Abstract
Apicomplexa parasites use complex cell cycles to replicate that are not well understood mechanistically. We have established a robust forward genetic strategy to identify the essential components of parasite cell division. Here we describe a novel temperature sensitive Toxoplasma strain, mutant 13-20C2, which growth arrests due to a defect in mitosis. The primary phenotype is the mis-segregation of duplicated chromosomes with chromosome loss during nuclear division. This defect is conditional-lethal with respect to temperature, although relatively mild in regard to the preservation of the major microtubule organizing centers. Despite severe DNA loss many of the physical structures associated with daughter budding and the assembly of invasion structures formed and operated normally at the non-permissive temperature before completely arresting. These results suggest there are coordinating mechanisms that govern the timing of these events in the parasite cell cycle. The defect in mutant 13-20C2 was mapped by genetic complementation to Toxoplasma chromosome III and to a specific mutation in the gene encoding an ortholog of nuclear actin-related protein 4. A change in a conserved isoleucine to threonine in the helical structure of this nuclear actin related protein leads to protein instability and cellular mis-localization at the higher temperature. Given the age of this protist family, the results indicate a key role for nuclear actin-related proteins in chromosome segregation was established very early in the evolution of eukaryotes.
Keywords: Apicomplexa, Toxoplasma gondii, cell cycle, mitosis, chromosome segregation, actin-related protein 4a
1.0 Introduction
Apicomplexan parasites replicate by strategies that differ in nuclear division, but are similar with respect to the formation of new daughter parasites by a unique process of internal budding. Apicomplexans are remarkably accomplished specialists of multinuclear replication. In schizogony, parasites form multiple nuclei by alternating rounds of DNA synthesis and mitosis with cytokinesis delayed until the last round of nuclear division [1]. Endopolygenic division is a variation on schizogony where multiple rounds of chromosome replication occur within a single (polyploid) nucleus that undergoes concerted nuclear division and parasite budding at the end of the cycle [2, 3]. By contrast, endodyogeny is a simple binary division with a single chromosome replication followed by concurrent mitosis and parasite budding [4]. Cytokinesis in endodyogeny occurs through a unique internal assembly of daughters that is very similar to the final cycle of schizogony in other Apicomplexans suggesting that there is a common set of cell cycle controls that operate in both division strategies [5, 6].
The comparative simplicity of endodyogenic division and strengths of the Toxoplasma experimental model system have allowed the major phases of the tachyzoite cell cycle and the basic order and timing of nuclear and organelle division to be established [7]. Toxoplasma tachyzoites divide using a three-phase cycle comprised of a G1 (60%), S (30%) and mitosis (10%) with the classic G2 period between S phase and mitosis either very short or absent [8, 9]. The length of the S phase period in the tachyzoite cell cycle has been independently verified by [H3]-thymidine radiolabeling, time-lapse microscopy of transgenic parasites expressing TgPCNA1-GFP (monitoring replication foci), and by calculating the length of S phase from genomic DNA analysis of synchronized populations [8]. It is not yet possible to pinpoint the start of mitosis, although conoids appear to form at this time [8, 10] and replication foci dissolve <1 h before nuclear division consistent with a minimal or absent G2 period [8]. This finding parallels similar observations in Theileria and other protozoa [11, 12]. S phase distributions in tachyzoites are peculiar [8, 13], with late S phase parasites (~1.8N) more numerous than parasites in early S. It is intriguing that internal daughter budding also appears to initiate in late S phase [13], and therefore, a novel apicomplexan mechanism has been proposed that safeguards the proper timing of budding with mitosis might be responsible for the 1.8N subpopulation [8]. Establishing the molecular basis for these observations is needed to understand these unusual cell cycle distributions.
Checkpoint control of apicomplexan cell division is poorly understood, although factors commonly associated with eukaryotic cell cycle checkpoints are present in apicomplexan parasites including cyclins, CDKs, MAPKs, and components of the anaphase promoting complex [14] that regulates mitosis. While these findings predict that cell cycle controls exist in these parasites, they do not provide information about where checkpoints function or how these controls operate to coordinate the diverse strategies utilized by these parasites to proliferate. Experimental proof for active checkpoint control in Toxoplasma has been controversial [15, 16], however, there is now growing evidence for a model that proposes specific transitions in the tachyzoite cell cycle where parasites naturally halt during progression through the G1, S and mitotic phases [8, 9, 17, 18]. In this paper, we describe a new Toxoplasma mitotic mutant and identify the essential defect responsible for conditional growth through genetic complementation. The fundamental nuclear factor revealed by these studies is required for chromosome segregation in Toxoplasma and other eukaryotes establishing an ancient cell cycle mechanism conserved over hundreds of million years of eukaryotic cell evolution. The primary defect in this mutant also provides insight into the mechanistic restrictions that coordinate mitosis and the internal assembly of daughter parasites in the Apicomplexa.
2.0 Material and Methods
2.1 Cell Culture
Parasites were grown in human foreskin fibroblasts (HFF) as described [19]. All transgenic and mutant parasite lines are derivatives of the RHΔhxgprt parasite strain. Temperature sensitive clone 13-20C2 was obtained by chemical mutagenesis of the RHΔhxgprt strain [18]. Parasite growth measurements were obtained using parasites pre-synchronized by limited invasion [20]. Briefly, a seed parasite stock was prepared using freshly lysed parasites inoculated into HFF monolayers (MOI 1:5) at 34°C. Following 30 min the monolayers were washed twice to remove parasites that had not invaded. The partially synchronized population was harvested a 36 h post-inoculation at 34°C (average size of vacuoles of 32-64) and the limited invasion process repeated once more. The resulting second synchronized population was used to inoculate experimental plates in triplicate. Average vacuole sizes were determined at selected time points from 50-100 randomly selected vacuoles.
2.2 Genetic complementation of mutant 13-20C2
Mutant 13-20C2 was complemented using the ToxoSuperCos cosmid genomic library described previously [18]. Mutant parasites were transfected with cosmid library DNA (50 μg DNA/5 × 107 parasites/transfection) in twenty independent electroporations. Each transfection was plated in a single T175cm2 flask, the parasites were recovered for 24h at 34°C, and then shifted to 40°C to select for temperature resistance. Populations that emerged from high temperature cultures were further selected with 1 μM pyrimethamine at 40°C to ensure integration of the cosmid backbone. Double resistant parasites (high temperature and drug) were passed four times before extraction of genomic DNA from the polyclonal population. Cosmid tags were recovered from the genomic DNA by plasmid-rescue protocols [18]. Rescued genomic inserts were sequenced using a T3 primer and the sequences mapped to the Toxoplasma genome to identify the rescue locus (ToxoDB: http://www.toxodb.org/toxo/).
To confirm the rescue locus, mutant 13-20C2 was complemented with individual cosmid clones from a cosmid collection mapped to the Toxoplasma genome (toxomap.wustl.edu/cosmid.html). Specific cosmids were transfected into 1×107 parasites using 10 μg of purified DNA. Cosmids with a Toxo-backbone were selected with 1 μM pyrimethamine, while those with a PSB-backbone were selected by phleomycin treatment of extracellular parasites (protocols can be downloaded at toxomap.wustl.edu/cosmid.html). A standard plaque assay performed in triplicate was utilized to quantify genetic rescue of the drug-resistance populations at the high temperature [21]. To resolve the contribution of individual genes carried by cosmids, we deleted specific ORFs in the cosmid insert by replacement with a cassette containing chloramphenicol acetyl transferase (CAT) and gentamycin acetyltransferase-3-I (AAC1). The replacement was performed by recombineering in E. coli as recently described [22]. Cosmid PSBLQ51, which spans the locus containing TgARP4a, provided the insert context for deletion of genes TgG2a or TgARP4a. Pairs of primers were first used to PCR the CAT/ AAC1 cassette followed by recombination of the PCR fragment into the cosmid insert to replace each gene (TgG2a: Forward primer, 5′ TTCAGGTTGTCTGCGTGTTTCGCGCCCATGTTTTTCCCCAGGACGTCAAGCCTCGACTACGGCTTCCATTGGCAA 3′ and reverse primer, 5′ ACGAGTCGAACTTTCCGCGCTCCTCGAATCCTCCCGCGAGACCTGCGAACATACGACTCACTATAGGGCGAATTG. TgARP4a: forward primer 5′CTCTCCCGTCTCTTTCTTCTCCTTCTTTTCCCCGTCCTTCTTCGTT GGCACCTCGACTACGGCTTCCATTGGCAA 3′ and reverse primer, 5′ CCTGGAGCGTCGTTCGCCGCCTGGAGCAGATGGCGCCACCGACGGTCTCATACGACTCACTATAGGGCGAATTG 3′). For direct complementation with DNA fragments, the TgARP4a genomic locus was amplified from genomic DNA (5,846 bp) prepared from parent and mutant DNA using a forward primer located at −956bp in the 5′ UTR of TgARP4a gene (5′ GACTTGTCTCGCACTAGAATGACCG) and a reverse primer located in 3′UTR, 978bp downstream of the TgARP4a stop codon (5′ GTGAAGCAGAGGAACCTTCCAGC). Complementation of mutant 13-20C2 with genomic fragments (5 μg DNA/1.5 × 107 parasites) carrying either the wt- or ts-allele of TgARP4a was performed as previously described [18].
