Abstract
Objectives
Vivax malaria has reemerged and become endemic in Korea. Our study aimed to analyze by both longitudinal and cross-sectional genetic diversity of this malaria based on the P vivax Merozoite Surface Protein (PvMSP) gene parasites recently found in the Korean peninsula.
Methods
PvMSP-1 gene sequence analysis from P vivax isolates (n = 835) during the 1996-2010 period were longitudinally analyzed and the isolates from the Korean peninsula through South Korea, the demilitarized zone and North Korea collected in 2008-2010 were enrolled in an overall analysis of MSP-1 gene diversity.
Results
New recombinant subtypes and severe multiple-cloneinfection rates were observed in recent vivax parasites. Regional variation was also observed in the study sites.
Conclusion
This study revealed the great complexity of genetic variation and rapid dissemination of genes in P vivax. It also showed interesting patterns of diversity depending, on the region in the Korean Peninsula. Understanding the parasiteninsula. Under genetic variation may help to analyze trends and assess the extent of endemic malaria in Korea.
Keywords: genetic diversity, Korea, Plasmodium vivax, PvMSP-1
1. Introduction
Plasmodium vivax is the most widespread human malarial species [1] and has reemerged in many regions of the world where malaria was eliminated in the 1950 and 1960s [1,2]. Until the late 1970s, P vivax malaria (vivax malaria) was highly endemic in the Korean Peninsula [3-5]. Subsequently, malaria was eradicated in South Korea (SK) [6], but reemerged in 1993 [7]. Vivax malaria outbreaks in SK and have been correlated to those of North Korea (NK) [3,4]. The fraction of soldiers with malaria during an outbreak has usually been >50% in SK in the past. The patient population has now shifted to include a majority of non-military personnel. It is obvious that malaria is endemic and is once again a serious public health threat in SK.
Previously we reported that the recently collected isolates were completely different from those in the initial period (1996-2000) and that this diversity showed rapid insemination in the recent samples [8]. In the present study, we have further analyzed recent vivax malaria parasites using both longitudinal (1996-2010) and cross-sectional genetic analysis. For regional characterization,civilian patients from SK, military patients from the DMZ (demilitarized zone) and patients from NK were enrolled. Within SK, the major malaria risk regions-Gangwha and Chelwon counties and Paju and Gimpo cities adjacent to the southern DMZ-were analyzed. In addition, the multiple-clone infection rates in Korean vivax malaria patients were investigated. In this study, the MSP-1 gene polymorphism of recent vivax isolates in the Korean Peninsula was demonstrated.
2. Materials and Methods
2.1. Malaria incidence and burden in Korea
The methods of spot map generation and malaria incidence rates in SK and NK were described in supplementary methods online.
2.2. Sample preparation
From 1996-2010 (except for 1997), blood was collected by venipuncture from 835 patients with confirmed vivax malaria who had never been abroad. DNA was extracted from 200 μl of each blood sample using a QIAamp® DNA Mini Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. Eight-hundred-thirty-five patient samples were used for longitudinal analysis based on the MSP-1 gene. For the regional analysis based on MSP-1 gene diversity, the following were included:
208 civilian samples from endemic areas in SK collected in 2007-2010;
111 samples taken from soldiers in the DMZ in 2008; and
28 isolates from NK that were collected between 2007 and 2010.
2.3. PCR amplification and sequencing of MSP-1 gene
Genomic DNAs obtained from the 835 patients during 1996-2010 were analyzed and compared to our previous study [8]. The amplified region of the MSP-1 gene included the sequence between Internal conserved block (ICB) 5 and 6 [9]. The products from the polymerase chain reaction (PCR) were purified and sequenced using an ABI 3730XL DNA Analyzer automated sequencer (Applied Biosystems, USA). For the multiple clone infection analysis, each PCR product was cloned using a TOPO-TA cloning kit (Invitrogen Corp., USA) and 10 colonies of each PCR product were purified and sequenced. Detailed sequence analysis information is described in the supplementary methods online at doi:10.1016/j.phrp.2011.11.039.
3. Results
3.1. Prevalence of the reemerged P vivax malaria in Korea
A spot map was made of a malaria outbreak including three patient groups in SK (civilians, soldiers in the DMZ and discharged soldiers) and the risk areas in SK (supplementary Figure 1). The prevalence of malaria in SK and NK during 1999-2009 is presented in the supplementary Figure 2.
