Abstract
The envelope glycoprotein, GP64, of Autographa californica nucleopolyhedrovirus (AcMNPV) is necessary and sufficient for the acid-induced membrane fusion activity that is required for fusion of the budded virus (BV) envelope and the endosome membrane during virus entry. Infectivity of the budded virus (BV) is neutralized by AcV1, a monoclonal antibody (MAb) directed against GP64. Prior studies indicated that AcV1 recognizes a conformational epitope and does not inhibit virus attachment to the cell, but instead inhibits entry at a step following virus attachment. We found that AcV1 recognition of GP64 was lost upon exposure of GP64 to low pH (pH 4.5) and restored by returning GP64 to pH 6.2. In addition, the AcV1 epitope was lost upon denaturation of GP64 in SDS but the AcV1 epitope was restored by refolding the protein in the absence of SDS. Using truncated GP64 proteins expressed in insect cells, we mapped the AcV1 epitope to a 24 amino acid region in the central variable domain of GP64. When sequences within the mapped AcV1 epitope were substituted with a c-Myc epitope and the resulting construct was used to replace wt GP64 in recombinant AcMNPV viruses, the modified GP64 protein appeared to function normally. However, an anti c-Myc monoclonal antibody did not neutralize infectivity of those viruses. Because binding of the c-Myc MAb to the same site in the GP64 sequence did not result in neutralization, these studies suggest that AcV1 neutralization may result from a specific structural constraint caused by AcV1 binding and not simply by steric hindrance caused by antibody binding at this position in GP64.
Keywords: Baculovirus, GP64, AcV1, Epitope, Monoclonal antibody, Neutralizing
Introduction
The Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) is a large double-stranded DNA virus (approximately 134 kbp) that produces two virion phenotypes during the infection cycle (Miller, 1997). One virion phenotype, the occlusion-derived virion (ODV), is adapted for stability in the environment and propagation of infection from animal to animal through oral transmission and infection of the midgut epithelial cells. In contrast, the other virion phenotype, the budded virion (BV), is adapted for propagation of infection from cell to cell throughout the animal after infection is established in the midgut (Granados and Lawler, 1981; Keddie, Aponte, and Volkman, 1989; Keddie and Volkman, 1985; Monsma, Oomens, and Blissard, 1996; Volkman and Goldsmith, 1984; Volkman et al., 1984). The major envelope protein of the BV is known as GP64 (Hohmann and Faulkner, 1983; Volkman and Goldsmith, 1984; Volkman et al., 1984). GP64 is essential for cell-to-cell transmission of the virus in cell culture and in the infected animal (Monsma, Oomens, and Blissard, 1996). GP64 serves two major roles during virus entry. First, GP64 is involved in host cell receptor binding (Hefferon et al., 1999) although the cellular receptor has not yet been identified. Second, GP64 mediates the low-pH-triggered membrane fusion activity necessary for release of the nucleocapsid into the cytosol during entry by endocytosis (Blissard and Wenz, 1992; Kingsley et al., 1999; Markovic et al., 1998; Monsma and Blissard, 1995; Plonsky et al., 1999; Volkman and Goldsmith, 1985). GP64 is found in the membrane as a homotrimer (Oomens, Monsma, and Blissard, 1995) and it has been hypothesized that GP64 changes conformation in response to low pH. Aggregates of multiple GP64 trimers have been identified in the membrane during membrane fusion (Markovic et al., 1998). For many other well-studied viral membrane fusion proteins, triggering of conformational changes in the fusion protein is believed to initiate the membrane fusion process. In one of the best current models, conformational changes in the trimeric fusion protein are believed to result in translocation and subsequent interaction of two separate alpha helical domains, forming a six-helix bundle structure in the trimeric fusion protein (Kielian and Rey, 2006). While it is not known whether such a structure forms during fusion mediated by GP64, it was previously demonstrated that a predicted alpha helix and an upstream hydrophobic region in the GP64 protein were important for membrane fusion activity of GP64 (Kingsley et al., 1999; Monsma and Blissard, 1995).
A monoclonal antibody, AcV1 (Hohmann and Faulkner, 1983), that neutralizes infectivity of AcMNPV BV is important as a tool for understanding the function of the GP64 protein. AcV1 is directed against the GP64 protein and pre-incubation of BV with AcV1 results in reductions in BV infectivity by 3–4 orders of magnitude (Hohmann and Faulkner, 1983; Volkman and Goldsmith, 1985; Volkman et al., 1984). Studies of BV attachment to host cells in the presence of AcV1 suggested that AcV1 neutralization did not result from inhibition of BV attachment but rather from a block in the ability of BV virions to use the endocytic pathway, at a stage after initial viral adsorption (Volkman and Goldsmith, 1985). AcV1 binds to native GP64 but does not recognize denatured GP64 by standard Western blot analysis (Hohmann and Faulkner, 1983), suggesting that AcV1 recognizes a conformational epitope. The purpose of the current study was to begin to dissect the antigenic, structural, and functional topography of GP64 using this conformation-dependent monoclonal antibody. To understand possible changes in GP64 conformation, we examined AcV1 binding to GP64 under conditions of normal and low pH, and using a series of truncated GP64 constructs. We found that AcV1 binding to GP64 was pH dependent, suggesting that GP64 conformation is modulated in response to pH. To identify the region of the GP64 protein recognized by AcV1, we generated a series of truncated recombinant GP64 proteins in insect cells, and used them to map the AcV1 epitope. AcV1 binding was mapped to a sequence of 24 amino acids located in the central variable domain of the GP64 protein. Substitution of 11 amino acids in the AcV1 epitope with a c-Myc epitope sequence abrogated binding of AcV1 to GP64, confirming the AcV1 epitope. GP64 proteins containing the c-Myc substitution were capable of fully rescuing an AcMNPV gp64-knockout virus, and the resulting protein was recognized by an anti c-Myc MAb. However, the anti c-Myc MAb did not neutralize infectivity.
Materials and Methods
Construction of plasmids and baculoviruses encoding C-terminally truncated GP64 proteins
A series of plasmids encoding C-terminally truncated GP64 proteins was generated by the following strategy: First, DNA fragments containing C-terminally truncated GP64 open reading frames were PCR amplified from a wild type (wt) AcMNPV DNA template. A single forward primer with an EcoRI restriction site engineered into the 5′ end (5′-CGG AAT TCC AAG GCT TCA ATA AGG AAC-3′; which included 31 bp of wt gp64 sequence from upstream of the gp64 translation initiation site), was used in combination with a downstream primer specific for each truncation (Table 1). Each downstream primer contained an XbaI site engineered for in-frame insertion of the AcMNPV gp64 gene into vector pIZ/V5-His (Invitrogen). Thus, EcoRI and XbaI restriction sites were engineered into to the 5′ and 3′ ends, respectively, of each PCR product. Each PCR product was digested with EcoRI and XbaI, purified, and ligated into the EcoRI and XbaI sites of vector pIZ/V5-His, to generate a truncated GP64 ORF fused in-frame at the C-terminus with a V5 and 6xHis tag.
Table 1.