2.3 Ectopic expression of DD-myc3-TgARP4a
The coding sequence of TgARP4a was amplified from cDNA produced from parent RHΔhxgprt strain and the mutant 13-20C2 using a forward primer, 5′ TATCAATTGTCTTCGTTGGCAATGCCGCCTCCGACTG 3′, and a reverse primer, 5′ ATACCTGCAGGTCACCGAC ACCGGCGGTTGATGATGTCGAC 3′ with incorporated restriction sites. PCR products were cloned into expression vector pdhfr- DDL106P-myc3X/sag-CAT or ptub- DDL106P-myc3X/sag-CAT using unique MfeI and SbfI cloning sites. The resulting construct design fused the FKBP destabilizing domain (DDL106P) [23] and three copies of the myc epitope tag to the N-terminus of the wt- or ts-TgARP4 coding region in an expression context driven by DHFR-TS or α-tubulin promoters. Plasmids were transfected into either RHΔhxgprt parental strain or the mutant 13-20C2 parasites by electroporation and stable transgenic clones were selected with 20 μM chloramphenicol.
2.4 Immunofluorescence microscopy
Confluent HFF cultures on the glass coverslips were infected with parasites for the indicated time. Cells were fixed in 3.7% paraformaldehyde, permeabilized in 0.25% Triton X-100 and blocked in 1% BSA in PBS. Incubations with primary antibody followed by the corresponding secondary antibody were performed for 1h each at room temperature with DAPI (0.5 μg/ml) added in the final incubation to stain genomic DNA. The following primary antibodies were used at the indicated dilutions: mouse monoclonal anti-myc (Santa Cruz Biotechnology, Santa Cruz, CA) at 1:500; anti-Atrx1, anti-ROP7 (kindly provided by Dr. Peter Bradley, UCLA, Los Angeles, CA) and anti-centrin 26-14.1 (kindly provided by Dr. Jeffrey Salisbury, Mayo clinic, Rochester, NY) at 1:1000; rabbit polyclonal anti-MORN1 (kindly provided by Dr. Mark-Jan Gubbels, Boston College, MA) at 1:2000, anti-IMC1 (kindly provided by Dr. Gary Ward, University of Vermont, VT) at 1:1000, and rat polyclonal anti-IMC3 (kindly provided by Dr. Mark- Jan Gubbels, Boston College, MA) at 1:2000. All Alexa-conjugated secondary antibodies (Invitrogen Life Technologies, Carlsbad, CA) were used at dilution 1:1000. After several washes with PBS, coverslips were mounted with Aquamount (Thermo Fisher Scientific, Waltham, MA), dried overnight at 4°C, and viewed on Zeiss Axiovert Microscope equipped with 100x objective. Images were collected both as wide-field and as z-stacks in 0.250μm steps, processed in Adobe Photoshop CS v4.0 using linear adjustment for all channels.
2.5 Western Blot analysis
Purified parasites obtained by needle passage, filtering through 3μm filters, and collected by centrifugation were washed in PBS. Total lysates were obtained by mixing isolated parasites with Leammli loading dye. Nuclear proteins were extracted using Pierce NE-PER kit (Thermo Fisher Scientific, Waltham, MA). All the buffers were supplied with 1x protease inhibitor cocktail (Thermo Fisher Scientific, Waltham, MA). Protein samples were mixed with Leammli loading dye, heated at 95°C for 10 min and separated on the SDS-PAGE gels. Proteins were transferred onto nitrocellulose membrane and probed with polyclonal anti-myc (Cell Signaling Technology, Danvers, MA) or anti-TgPCNA1 antibody [24]. After incubation with secondary HRP-conjugated anti-rabbit antibody, proteins were visualized in enhanced chemiluminescence reaction.
3.0 Results
3.1 Chemical mutant 13-20C2 is a temperature sensitive strain with defects in nuclear division
Methods to generate conditional growth mutants by chemical mutagenesis in Toxoplasma gondii were established three decades ago [25], although only recent genetic strategies permit the affected genes to be identified [18]. These advances opened investigations into cell cycle mechanisms of apicomplexan parasites through the generation of an expanded collection of conditional growth strains in T. gondii [18]. The clone 13-20C2 is one of the isolates from a recent screen of >60,000 chemical mutants that displayed restricted growth at the non-permissive temperature of 40°C, while growing at a similar rate to the parental RHΔhxgprt strain at the permissive temperature of 34°C (Fig. 1A). Growth arrest of mutant 13-20C2 at 40°C was rapid and vacuole distributions were consistent with a defect in a time-limited event. Mutant 13-20C2 parasites arrested immediately in cycle or advanced to the next cell cycle before stopping depending on their relative position (before or behind) the critical cell cycle defect at the time of the shift to higher temperature. Like many of the growth mutants isolated in this screen [18], mutant 13-20C2 has a conditional-lethal phenotype. Mutant 13-20C2 parasites lost the ability to form plaques when exposed to progressively longer times at the higher temperature with only 6% of parasites (15 plaques) viable after a 48 h exposure to 40°C (Fig. 1B). Longer incubations at the restrictive temperature resulted in the complete loss of population viability (not shown).
Figure 1. Tachyzoite strain 13-20C2 carries a lethal mutation that is conditional with respect to temperature.
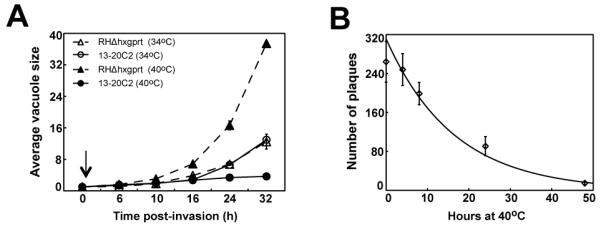
(A) The growth of parental RHΔhxgprt and mutant 13-20C2 parasites were monitored in populations partially synchronized by limited invasion over a 32 h period at 34°C and 40°C. The arrow indicates a post-invasion time when cultures were shifted to 40°C. To determine average vacuole sizes, randomly selected vacuoles (>50) in three independent cultures per strain and temperature condition were directly monitored by light microscopy. (B) The lethality of the temperature defect was determined by plaque assay. Freshly invaded mutant parasites were incubated at 40°C for the indicated times before the cultures were shifted to the permissive temperature (34°C) to allow for plaque development. Plaque numbers represent the average of three independent flasks.
To understand the terminal growth phenotype of mutant 13-20C2, we examined the cell biology of 13-20C2 parasites cultured at permissive or non-permissive temperatures. The general cell and nuclear appearance of mutant 13-20C2 parasites at 34°C revealed they were indistinguishable from the parent strain (not shown), whereas morphological defects and DNA loss was predominant at 40°C (Fig. 2). The degree of chromosome loss was variable and this appeared to influence the timing of subsequent cell cycle arrest at the higher temperature. A slight decrease in DNA (based on DAPI estimates) in daughter parasites appeared to allow progression to the next division cycle, while significant DNA loss was responsible for immediate arrest of cell division. Flow cytometric analysis of mutant 13-20C2 grown at 40°C showed a large subpopulation of parasites possessed <1N DNA content (data not shown) consistent with the defects in chromosome segregation and nuclear division observed in the microscopic images.