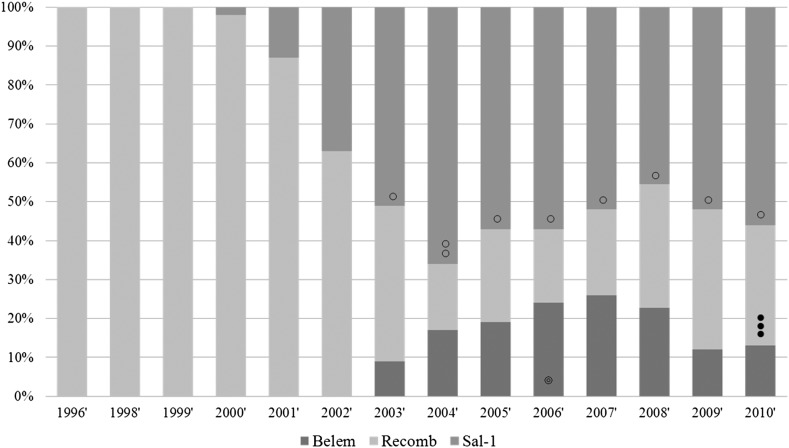
Figure 1. Frequency of PvMSP strains in Korea during the study period. Between 1996 and 2000, the majority of the Korean isolates have changed from recombinant to Sal-1. Black spots indicate the recombinant subtypes, white spots indicate the Sal-1 subtype and white spots with a smaller spot inside indicate the Belem subtypes. No spot region indicates only one subtype (S-b, B-1 and R1) without variation; ○: S-a subtype, ◌◌: S-a and S-c subtypes; ◎: B-2 subtype); ●●●: R2, R3 and R4 subtypes.
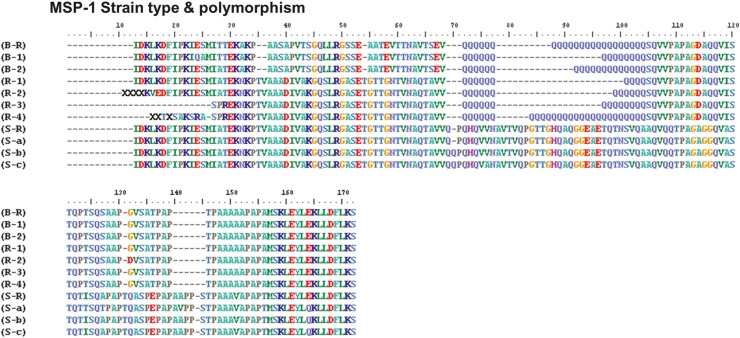
Figure 2. Comparison of amino acid sequences of the PvMSP-1 protein found in SK with worldwide strains. The amino acid positions indicated by the shaded regions depict non-synonymous substitutions and poly-Q residues in the Korean isolates.
3.2. Longitudinal analysis on MSP-1gene diversity
The block 5 of PvMSP-1 polymorphism among 835 isolates including the previous data [8] was analyzed. Three MSP-1 types (Sal-1, Belem and Recombinant) have been appeared in Korean vivax isolates. Sal-1 showed major frequency (over 50%) since 2003 (see supplementary Table 2). In the subtype analysis, S-a and S-b were the major Sal-1 subtypes. Only B1 subtype has been observed except 2006 since its appearance (Figure 1). New recombinant subtypes with different numbers of Q-repeats (10×Q, 12×Q, 14×Q and 27×Q) were found in patients in 2010 and named R1 R2, R3 and R4, respectively (Figure 2).
3.3. Comparison of MSP-1 genotypes in the Korean peninsula: SK, DMZ and NK
MSP-1 gene diversity of civilians in SK, soldiers in the DMZ and the NK samples collected over 3 years (2007-2010) were analyzed. The variation in MSP-1 types in soldiers was similar to that seen in SK. In NK, however, the recombinant type was predominant (53.6%), followed by Sal-1 (32.1%) and Belem (14.3%), see Figure 3A. The frequency of the S-b subtype (28.9%) was slightly higher than the S-a subtype (23.6%) in SK.
Figure 3. (A) The portions of MSP-1 types; and (B) its subtypes in patients from SK, the DMZ and NK.