Primers for N- and C- terminal GP64 truncations
| C-terminal GP64 truncations | N-terminal GP64 Truncations |
|---|---|
| A single forward primer: | A single reverse primer: |
| 5′-CGGAATTCCAAGGCTTCAATAAGGAAC-′ | 5′-GAGAAGCTTCATTAATATTGTCTATTACGGTT-3′ |
| Reverse primers: | Forward primers: |
| CTr1(21-482): 5′-GCTCTAGAGAAGTCAATTTAGCGGCC-3′ | NTr1(21-512): 5′-GAGGAATTCGCGGAGCACTGCAACGCGCAA-3′ |
| CTr2(21-435): 5′-GCTCTAGATCTTTCCAACTGTCGTG-3′ | NTr2(125-512): 5′-GAGGAATTCAGCGACGACTGTTTTCGCGAC-3′ |
| CTr3(21-294): 5′-GCTCTAGACCCTCTGTGTACTTGGCTC-3′ | NTr3(186-512): 5′-GAGGAATTCGTATACATTTTGGACGCTGAG-3′ |
| CTr4(21-273): 5′-GCTCTAGACTCTCGACTTTGCGTTTAATGC-3′ | NTr4(264-512): 5′-GAGGAATTCAAAGTCGAGCACCGAGTCAAG-3′ |
| CTr5(21-217): 5′-GCTCTAGAGACTTTTGTTTGAGAATC-3′ | NTr5(271-512): 5′-GAGGAATTCCTGATGCATGCGCACATCAAC-3′ |
| CTr6(21-157): 5′-GCTCTAGAGTGTGGTGCGCAAAGTG-3′ | NTr6(377-512): 5′-GAGGAATTCGATTTTAGCAACTACAAGGAA-3′ |
| CTr7(21-131): 5′-GCTCTAGATCGCGAAAACAGTCGTCGC-3′ | NTr7(404-512): 5′-GAGGAATTCAGTTGGAAAGATGCCAGCGGC-3′ |
| CTr8(21-98): 5′-GCTCTAGACCCACATTCAGCGTTTTC-3′ | NTr8(459-512): 5′-GAGGAATTCGGCGGCGTCGGCACCAGTCTG-3′ |
| CTr9(21-58): 5′-GCTCTAGAGTCTCCACGATGGTGATTTC-3′ | NTr9(481-512): 5′-GAGGAATTCACTTCGTTCATGTTTGGTCAT-3′ |
To generate recombinant baculoviruses expressing the truncated GP64 proteins, GP64 constructs in pIZ/V5-His were linearized with ClaI and the ends made blunt with Klenow (Promega), then digested with EcoRI to excise the fragment containing the GP64 gene. Truncation-containing fragments were subcloned into the EcoRI and StuI sites of the pFastBac1 plasmid (Invitrogen). All constructs were confirmed by DNA sequencing. DH10Bac cells (Invitrogen) were transformed with the pFastBac1 constructs, then selected in kanamycin, gentamicin and tetracycline; and recombinant bacmids were selected by a color screen. Bacmid DNA was isolated from the single colony isolates of each construct and PCR amplification was used to verify each truncated GP64 construct. Sf9 cells were subsequently transfected with the bacmid DNAs and the resulting viruses were harvested and amplified as described in the Bac-to-Bac baculovirus expression system manual (Invitrogen). Infectious BV titres were determined using an endpoint dilution assay (O’Reilly, Miller, and Luckow, 1992).
Construction of plasmids and baculoviruses encoding N-terminally truncated GP64 proteins
N-terminally truncated GP64 constructs were generated in the following manner. Portions of the GP64 protein (downstream from the signal peptide) were PCR amplified using primers that contained either an EcoRI site (upstream primer) or a HindIII site (downstream primer) (Table 1). The resulting PCR products were digested with EcoRI and HindIII, then subcloned into the EcoRI/HindIII sites of pdFB-gp64sig-cMyc, a pFastBac-derived plasmid containing the gp64 promoter, the signal peptide and cleavage site, followed by a cMyc epitope tag and a cloning site (Figure 2A). Thus, N-terminally truncated forms of the GP64 gene that are cloned into vector pdFB-gp64sig-cMyc express a protein that contains the GP64 signal peptide and cleavage site plus a c-Myc tag. GP64 proteins are therefore truncated at the N-terminus of the mature GP64 protein and all constructs contain an N-terminal c-Myc epitope tag linked by a Phe residue to a portion of the GP64 protein. Transpositions of inserts from donor plasmids into the gp64-null bacmid (Lung et al., 2002) were confirmed by PCR analysis and by DNA sequencing. Cells stably expressing OpMNPV GP64 (cell line Sf9Op1D) (Plonsky et al., 1999) were transfected with each bacmid DNA and the resulting viruses were harvested from cell supernatants and titred on Sf9Op1D cells.
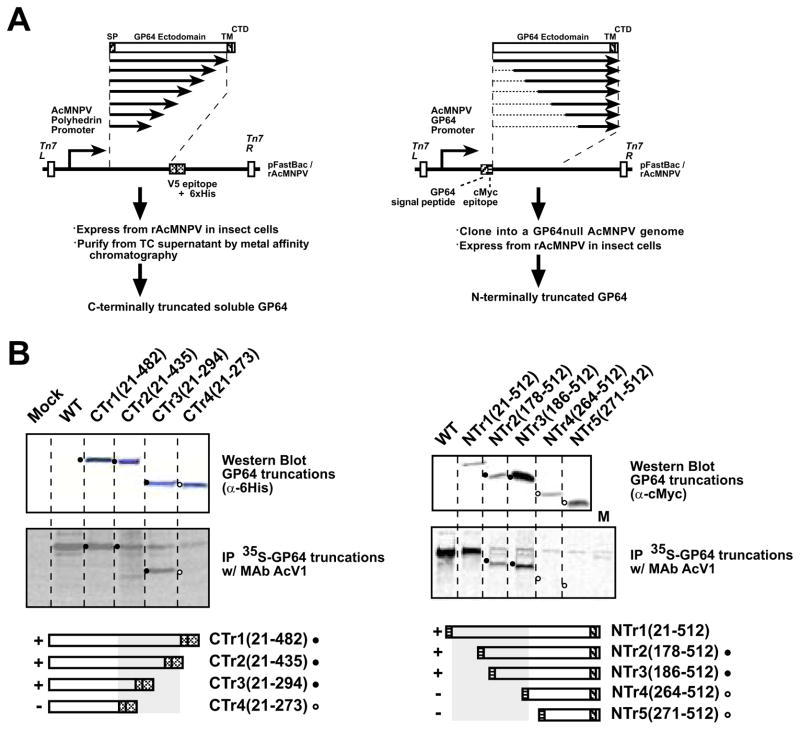
FIGURE 2.
A. Construction of N- and C-terminally truncated GP64 proteins. The strategy for PCR amplification, subcloning, and expression of portions of the GP64 coding region are shown as diagrams. The strategy for construction of C-terminally truncated constructs is shown on the left, and that for construction of N-terminally truncated constructs on the right
B. Western blot analysis and immunoprecipitation of C- and N-terminally truncated GP64 proteins
(SP, signal peptide; cMyc, cMyc tag; V5, V5 epitope; 6His, 6His tag; TM, transmembrane domain; CTD, cytoplasmic tail domain)
Left panel: Cells were infected with recombinant AcMNPV expressing secreted soluble C-terminally truncated GP64 constructs (CTr1(21-482), CTr2(21-435), CTr3(21-294) and CTr4(21-273)). Cells were metabolically labeled with 35S-Methionine for 8 hours and labeled soluble GP64 proteins were harvested from cell supernatants at 72 h pi and prepared as describe in the Materials and Methods section. GP64 was immunoprecipitated with MAb AcV1 and examined by SDS-PAGE (center panel). The top panel shows a Western blot of the cell supernatant preparations used for Immunoprecipitation experiments. GP64 constructs were detected on Western blots with an anti-6His antibody. Circles (filled or open) represent the positions of corresponding bands in upper and center panels. Open circles represent the positions of proteins that were not immunoprecipitated by AcV1 (center panel). Grey boxes (bottom panels) indicate constructs immunoprecipitated by AcV1 and containing the AcV1 epitope
Right panel: Cells infected with wild type or recombinant AcMNPV constructs and expressing wild-type AcMNPV GP64 or recombinant N-terminally truncated GP64 constructs (NTr1(21-512), NTr2(178-512), NTr3(186-512), NTr4(264-512) and NTr5(271-512)) were metabolically labeled with 35S-Methionine for 8 hours and cells were lysed at 72 h pi. GP64 was immunoprecipitated with MAb AcV1 and examined by SDS-PAGE (center panel). The top panel shows a Western blot of extracts used for Immunoprecipitation experiments. On Western blots, GP64 constructs were detected with an anti c-Myc antibody.
Immunofluorescence assay
Sf9 cells (2 × 105 cells/well) were plated in app. 150 mm diameter wells of 24 well plates (Corning Inc.) and cells were allowed to attach for 1 h, then infected at an MOI of 10 with either wt AcMNPV or the recombinant viruses expressing N-terminally truncated GP64 and incubated for 48 h. Cells were then fixed with 3% paraformaldehyde, a non-permeabilizing fixative. Cells were washed with Phosphate Buffered Saline (PBS, pH 7.4) and then incubated with a blocking buffer (1% gelatin in PBS, pH 7.4) at 27 °C for 2 h. After washing with PBS, the cells were incubated with a primary anti c-Myc monoclonal antibody (ATCC CRL-1729, MYC 1-9E10.2; 1:75 dilution in PBS) at 27 °C for 1 h. Cells were washed 3 times with blocking buffer and incubated with Alexa fluor 488-anti-mouse (Molecular Probes) at a 1:1000 dilution in PBS at 27 °C for 1 h. After washing 3 times with PBS (pH 7.4), florescence was observed with an Olympus IX70 fluorescence microscope.