Figure 2. Immunofluorescent analysis of mutant 13-20C2 reveals severe defects in chromosome segregation and nuclear division.
Immunofluorescent microscopy of parental strain RHΔhxgprt (a, b, c) and mutant 13-20C2 was performed 16 h post-infection at 34°C (d, e, f) and 40°C (g, h, I, k, l, m) to determine the primary temperature defect. Representative images showing the co-staining with anti-IMC1 (green) antibodies and DAPI (blue) revealed the primary retention of genomic DNA in the mother residual mass (dashed circled) occurred in parasites grown at the non-permissive temperature (40°C). The loss of chromosome material was variable with some parasites retaining significant DNA (middle panels), while other newly formed parasites lacked detectable DAPI staining (lower panels). Magnification bar (5 μm) is indicated in the lower right hand Nomarski image.
3.2 Nuclear architecture, budding and organellar biogenesis in mutant 13-20C2
There is an intimate relationship between mitosis and biogenesis of internal daughter structures that comprise the invasion apparatus in Toxoplasma (and other members of Apicomplexa) [4, 7]. To determine how the defect in mutant 13-20C2 might affect assembly of the invasion apparatus during mitosis, we examined various cellular compartments and organelles that undergo biosynthetic changes in the tachyzoite cell cycle. As parasites progress into S phase from G1 they duplicate their centrosomes [8, 26], which migrate to an apical location where they are thought to physically connect to the developing bud scaffolds by unknown mechanisms [7]. Utilizing anti-centrin antibody to visualize centrosomes in mutant 13-20C2, we determined the duplication of the centrosome was not affected by temperature, although there were differences in centrosome orientation (Fig. 3A and B, panels a-d). The faithfully replication of centrosomes indicated that passage through the G1/S phase checkpoint known as START had occurred before growth arrest [27], while changes in migration patterns may be related to the disturbed nucleus architecture that ultimately results in chromosome mis-segregation in this mutant. In apicomplexan replication the spindle pole (also called the centrocone) undergoes significant changes in S phase and mitosis that can be monitored with anti-MORN1 antibodies (membrane occupation and recognition nexus protein 1, [28]). Like the centrosomes, we found morphological transitions of the spindle pole also occurred in mutant parasites grown at high temperature (Fig. 3A and B, panels e-h). In most parasites grown at 40°C, MORN1 structures were duplicated (with evidence of ring formation, see inset Fig. 3B, e panel) and segregated into daughters. Interesting, the residual basal complex from early cell cycle did not resolve fully at high temperature (Fig. 3B, panels e-h). Consistent with the preservation of microtubule organizing centers in this mutant (i.e. MTOCs apical complex and spindle), organelle biogenesis and segregation into daughters unfolded with similar timing regardless of temperature (compare Figs 3A and B, panels i-q). Thus, the apicoplast was duplicated and segregated into daughter parasites, and newly synthesized rhoptry, secretory organelles were also packaged into internal daughter buds in parasites whose subsequent growth was stopped by the high temperature.
Figure. 3. The mitotic defect in mutant 13-20C2 does not prevent daughter formation or organellar duplication and targeting.
Cell biologic analyses of mutant 13-20C2 parasites cultured for 24 h at 34°C (A) or 40°C (B). Co-stains: Green stains are anti-centrin (centrosome), anti-MORN1 (centrocone and basal complex), anti-Atrx1 (apicoplast) or anti-ROP7 (Rhoptries). Red stain is anti-IMC1. Blue stain is DAPI. Marker guides are decolorized/inverted green/red merged images. Note the consistent posterior location of centrosomes in daughter parasites formed at 40°C (d image panel). Different MORN1 patterns (see guides in h image panels) mark the sequence of cell cycles with the larger posterior body (arrow) retained from the previous basal complex, while the duplicated MORN1 centrocone with ring (star) was formed in the current cell cycle. It is interesting to note that in this example of parasites grown at high temperature (a four vacuole trying to make eight parasites) showed evidence of all three cell cycles with two residual basal complexes evident (total of six basal MORN1 bodies). The two basal MORN1 bodies (arrows) from parasites originally dividing 1-to-2 is still present along with the four MORN1 basal bodies from the 2-to-4 division and the newly formed centrocone/ring structure (star). Marker guides for apicoplast/daughters are indicated in m-panels, and for rhoptries/daughters in q-panels. Dashed circles denote the chromosome material lost in the residual mass. Magnification is indicated in the lower marker guide panels with a 5 μm black bar. (C) Comparison of two chromosome mis-segregation mutants showing distinct severity of defect. Mutant 12-42D6 has an uncoupling phenotype leading to relaxed chromosomes (expanded blue DAPI staining) and severely degenerated bud structures (anti-IMC1, red). By contrast, mutant 13-20C2 has compact, if fragmented nuclei, and well developed buds.
The ability of mutant 13-20C2 to form bipolar spindles and new internal buds along with the proper packaging of invasion organelles into daughter parasites suggests the defect in mutant 13-20C2 has a restricted cell cycle context involving specific intranuclear mechanisms required for chromosome segregation and nuclear division. This level of biosynthetic coordination is lost when parasites are treated with dinitroanalines [29] and in other mitotic mutants where more dramatic uncoupling phenotypes predominate [18]. The recently described mitotic mutant of T. gondii 12-42D6 is rescued by a RCC1 domain-containing protein [18], which may have a role in chromosome condensation. A direct comparison of mutants 13-20C2 and 12-42D6 at the non-permissive temperature demonstrated a pronounced defect in the nuclear division (Fig. 3C) that in mutant 12-42D6 leads to a loss of DNA compaction and severe breakdown in the DNA and IMC1 containing structures (centrosomes are mostly retained in the DNA, Fig. 3C, panel b). By contrast, mutant 13-20C2 (Fig. 3C, panels c and d) preserves many of the critical mitotic structures at much higher frequency resulting in a milder phenotype despite the lethal growth arrest.
3.3 Genetic rescue of the conditional temperature defect of mutant 13-20C2
In order to link the fatal chromosome mis-segregation of mutant 13-20C2 with a specific protein defect, we used a forward genetic approach based on complementation of the mutant with a T. gondii cosmid genomic library (http://toxomap.wustl.edu/cosmid.html). Mutant parasites grown at 34°C were transfected with the ToxoSuperCos library in 20 independent electroporations [18]. Temperature resistant populations were obtained from two of these transfections that underwent further pyrimethamine (1 μM) selection to promote more complete integration of cosmid DNA into parasite chromosomes. Identification of the integrated cosmid DNA was accomplished by marker-rescue techniques and sequencing of the recovered cosmid inserts. Two distinct genomic inserts recovered by these methods were mapped to Toxoplasma chromosome III at position 665,366bp and 711,113bp where five genes were predicted to lie within the complemented locus (see diagram Fig. 4A). Two of the five genes were eliminated through additional genetic complementation with overlapping cosmid clones mapped to this locus. Transformation of mutant 13-20C2 with a cosmid carrying a insert that spanned the 3′ end of the locus (cosmid PSBMM94) did not rescue the temperature sensitive defect (Fig. 4A), while cosmid PSBLQ51 was able to confer resistance (Gene #1 is not fully encompassed), implicating three genes that encode, alpha glucosidase II (Gene #2, TgG2a), actin-related protein 4a (Gene #3, TgARP4a) or a hypothetical protein (Gene #4, TGGT1_002030).
Figure 4. Identification of the defective gene in mutant 13-20C2 by genetic complementation.