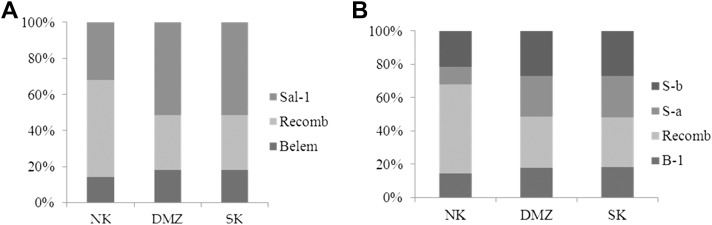
The percentage of S-a to S-b subtypes was almost equal in the DMZ (27% and 28.8%, respectively). In the North Korean isolates, a high portion of recombinant type (53.6%) was observed. The incidence of the S-b subtype (21.4%) was much higher than S-a (10.7%). In Belem subtypes, there were similar frequency rates (14.3-17.3%) in the three regions, see Figure 3B.
3.4. Cross-sectional genotyping in malaria risk regions in South Korea
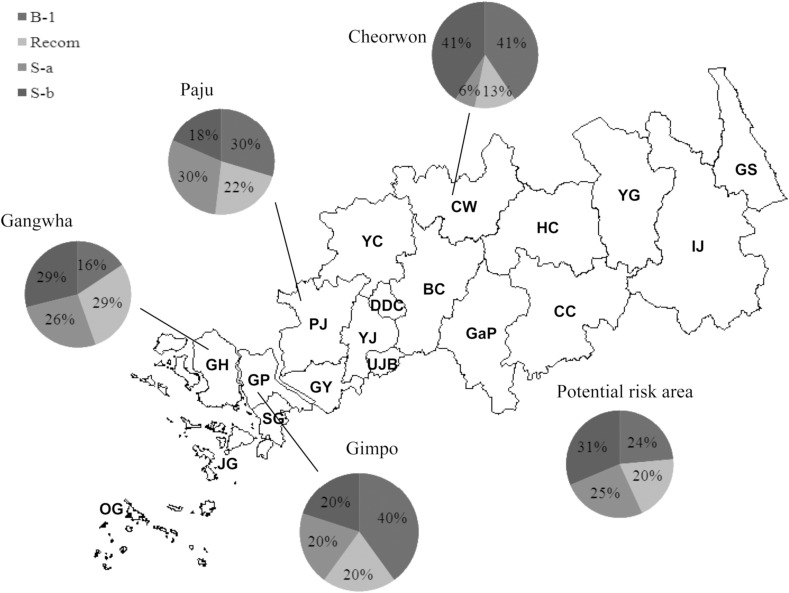
The 208 samples collected from Gangwha (N =83), Gimpo (N = 15), Paju (N = 27) and Cheorwon
(N = 32), and other regions including potential risk areas (N = 51) in 2006-2010 were enrolled. Gangwha, Gimpo and Paju are located in the western part of SK, while Cheorwon is located in the middle of the risk areas (see Figures 4 and supplementary Figure 1). Other regions include the potential risk regions and scattered samples in SK, as described in the spot map (supplementary Figure 1). A similar pattern of subtype variation was observed in Gang-wha, Paju and other areas with a risk of vivax malaria. A relatively higher portion (40%) of B-1 type was, however, shown in Gimpo isolates and only
Figure 4. The diversity in MSP-1 subtypes depending on the major malaria risk areas in Korea (Gangwha, Bucheon, Cheolwon, Gimpo, Paju, etc.).
6% of S-a type was found in patients from Cheorwon (Figure 4).
3.5. Prevalence of mixed-clone infection in P vivax
We invested the multiple clone infection rates in vivax patients. Eighty-eight isolates from SK, 22 isolates from DMZ soldiers and 23 isolates from NK that were collected during 2007 and 2008 were analyzed. We observed 56.5% mixed-clone infection rates in SK, 77%in the DMZ and 32% in NK. In further analysis of the major risk regions in SK, multiple infection rates varied from 41.7-73.3% and double, triple or even quadruple clones were observed, as described in Table 1.
Table 1.
