SUMMARY
Tumor cells metastasize to distant organs through genetic and epigenetic alterations, including changes in microRNA (miR) expression. Here we find miR-22 triggers epithelial-mesenchymal transition (EMT), enhances invasiveness and promotes metastasis in mouse xenografts. In a conditional mammary gland-specific transgenic (TG) mouse model, we show that miR-22 enhances mammary gland side-branching, expands the stem cell compartment, and promotes tumor development. Critically, miR-22 promotes aggressive metastatic disease in MMTV-miR-22 TG mice, as well as compound MMTV-neu or -PyVT-miR-22 TG mice. We demonstrate that miR-22 exerts its metastatic potential by silencing anti-metastatic miR-200 through direct targeting of the TET (Ten eleven translocation) family of methylcytocine dioxygenases, thereby inhibiting demethylation of the mir-200 promoter. Finally, we show that miR-22 overexpression correlates with poor clinical outcomes and silencing of the TET-miR-200 axis in patients. Taken together, our findings implicate miR-22 as a crucial epigenetic modifier and promoter of EMT and breast cancer stemness towards metastasis.
INTRODUCTION
Metastasis remains one of most complex and challenging problems of contemporary oncology. During metastatic dissemination, a cancer cell from a primary tumor executes a multi-step process: intravasation, survival in the circulation, extravasation, micrometastases and colonization (Fidler, 2003). Recent studies have demonstrated that as tumors progress, genetic and epigenetic mechanisms may result in the emergence of a self-renewing metastatic cancer stem cell (CSC), also referred to as cancer initiating cell (CIC) (Ito et al., 2009; Visvader and Lindeman, 2008). This metastatic CSC eventually enters the blood stream and seeds a secondary tumor in a distinct organ. CSCs/CICs are thought to exist in primary tumors from the very early stages of tumorigenesis and may be the oncogenic derivatives of normal-tissue stem or progenitor cells.
CSCs may also arise as a consequence of the epithelial-mesenchymal transition (EMT), a developmental program that is instrumental to the acquisition of stemness by nontransformed and tumor cells (Chaffer and Weinberg, 2011; Gupta and Massague, 2006; Thiery, 2009). While much progress has been garnered in elucidating the molecular pathways that trigger EMT and metastasis, a number of key mechanistic gaps remain to be explained, suggesting that critical players in the process have yet to be identified.
MicroRNAs (miRNAs) are small non-coding RNAs that lead to silencing of their cognate target genes by either degrading mRNA molecules or inhibiting their translation (Bartel, 2004). Indeed, miRNAs have been implicated in the regulation of a variety of cellular processes, including stemness and metastasis, implying that they can function either as oncogenes or tumor suppressors (Calin et al., 2005; He et al., 2005; Ma et al., 2007). For instance, the miR-200 family members (miR-200s) are known as tumor suppressive miRNAs that regulate the EMT and control stemness by directly targeting transcriptional repressor Zeb1/2 (zinc finger E-box-binding homeobox 1/2) and polycomb repressor complex proteins, such as Bmi1 (B lymphoma Mo-MLV insertion region 1 homolog) and Suz12 (suppressor of zeste 12 homolog) (Iliopoulos et al., 2010; Shimono et al., 2009). Interestingly, miR-200s are downregulated in different types of cancer (Adam et al., 2009; Bendoraite et al., 2010; Olson et al., 2009), and they are specifically downregulated in breast CSCs in comparison with non-tumorigenic cancer cells (Shimono et al., 2009).
The selective deregulation of miRNA gene expression may be due to deletion, amplification or mutations targeting the miRNAs themselves, as well as deregulation of transcription factors and epigenetic regulators targeting the genes encoding them (Breving and Esquela-Kerscher, 2010; Calin et al., 2005). In particular, while, promoter hypermethylation and associated inactivation of tumor suppressor genes is an established molecular hallmark of human cancer (Esteller, 2008; Jones and Baylin, 2007), this epigenetic lesion has recently been extended to the DNA methylation-mediated silencing of miRNAs (Lujambio et al., 2007; Saito et al., 2006; Toyota et al., 2008). DNA methylation can be heritably maintained across cell division, but can also be reversibly/dynamically altered to establish new epigenetic programs.
The recent discovery of members of the TET (Ten eleven translocation) family that can specifically modify DNA by hydroxylating 5-methylcytosine (5mC) may explain how cells can erase existing methylation marks (Ito et al., 2010; Ko et al., 2010; Tahiliani et al., 2009). Within the TET family of proteins, TET1, TET2, and TET3 have been shown to convert 5mC to 5-hydroxymethylcytosine (5hmC), while TET2 inactivating deletion and mutations have been frequently observed in hematopoietic malignancies (Ko et al., 2010; Tahiliani et al., 2009). Notably, the level of 5hmC, so tightly associated with TET gene expression, is found increased in differentiated cells and is profoundly reduced in many types of tumors, identifying 5hmC as a biomarker associated with tumor development (Haffner et al., 2011; Yang et al., 2012).