Western blot analysis
Cell lysates were prepared by washing cultured cells with PBS and resuspending cells in NET buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.5% deoxycholate, 1.0% Nonidet P-40, 1mM EDTA) to which a protease inhibitor cocktail (Complete; Roche Applied Science) was added according to the manufacturer’s instructions. NET buffer (50 μl) was added to 1 × 106 cells and incubated for 30 min at 4 °C, and then nuclei were removed by pelleting at 4 °C for 10 min at 18,000 x g. For Western blot analysis, 10 μl of the above cell lysate were mixed with 10 μl of 2x Laemmli buffer (125 mM Tris, 2% sodium dodecyl sulfate, 5% 2-mercaptoethanol, 10% glycerol, 0.001% bromophenol blue, pH 6.8) and heated to 100°C for 5 min prior to SDS–10% polyacrylamide gel electrophoresis (SDS-PAGE). Gels were blotted onto Immobilon-P membranes (Millipore) and blocked overnight at 4 °C in TBST (25 mM Tris, pH 7.6, 150 mM NaCl, 0.1% Tween 20, 5% powdered milk). Blots were incubated for 1 h at room temperature with the following primary antibodies diluted in TBST: AcV5 MAb (which recognizes a linear epitope within AcMNPV GP64) diluted 1:100; anti-His polyclonal antibody (MBL international) diluted 1:4000; or anti c-Myc MAb diluted 1:1000. After washing 3 times in TBST, blots were incubated with a secondary antibody consisting of a goat anti-mouse or goat anti-rabbit IgG-alkaline phosphatase conjugate (Promega) at a dilution of 1:10,000. Western blots were processed as described earlier (Blissard and Wenz, 1992).
Protein refolding
For refolding, proteins in the SDS-Polyacrylamide gels were incubated in transfer buffer I (0.01% Triton X-100, 48 mM Tris, 39 mM Glycine, 20%methanol, pH 9.2) twice for 15 min, and then transfer buffer II (48 mM Tris, 39 mM Glycine, 20%methanol, pH9.2) twice for 15 min, then transferred onto Immobilon-P membranes (Millipore) in transfer buffer II and processed by standard procedures for Western blots.
35S-methionine-labeled virions and immunoprecipitation
C-terminal and N-terminal GP64 truncations were labeled with 35S-Methionine and prepared for immunoprecipitation (IP) in the following manner. Sf9 cells (1×106 cells/well) were plated in app. 35 mm diameter wells of six-well plates (Corning Inc.). Cells were allowed to attach for 1 h, then infected with either wt AcMNPV or recombinant viruses expressing N-terminally or C-terminally truncated GP64 at an MOI of 10 or 20 for 1 h. At 29 hours post infection (h pi), the cells were starved by incubation in 1 ml methionine-free Grace’s medium (Invitrogen) for 1 hour, followed by addition of 35S-EasyTag Express protein labeling mix (1175.0 Ci/mmol, Perkin-Elmer) to a final concentration of 10 μCi/ml. At 37 h pi, unlabeled methionine was added to a final concentration of 10 mM and cells were incubated at 27 °C for an additional 48 hours. For C-terminal truncations, soluble truncated GP64 proteins were harvested from the supernatant by the following method: Cell debris were removed from the supernatant by centrifugation at 5000 x g for 10 min (Sorvall F20 rotor), then virus was pelleted by centrifugation at 100,000 x g for 2 hours. Supernatants containing soluble secreted GP64 proteins were retained for analysis. For N-terminally truncated GP64 proteins, cell lysates were prepared by the following methods: Cells were washed once with PBS, then 300 μl NET buffer was added and incubated for 20 min on ice to lyse cells. Cell lysates (50 μl) in NET buffer were precleared by adding 50 μl of 10% rabbit serum (Sigma) and 50 μl of a 10% slurry of Protein A Sepharose (Sigma) in NET buffer and incubating with agitation for 1 h at 4 °C. After centrifuging (14,000 x g, 5 min), the supernatant was retained and MAb AcV1 (100 μl) was added to each precleared lysate along with 50 μl of a 50% slurry of Protein G Agarose (Pierce). The mixture was incubated overnight at 4 °C with continuous gentle rocking. Immunoprecipitates were collected by centrifugation (14,000 x g, 5 min) and washed twice with NET buffer. The precipitated material was solubilized by heating for 5 min at 95 °C in Laemmli buffer and analyzed by electrophoresis on 10% SDS-PAGE gels. Labeled proteins were visualized by imaging on phosphorImager screens.
Low pH treatments
To examine AcV1 binding to GP64 under various conditions, wild type or recombinant GP64 constructs were metabolically labeled with 35S-Methionine as described above, exposed to various pH treatments, then immunoprecipitated. The Sf9 cells (1×106 cells) were lysed in 0.5 ml of NET buffer followed by a 30 min agitation at 4 °C. The lysate was cleared of the detergent-insoluble material by centrifugation for 30 min at 90,000 x g at 4 °C. The cell lysates were subjected to the following treatments: 1) low pH only; Lysates were adjusted to pH 4.5 by addition of PBS (pH 1.7), 2) no pH change (pH6.2) and 3) low pH treatment followed by readjusting to pH 6.2 [(pH was shifted to pH 4.5 by adding PBS (pH 1.7), incubating for 30 min at pH 4.5, then readjusting the pH to 6.2 by adding PBS (pH 12)]. GP64 proteins were then immunoprecipitated from lysates directly using AcV1 (as described above) or a control anti c-Myc MAb.
To examine the effects of low pH treatment on BV infectivity, AcMNPV BV were exposed to various pH values, and then examined for changes in infectivity. Wild type AcMNPV BV were adjusted to pH 4.5 by adding 500 μl PBS (pH 1.7) or Grace’s medium (pH 2.1) to 1 ml of a virus (BV) stock supernatant (pH 6.2, 2.18×108 pfu/ml). The virus was incubated for 30 min at the adjusted pH at room temperature. Virus preparations were then readjusted to pH 6.2 by the addition of 600 μl PBS (pH 12.3) or Grace’s medium (pH9.6), respectively. The titre of the virus from each treatment was determined by end-point dilution assay.
Site-directed mutagenesis
c-Myc substitution mutations were made by replacing 33 nt in the GP64 ORF with 33 nt of sequence encoding the c-Myc epitope: 5′-GAA CAA AAA CTC ATC TCA GAA GAG GAT CTG AAT-3′. The substitutions were generated by the site-directed, ligase-independent mutagenesis method (Chiu et al., 2004). First, a DNA fragment containing the AcMNPV GP64 promoter and open reading frame was PCR amplified from a wild type AcMNPV DNA template using primers that contained a KpnI site (upstream primer) or a HindIII site (downstream primer). The PCR product was digested with KpnI and HindIII, and subcloned into the KpnI and HindIII sites of pGEM-3Z (Promega). The resulting plasmid DNA contained 211 nt upstream of the gp64 ORF, the gp64 ORF, and 13 nt downstream of the gp64 ORF, and was designated pGEM-gp64 and used for the subsequent inverse PCR. Each c-Myc substitution required four primers for a single inverse PCR reaction. Two primers were designed to introduce c-Myc sequence into the 5′-adapter tail of primers (Table 3.). Following amplification, the resulting PCR mixtures were purified with Qiaex II resins (Qiagen), digested with Dpn I and then hybridized using two cycles of 65 °C for 5 min and 30 °C for 15 min, to generate the plasmid containing the substitution mutation. An aliquot of 3 μl of the purified PCR reaction was used to transform electrocompetent E.coli. TOP10 cells. Clones were screened and recombinants identified by colony PCR analysis. The mutated gp64 constructs were excised from the pGEM-gp64 by digestion with KpnI and HindIII and subcloned into the KpnI and HindIII sites of the pFastBac1 plasmid (Invitrogen). All constructs were confirmed by DNA sequencing then used for transposition into a gp64null AcMNPV genome. Bacmid DNAs were transfected into Sf9 cells and the resulting viruses, vAc-Glu273/cmyc and vAc-Lys277/cmyc, were harvested and amplified as described in the Bac-to-Bac baculovirus expression system manual (Invitrogen). Infectious BV titres were determined on Sf9 cells using an endpoint dilution assay (O’Reilly, Miller, and Luckow, 1992).
Table 3.