(A) Results of mutant complementation with cosmid libraries and individual cosmids are diagrammed. Mutant 13-20C2 was complemented with cosmid genomic library DNA. Sequence tags (black arrows) were recovered from temperature and drug resistant populations and mapped to a single locus on chromosome III (665,366bp to 711,113bp). To confirm the complementing locus, the original mutant 13-20C2 was complemented with DNA from two cosmid clones, PSBMM94 (no growth at 40°C) that spans the right side of the locus and cosmid PSBLQ51 (100% growth at 40°C compared to growth at 34°C), which spans the center of the locus and includes genes TGGT1_002010 (Gene #2, TgG2a), TGGT1_002020 (Gene #3, TgARP4a), and TGGT1_002030 (Gene #4, hypothetical). To narrow the gene list of cosmid PSBLQ51 further, mutant 13-20C2 was complemented with cosmid DNA bearing either a deletion in Gene #2 (TgG2a) or Gene #3 (TgARP4a). Mutant parasites transfected with individual cosmids were first selected in bleomycin and the drug resistant populations tested for temperature resistant growth by plaque assay. Ability to form plaques is expressed as a percentage of the plaques formed at 40°C versus parallel controls grown at 34°C. To test the ability of genomic fragments to complement, DNA fragments spanning the TgARP4a locus including promoter and 3′-UTR sequences were amplified from mutant 13-20C2 or parental genomic DNA. (B) Sequencing of TgARP4a cDNA from mutant 13-20C2 and parental RHΔhxgprt parasites identified a single transition mutation at 1,862bp (T/C) in the coding sequence of the tsTgAPR4a resulting in a change of isoleucine to threonine at residue 621 of the predicted TgARP4a coding sequence. Mutated codon is shown boxed. (C) Alignment of the helical region that contains the I621T mutation of tsTgARP4a with selected orthologs (Plasmodium falciparum PF14_0218; Saccharomyces cereviseae SacArp4p; Homo sapiens HmALP6B). Hydrophobic residue corresponding to isoleucine 621 (starred column) in TgARP4a is conserved in the analyzed orthologs. Note that the 621-residue in the helix is preserved by conservative substitutions isoleucine and valine. Identical residues as compared to TgARP4a are indicated in the ortholog sequences as a period. (D, E) Predicted folding of the polypeptides I383-L432 (Sc_Arp4) and F614-L665 (TgARP4a). Crystal structure of yeast protein (pdb:3QB0) was used to model a part of the Subdomain 3 of the actin fold of TgARP4a contain the temperature sensitive mutation. The alpha helix is shown in blue and the corresponding residue I621 in TgARP4a to the V385 in yeast Arp4p are shown in red. Bulky side chains of the residues of three α-helixes and one β-sheet that form a hydrophobic pocket are shown and corresponding residues are labeled on the outside. Note the position of I621 in TgARP4a may interact with a hydrophobic pocket that would like be altered by the mutation to threonine in the ts-TgARP4a protein. (F) Ribbon drawing of the helix I617-D626 showing the precise location of the I621T change.
To further resolve the locus, we modified cosmid PSBLQ51 by recombineering in E.coli [22] to individually delete the TgG2a and TgARP4a genes in the cosmid insert by replacement with a chloramphenycol acetyltransferase (CAT) cassette (Fig. 4A). Transformation of the cosmid PSBLQ51-ΔTgG2a into mutant 13-20C2 successfully rescued mutant 13-20C2 parasites grown at 40°C, whereas cosmid PSBLQ51-ΔTgARP4a failed to rescue the temperature defect indicating that the wtTgARP4a allele was responsible for conferring temperature resistance. To resolve whether wtTgARP4a was rescuing a defective tsTgARP4a allele in mutant 13-20C2 or was a generalized suppressor of the defect, we first tested the ability of locus-specific DNA amplified from parental RHΔhxgprt or the mutant 13-20C2 strains to rescue the temperature defect. Mutant 13-20C2 parasites transfected with DNA fragments encompassing wtTgARP4a, but not fragments carrying the tsTgARP4a gene formed viable plaques at 40°C (Fig. 4A), which provided additional evidence that the defect in mutant 13-20C2 is due to a mutation in TgARP4a. Finally, sequencing TgARP4a cDNA fragments transcribed from parental strain and mutant 13-20C2 mRNAs identified a single pyrimidine transition (T to C) that introduces a non-synonymous mutation (I621T) in the coding sequence of tsTgARP4a (Fig. 4B).
Structural alignment places the isoleucine at position 621 in the middle of an alpha helixal region (I617-D626) that is conserved not only in several apicomplexan ARP4a orthologs but also in mammalian and yeast ARP4a (Fig. 4C). We modeled TgARP4a protein into a template of the recently resolved crystal structure of the yeast Arp4p [30]. Six major insertions can be identified. (Fig. S1). The alpha helix 617-626 is a part of the subdomain 3 of the actin fold. It forms a hydrophobic pocket with two other alpha helixes and one beta sheet (Fig. 4D & E). Side-chains of hydrophobic residues face the internal chamber and stabilize a folded state of this region. Based on the resolved structure of the yeast Arp4p this part of subdomain 3 contributes to the formation of the interdomain cleft that holds adenosine group of ATP [31]. It is likely that the mutation that changes a bulky hydrophobic residue (isoleucine 621) to a polar residue threonine in the TgARP4a-ts allele causes a distortion to the bundle structure and my affect the positioning of ATP (Fig. 4F).
3.4 Nuclear localization of TgARP4a is required to complete mitosis in Toxoplasma
Actin-related proteins have been studied in animal and yeast models with as many as ten family members found in higher eukaryotes. A subset of actin-related proteins (ARP4-10) are localized to the nucleus, and it is this class whose function is least understood. Diverse mechanisms involving chromatin remodeling, histone packaging, transcriptional regulation, and spindle assembly have been genetically linked to actin-related proteins, although the precise molecular role of actin-related factors in these processes remains to be fully explained. In the Apicomplexa, three conventional actin-related genes have been identified, ARP1, 4, and 6 [32]. To further investigate the role of TgARP4a in T. gondii replication, we explored the changes in expression and cellular localization of the ts mutant and wt isoforms of TgARP4a via ectopic protein expression utilizing an epitope tag for visualization. Fusion of FKBP-3xmyc (DDmyc) domain to the N-terminus of full lengthTgARP4a was chosen for these experiments because C-terminal additions to TgARP4a were not tolerated by tachyzoites. FKBP fusion destabilizes cytoplasmic proteins and targets them to the proteasome [33, 34]. Addition of the small molecule ligand Shield-1 rescues the protein from this fate. To confirm DDmyc-wtTgARP4a was functional, we transfected mutant 13-20C2 parasites with this construct and demonstrated the wild type fusion protein could complement the temperature defect of this mutant strain (data not shown). Interestingly, modulation of the recombinant protein stability with ligand Shield 1 did not prevent rescue at 40°C because the lower levels of DDmyc-wtTgARP4a (without Shield1) were sufficient to meet the cell cycle requirement for this essential factor as was easily detected by Western blot (Fig. 5 Western total lysate). This was not a surprising result as native TgARP4a mRNA is expressed in the lower ~30% abundance class, while the DHFR-TS or tubulin promoter driving these ectopic constructs flanks a genetic locus that encodes an mRNA in the >80% class (and necessary to detect TgARP4a in IFAs). Despite significant overexpression of DDmyc-wtTgARP4a, the fusion protein was cell cycle regulated (Fig. S2) with peak expression in S and mitotic phases. The protein was detectable in G1 parasites, although at much lower levels (Fig. S2).
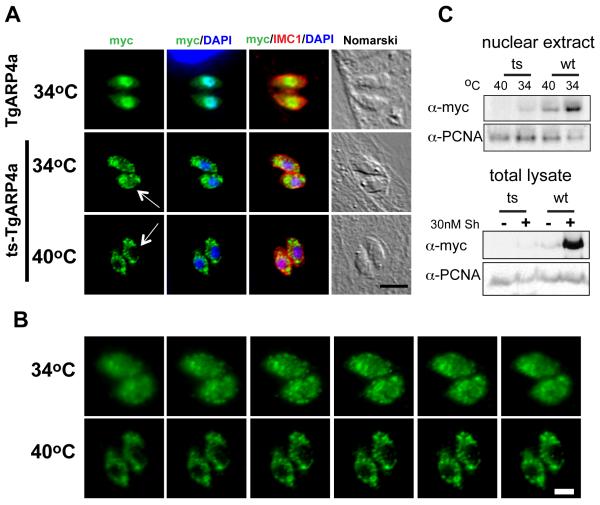
Figure 5. The I621T mutation in tsTgARP4a results in mis-localization.