Here we show that miR-22 triggers enhanced mammary gland hyperplasia as well as a marked expansion of the mammary stem cell compartment, hence triggering tumor initiation. Importantly, we demonstrate that miR-22 promotes the metastatic process and EMT through its ability to repress the expression of miR-200s as well as 5hmC by directly targeting members of the TET family.
RESULTS
miR-22 promotes EMT and tumor invasion and metastasis
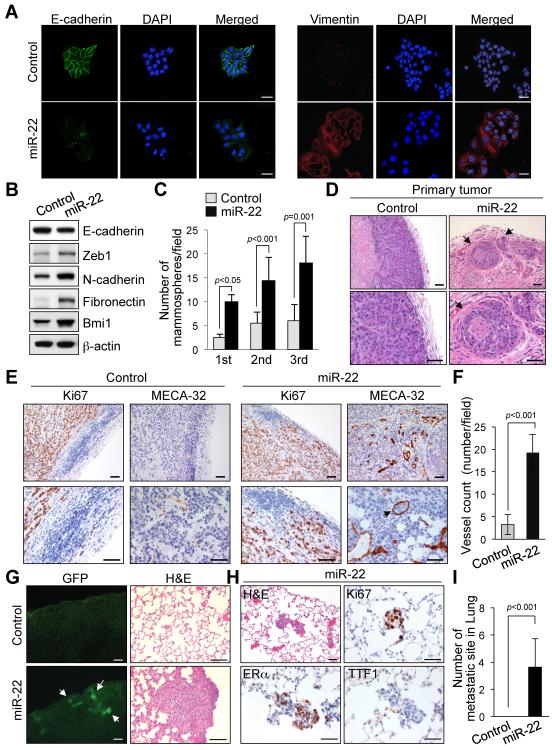
To investigate the potential oncogenic effects of miR-22 expression, we first transduced a retroviral vector encoding the human mir-22 gene into immortalized human breast epithelial cells, HMEC and MCF-10A, and unexpectedly found that ectopic expression of miR-22 results in an increased cell motility in an in vitro assay (Figure S1A and data not shown). A dramatic morphological change was also observed in miR-22-overexpressing cells, in which the typical cobblestone-like appearance of normal epithelium was replaced by a spindle-like, fibroblastic morphology (Figure S1B). In agreement with these observations, we found that miR-22-overexpressing cells displayed a silenced expression of the key epithelial marker E-cadherin, and the upregulations of the mesenchymal markers N-cadherin, Vimentin and Fibronectin (Figures 1A and 1B and Figure S1C). The epithelial-mesenchymal transition (EMT) is known to be a central mechanism responsible for invasiveness and metastasis of breast cancer and is also associated with normal and malignant mammary stem cell function (Gupta and Massague, 2006). Therefore we speculated that miR-22 could also induce stemness. Noticeably, miR-22 expression increased the ability of MCF-10A cells to develop into mammosphere structures, suggesting that miR-22 triggers EMT and stemness (Figure 1C and Figures S1D and S1E).
Figure 1. miR-22 enhances EMT and tumor invasion and metastasis.
(A and B) MCF-10A cells infected with the miR-22 expressing or empty vector were subjected to immunofluorescence (A) and Western blot (B) analyses for the indicated proteins. Scale bars, 20 μm.
(C) Replating efficiency of mammospheres derived from MCF-10A cells expressing miR-22 was measured. The number of mammospheres per 1000-plated cells in each culture was then quantified. The data are represented as mean ± SD from three independent experiments.
(D–F) H&E-stained sections of primary mammary tumors formed by MCF-7 cells expressing miR-22, at 12 weeks after orthotopic transplantation. Arrows indicate areas of lymphatic invasion. Scale bars, 50 μm.
(E and F) Ki67- and MECA-32–stained sections of primary mammary tumors formed by MCF-7 cells expressing miR-22 (E). Arrows indicate tumor cells within a vessel. Scale bars, 50 μm. The quantification of vessel numbers (using MECA-32–stained sections) at the center and the edge of the primary mammary tumors is also shown (F) (n=6).
(G) GFP images (left) and H&E staining (right) of lungs isolated from mice that received orthotopic injection of MCF-7 cells expressing miR-22, at 12 weeks after transplantation. Arrow indicates clusters of metastatic cells. Scale bars, 100 μm.
(H and I) H&E-, Ki67-, ERα- and TTF1-stained sections of lungs isolated from mice that received orthotopic injection of MCF-7 cells expressing miR-22, at 12 weeks after transplantation (H). Scale bars, 50 μm. The number of lung micrometastases (micromets) per section in individual mice was also quantified (I) (n=6).