Primers for c-Myc substitutions
| Glu273/cmyc | Glu273/cmyc-F: (5′-GAAGAGGACCTGAATCGCCACAACGTTAGAGCC-3′) |
| Glu273/cmyc-R: (5′-TGAGATGAGTTTTTGTTCGACTTTGCGTTTAATGCA-3′) | |
| Glu273-F: (5′-CGCCACAACGTTAGAGCC-3′) | |
| Glu273-R: (5′-GACTTTGCGTTTAATGCA-3′) | |
| Lys277/cmyc | Lys277/cmyc-F: (5′-GAAGAGGACCTGAATAGAGCCAAGTACACAGAG-3′) |
| Lys277/cmyc-R: (5′-TGAGATGAGTTTTTGTTCGACTCGGTGCTCGACTTT-3′) | |
| Lys277-F: (5′-AGAGCCAAGTACACAGAG-3′) | |
| Lys277-R: (5′-GACTCGGTGCTCGACTTT-3′) |
Overlapping c-Myc sequences are underlined
Neutralization assay
Previously, it was demonstrated that concentrations of ≥ 2 μg AcV1 per 106 PFU of AcMNPV BV were sufficient for neutralization. In the current study we used excess concentrations of Mab (4.87 – 71 μg MAb per 106 PFU AcMNPV BV). A volume of 30 μl wt AcMNPV budded viruses (1.23 × 108 PFU/ml), NTr1(21-512) (3 × 107 PFU/ml), vAc-Glu273/cmyc (1.93 × 107 PFU/ml), vAc-Lys277/cmyc (1.73×107 PFU/ml) were diluted to a total volume of 40 μl with PBS (pH 6.2) containing either AcV1 (24 μg), anti c-Myc (18 μg), B12D5 (antibody ascites, 37 μg) or normal mouse IgG (25 μg), and incubated for 1 h at 27 °C. Titres of the treated and untreated virus mixtures were determined by endpoint dilution assay as described previously (O’Reilly, Miller, and Luckow, 1992; Volkman and Goldsmith, 1985).
Results and Discussion
The AcMNPV GP64 protein is a membrane fusion protein that mediates viral entry by receptor-mediated endocytosis (Blissard and Wenz, 1992; Hefferon et al., 1999; Hohmann and Faulkner, 1983; Monsma, Oomens, and Blissard, 1996; Volkman et al., 1984). In the endosome, GP64 is believed to change conformation in response to low pH, and that change is believed to trigger GP64-mediated membrane fusion activity. The anti-GP64 monoclonal antibody, AcV1, neutralizes infectivity of AcMNPV BV and recognizes a conformational epitope in the native but not denatured GP64 protein (Hohmann and Faulkner, 1983; Volkman et al., 1984). In prior studies using AcV1, low pH treatments of GP64 on infected cells inhibited immunofluorescent detection and fusion activity (Chernomordik et al., 1995). To confirm and extend studies of GP64 conformation, and to characterize AcV1 binding and inhibition of GP64 function, we used immunoprecipitation assays in combination with truncated and substituted forms of GP64 to examine AcV1 binding under various conditions and we mapped the AcV1 epitope.
Low pH treatment of GP64 results in loss of the AcV1 epitope
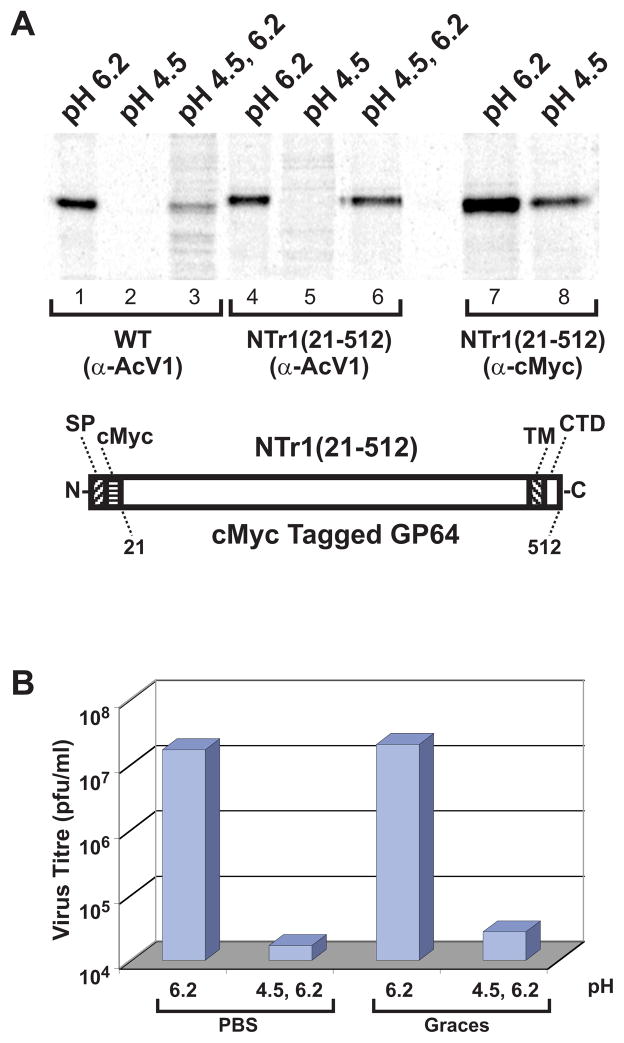
To examine AcV1 binding to GP64 under conditions of neutral and low pH, wild type and modified GP64 proteins were immunoprecipitated under conditions of normal or low pH, pH 6.2 or 4.5, respectively. Because Sf9 cells are typically cultured in media at pH 6.0 – 6.2 and AcMNPV infections are typically performed at the same pH values, the pH value of 6.2 was selected as the “normal” or “neutral” pH for GP64 conformation studies. Cells expressing either wt GP64 or a GP64 protein with an N-terminal c-Myc epitope tag (NTr1/21-512), were lysed and GP64 was immunoprecipitated with either MAb AcV1 or a control anti c-Myc MAb (Fig. 1A, lanes 1, 4, and 7). To determine if the AcV1 epitope was present and available in various conformations, the GP64 protein was incubated at various pH values prior to antibody binding and immunoprecipitation.
FIGURE 1.
A. Immunoprecipitation of 35S-Methionine-labeled GP64 constructs with MAb AcV1 (lanes 1–6) and anti c-Myc (lanes 7–8). Cells expressing wild-type AcMNPV or NTr1(21-512) GP64 proteins were metabolically labeled with 35S-Methionine. Cell lysates containing wild type GP64 or NTr1(21-512) proteins were subjected to various treatments (pH 6.2, pH 4.5, or pH 4.5 followed by a shift to 6.2), then subjected to immunoprecipitation with MAb AcV1 or anti c-Myc, and analyzed on an SDS-10% polyacrylamide gel. The diagram (bottom) shows the construction of a control protein, N-terminally cMyc tagged GP64, NTr1(21-512). (SP, signal peptide; cMyc, cMyc tag; TM, transmembrane domain; CTD, cytoplasmic tail domain)
B. Effect of low pH treatment on BV infectivity. Wild type AcMNPV budded viruses at pH 6.2 were adjusted to pH 4.5 with either PBS (pH 1.7) or Grace’s medium (pH 2.1). The low pH adjusted virus was incubated for 30 min at room temperature, then returned to pH 6.2 by addition of either PBS (pH 12.3) or Grace’s medium (pH 9.6). Titres were then determined by end-point dilution. Control BV preparations were similarly treated with PBS or Grace’s medium but at pH 6.2 only. The results represent the average value of three separate replicate experiments.
When preincubated at pH 6.2, both wild type GP64 and c-Myc tagged GP64 were efficiently immunoprecipitated with MAb AcV1. However, when preincubated at pH 4.5, neither construct was recognized and immunoprecipitated by AcV1 (Fig. 1A, lanes 2 and 5). To determine if the observed effect resulted from a general inability of monoclonal antibodies to bind and immunoprecipitate GP64 at pH 4.5, an anti c-Myc MAb was used to immunoprecipitate the N-terminally c-Myc tagged GP64 protein in a parallel experiment. The anti c-Myc MAb bound and immunoprecipitated the N-terminally tagged GP64 protein at both pH 6.2 and 4.5 (Fig. 1A, lanes 7 and 8). Thus, failure of AcV1 to bind GP64 at low pH (pH 4.5) is not due to an intrinsic property of monoclonal antibody binding at pH 4.5, but appears to be due to a change in the availability of the conformation-specific AcV1 epitope. To determine if the loss of the AcV1 epitope at low pH was reversible, we exposed wt and c-Myc tagged GP64 constructs to pH 4.5 for 30 min, and then returned the proteins to pH 6.2 and immunoprecipitated GP64 with AcV1. Although the efficiency of GP64 detection by this method was slightly reduced, AcV1 recognition was restored by returning GP64 to pH 6.2 after low pH treatment (Fig. 1A, lanes 3 and 6). Thus, the conformation-specific AcV1 epitope that is lost when GP64 is denatured by SDS and beta-mercaptoethanol treatment was also lost upon exposure to low pH. However, the pH-induced change was reversible as the AcV1 epitope was restored when the pH was returned to pH 6.2.