Proportion of mixed-clone infections in P vivax isolates from the risk areas in South Korea
| Area | Gangwha | Cheorwon | Gimpo | Paju | Potential risk area |
|---|---|---|---|---|---|
| Multiplicity | n (%) | n (%) | n (%) | n (%) | n (%) |
| Single | 10 (35.7) | 4 (26.7) | 7 (58.3) | 14 (46.7) | 11 (50.0) |
| Double | 10 (35.7) | 10 (66.7) | 2 (16.7) | 13 (43.3) | 8 (36.4) |
| Triple | 7 (25.0) | 1 (6.7) | 2 (16.7) | 3 (10.0) | 2 (9.1) |
| Quadruple | 1 (3.6) | 1 (8.3) | 1 (4.5) | ||
| Total no (n) | 28 | 15 | 12 | 30 | 22 |
| Mixed infection rate (%) | 64.3 | 73.3 | 41.7 | 53.3 | 50 |
4. Discussion
The present longitudinal (1996-2010) and crosssectional analysis of three regions in the Korean Peninsula provided more evidence of the complexity of genetic variation and supports the previous study [8]. As described in the supplementary Figures 1 and 2, the prevalence of vivax malaria NK is slightly different to that in SK. The border region is the most important region for malaria control in the Korean Peninsula. Since 2 years of military duty is mandatory for Korean men and soldiers make up about half of the long-term latent patients in Korea [10], patients are usually grouped into three bands: soldiers, discharged soldiers
(DS) and civilians, as shown in the spot map (supplementary Figure 1A).
Park et al [5] showed that the portion of civilians with vivax malaria has increased. Until recently, the proportion of soldiers including DS has made up almost half of the total number of vivax malaria cases [6]. In this regard, this study including three groups-SK, NK samples and DMZ soldiers-provides valuable data for genetic research into vivax malaria
The PvMSP-1 protein expressed on the surface of the malaria merozoite [11] is organized in several variable blocks flanked by 10 conserved sections [12]. Block 5 shows a dimorphic pattern of sequences that have little homology [13]. PvMSP-1 has been used as a polymorphic marker to study genetic structure of P vivax populations and is also a valuable epidemiological marker in endemic areas [12,14-16]. In this study, we have reviewed the block 5 PvMSP-1 polymorphism among isolates collected in SK from 1996-2010. The result of the variation pattern was not very different to the previous study [8]. Interestingly, however, we found new recombinant subtypes with different numbers of Q residue repeats (12×Q, 14×Q and 27×Q) from indigenous Korean patients in 2010 (Figure 2). 12×Q, and 27×Q subtypes were found in Ganghwa and 14×Q subtype was found in Gimpo. The 12×Q subtype has only ever been found in Azerbaijan [17] but recombinant 14×Q and 27×Q subtypes have been newly found in the present study. The origination of these new recombinant subtypes is unclear, but their appearance might be further evidence of the rapid dissemination of newly introduced P vivax, as described in our previous study [8], and they show the complexity of genotypes in Korean vivax malaria. Choi et al [18] described an imported case of vivax case that broke out in an endemic area in Korea like an indigenous case. They suggested that imported cases might increase the diversity of vivax genotypes in Korean peninsula. These results suggest that there is great need for further surveillance of genetic diversity in P vivax.
In this study we tried to investigate the three groups of vivax patients; civilian patients in SK and NK and soldiers with malaria in the DMZ. Several researchers have suggested that the P vivax malaria situation in SK has been directly influenced by infected mosquitoes originating from NK [19,20]. The relative risk on vivax prevalence of NK to SK is very high (supplementary Figure 2). Since the number of NK isolates was relatively small, we were unable to find the direct relationship of genotypes found in SK and NK. It was, at least, was very clear that DMZ isolates were closely related the isolates from SK patients. It seems that SK has its own malaria transmission cycle. Moreover, it is meaningful that the present study showed the recent change in the diversity of isolates from soldiers in Korea. We also carried out a cross-sectional study in SK. As described in Figure 4, the diversity of isolates in Cherwon was unique compared to other areas. The portion of the S-b subtype was higher than S-a, and a very high frequency of B-1 was observed. There were also a relatively lower number of recombinant subtypes found. Cherwon is located in the middle of the risk areas(Figure 4) and has different geographical features. Jun et al [3] suggested that the west part of malaria risk areas in SK can be influenced by the malaria situation in NK. It seems that a different transmission cycle correlates with NK and in Cheorwon area. This study suggests that there we need to survey the genotypes of NK isolates, using a large sample size to demonstrate the genotyping status in the Korean Peninsula.