“see also Figures S1A–S1G”.
To determine whether miR-22 could induce similar effects in vivo, we next established a xenograft model using the non-metastatic breast cancer cell line MCF-7. We used a miR-22/GFP expressing retroviral vector to transduce MCF-7 cells and implanted them into cleared mammary fat pads of Nu/Nu immunodeficient mice (Figures S1F and S1G). Within three weeks of injection, recipient mice developed macroscopic GFP+ tumors with expansive and infiltrative growth patterns; by 12 weeks, in 2 of 6 cases massive lymphatic invasions were also observed in the adjacent normal breast tissues (Figure 1D). In comparison, at the same time point, MCF-7 control tumors were strictly non-invasive. The distribution, but not the total number of Ki67+ (proliferating) cells in miR-22-overexpressing tumors, was also distinct from that seen in the control tumors: in the former, large necrotic centers were apparent and Ki67+ cells were enriched in the highly vascularized invasive regions, whereas in the latter the vascularization was poor and the Ki67+ cells were evenly distributed (Figure 1E and Figure S1F). Furthermore, the vessels associated with the invasive regions of miR-22– overexpressing tumors were observed not only in the stroma (peritumoral), but also within the epithelial tumor masses (intratumoral) (Figures 1E and 1F).
To determine whether miR-22–overexpressing MCF-7 cells could metastasize to distant sites, we undertook a detailed pathological analysis of the major organs of our implanted recipients. Strikingly, 6 out of 6 mice that received the orthotopic implantation of miR-22–overexpressing cells displayed numerous lung metastases that were readily detectable by both GFP fluorescence and histological analysis (Figure 1G). These clusters of cells were stained positive for Ki67 and estrogen receptor α (ERα), and negative for thyroid transcription factor 1 (TTF1; a marker of primary lung adenocarcinoma), further confirming their origin (Figure 1H). In contrast, no metastases were found in control animals (Figure 1I). Taken together, our data suggest that miR-22 can contribute to cell proliferation, invasion and angiogenesis, and importantly, can impart metastatic properties on otherwise non-metastatic cells.
miR-22 increases mammary stem population, and promotes tumorigenesis and metastasis in vivo in transgenic mice
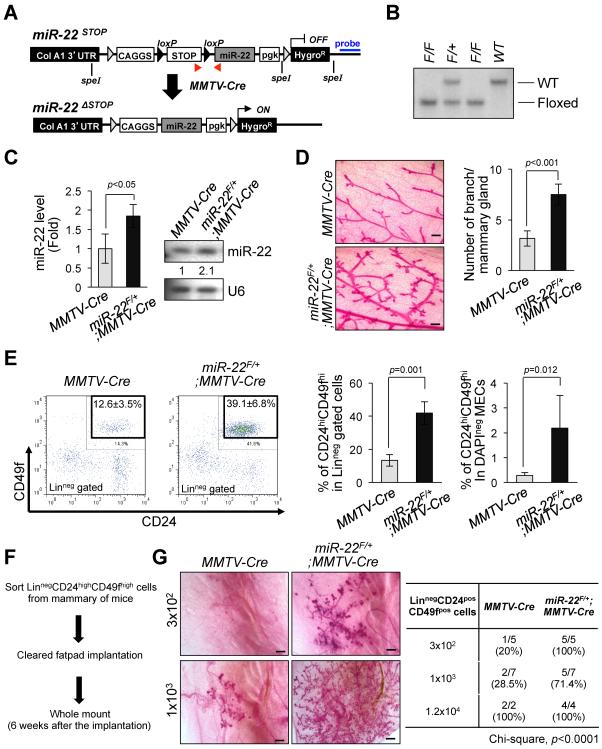
Although these cell line studies were highly suggestive, we attempted to confirm and expand on our findings using a conditional transgenic mouse model. To this end, we engineered mice that harbored within their Collagen A1 locus the CAGGS promoter and the mir-22 genomic sequence separated by a LoxP flanked transcriptional STOP cassette (hereafter referred to as miR-22F/+) (Figures 2A and 2B). By crossing these mice with mice that express the Cre recombinase under the control of the mouse mammary tumor virus promoter (MMTV-Cre), we could induce targeted expression of miR-22 in the mammary glands. Real-time quantitative PCR and Northern blot analyses revealed that miR-22 expression was increased 2~3 fold in miR-22F/+;MMTV-Cre mice compared to littermate controls (Figure 2C).
Figure 2. miR-22 increases mammary gland side-branching and stemness in vivo in transgenic mice.
(A) Schematic representation of the strategy for generation of floxed miR-22 mouse embryonic stem cells. Red arrows indicate the positions of primers used for genotyping the miR-22 transgenic mice. Blue line indicates the position of probe for the Southern blot analysis. The F1 floxed miR-22 founder mice were bred to MMTV-Cre strain to delete LoxP site.