(A) In order to better understand the temperature defect caused by tsTgARP4a, we compared the subcellular localization of this mutant protein to the wild type isoform over expressed in the parental strain. Transgenic clones expressing DDmyc-wtTgARP4a showed tight nuclear localization at either temperature (34°C expression shown only in top panels); green=anti-myc stain. By contrast, DDmyc-tsTgARP4a was primarily cytoplasmic with some nuclear localization at the non-permissive temperature of 34°C (middle panels). Fluorescent micrographs of DDmyc-tsTgARP4a transgenic parasites were taken using >10-fold longer time exposures. Magnification bar (5 μm) is indicated. (B) Image series scanning through representative 34°C and 40°C vacuoles demonstrated the absence of detectable nuclear DDmyc-tsTgARP4a at the non-permissive temperature. Magnification bar (2 μm) is indicated. (C) Upper panels: To assess the affect of temperature on the levels of nuclear ts or wtTgARP4a, the transgenic clones were induced with 30nM Shield1 at 34°C or 40°C and nuclear extracts prepared as described in the Material and Methods. Nuclear levels of the fusion proteins were detected by Western blots stained with anti-myc antibody. Note the levels of either TgARP4a isoform are affected by higher temperature with the tsTgARP4a undetectable in the parasite nuclear extracts at 40°C similar to the IFA results above. Lower panels: Protein levels in total lysates of transgenic parasites expressing DDmyc-wtTgARP4a or DDmyc- tsTgARP4a fusion proteins grown in the presence or absence of 30 nM Shield1 (Sh) at 34°C were subjected to the western blot analysis using anti-myc antibody. The Western blot membrane was secondarily probed with anti-PCNA1 antibody to ensure equal loading.
Nuclear localization of eukaryotic ARP4 has been well documented in numerous studies [35-37] and similar to other eukaryotic ARP4 orthologs, we found DDmyc-wtTgARP4a was predominantly nuclear (Fig. 5A and S2). By contrast, DDmyc-tsTgARP4a was mostly detected in the cytoplasm (Fig. 5A and B) with some DDmyc-tsTgARP4a detected in the nucleus at 34°C (see arrows Fig. 5A). These observations indicated under the permissive temperature sufficient mutant protein might be properly folded and delivered to the nucleus. The shift to 40°C abolished even this residual nuclear transfer of the mutated protein. Serial image stacks through the nucleus confirmed the mutant protein was not detectable in the nucleus at the restricted temperature (Fig. 5B) and the subcellular difference could be established in nuclear extracts (Fig. 5C). Western analysis of these two fusion proteins in nuclear extracts showed levels of DDmyc-tsTgARP4a were also affected by temperature with higher protein present in total lysates at 34°C as compared to parasites grown at 40°C (Fig. 5C).
4.0 Discussion
Here we describe a novel conditional mutant of T. gondii that rapidly growth arrests at higher temperature. Chromosome mis-segregation and abnormal nuclear division is the predominant phenotype of this mutant, which classifies this clone in a broad category of conditional mutants with defects in mitosis isolated in a recent chemical mutagenesis screen [18]. A nucleotide transition that alters a single amino acid within the helix adjacent the conserved actin domain of TgARP4a appears to be responsible for the mitotic failure in this mutant. This mutation dramatically reduces overall protein levels and causes a mis-localization of the essential TgARP4a factor at higher temperatures leading to a lethal result for this strain. Protein alignment of apicomplexa ARP4a orthologs with yeast and human revealed the introduction of extended peptide inserts in the Toxoplasma and Neospora proteins that increased the TgARP4a to 727 amino acids from the smaller yeast Arp4p (489aa). The function of these inserts is not known, although analysis of TgARP4a structure by PONDR VLXT, VL3 and PONDR-FIT algorithms predicts the extra peptide sequence might increase the disorder of TgARP4a tertiary structure (data not shown). One protein domain (491-547aa) is of a particular interest where the combination of disorder with clustering of positive charged residues indicates the presence of nuclear localization signals (NLS) similar to the NLS recently described in Toxoplasma histone acetyltransferase TgGCN5 [38]. Placement of the I621T mutation in the protein structure revealed a location in a conserved domain of the protein with the isoleucine hydrophobic residue preserved from yeast to human. The loss of hydrophobicity in this short a-helix might affect the overall stability of the protein or it could affect the accessibility of the putative NLS. Additional studies will be required to determine whether the folding defect affects nuclear import or results in the inability of the protein to pass the ER quality control system.
TgARP4 belongs to the actin family of proteins that have an unusual constitution in the Apicomplexa. In most eukaryotes a core set of eight ARP factors are recognized by their overall similarity to actin and the presence of an ATP/ADP binding pocket termed the actin fold [39, 40]. While similar in sequence, ARP factors serve diverse functions in cells that involve regulating the actin cytoskeleton, controlling movement of intracellular cargo, or altering the topology of chromosome structure (for review see refs. [41, 42]). The prototypical actin-related protein, ARP1, which serves an essential role in the dynactin complex responsible for vesicular trafficking, is conserved across all eukaryotes including the Apicomplexa [40]. Similarly other ARP family members (e.g. ARP4-6, 8) that have nuclear functions are also conserved in Apicomplexa notwithstanding there are fewer numbers; ARP4 and 6 are present in the Apicomplexa, while ARP5 and 8 are missing [32]. Most surprising is the lack of genes encoding the ARP2/3 pair known to control actin polymerization in many eukaryotes [40]. In the absence of conventional ARPs, apicomplexa parasites may rely on a novel family of actin-like proteins (ALP) to fulfill specific roles in actin motility and to serve tasks more specific to apicomplexan cellular biology [32, 43, 44].
The occurrence of conventional nuclear ARP factors, (ARP4/6) in the Apicomplexa suggests the role for ARPs in regulating parasite chromatin may have ancient origins. In yeast, ARP4 is associated with two chromatin-remodeling complexes INO80 and SWR-C where interaction is primarily with the major ATPase subunit of the SWI2/SNF2 family and conventional actin [41]. ARP4 also participates in the covalent modification of histones as a component of NuA4 histone acetyltransferase complex. Epigenetic control of gene expression is now well established in the Apicomplexa [45, 46] and several histone modifying enzymes of GNAT and MYST families along with a complex containing histone diacetyltransferase (HDAC) have recently been described [47-50]. Less studied is chromatin modifying complexes where ARP4 is known to participate, although components of the NuA4, INO80 and SWR-C complexes are encoded in the Apicomplexa. In Toxoplasma and Plasmodium all core subunits of the complexes including SWI/SNF2 family ATPases (INO80complex and SWR-C complex), RuvB helicases and histone acetyltransferases (NuA4 complex) are present (see Fig. S3 for a full list). Interestingly, there is some variation in nuclear ARP components with Toxoplasma (also Neospora and Theileria) encoding two isoforms, ARP4a and ARP4b, while a single ARP4 is present in Plasmodium spp and many other Apicomplexa [32]. Dual ARP4 isoforms are less common, and when they occur they can have redundant function a description that applies to tissue-specific isoforms of ARP4 in human cells (BAF53A and BAF53B) with similar roles in the different tissues [51]. Toxoplasma ARP4a (chrIII) and ARP4b (chrVIIb) are highly similar (31% identity/45% similarity), and are expressed by equal mRNA levels (not shown), however, our genetic experiments indicate these isoforms likely perform different, non-overlapping functions as the presence of the wt- TgARP4b allele was not sufficient to overcome the lethal affects of a mutant TgARP4a. It is possible the wealth of chromatin remodeling components in Toxoplasma that exceeds the typical complement of other eukaryotes could relate to the retention of independent APR4s. Toxoplasma encodes three ATPases of the SWI2/SNF2 family and three RuvB helicases, while only one for each requires for INO80 and SWR-C complexes (Fig. S3).