Low pH treatment of AcMNPV BV neutralizes infectivity
Because recognition of the AcV1 epitope was lost upon exposure to pH 4.5 but was restored by incubation at pH 6.2, we next examined the effect of low pH treatment on baculovirus infectivity. We asked whether the previously reported inactivation of AcMNPV by low pH, could be reversed by returning the virus to neutral pH. Wild type AcMNPV BV was treated for 30 min with PBS or Grace’s medium adjusted to pH 4.5, then the pH was readjusted to pH 6.2 and the virus infectivity was determined. As a control, the same virus preparation was exposed to PBS or Grace’s media at pH 6.2 (instead of pH 4.5), but otherwise treated identically. Exposure of AcMNPV BV to pH 4.5 resulted in an approximately 1000-fold reduction in the infectious titre (Fig. 1B). Similar results were obtained in three replicate experiments. Thus, although the low pH induced conformational change associated with AcV1 binding was reversible, exposure to pH 4.5 appears to irreversibly neutralize virus infectivity.
AcV1 epitope mapping with truncated GP64 proteins
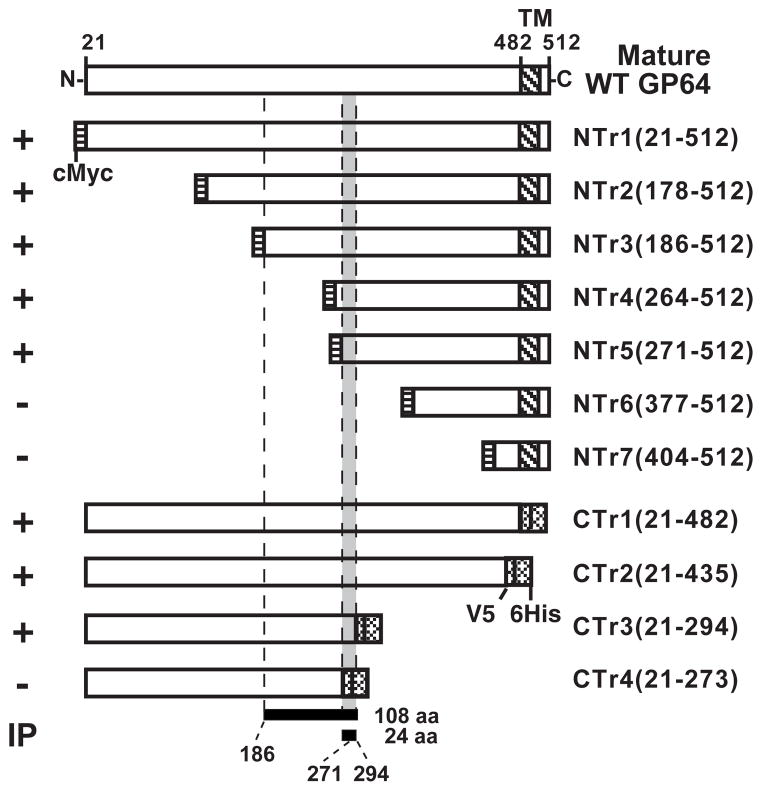
To map the epitope recognized by AcV1, we generated a series of truncated GP64 constructs for expression in insect cells. We first constructed 9 baculovirus expression vectors that encode and express soluble forms of GP64 that are C-terminally truncated and tagged at the C-terminus with a V5 epitope and 6His. The construct names (CTr1(21-482), CTr2(21-435), CTr3(21-294), CTr4(21-273); Table 2) identify the GP64 amino acids present in each construct (Table 2 and Fig. 2A, left panel). GP64 is a 512 amino acid protein that is processed by signal peptide cleavage after amino acid 20 and the mature form of wild type GP64 consists of amino acids 21–512. Thus, the mature form of C-terminal deletion construct CTr1(21-482) is comprised of amino acids 21–482 from GP64, plus a C-terminal V5 epitope and 6His tag sequence. The C-terminally truncated GP64 protein constructs were metabolically labeled with 35S-methionine and collected as secreted protein from cell supernatants. Truncated GP64 proteins were identified directly by SDS-PAGE of cell supernatants, autoradiography (data not shown), and by Western Blot analysis of cell supernatants with an anti-His antibody (Fig. 2B, left panels, top). All constructs were subjected to immunoprecipitation by AcV1. Representative results are shown in Figure 2B (left panels, center) and additional results are indicated in Table 2. Constructs containing at least amino-acids 21–294 of AcMNPV GP64 were immunoprecipitated by AcV1 whereas constructs containing amino acids 21–273 or fewer were not (Fig. 2B, left panel, center; lane 5 and 6; Table 2).
Table 2.
Summary of the properties of GP64 truncations
| C-terminally truncations detected by:
|
N-terminally truncations detected by
|
||||||
|---|---|---|---|---|---|---|---|
| WB anti-His | IP AcV1 | Refolding AcV1 | WB anti-His | IP AcV1 | Refolding AcV1 | ||
| CTr1(21-482) | + | + | + | NTr1(21-512) | + | + | + |
| CTr2(21-435) | + | + | + | NTr2(125-512) | + | + | + |
| CTr3(21-294) | + | + | + | NTr3(186-512) | + | + | + |
| CTr4(21-273) | + | − | − | NTr4(264-512) | + | − | + |
| CTr5(21-217) | + | − | − | NTr5(271-512) | + | − | + |
| CTr6(21-157) | + | − | − | NTr6(377-512) | + | − | − |
| CTr7(21-131) | + | − | − | NTr7(404-512) | + | − | − |
| CTr8(21-98) | + | − | − | NTr8(459-512) | + | − | − |
| CTr9(21-58) | + | − | − | NTr9(481-512) | + | − | − |
WB, Western Blot. IP, Immunoprecipitation. Refolding, Refolding Assay.
The numbers in parentheses indicate the positions of amino acids covering the peptides.
Next, we generated a series of N-terminally truncated GP64 protein constructs. Each N-terminally truncated GP64 protein is truncated from the N-terminus of the mature (cleaved) GP64 protein and contains a c-Myc epitope tag at the N-terminus of the truncated GP64 protein (Fig. 2A, right panel). Each N-terminally truncated GP64 construct was cloned under the control of the AcMNPV GP64 promoter and inserted into an AcMNPV Bacmid containing a gp64 deletion (Lung et al., 2002). Viruses were propagated in a stable cell line that constitutively expresses the wild type OpMNPV GP64 protein as described previously (Lung et al., 2002; Plonsky et al., 1999). To confirm that the N-terminal GP64 truncations were transported through the secretory pathway and localized on the cell surface, the presence of each truncated GP64 construct at the cell surface was confirmed by immunofluorescence microscopy. All truncated proteins indicated in Figure 4 were detected at the cell surface (data not shown). N-terminally truncated GP64 proteins were metabolically labeled with 35S-methionine and GP64 constructs containing the AcV1 epitope were immunoprecipitated from cell lysates with AcV1 (Fig. 2B, right panels, center). To confirm the presence of each GP64 construct, cell extracts were examined by Western blot analysis with an anti c-Myc antibody (Fig. 2B, right panels, top). N-terminally truncated constructs that contained at least amino acids 186–512 were immunoprecipitated by AcV1, while constructs containing fewer GP64 sequences were not (Table 2; Fig. 2B). Thus, by immunoprecipitating native N- and C- terminally truncated GP64 constructs expressed in insect cells, the AcV1 epitope was mapped to a 108 amino acid sequence from 186–294 (Fig. 4).
FIGURE 4.
Summary of AcV1 epitope mapping. The diagram shows a schematic representation of the wild type GP64 ectodomain (top bar) and deletion mutants of GP64 combined with the results of immunoprecipitation (IP) experiments. The numbers at the top represent the amino acid positions in the wild type GP64 protein. Names of the truncated GP64 constructs are indicated on the right and the results of IP analyses are summarized on the left (+ or −). The binding site (epitope) of MAb AcV1 was mapped to a sequence of 108 amino acids (bottom; amino acids186 –294) with native GP64 proteins and to a sequence of 24 amino acids (271–294) in protein refolding assays. (cMyc, cMyc tag; TM, transmembrane domain; V5, V5 epitope; 6His, 6His tag; CTD, cytoplasmic tail domain).