(B) Genomic DNAs were isolated from the tails of miR-22-LoxP mice were digested by Spe I and subjected to Southern blot analysis.
(C) Total RNAs isolated from mammary gland tissues of miR-22F/+;MMTV-Cre mice were subjected to real-time qPCR (left) (n=4) or Northern blot analysis (right) to evaluate miR-22 expression.
(D) Whole mount analyses were conducted on 7-weeks old miR-22F/+;MMTV-Cre mice and MMTV-Cre littermates (left) and the number of mammary gland side-branches was quantified (right) (n=3). Scale bars, 500 μm.
(E) Distribution of CD45negCD31negCD140anegTer119neg mouse mammary cells according to their expression of CD24 and CD49f were analyzed on 7-weeks old miR-22F/+;MMTV-Cre mice and littermate controls (left). Mouse mammary stem cells (MSCs) according to their expression of CD24highCD49high in Lineagenegative (middle) or total mammary epithelial cells (right) were quantified by a flow cytometric analysis (n=4).
(F and G) Schematic representation of limiting dilution transplantation experiments with CD24highCD49fhigh MSCs (F). 3 × 102, 1 × 103 or 1.2 × 104 LinnegCD24highCD49high MSCs isolated from 7-weeks old miR-22F/+;MMTV-Cre mice and littermate controls were injected into the cleared fat-pad of 3-weeks old FVB/NJ female mice and whole mount analyses were then conducted at 6 weeks after injection (G). Representative images of mammary gland side-branches are shown in the left panel. The resulting data were also analyzed by Chi-square test (right).
“see also Figures S1H–S1J”.
Noticeably, in agreement with our observation in cell line studies, epithelial cell migration and Western blot analyses in breast epithelium obtained from miR-22F/+;MMTV-Cre and control mice revealed a marked increase in EMT features in cells from the transgenic animals at young age (Figures S1H and S1I). Strikingly, by 7 weeks of age miR-22 transgenic mice demonstrated an increased ductal side-branching (p<0.001, Figure 2D), higher numbers of mammary stem cells (MSCs; defined as CD24highCD49fhigh) (p=0.001, Figure 2E), and cells from these mice displayed an increased ability to form mammospheres - three-dimensional structures in culture (p=0.019, Figure S1J). In agreement with these findings, it has been previously reported that miR-22 is highly expressed in mouse mammary progenitor cells (Ibarra et al., 2007). Importantly, limiting dilution transplantation experiments with CD24highCD49fhigh MSCs revealed that the increase in lobuloalveolar structures in miR-22 transgenic mice directly relates to MSCs, but not to aberrant hormonal signaling during the oestrus cycle (Figures 2F and 2G).
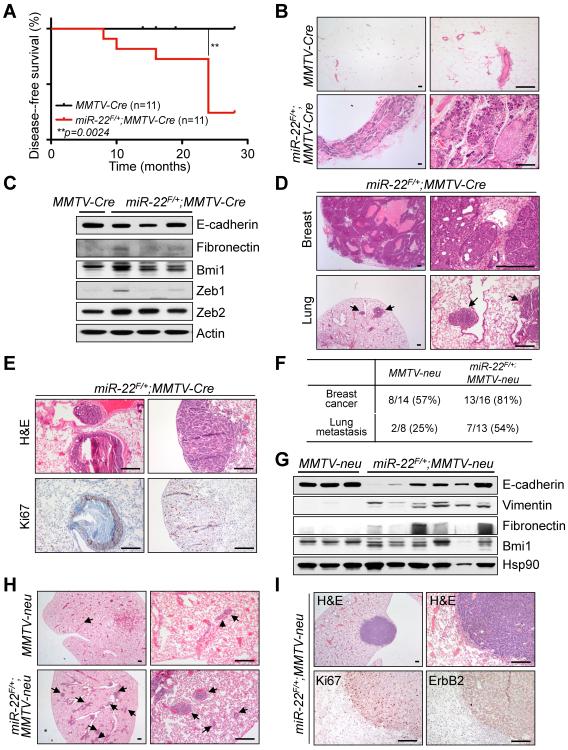
Next, we performed a long-term evaluation of spontaneous mammary tumorigenesis and metastasis in vivo in miR-22 transgenic mice. Remarkably, miR-22 overexpression in mice resulted in a progressive decrease in the disease-free survival rate at the age of 8~24 months (n=11) (p=0.0024, Figure 3A). Diffuse alveolar and ductal hyperplasia, focal mammary ductal ectasia, impaction of thick materials, and more importantly, focal ductal carcinomas were observed in virgin female miR-22F/+;MMTV-Cre mice (at approximately 60% incidence), whereas no hyperplasia nor tumor was observed in control mice (Figure 3B). Noticeably, miR-22F/+;MMTV-Cre mice displayed EMT-related breast tumor phenotypes (Figure 3C) and their primary tumors represented both luminal (cytokeratin 8-positive and cytokeratin 14-negative) and mixed luminal-basal (cytokeratin 8-positive and cytokeratin 14-positive) cell fates, although the intensities of positivity of cytokeratin 8 are partially weaker than those of normal mammary glands (Figure S2A). Most strikingly, post-pregnant miR-22F/+;MMTV-Cre mice developed lung metastases at 8 to 10 months of age (at 100% incidence) mostly in the peri-bronchiolar lymphatic space (n=3) (Figures 3D and 3E).