It is documented that ARP4 forms a heterodimer with β-actin and is postulated to bring about association of the complex with chromatin to generate appropriate chromatin architecture. This activity is thought to influence the state of gene expression [52-55]. Another potential function for ARP4-complexes appears to involve kinetochore attachment uncovered by forward genetic studies in yeast [56-59]. Recent studies of temperature-sensitive arp4 mutants demonstrate primary defects in G2/M phase functions that are associated with spindle abnormalities [56]. This role for ARP4 appears to have a restricted chromosome context as this factor is associated with centromeres throughout the cell cycle. Based on the loss of protein interaction in arp4 mutants, it is speculated that ARP4-containing complexes are required to achieve the proper chromatin structure at centromeres for efficient binding of the inner kinetochore proteins [56] and that this role is independent of histone acetylation [60]. Clarifying the competing functions for ARP4 in gene expression and cell cycle will require more study, however, the discoveries described here provide solid evidence that a vital role in chromosome segregation is a conserved function for ARP4-containing complexes in the early branching Apicomplexa.
Mitosis in the Apicomplexa has novel features stemming from an intimate relationship with daughter budding that initiates in late S-phase and continually builds while mitotic events unfold. To orchestrate a process of such complexity with timely and spatial precision, the idea of physical constrains connecting the nuclear division apparatus with daughter bud scaffold had been proposed (for review see [7]). Thus, structural integrity of the parasite budding cytoskeleton is suggested to be an important factor to the feedback mechanisms that coordinate replication in apicomplexan parasites. We do not yet understand the details of these mechanisms or the critical points of interdependence. Tachyzoites have two microtubule organizing centers (MTOC), associated with subpellicular microtubules that shape the internal daughter parasite, and the spindle apparatus (nucleus), which serves chromosome segregation. The existence of multiple MTOC makes possible the division of labor between the tasks associated with nuclear division and budding events. Experiments with microtubule depolymerization agent oryzalin and stabilizing agent taxol call into question the level of coordination between the principal tasks of these organizing centers [7, 29, 61]. When tachyzoites are exposed to oryzalin both cycles of chromosome replication and budding are abnormal with evidence of uncontrolled reduplication [29]. In the pursuit of answers to how the parasite cell cycle is regulated, it is clear conditional cell cycle mutants will be an important resource to help establish the framework of these relationships. Mitotic mutant 13-20C2 with more subtle defect in the innermost workings of chromosome segregation offers some insight to these processes. Unlike oryzalin treatment, the orientation between the centrosome and budding center is maintained in this mutant during growth arrest at the non-permissive temperature. As a consequence, centrosome numbers are normal and organellar biogenesis and segregation follows the regular path even if packaging of nuclear material into daughter parasites fails. Key to the residual cell cycle coordination is the preservation of the spindle pole, which duplicates and segregates at the higher temperature and remains oriented to the two MTOC, centrosome and apical complex. Similar evidence for the physical context of cell cycle coordination has been reported in other mitotic mutants [18, 62] suggesting we might expect the biochemical components of cell cycle coordination to be tethered to these structures. The continued characterization of conditional cell cycle mutants as well as studies of the critical checkpoint factors that govern mitosis in these parasites should help us further resolve the key strategies the Apicomplexa employ to coordinate their peculiar cell cycles.
Supplementary Material
Figure S1. Structural alignment of TgARP4a and S. cereviseae Arp4p.
Amino acid sequence of TgARP4a was modeled onto the resolved crystal structure of the yeast ortholog Arp4p (3QB0 chain A) using SWISS-PRO software [63]. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modeling [63]. Predicted folding into beta-sheet structure is indicated by arrow and alpha helix is shown by sequence of letter h. Modeling revealed a high structural resemblance between the two orthologs despite only 17.8% similarity in primary amino acid sequence. The major differences between the two proteins are due to multiple inserts (loops) that are shown in the boxes.
Figure S3. Putative orthologs of known ARP4-associated complexes.
Genomes of T. gondii and P. falciparum were examined for a presence of histone modifying complex Nu4A and chromatin remodeling complexes INO80 and SWR-C found in yeast. Results of the pBLAST search using yeast complex subunits as a query are summarized in the table. E-values higher then -16 were considered non-significant and indicated as none. Footnotes a=Abbreviation of S. cerevisiae subunit obtained from http://yeastgenome.org and corresponding accession number in NCBI database, b=Gene accession number for T. gondii TGME49 strain obtained from http://toxodb.org, c=E-value of T. gondii versus S. cerevisiae protein, d=Gene accession number for P. falciparum 3D7 strain obtained from http://plasmodb.org, e=E-value of P. falciparum versus S. cerevisiae protein, f=Gene product description from http://toxodb.org
Figure S2. Cell cycle regulation of DDmyc-wtTgARP4a fusion protein.
A transgenic clone expressing DDmyc-wtTgARP4a was evaluated for cell cycle expression. The promoter driving this construct was derived from the dihydrofolate reductase gene promoter (>80% in abundance class for mRNA expression). Parasites were grown in the presence of 30nM Shield1, fixed and stained with anti-myc (green), anti-IMC3 (red) and stained with DAPI (blue). Cell cycle phases are estimated by the relative parasite size, presence or absence of internal daughter, nuclear size and DAPI staining intensity. Note the concentration of the fusion protein in the parasite nucleus with minimal cytoplasmic staining. The fusion protein was nearly undetectable in G1 phase parasites indicating a dynamic post-transcriptional mechanism may be involved in the cell cycle regulation of this protein.
Acknowledgements
This work was supported by grants from the National Institutes of Health to MWW (R01-AI077662 and R01-AI089885. T. gondii genomic and/or cDNA sequence data were accessed via http://ToxoDB.org.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6.0 References
- 1.Dubey JP, Carpenter JL. Toxoplasma gondii-like schizonts in the tracheal epithelium of a cat. J Parasitol. 1991;77:792–6. [PubMed] [Google Scholar]
- 2.Canning EU, Sinden RE. The organization of the ookinete and observations on nuclear division in oocysts of Plasmodium berghei. Parasitology. 1973;67:29–40. doi: 10.1017/s0031182000046266. [DOI] [PubMed] [Google Scholar]
- 3.Speer CA, Dubey JP. An ultrastructural study of first- and second-generation merogony in the coccidian Sarcocystis tenella. J Protozool. 1981;28:424–31. doi: 10.1111/j.1550-7408.1981.tb05314.x. [DOI] [PubMed] [Google Scholar]
- 4.Striepen B, Jordan CN, Reiff S, van Dooren GG. Building the perfect parasite: Cell division in apicomplexa. PLoS Pathog. 2007;3:e78. doi: 10.1371/journal.ppat.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheffield HG, Melton ML. The fine structure and reproduction of Toxoplasma gondii. J Parasitol. 1968;54:209–26. [PubMed] [Google Scholar]
- 6.Ogino N, Yoneda C. The fine structure and mode of division of Toxoplasma gondii. Arch Ophthalmol. 1966;75:218–27. doi: 10.1001/archopht.1966.00970050220015. [DOI] [PubMed] [Google Scholar]