Analysis of refolded GP64 proteins
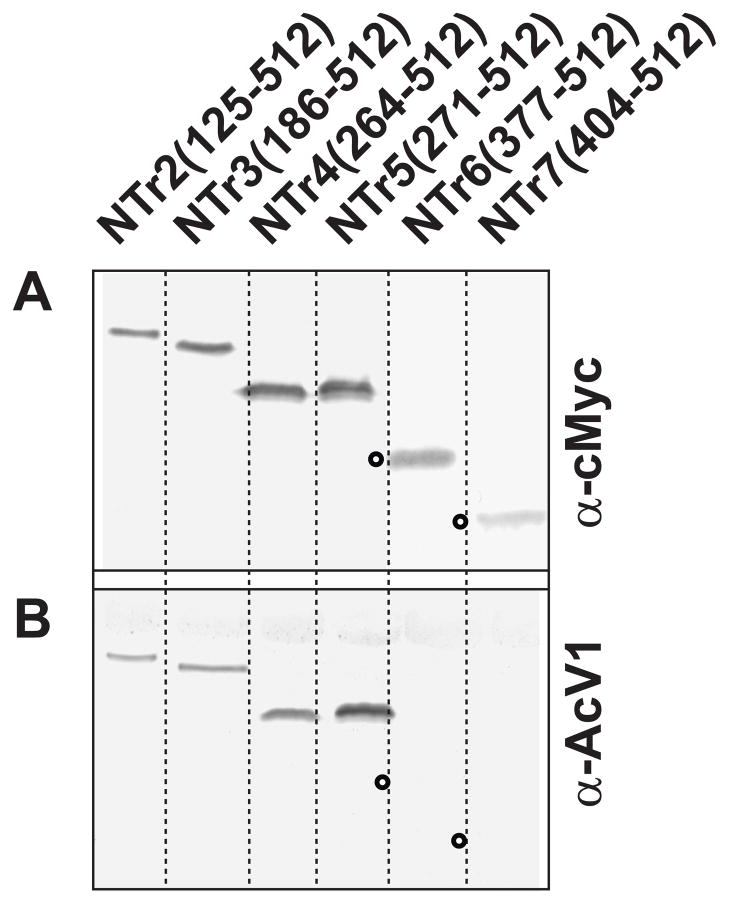
Because the AcV1 epitope is lost upon denaturation in SDS and beta-mercaptoethanol, AcV1 does not identify GP64 by traditional Western blot analysis. However, our prior analysis showed that loss of AcV1 binding at low pH was restored by returning the pH to 6.2. We therefore examined the possibility that the loss of the AcV1 epitope by denaturation in SDS, may be restored by refolding the protein in the absence of SDS. To address this question, we performed in-gel refolding experiments. Truncated and wild type GP64 proteins were denatured in SDS and reduced, then electrophoresed in standard polyacrylamide gels. GP64 proteins were then refolded by incubating the gels in a series of buffers containing no SDS (see Materials and Methods) for 30 min, then transferring the protein to Immobilon-P membranes (Millipore) and processing for Western blots by standard procedures. Blots from gels containing refolded proteins were challenged with either MAb AcV1 or a control antibody (anti c-Myc or anti-6His). Using this in-gel refolding technique, AcV1 detected N-terminal truncated GP64 proteins NTr4 (264-512), NTr5 (271-512), but not NTr6 (377-512) and NTr7 (404-512) (Fig. 3 and Table 2). Thus, by refolding GP64, we were able to map the AcV1 binding site further, to a 24 amino acid region between amino acids 271 and 294 (Fig. 4).
FIGURE 3.
Western blot analysis of refolded N-terminally truncated GP64 constructs. Protein lysates from cells expressing N-terminal GP64 truncations were subjected to 10% SDS-PAGE. Proteins were refolded in the acrylamide gel by incubation in a transfer buffer containing no SDS (see Materials and Methods) for 30 min, then transferred onto Immobilon-P membranes (Millipore) and processed for Western blots by standard procedures. Blots from gels containing refolded proteins were challenged with either anti c-Myc (Top panels) or MAb AcV1 (Bottom panels). Open circles represent the positions of proteins that were not detected by MAb AcV1.
c-Myc substitutions in the AcV1 epitope
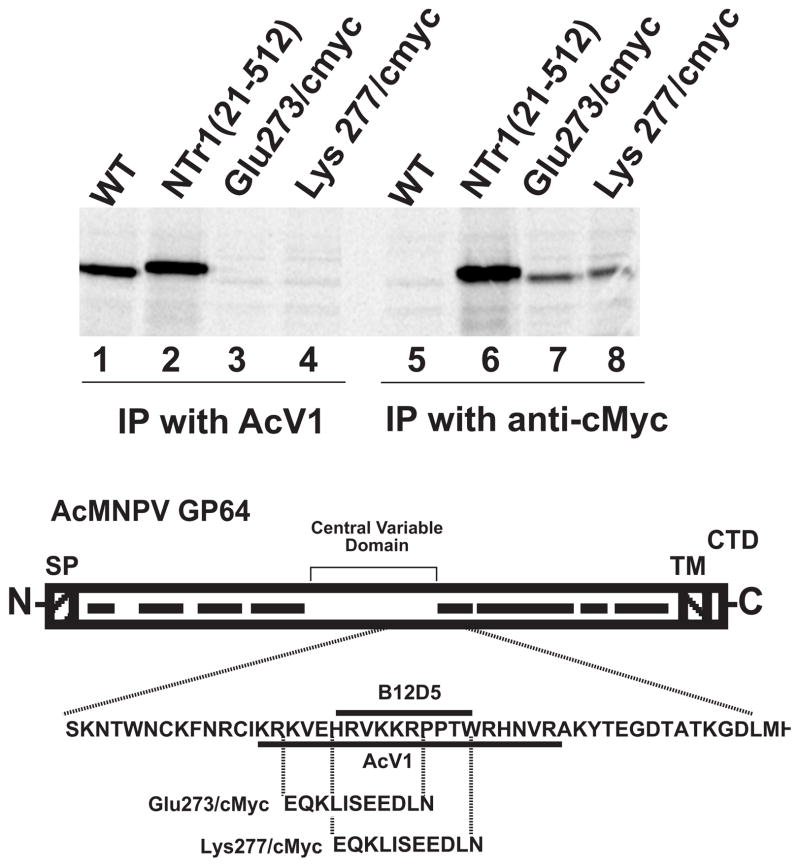
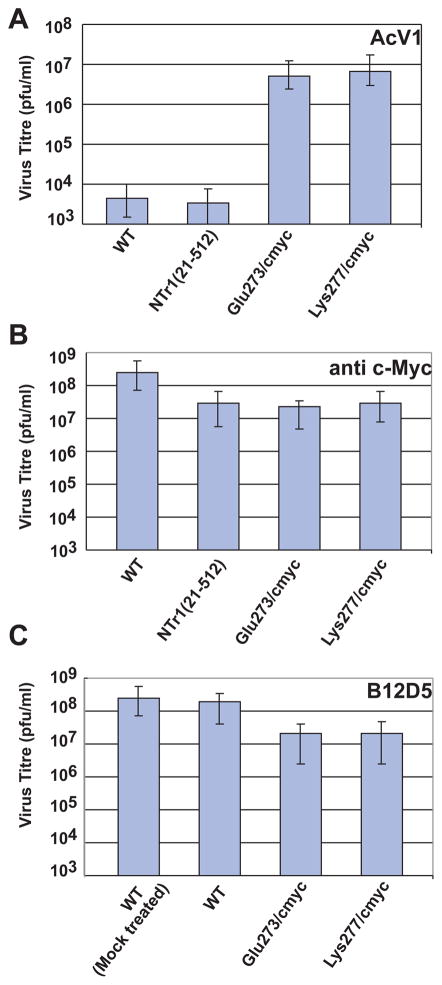
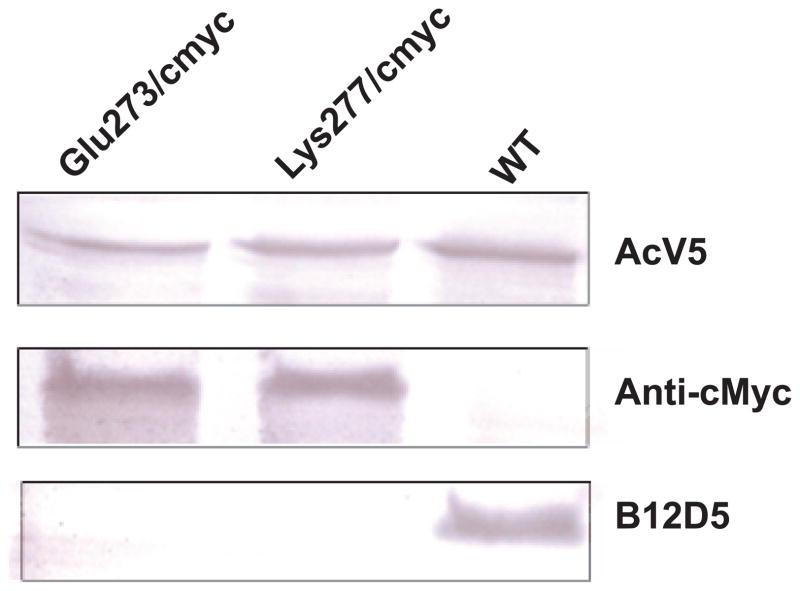
To confirm the location of the AcV1 epitope, we asked whether substitution mutations in amino acid positions between 271 and 294 would abolish binding of AcV1 to GP64. Two substitutions of 11 amino acids each were introduced into amino acid positions 273–284 (substitution Glu273/cmyc) and 277–288 (substitution Lys277/cmyc), respectively (Fig. 5). In each case, an 11 amino acid c-Myc epitope sequence was substituted for the corresponding 11 amino acids from the GP64 protein. GP64 constructs that contained substitution mutations Glu273/cmyc and Lys277/cmyc were then used to generate two recombinant AcMNPV viruses in which the wild type GP64 gene was deleted and replaced by a GP64 gene containing a substitution in the AcV1 epitope region. GP64 genes encoding substitution mutations were inserted into the polyhedrin locus of a gp64null AcMNPV genome (Lung et al., 2002). The GP64 proteins containing the Glu273/cmyc and Lys277/cmyc substitutions were able to functionally substitute for the wt GP64 protein and were incorporated into virions at levels similar to that of the wt GP64 protein (data not shown). Viruses encoding and expressing only the Glu273/cmyc or Lys277/cmyc GP64 substitutions replicated efficiently in Sf9 cells and required no wild type GP64 for virus propagation. To determine if the Glu273/cmyc and Lys277/cmyc substitutions abolished AcV1 binding, GP64 proteins were metabolically labeled with 35S-Methionine and immunoprecipitated with AcV1. While wt GP64 and a control GP64 (N-terminally c-Myc tagged GP64 NTr(21-512), Fig. 1A) were efficiently immunoprecipitated by AcV1, GP64 proteins containing the Glu273/cmyc and Lys277/cmyc substitutions were not (Fig. 5, lanes 1 and 2 vs. 3 and 4). As a control, similar immunoprecipitation experiments were performed using an anti c-Myc antibody. GP64 control, Glu273/cmyc, and Lys277/cmyc substitution constructs were immunoprecipitated by the anti c-Myc antibody while wt GP64 was not, as expected (Fig. 5, lanes 6, 7 and 