Figure 3. miR-22 induces mammary tumorigenesis and metastasis in vivo in transgenic mice.
(A) Cumulative disease-free survival analysis. A statistically significant decrease in lifespan for the miR-22F/+;MMTV-Cre cohort was found compared with the MMTV-Cre cohort (p=0.0024, n=11).
(B) H&E-stained sections of diffuse alveolar and ductal hyperplasia isolated from 12-months old miR-22F/+;MMTV-Cre mice. Scale bars, 200 μm.
(C) Lysates from mammary tumors of miR-22F/+;MMTV-Cre mice were subjected to Western blot analysis for the indicated proteins.
(D and E) H&E-stained sections of primary mammary tumors and lungs isolated from 8-months old post-pregnant miR-22F/+;MMTV-Cre mice (D). Arrows indicate clusters of metastatic cells in the lung. Scale bars, 200 μm. H&E- and Ki67-stained sections of lungs isolated from miR-22F/+;MMTV-Cre mice are also shown (E). Scale bars, 200 μm.
(F) The incidence of metastases to the lung in MMTV-neu;miR-22F/+;MMTV-Cre (represented as miR-22F/+;MMTV-neu) and MMTV-neu littermates (8~16 months old) was quantified.
(G) Lysates from mammary tumors of miR-22F/+;MMTV-neu were subjected to Western blot analysis for the indicated proteins.
(H and I) H&E-stained sections of the lungs isolated from miR-22F/+;MMTV-neu mice and littermate controls (H). Arrows indicate clusters of metastatic cells in the lung. Scale bars, 200 μm. H&E-, Ki67- and ErbB2-stained sections of lungs isolated from miR-22F/+;MMTV-neu mice are also shown (I). Scale bars, 200 μm.
“see also Figure S2”.
In addition, by utilizing a series of genetically engineered mouse models, we further explored whether miR-22 can promote breast cancer and metastasis in vivo. As MMTV-PyVT transgenic mice are known to develop multifocal mammary tumors that spontaneously metastasize to the lung (Guy et al., 1992a), we first crossed these mice with our miR-22F/+;MMTV-Cre mice and analyzed the resultant compound mutants for development of primary tumors and lung metastases. Although miR-22 overexpression had no significant effect on the development or size of primary mammary gland lesions in this model, the penetrance of metastatic cancer, analyzed at different time points (8 to 12 weeks), was increased in MMTV-PyVT;miR-22F/+;MMTV-Cre mice when compared to MMTV-PyVT controls (Figures S2B and S2C).
As MMTV-PyVT animals are difficult to compare precisely due to their very early onset of lung metastases, we next generated and analyzed MMTV-neu;miR-22F/+;MMTV-Cre mice, taking advantage of the fact that MMTV-neu mice express unactivated neu (c-ErbB2) and show less aggressive mammary tumors and lung metastases (Guy et al., 1992b). Remarkably, the development of primary mammary gland lesions as well as the incidence of lung metastases was significantly increased in MMTV-neu;miR-22F/+;MMTV-Cre mice, by ~19 months of age, when compared to MMTV-neu controls (Figure 3F and Figure S2D). Importantly, MMTV-neu;miR-22F/+;MMTV-Cre mice exhibited EMT-related breast tumor phenotypes (Figure 3G and Figure S2E). Interestingly, a significant number of MMTV-neu;miR-22F/+;MMTV-Cre breast tumors were both cytokeratin 8-positive and cytokeratin 14-positive, whereas MMTV-neu control primary tumors were cytokeratin 8-positive and cytokeratin 14-negative, suggesting that miR-22 induces a significant luminal-to-basal cell fate change in breast tumor cells in the presence of neu oncogene insult (Figure S2F). Notably, MMTV-neu;miR-22F/+;MMTV-Cre breast tumors also presented a significant increase in the population of CD24posCD90pos cancer stem cells (CSCs) compared to MMTV-neu control tumors (Figure S2G and Ref. (Malanchi et al., 2012)). Finally, the penetrance of metastases to the lung were greatly increased in MMTV-neu;miR-22F/+;MMTV-Cre mice when compared to MMTV-neu control mice of the same age (Figures 3H and 3I).