- 7.Gubbels MJ, White M, Szatanek T. The cell cycle and Toxoplasma gondii cell division: Tightly knit or loosely stitched? Int J Parasitol. 2008;38:1343–58. doi: 10.1016/j.ijpara.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Radke JR, Striepen B, Guerini MN, Jerome ME, Roos DS, White MW. Defining the cell cycle for the tachyzoite stage of Toxoplasma gondii. Mol Biochem Parasitol. 2001;115:165–75. doi: 10.1016/s0166-6851(01)00284-5. [DOI] [PubMed] [Google Scholar]
- 9.Radke JR, White MW. A cell cycle model for the tachyzoite of Toxoplasma gondii using the herpes simplex virus thymidine kinase. Mol Biochem Parasitol. 1998;94:237–47. doi: 10.1016/s0166-6851(98)00074-7. [DOI] [PubMed] [Google Scholar]
- 10.Swedlow JR, Hu K, Andrews PD, Roos DS, Murray JM. Measuring tubulin content in Toxoplasma gondii: A comparison of laser-scanning confocal and wide-field fluorescence microscopy. Proc Natl Acad Sci U S A. 2002;99:2014–9. doi: 10.1073/pnas.022554999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irvin AD, Ocama JG, Spooner PR. Cycle of bovine lymphoblastoid cells parasitised by Theileria parva. Res Vet Sci. 1982;33:298–304. [PubMed] [Google Scholar]
- 12.Fujishima M. Microspectrophotometric and autoradiographic study of the timing and duration of pre-meiotic DNA synthesis in Paramecium caudatum. J Cell Sci. 1983;60:51–65. doi: 10.1242/jcs.60.1.51. [DOI] [PubMed] [Google Scholar]
- 13.Hu K, Roos DS, Angel SO, Murray JM. Variability and heritability of cell division pathways in Toxoplasma gondii. J Cell Sci. 2004;117:5697–705. doi: 10.1242/jcs.01494. [DOI] [PubMed] [Google Scholar]
- 14.Baker DJ, Dawlaty MM, Galardy P, van Deursen JM. Mitotic regulation of the anaphase-promoting complex. Cell Mol Life Sci. 2007;64:589–600. doi: 10.1007/s00018-007-6443-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrissette NS, Sibley LD. Disruption of microtubules uncouples budding and nuclear division in Toxoplasma gondii. J. Cell Sci. 2002;115:1017–1025. doi: 10.1242/jcs.115.5.1017. [DOI] [PubMed] [Google Scholar]
- 16.Shaw MK, Roos DS, Tilney LG. DNA replication and daughter cell buddingare not tightly linked in the protozoan parasite Toxoplasma gondii. Microbes Infect. 2001;3:351–62. doi: 10.1016/s1286-4579(01)01392-2. [DOI] [PubMed] [Google Scholar]
- 17.de Felipe MM Conde, Lehmann MM, Jerome ME, White MW. Inhibition of Toxoplasma gondii growth by pyrrolidine dithiocarbamate is cell cycle specific and leads to population synchronization. Mol Biochem Parasitol. 2008;157:22–31. doi: 10.1016/j.molbiopara.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gubbels MJ, Lehmann M, Muthalagi M, Jerome ME, Brooks CF, Szatanek T, Flynn J, Parrot B, Radke J, Striepen B, White MW. Forward genetic analysis of the apicomplexan cell division cycle in Toxoplasma gondii. PLoS Pathog. 2008;4:e36. doi: 10.1371/journal.ppat.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roos DS, Donald RG, Morrissette NS, Moulton AL. Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods in cell biology. 1994;45:27–63. doi: 10.1016/s0091-679x(08)61845-2. [DOI] [PubMed] [Google Scholar]
- 20.Gaji RY, Behnke MS, Lehmann MM, White MW, Carruthers VB. Cell cycle-dependent, intercellular transmission of Toxoplasma gondii is accompanied by marked changes in parasite gene expression. Molecular microbiology. 2011;79:192–204. doi: 10.1111/j.1365-2958.2010.07441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Striepen B, Soldati D. Genetic manipulation of Toxoplasma gondii. In: Weiss LM, Kim K, editors. Perspectives and methods. Academic Press; London: 2007. pp. 391–418. [Google Scholar]
- 22.Brooks CF, Johnsen H, van Dooren GG, Muthalagi M, Lin SS, Bohne W, Fischer K, Striepen B. The Toxoplasma apicoplast phosphate translocator links cytosolic and apicoplast metabolism and is essential for parasite survival. Cell Host Microbe. 2010;7:62–73. doi: 10.1016/j.chom.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herm-Gotz A, Agop-Nersesian C, Munter S, Grimley JS, Wandless TJ, Frischknecht F, Meissner M. Rapid control of protein level in the apicomplexan Toxoplasma gondii. Nature methods. 2007;4:1003–5. doi: 10.1038/nmeth1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guerini MN, Que X, Reed SL, White MW. Two genes encoding unique proliferating-cell-nuclear- antigens are expressed in Toxoplasma gondii. Molecular and biochemical parasitology. 2000;109:121–31. doi: 10.1016/s0166-6851(00)00240-1. [DOI] [PubMed] [Google Scholar]
- 25.Pfefferkorn ER, Pfefferkorn LC. Quantitative studies of the mutagenesis of Toxoplasma gondii. J Parasitol. 1979;65:364–70. [PubMed] [Google Scholar]
- 26.White MW, Jerome ME, Vaishnava S, Guerini M, Behnke M, Striepen B. Genetic rescue of a Toxoplasma gondii conditional cell cycle mutant. Molecular microbiology. 2005;55:1060–71. doi: 10.1111/j.1365-2958.2004.04471.x. [DOI] [PubMed] [Google Scholar]
- 27.Nasmyth K. A prize for proliferation. Cell. 2001;107:689–701. doi: 10.1016/s0092-8674(01)00604-3. [DOI] [PubMed] [Google Scholar]
- 28.Gubbels MJ, Vaishnava S, Boot N, Dubremetz JF, Striepen B. A morn-repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. Journal of cell science. 2006;119:2236–45. doi: 10.1242/jcs.02949. [DOI] [PubMed] [Google Scholar]
- 29.Morrissette NS, Sibley LD. Disruption of microtubules uncouples budding and nuclear division in Toxoplasma gondii. J Cell Sci. 2002;115:1017–25. doi: 10.1242/jcs.115.5.1017. [DOI] [PubMed] [Google Scholar]
- 30.Fenn S, Breitsprecher D, Gerhold CB, Witte G, Faix J, Hopfner KP. Structural biochemistry of nuclear actin-related proteins 4 and 8 reveals their interaction with actin. The EMBO journal. 2011;30:2153–66. doi: 10.1038/emboj.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kabsch W, Holmes KC. The actin fold. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 1995;9:167–74. doi: 10.1096/fasebj.9.2.7781919. [DOI] [PubMed] [Google Scholar]
- 32.Gordon JL, Sibley LD. Comparative genome analysis reveals a conserved family of actin-like proteins in apicomplexan parasites. BMC Genomics. 2005;6:179. doi: 10.1186/1471-2164-6-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daher W, Plattner F, Carlier MF, Soldati-Favre D. Concerted action of two formins in gliding motility and host cell invasion by Toxoplasma gondii. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herm-Gotz A, Agop-Nersesian C, Munter S, Grimley JS, Wandless TJ, Frischknecht F, Meissner M. Rapid control of protein level in the apicomplexan Toxoplasma gondii. Nat Methods. 2007;4:1003–5. doi: 10.1038/nmeth1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harata M, Oma Y, Tabuchi T, Zhang Y, Stillman DJ, Mizuno S. Multiple actin-related proteins of Saccharomyces cerevisiae are present in the nucleus. J Biochem. 2000;128:665–71. doi: 10.1093/oxfordjournals.jbchem.a022799. [DOI] [PubMed] [Google Scholar]
- 36.Kandasamy MK, McKinney EC, Meagher RB. Cell cycle-dependent association of arabidopsis actin-related proteins atarp4 and atarp7 with the nucleus. Plant J. 2003;33:939–48. doi: 10.1046/j.1365-313x.2003.01691.x. [DOI] [PubMed] [Google Scholar]
- 37.Meagher RB, Kandasamy MK, McKinney EC, Roy E. Chapter 5. Nuclear actin-related proteins in epigenetic control. Int Rev Cell Mol Biol. 2009;277:157–215. doi: 10.1016/S1937-6448(09)77005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dixon SE, Stilger KL, Elias EV, Naguleswaran A, Sullivan WJ., Jr A decade of epigenetic research in Toxoplasma gondii. Mol Biochem Parasitol. 2010 doi: 10.1016/j.molbiopara.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poch O, Winsor B. Who’s who among the Saccharomyces cerevisiae actin-related proteins? A classification and nomenclature proposal for a large family. Yeast. 1997;13:1053–8. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1053::AID-YEA164>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 40.Frankel S, Mooseker MS. The actin-related proteins. Current opinion in cell biology. 1996;8:30–7. doi: 10.1016/s0955-0674(96)80045-7. [DOI] [PubMed] [Google Scholar]