8). Thus, the AcV1 epitope was mapped to a position between amino acids 271 and 294 and substitutions of either amino acids 273–284 or 277–288 abolished AcV1 binding. Interestingly, the two viruses containing the AcV1 substitutions were almost as infectious as wt AcMNPV on Sf9 cells, with titres of 1.93×107 PFU/ml (vAc-Glu273/cmyc) and 1.73×107 PFU/ml (vAc-Lys277/cmyc), as compared to 1.23×108 PFU/ml for wt AcMNPV on Sf9 cells. However, viruses carrying GP64 proteins with the two substitutions (Glu273/cmyc and Lys277/cmyc) could not be neutralized by antibody AcV1 (Fig. 7A). It is also of interest that another monoclonal antibody known as B12D5 (Keddie, Aponte, and Volkman, 1989) was previously mapped to a linear epitope in the same region (amino acids KKRPPTWRHNV at 277–287) (Monsma and Blissard, 1995) and the B12D5 epitope overlaps that of AcV1 (Fig. 5). The two GP64 substitutions (Glu273/cmyc and Lys277/cmyc) that abolish AcV1 binding (Fig. 5) also abolish binding of B12D5 to GP64 (Fig. 6). However unlike AcV1, we found that B12D5 was not able to neutralize infectivity of wt AcMNPV BV (Fig. 7C).
FIGURE 5.
Immunoprecipitation of GP64 protein constructs containing substitutions in the AcV1 epitope region. The c-Myc epitope sequence was substituted for GP64 sequences at amino acid positions 273–284 (Glu273/cmyc) and 277–288 (Lys277/cmyc). Cells infected with recombinant AcMNPV viruses expressing the c-Myc substituted forms of GP64 (replacing wt GP64) were 35S-Methionine labeled and the GP64 constructs examined by immunoprecipitation with either AcV1 (lanes 1–4) or the anti c-Myc MAb (lanes 5–8). The diagram (bottom) shows the locations and sequences of the c-Myc substitutions relative to the GP64 coding sequence and the mapped AcV1 and B12D5 epitopes. The central variable domain and conserved domains (black bars) are indicated on the diagram of the GP64 coding region. (SP, signal peptide; TM, transmembrane domain; CTD, cytoplasmic tail domain).
FIGURE 7.
Neutralization of AcMNPV viruses displaying GP64 protein constructs with either a wild type or modified AcV1 epitope region. The graphs show titres of viruses displaying wild-type AcMNPV GP64 (WT), N-terminally c-Myc tagged GP64 (NTr1(21-482)), or GP64 proteins with c-Myc substitutions (Glu273/cmyc and Lys277/cmyc) in the mapped AcV1 epitope. The titre of each BV preparation was determined by end-point dilution analysis after incubation with MAb AcV1 (Panel A), anti c-Myc MAb (Panel B), or MAb B12D5 (Panel C). Each bar represents the average titre obtained from three separate experiments. (Mock treated represents no antibody treatment.)
FIGURE 6.
Western blot analysis of GP64 protein constructs containing c-Myc substitutions in the AcV1 epitope region. Lysates of Sf9 cells infected with either wt AcMNPV (WT) or recombinant viruses expressing GP64 c-Myc substitution constructs (Glu273/cmyc and Lys277/cmyc) were examined by Western blot analysis with antibodies AcV5 (top panel), Anti c-Myc (center panel), or B12D5 (lower panel).
The mechanism of AcV1-mediated inhibition of infectivity is not known although prior studies indicate that AcV1 does not interfere with virion binding at the cell surface (Volkman and Goldsmith, 1985). If AcV1 binding results in neutralization by steric hindrance as a result of an antibody molecule occupying this specific site, we would predict that substitution of this site by the c-Myc epitope in constructs Glu273/cmyc and Lys277/cmyc would result in GP64 proteins that would no longer be neutralized by AcV1 but could be neutralized by an anti c-Myc monoclonal antibody. However, in neutralization studies with viruses carrying the wild type or substituted forms of GP64 (Fig. 7), we found that while AcV1 neutralization was indeed abolished by the Glu273/cmyc and Lys277/cmyc substitutions, the GP64 proteins carrying the Glu273/cmyc and Lys277/cmyc substitutions were not neutralized by the anti c-Myc monoclonal antibody (Fig. 7, panel A and B). Thus, the mechanism of AcV1 mediated neutralization of GP64 function does not appear to occur through simple steric hindrance caused by antibody binding at this site but likely involves a more complex interaction between AcV1 and GP64 proteins.