Taken together, these data indicate that miR-22 plays a crucial role in breast stem cell biology, tumorigenesis and metastasis in vivo.
miR-22 triggers methylation-dependent silencing of miR-200
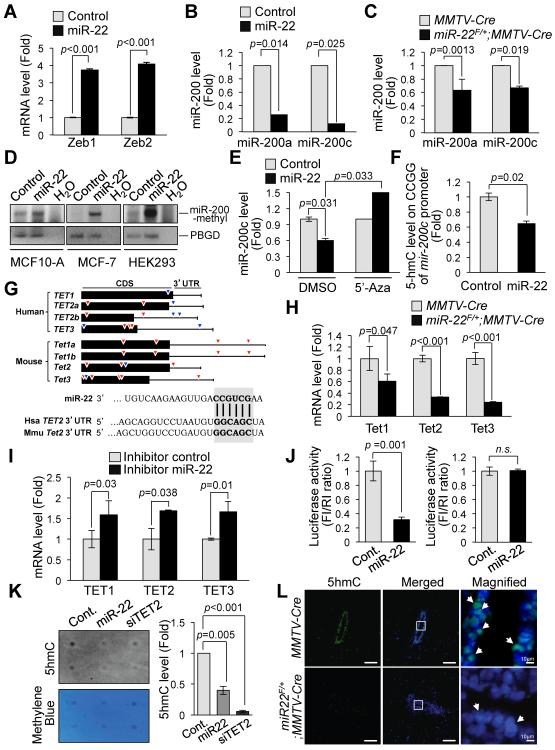
The E-box-binding transcription factors Zeb1 (also known as TCF8 and δEF1) and Zeb2 (also known as ZFXH1B and SMAD-interacting protein 1 [SIP1]), and the miR-200 family members have been shown to regulate EMT in multiple cancer types (Gregory et al., 2008; Park et al., 2008; Peinado et al., 2007). Zeb1 and Zeb2 are key transcriptional repressors of E-cadherin, while the miR-200 family is known as important regulators of both EMT and mammary stem cell function through direct targeting of the mRNA of both Zeb1/Zeb2 and Bmi1 (Iliopoulos et al., 2010; Shimono et al., 2009). Strikingly, we found that miR-22 is able to repress miR-200a and miR-200c expression, leading to upregulation of Zeb1, Zeb2 and Bmi1 in both human breast epithelial cells and mouse mammary epithelium prepared from miR-22F/+;MMTV-Cre mice (Figures 1B and 4A–4C and Figures S1E and S1I). Taken together, these data suggest that in mammary epithelial cells miR-22 induces EMT and stemness by suppressing the expression of miR-200 family.
Figure 4. miR-22 regulates epigenetic inactivation of miR-200 and directly targets TET methylcytosine dioxygenases.
(A and B) Real-time qPCR analysis of Zeb1/Zeb2 (A) or miR-200a/miR-200c (B) with RNAs from MCF-10A cells infected with the miR-22 expressing or empty vector. The data are represented as mean ±SD from three independent experiments.
(C) Real-time qPCR analysis of miR-200a/miR-200c with RNAs obtained from miR-22F/+;MMTV-Cre mice and littermate controls. The data are represented as mean ±SD (n=3).
(D) Methylation-specific PCR analysis of mir-200c CpG islands with genomic DNAs purified from the indicated cells infected with the miR-22 expressing vector.
(E) Restored miR-200c expression upon treatment with DNA demethylating agent 5'-aza-2'-deoxycytidine (5'-Aza) in MCF-10A cells infected with the miR-22 expressing vector. The data are represented as mean ±SD.
(F) GlucMS-qPCR analysis of CpG islands within the mir-200c promoter regions specifically enriched for 5-hydroxymethylcytosine (5hmC) in MCF-10A cells infected with the miR-22 expressing vector. The data are represented as mean ±SD.
(G) Representative seed sequences for miR-22 on the TET family: 7 base pairs (red-colored) and 8 base pairs (blue-colored) on human and mouse TET family (top) and seed match sequences of miR-22 within 3'UTR of human and mouse TET2b as an example (bottom) are shown.
(H) Total RNAs isolated from primary mammary epithelial cells of miR-22F/+;MMTV-Cre mice or littermate controls were subjected to real-time qPCR for Tet1, Tet2 and Tet3 mRNA. The data are represented as mean ±SD (n=3).
(I) Total RNAs isolated from MCF-10A cells transfected with the inhibitor (decoy) of miR-22 were subjected to real-time qPCR for TET1, TET2 and TET3 mRNA. The data are represented as mean ±SD.
(J) Luciferase assay of the luciferase gene linked to the 3'UTR of TET2. HEK293 cells were transiently transfected with a combination of pGL3 firefly luciferase reporter plasmids encoding wild-type (left) or mutated (right) 3'UTR sequences of human TET2b, miR-22 and a Renilla luciferase reporter for normalization. The data are represented as mean ±SD.