- 41.Blessing CA, Ugrinova GT, Goodson HV. Actin and arps: Action in the nucleus. Trends Cell Biol. 2004;14:435–42. doi: 10.1016/j.tcb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 42.Dion V, Shimada K, Gasser SM. Actin-related proteins in the nucleus: Life beyond chromatin remodelers. Curr Opin Cell Biol. 2010;22:383–91. doi: 10.1016/j.ceb.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 43.Gordon JL, Beatty WL, Sibley LD. A novel actin-related protein is associated with daughter cell formation in Toxoplasma gondii. Eukaryot Cell. 2008;7:1500–12. doi: 10.1128/EC.00064-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gordon JL, Buguliskis JS, Buske PJ, Sibley LD. Actin-like protein 1 (alp1) is a component of dynamic, high molecular weight complexes in Toxoplasma gondii. Cytoskeleton (Hoboken) 2010;67:23–31. doi: 10.1002/cm.20414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hakimi MA, Deitsch KW. Epigenetics in apicomplexa: Control of gene expression during cell cycle progression, differentiation and antigenic variation. Curr Opin Microbiol. 2007;10:357–62. doi: 10.1016/j.mib.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 46.Sullivan WJ, Jr., Hakimi MA. Histone mediated gene activation in Toxoplasma gondii. Mol Biochem Parasitol. 2006;148:109–16. doi: 10.1016/j.molbiopara.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 47.Bhatti MM, Sullivan WJ., Jr Histone acetylase gcn5 enters the nucleus via importin-alpha in protozoan parasite Toxoplasma gondii. J Biol Chem. 2005;280:5902–8. doi: 10.1074/jbc.M410656200. [DOI] [PubMed] [Google Scholar]
- 48.Dixon SE, Bhatti MM, Uversky VN, Dunker AK, Sullivan WJ., Jr Regions of intrinsic disorder help identify a novel nuclear localization signal in Toxoplasma gondii histone acetyltransferase tggcn5-b. Mol Biochem Parasitol. 2011;175:192–5. doi: 10.1016/j.molbiopara.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saksouk N, Bhatti MM, Kieffer S, Smith AT, Musset K, Garin J, Sullivan WJ, Jr., Cesbron-Delauw MF, Hakimi MA. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol Cell Biol. 2005;25:10301–14. doi: 10.1128/MCB.25.23.10301-10314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith AT, Tucker-Samaras SD, Fairlamb AH, Sullivan WJ., Jr Myst family histone acetyltransferases in the protozoan parasite Toxoplasma gondii. Eukaryot Cell. 2005;4:2057–65. doi: 10.1128/EC.4.12.2057-2065.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olave IA, Reck-Peterson SL, Crabtree GR. Nuclear actin and actin-related proteins in chromatin remodeling. Annu Rev Biochem. 2002;71:755–81. doi: 10.1146/annurev.biochem.71.110601.135507. [DOI] [PubMed] [Google Scholar]
- 52.Jiang YW, Stillman DJ. Epigenetic effects on yeast transcription caused by mutations in an actin- related protein present in the nucleus. Genes & development. 1996;10:604–19. doi: 10.1101/gad.10.5.604. [DOI] [PubMed] [Google Scholar]
- 53.Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the swi/snf-like baf complex to chromatin after t lymphocyte receptor signaling. Cell. 1998;95:625–36. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 54.Gorzer I, Schuller C, Heidenreich E, Krupanska L, Kuchler K, Wintersberger U. The nuclear actin-related protein act3p/arp4p of Saccharomyces cerevisiae is involved in transcription regulation of stress genes. Molecular microbiology. 2003;50:1155–71. doi: 10.1046/j.1365-2958.2003.03759.x. [DOI] [PubMed] [Google Scholar]
- 55.Harata M, Zhang Y, Stillman DJ, Matsui D, Oma Y, Nishimori K, Mochizuki R. Correlation between chromatin association and transcriptional regulation for the act3p/arp4 nuclear actin-related protein of Saccharomyces cerevisiae. Nucleic acids research. 2002;30:1743–50. doi: 10.1093/nar/30.8.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogiwara H, Ui A, Kawashima S, Kugou K, Onoda F, Iwahashi H, Harata M, Ohta K, Enomoto T, Seki M. Actin-related protein arp4 functions in kinetochore assembly. Nucleic acids research. 2007;35:3109–17. doi: 10.1093/nar/gkm161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang H, Richardson DO, Roberts DN, Utley R, Erdjument-Bromage H, Tempst P, Cote J, Cairns BR. The yaf9 component of the swr1 and nua4 complexes is required for proper gene expression, histone h4 acetylation, and htz1 replacement near telomeres. Molecular and cellular biology. 2004;24:9424–36. doi: 10.1128/MCB.24.21.9424-9436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Le Masson I, Yu DY, Jensen K, Chevalier A, Courbeyrette R, Boulard Y, Smith MM, Mann C. Yaf9, a novel nua4 histone acetyltransferase subunit, is required for the cellular response to spindle stress in yeast. Molecular and cellular biology. 2003;23:6086–102. doi: 10.1128/MCB.23.17.6086-6102.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krogan NJ, Baetz K, Keogh MC, Datta N, Sawa C, Kwok TC, Thompson NJ, Davey MG, Pootoolal J, Hughes TR, Emili A, Buratowski S, Hieter P, Greenblatt JF. Regulation of chromosome stability by the histone h2a variant htz1, the swr1 chromatin remodeling complex, and the histone acetyltransferase nua4. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13513–8. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Babiarz JE, Halley JE, Rine J. Telomeric heterochromatin boundaries require nua4-dependent acetylation of histone variant h2a.Z in Saccharomyces cerevisiae. Genes & development. 2006;20:700–10. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shaw MK, Compton HL, Roos DS, Tilney LG. Microtubules, but not actin filaments, drive daughter cell budding and cell division in Toxoplasma gondii. J Cell Sci. 2000;113(Pt 7):1241–54. doi: 10.1242/jcs.113.7.1241. [DOI] [PubMed] [Google Scholar]
- 62.Radke JR, Gubbels MJ, Jerome ME, Radke JB, Striepen B, White MW. Identification of a sporozoite-specific member of the Toxoplasma sag superfamily via genetic complementation. Molecular microbiology. 2004;52:93–105. doi: 10.1111/j.1365-2958.2003.03967.x. [DOI] [PubMed] [Google Scholar]
- 63.Arnold K, Bordoli L, Kopp J, Schwede T. The swiss-model workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Structural alignment of TgARP4a and S. cereviseae Arp4p.
Amino acid sequence of TgARP4a was modeled onto the resolved crystal structure of the yeast ortholog Arp4p (3QB0 chain A) using SWISS-PRO software [63]. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modeling [63]. Predicted folding into beta-sheet structure is indicated by arrow and alpha helix is shown by sequence of letter h. Modeling revealed a high structural resemblance between the two orthologs despite only 17.8% similarity in primary amino acid sequence. The major differences between the two proteins are due to multiple inserts (loops) that are shown in the boxes.
Figure S3. Putative orthologs of known ARP4-associated complexes.
Genomes of T. gondii and P. falciparum were examined for a presence of histone modifying complex Nu4A and chromatin remodeling complexes INO80 and SWR-C found in yeast. Results of the pBLAST search using yeast complex subunits as a query are summarized in the table. E-values higher then -16 were considered non-significant and indicated as none. Footnotes a=Abbreviation of S. cerevisiae subunit obtained from http://yeastgenome.org and corresponding accession number in NCBI database, b=Gene accession number for T. gondii TGME49 strain obtained from http://toxodb.org, c=E-value of T. gondii versus S. cerevisiae protein, d=Gene accession number for P. falciparum 3D7 strain obtained from http://plasmodb.org, e=E-value of P. falciparum versus S. cerevisiae protein, f=Gene product description from http://toxodb.org
Figure S2. Cell cycle regulation of DDmyc-wtTgARP4a fusion protein.
A transgenic clone expressing DDmyc-wtTgARP4a was evaluated for cell cycle expression. The promoter driving this construct was derived from the dihydrofolate reductase gene promoter (>80% in abundance class for mRNA expression). Parasites were grown in the presence of 30nM Shield1, fixed and stained with anti-myc (green), anti-IMC3 (red) and stained with DAPI (blue). Cell cycle phases are estimated by the relative parasite size, presence or absence of internal daughter, nuclear size and DAPI staining intensity. Note the concentration of the fusion protein in the parasite nucleus with minimal cytoplasmic staining. The fusion protein was nearly undetectable in G1 phase parasites indicating a dynamic post-transcriptional mechanism may be involved in the cell cycle regulation of this protein.