Comparison of predicted GP64 proteins from group I NPV baculoviruses and the related GP75 proteins of the Thogoto-like viruses revealed 8 conserved domains along the entire length of the approximately 462 amino acid GP64 ectodomain. In addition, a non-conserved “central variable domain” of approximately 118 amino acids is located in the central portion of the ectodomain (Fig. 5). Epitopes for two monoclonal antibodies (B12D5 and AcV5) directed against AcMNPV GP64 were mapped in a prior study (Monsma and Blissard, 1995) (Fig. 5). Because those antibodies recognized linear epitopes exposed on either native (B12D5) or denatured (B12D5 and AcV5) GP64, a simple epitope library screening approach was used previously. In the current study, we used several approaches to map the epitope of the neutralizing AcV1 monoclonal antibody. N- and C- terminal deletion constructs were used to map the AcV1 conformational epitope to a 24 amino acid sequence in the central variable domain of GP64. Because N- and C- terminally truncated forms of GP64 were recognized, these data indicate that additional sequences within the GP64 protein are not required and that the epitope is included within the 24 amino acid sequence. Why does AcV1 bind only to the native GP64 protein and not to denatured GP64? One possible explanation is that the native structure of GP64 may contain a localized pH-sensitive conformation within this 24 amino acid region, and this conformation is required for AcV1 binding. Thus, because AcV1 does not bind in the presence of SDS or at low pH (4.5), yet maps to a very limited 24 amino acid sequence, we speculate that AcV1 binding must require a highly localized conformation of the epitope sequence. Substitution of a c-Myc epitope in two 11 amino acid positions within the 24 amino acid AcV1 epitope, resulted in GP64 proteins that could be functionally substituted for wild type GP64, but were not recognized by AcV1. Thus, the AcV1 epitope sequence per se is not necessary for the function of GP64. The observation that AcV1 neutralizes infectivity but does not bind the low pH conformation of GP64, suggests that AcV1 binding somehow interferes with the low pH activated conformational change. Interestingly, when the c-Myc epitope was substituted into the AcV1 epitope region, anti c-Myc did not neutralize infectivity of the virus. This suggests that the AcV1 mediated neutralization does not result simply from steric hindrance, but rather from inhibition of a specific conformational change in the GP64 protein.
In a contemporary model for the function of viral membrane fusion proteins such as Influenza HA and HIV GP41, the triggering of membrane fusion results in a refolding of the protein such that a “hairpin” is formed, bringing the distal end of the fusion protein (containing the hydrophobic fusion domain) in close proximity to the transmembrane domain (which spans the viral envelope). Thus, the cellular membrane is placed in close proximity to the viral envelope membrane. In some cases, hairpin formation is mediated by the interaction of two alpha helices. Thus, for trimeric fusion proteins, a “trimer of hairpins” is believed to form and to generate a six-helix bundle. Experimental evidence shows that in such cases, fusion can be disrupted by soluble peptides that encode one of the helices – presumably inhibiting fusion by competing for the helix-helix interaction and preventing stable hairpin formation. The AcMNPV GP64 protein contains a predicted (highly conserved) amphipathic alpha helix at amino acids 302–345, immediately downstream of the central variable domain. Prior studies show that mutations that would disrupt formation of the predicted helix or the amphipathic nature of the helix also disrupt membrane fusion by GP64 (Kingsley et al., 1999; Monsma and Blissard, 1995). In addition, a 29-mer peptide consisting of amino acids 301 to 329 also disrupted membrane fusion (Kingsley et al., 1999). These data provide preliminary evidence for possible participation of the amphipathic alpha helix at 301 in membrane fusion. The AcV1 epitope is located at amino acid position 271 – 294, immediately upstream of the predicted 301 alpha helix. Thus, we would speculate that while AcV1 binding requires a local conformation found in the neutral pH conformation of GP64, neutralization of GP64 function (and viral infectivity) by AcV1 may result from inhibition of a required conformation change associated with repositioning the alpha helix or its interacting partner.
Acknowledgments
The authors thank Gretchen Hoffmann, Josh Huffer and Jodie Mangor for assistance with construction of GP64 truncation mutants. We also gratefully acknowledge Loy Volkman for providing monoclonal antibody B12D5 and Peter Faulkner for hybridoma lines AcV1 and AcV5. This work was supported by NIH grant AI33657 and project 1255 of the Boyce Thompson Institute.
References
- Blissard GW, Wenz JR. Baculovirus GP64 envelope glycoprotein is sufficient to mediate pH dependent membrane fusion. J Virol. 1992;66:6829–6835. doi: 10.1128/jvi.66.11.6829-6835.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik L, Leikina E, Cho M, Zimmerberg J. Control of baculovirus GP64-induced syncytium formation by membrane lipid composition. J Virol. 1995;69(5):3049–3058. doi: 10.1128/jvi.69.5.3049-3058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu J, March PE, Lee R, Tillett D. Site-directed, Ligase-Independent Mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 2004;32(21):e174. doi: 10.1093/nar/gnh172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados RR, Lawler KA. In vivo pathway of Autographa californica baculovirus invasion and infection. Virology. 1981;108:297–308. doi: 10.1016/0042-6822(81)90438-4. [DOI] [PubMed] [Google Scholar]
- Hefferon K, Oomens A, Monsma S, Finnerty C, Blissard G. Host cell receptor binding by baculovirus GP64 and kinetics of virion entry. Virology. 1999;258:455–468. doi: 10.1006/viro.1999.9758. [DOI] [PubMed] [Google Scholar]
- Hohmann AW, Faulkner P. Monoclonal antibodies to baculovirus structural proteins: Determination of specificities by Western blot analysis. Virology. 1983;125(2):432–444. doi: 10.1016/0042-6822(83)90214-3. [DOI] [PubMed] [Google Scholar]
- Keddie BA, Aponte GW, Volkman LE. The pathway of infection of Autographa californica nuclear polyhedrosis virus in an insect host. Science. 1989;243(4899):1728–1730. doi: 10.1126/science.2648574. [DOI] [PubMed] [Google Scholar]
- Keddie BA, Volkman LE. Infectivity difference between the two phenotypes of Autographa californica nuclear polyhedrosis virus: Importance of the 64K envelope glycoprotein. J Gen Virol. 1985;66(5):1195–1200. [Google Scholar]
- Kielian M, Rey FA. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat Rev Microbiol. 2006;4(1):67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley DH, Behbahani A, Rashtian A, Blissard GW, Zimmerberg J. A discrete stage of baculovirus GP64-mediated membrane fusion. Molecular Biology of the Cell. 1999;10(12):4191–4200. doi: 10.1091/mbc.10.12.4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung O, Westenberg M, Vlak JM, Zuidema D, Blissard GW. Pseudotyping Autographa californica multicapsid Nucleopolyhedrovirus (AcMNPV): F proteins from Group II NPVs are functionally analogous to AcMNPV GP64. J Virol. 2002;76:5729–5736. doi: 10.1128/JVI.76.11.5729-5736.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovic I, Pulyaeva H, Sokoloff A, Chernomordik LV. Membrane fusion mediated by baculovirus gp64 involves assembly of stable gp64 trimers into multiprotein aggregates. J Cell Biol. 1998;143(5):1155–66. doi: 10.1083/jcb.143.5.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LK, editor. The Baculoviruses. New York: Plenum Press; 1997. [Google Scholar]
- Monsma SA, Blissard GW. Identification of a membrane fusion domain and an oligomerization domain in the baculovirus GP64 Envelope Fusion Protein. J Virol. 1995;69(4):2583–2595. doi: 10.1128/jvi.69.4.2583-2595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsma SA, Oomens AGP, Blissard GW. The GP64 Envelope Fusion Protein is an essential baculovirus protein required for cell to cell transmission of infection. J Virol. 1996;70:4607–4616. doi: 10.1128/jvi.70.7.4607-4616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly DR, Miller LK, Luckow VA. Baculovirus expression vectors, a laboratory manual. W. H. Freeman and Co; New York: 1992. [Google Scholar]
- Oomens AGP, Monsma SA, Blissard GW. The baculovirus GP64 Envelope Fusion Protein: synthesis, oligomerization, and processing. Virology. 1995;209:592–603. doi: 10.1006/viro.1995.1291. [DOI] [PubMed] [Google Scholar]
- Plonsky I, Cho MS, Oomens AGP, Blissard GW, Zimmerberg J. An analysis of the role of the target membrane on the gp64-induced fusion pore. Virology. 1999;253:65–76. doi: 10.1006/viro.1998.9493. [DOI] [PubMed] [Google Scholar]
- Volkman LE, Goldsmith PA. Budded Autographa californica NPV 64K protein: further biochemical analysis and effects of postimmunoprecipitation sample preparation conditions. Virology. 1984;139:295–302. doi: 10.1016/0042-6822(84)90375-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman LE, Goldsmith PA. Mechanism of neutralization of budded Autographa californica nuclear polyhedrosis virus by a monoclonal antibody: Inhibition of entry by adsorptive endocytosis. Virology. 1985;143(1):185–195. doi: 10.1016/0042-6822(85)90107-2. [DOI] [PubMed] [Google Scholar]
- Volkman LE, Goldsmith PA, Hess RT, Faulkner P. Neutralization of budded Autographa californica NPV by a monoclonal antibody: Identification of the target antigen. Virology. 1984;133(2):354–362. doi: 10.1016/0042-6822(84)90401-X. [DOI] [PMC free article] [PubMed] [Google Scholar]