(K) Genomic DNA purified from HEK293 cells expressing control miRNA, miR-22 or TET2 siRNA was denatured and neutralized. Global 5hmC levels were then measured by using a dot blot assay with anti-5hmC antibody and normalized by methylene blue staining (left). The resulting 5hmC levels were also quantified (right).
(L) 5hmC- and DAPI-stained sections of the duct of mammary glands isolated from 7-weeks old miR-22F/+;MMTV-Cre mice or littermate controls. Scale bar, 100 μm. The arrows indicate the cells with strong 5hmC positive signals.
“see also Figure S3”.
Recent studies have likewise shown that CpG island hypermethylation-mediated epigenetic silencing of the miR-200 family is associated with upregulation of Zeb1/Zeb2 expression, EMT and metastasis (Davalos et al., 2011; Neves et al., 2010; Vrba et al., 2010). We therefore analyzed the promoter of mir-200 using the methyl-specific PCR (MSP) analysis and found that it is indeed hypermethylated upon miR-22 overexpression (Figure 4D). Notably, treatment of these cells with a DNA-demethylating agent, 5'-aza-2'-deoxycytidine (5'-Aza), reversed the effects of miR-22 and restored expression of miR-200 transcripts (Figure 4E and Figure S3A), suggesting that miR-22 may have a functional role in the dynamic control of DNA methylation.
The recent identification of 5-hydroxymethylcytosine (5hmC) as a novel demethylation standpoint and a biomarker associated with tumor development (Cimmino et al., 2011; Haffner et al., 2011; Nestor et al., 2012; Song et al., 2011; Wu and Zhang, 2010; Yang et al., 2012) prompted us to hypothesize that miR-22 may antagonize miR-200 transcription by controlling 5hmC levels on genomic DNA. Indeed, miR-22 overexpression significantly depleted 5hmC levels within mir-200 promoter CpG islands (Figure 4F). Taken together, these results suggest that miR-22 suppresses the demethylation of the promoter within the mir-200 gene and represses its expression, leading to upregulation of its cognate targets.
miR-22 regulates the level of 5-hydroxymethylcytosine by directly targeting TET family members
miR-22 has been shown to exert growth-suppressive functions in vitro in some cancer cell lines (Ling et al., 2012; Pandey and Picard, 2009); our investigation, however, revealed an unexpected oncogenic function for miR-22, which we further explored in order to identify the targets of its oncogenic activity in breast cancer. Potential molecular targets of miR-22 were predicted by prediction algorithms including TargetScan 6.0 (http://www.targetscan.org) (Lewis et al., 2005), microRNA.org (http://www.microrna.org) (Betel et al., 2008) and miRBase (http://www.mirbase.org) (Griffiths-Jones et al., 2006). Among the potential targets, we focused on the TET (Ten eleven translocation) family – enzymes that contribute to DNA demethylation by converting 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) and have recently been implicated in epigenetic reprogramming, cellular differentiation and cancer (Ito et al., 2010; Ko et al., 2010; Langemeijer et al., 2009; Tahiliani et al., 2009) (Figure 4G and Figure S3B). Remarkably, real-time qPCR and Western blot analyses revealed that miR-22 expression in MCF-10A, human embryonic kidney (HEK) 293 cells, and in vivo in the mammary glands of miR-22F/+;MMTV-Cre mice leads to a robust downregulation of TET family members (Figure 4H and Figures S1I, S3C and S3D). This was in agreement with the high-degree of cross species conservations of the predicted miR-22 microRNA recognition elements (MREs) in TET family members (Figure 4G and Figure S3B). In contrast, inhibiting miR-22 by using antisense oligonucleotides resulted in a significant elevation in expression of TET family members (Figure 4I). Notably, a luciferase reporter assay performed using the 3'UTR region of TET2 (which however contains the miR-22 MREs also identical within the 3'UTR of TET1, TET2 and TET3) (Figure 4G), and its mutant version in the miR-22 MREs, demonstrated that miR-22-mediated repression of TET is due to a direct interaction between miR-22 and the TET genes (Figure 4J and Figures S3E and S3F).
A reduction in the levels of 5hmC has been recently observed in cancer cells, along with inactivation of TET proteins (Haffner et al., 2011; Nestor et al., 2012; Yang et al., 2012). We therefore next assessed whether miR-22 can remodel the epigenetic landscape by altering 5hmC levels in the genome through the TET family. Dot blot assay and immunofluorescence analysis with anti-5hmC antibodies revealed that miR-22 overexpression in MCF-10A cells is indeed able to reduce global 5hmC levels (Figure 4K and Figure S3G). More importantly, 5hmC levels were drastically reduced in vivo in the mammary glands of miR-22F/+;MMTV-Cre mice compared to littermate controls (Figure 4L). Taken together, these data suggest that changes in this important epigenetic mark (e.g. 5hmC) may contribute to the ability of miR-22 to impact on EMT, stemness and metastasis.