Finally we investigated the proportion of mixed clone infection. A high proportion of mixed clone infection (77%) was observed in DMZ isolates. This might support the hypothesis that the DMZ is the pathway for malaria transmission in the Korean Peninsula. Although NK isolates showed the lowest mixed clone infection rates (33%), the small numbers of samples might be used to represent status of vivax malaria NK. In further analysis among SK isolates, Cheorwon isolates showed the highest proportion of mixed infection (73.3%), and was similar to that of DMZ isolates. Ganghwa, the highest risk area for Korean civilians followed, with 64.3% having mixed infection. Double-, triple- or fourclone infections were found in the study sites. It has been reported that 38% of mixed infections were observed in Papua New Guinea, 33.3% in Thailand, 0.7% in India and 44.8% in Colombia [21]. Korea is considered a hypoendemic area for malaria. Cui [22] reported that relatively low genetic diversity has been detected in the reemerging P vivax malaria in Korea. In this study, however, Korea showed the extremely high rates of mixed clone infection. This study contradicts the previous results [22] and provides evidence of the complexity of genetic vivax malaria diversity in Korea. Recently Han et al [23] reported that the allelic diversity of Pvmsp-1 was increased in Korean isolates.
5. Conclusion
Our overall study on vivax malaria in the Korean Peninsula revealed the high complexity of genetic variation and rapid dissemination of P vivax genes. It also showed interesting patterns of diversity, depending on the regions analyzed in the Korean Peninsula. Understanding the patterns of P vivax genetic may help us analyze trends and assess the extent of endemic malaria in Korea.
Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.phrp.2011.11.039.
Acknowledgments
This study was supported by an intramural fund provided by the Korea National Institute of Health (No. 4837-302-210-13).
References
- 1.Mendis K, Sina BJ, Marchesini P, et al. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001 Jan-Feb;64:97–106. doi: 10.4269/ajtmh.2001.64.97. [DOI] [PubMed] [Google Scholar]
- 2.Guerra CA, Hay SI, Lucioparedes LS, et al. Assembling a global database of malaria parasite prevalence for the Malaria Atlas Project. Malar J. 2007 Feb 16;6:17. doi: 10.1186/1475-2875-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jun G, Yeom JS, Hong JY, et al. Resurgence of Plasmodium vivax malaria in the Republic of Korea during 2006-2007. Am J Trop Med Hyg. 2009 Oct;81(4):605–10. doi: 10.4269/ajtmh.2009.09-0111. [DOI] [PubMed] [Google Scholar]
- 4.Yeom JS, Kim TS, Oh S, et al. Plasmodium vivax malaria in the Republic of Korea during 2004-2005: changing patterns of infection. Am J Trop Med Hyg. 2007 May;76(5):865–8. PubMed. [PubMed] [Google Scholar]
- 5.Park JW, Jun G, Yeom JS. Plasmodium vivax malaria: status in the Republic of Korea following reemergence. Korean J Parasitol. 2009 Oct;47(Suppl):S39–50. doi: 10.3347/kjp.2009.47.S.S39. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chai JY. Re-emerging Plasmodium vivax malaria in the Republic of Korea. Korean J Parasitol. 1999 Sep;37(3):129–43. doi: 10.3347/kjp.1999.37.3.129. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korean Center for Disease Control and Prevention Statistics. Commun Dis Rep. 2007;18(1):13. [Google Scholar]
- 8.Choi YK, Choi KM, Park MH, et al. Rapid dissemination of newly introduced Plasmodium vivax genotypes in South Korea. Am J Trop Med Hyg. 2010 Mar;82(3):426–32. doi: 10.4269/ajtmh.2010.09-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim CS, Kim SH, Kwon SI, et al. Analysis of Plasmodium vivax merozoite surface protein-1 gene sequences from resurgent Korean isolates. Am J Trop Med Hyg. 2000 Feb;62(2):261–5. doi: 10.4269/ajtmh.2000.62.261. [DOI] [PubMed] [Google Scholar]
- 10.Nishiura H, Lee HW, Cho SH, et al. Estimates of short- and long term incubation periods of Plasmodium vivax malaria in the Republic of Korea. Trans R Soc Trop Med Hyg. 2007 Apr;101(4):338–43. doi: 10.1016/j.trstmh.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Leclerc MC, Gauthier C, Villegas L, et al. Genetic diversity of merozoite surface protein-1 gene of Plasmodium vivax isolates in mining villages of Venezuela (Bolivar State). Acta Trop. 2005 Jul;95(1):26–32. doi: 10.1016/j.actatropica.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Del Portillo HA, Longacre S, Khouri E, et al. Primary structure of the merozoite surface antigen 1 of Plasmodium vivax reveals sequences conserved between different Plasmodium species. Proc Natl Acad Sci USA. 1991 May;88(9):4030–4. doi: 10.1073/pnas.88.9.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Putaporntip C, Jongwutiwes S, Tanabe K, et al. Interallelic recombination in the merozoite surface protein-1 (MSP-1) gene of Plasmodium vivax from Thai isolates. Mol Biochem Parasitol. 1997;84:49–56. doi: 10.1016/s0166-6851(96)02786-7. [DOI] [PubMed] [Google Scholar]
- 14.Kolakovich KA, Ssengoba A, Wojcik K, et al. Plasmodium vivax:favored gene frequencies of the Merozoite Surface Protein-1 and the multiplicity of infection in a malaria endemic region. Exp. Parasitol. 1996 Jun;83(1):11–8. doi: 10.1006/expr.1996.0044. [DOI] [PubMed] [Google Scholar]
- 15.Figtree M, Pasay CJ, Slade R, et al. Plasmodium vivax synonymous frequencies, evolution and population structure deduced from diversity in AMA-1 and MSP-1 genes. Mol Biochem Parasitol. 2000 Apr;108(1):53–66. doi: 10.1016/s0166-6851(00)00204-8. [DOI] [PubMed] [Google Scholar]
- 16.Cui L, Escalante AA, Imwong M, Snounou G. The genetic diversity of Plasmodium vivax populations. Trends Parasitol. 2003 May 19;(5):220–6. doi: 10.1016/s1471-4922(03)00085-0. [DOI] [PubMed] [Google Scholar]
- 17.Leclerc MC, Hugot JP, Durand P, et al. Evolutionary relationships between 15 Plasmodium species from new and old world primates (including humans): an 18S rDNA cladistic analysis. Parasitology. 2004 Dec;129(Pt 6):677–84. doi: 10.1017/s0031182004006146. [DOI] [PubMed] [Google Scholar]
- 18.Choi KM, Choi YK, Kang YA, et al. Study of the genetic discrimination between imported and autochthonous cases of malaria in SK. J Travel Med. 2011 Jan-Feb;18(1):63–6. doi: 10.1111/j.1708-8305.2010.00473.x. [DOI] [PubMed] [Google Scholar]
- 19.Lee JS, Lee WJ, Cho SH, et al. Outbreak of vivax malaria in areas adjacent to the demilitarized zone, SK, 1998. Am J Trop Med Hyg. 2002 Jan;66(1):13–7. doi: 10.4269/ajtmh.2002.66.13. [DOI] [PubMed] [Google Scholar]
- 20.Ree HI. Can malaria be endemic in SK? Korean J Infect Dis. 1998;4:397–400. [Google Scholar]
- 21.Joshi H, Prajapati SK, Verma A, et al. Plasmodium vivax in India. Trends Parasitol. 2008 May;24(5):228–35. doi: 10.1016/j.pt.2008.01.007. Epub 2008 Apr 9. Review. [DOI] [PubMed] [Google Scholar]
- 22.Cui L, Escalante AA, Imwong M, et al. The genetic diversity of Plasmodium vivax populations. Trends Parasitol. 2003 May;19(5):220–6. doi: 10.1016/s1471-4922(03)00085-0. Review. [DOI] [PubMed] [Google Scholar]
- 23.Han ET, Wang Y, Lim CS, et al. Genetic diversity of the malaria vaccine candidate merozoite surface protein 1 gene of Plasmodium vivax field isolates in Republic of Korea. Parasitol Res. 2011 May 10; doi: 10.1007/s00436-011-2413-5. [DOI] [PubMed] [Google Scholar]