Downregulation of miR-22 or the TET family members alters EMT, stemness and miR-200 expression levels
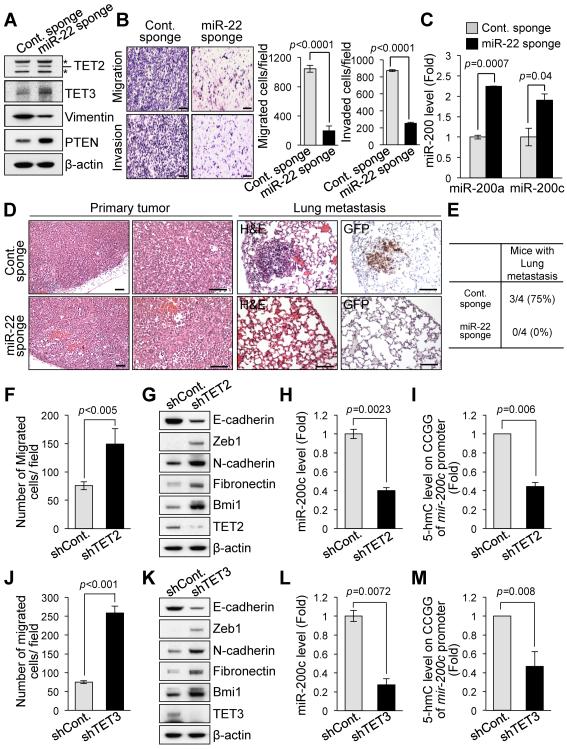
Our observations that ectopic expression of miR-22 promotes EMT and represses TET and miR-200 in turn prompted us to investigate whether miR-22 inhibition could suppress EMT, leading to the elevation of expression of TET and miR-200. To address this possibility, we first evaluated miR-22 expression rates in a large panel of breast cancer cells (including 2 non-tumorigenic cells, 7 ERα-positive and 4 ERα-negative breast cancer cell lines); we then focused on LM2 cells as these cells are highly metastatic and express a substantial amount of miR-22 compared to other breast cancer cell lines (Figure S4A). Next, we generated a retroviral polymerase II sponge by inserting miR-22 binding sites, arrayed in tandem, into the 3'UTR of a reporter gene encoding destabilized GFP driven by the CMV promoter (Figure S4B, top and Ref. (Ebert et al., 2007)). These miR-22 binding sites were perfectly complementary in the seed region, with a bulge at positions 9–12 to prevent degradation of the sponge RNA (Figure S4B, bottom). To examine the effects of miR-22 inhibition on EMT and miR-200 expression, we infected LM2 cells expressing abundant endogenous miR-22 with either the CXCR4 control sponge plasmid (Cont. Sponge) or the sponge plasmid complementary to miR-22 (miR-22 Sponge). Importantly, inhibition of miR-22 by the miR-22 sponge in LM2 cells led to a reduction of metastatic phenotypes, as measured by cell migration, invasion abilities and the expression of the mesenchymal marker Vimentin accompanied with elevation in the levels of TET proteins (Figures 5A and 5B). Noticeably, inhibiting miR-22 by the miR-22 sponge also resulted in a significant increase in the levels of miR-200 family members (Figure 5C). In agreement with these in vitro observations, miR-22 decoying by the miR-22 sponge drastically inhibited in vivo breast cancer metastases to the lung in xenograft models established with LM2 cells (Figures 5D and 5E).
Figure 5. Loss of miR-22 or TET family members alters EMT, stemness and miR-200 levels.
(A) Cell lysates from LM2 cells infected with the control or miR-22 sponge were subjected to Western blot analysis for the indicated proteins. PTEN protein was used as a verified miR-22 target to show the efficacy of the miR-22 sponge in this analysis.
(B) LM2 cells infected with the miR-22 sponge were subjected to the cell migration (top) and invasion assay (bottom). Representative fields of the cells are shown (left). Scale bars, 100 μm. The migrated or invaded cells were also quantified (right). The data are represented as mean ±SD from three independent experiments.
(C) Real-time qPCR analysis of miR-200a/miR-200c with RNAs from LM2 cells infected with the miR-22 sponge. The data are represented as mean ±SD.
(D and E) H&E-stained sections of primary mammary tumors (left), or H&E-stained and Ki67-stained sections of lungs isolated from mice that received orthotopic injection of control- or miR-22 sponge-infected LM2 cells (right) (D). Scale bars, 100 μm. The incidence of metastases to the lung in mice at 10 weeks after orthotopic injection is also shown (E).
(F and J) MCF-10A cells infected with the lentiviral vector expressing TET2 (F) or TET3 (J) shRNA were subjected to the cell migration assay and the migrated cells were then quantified. The data are represented as mean ±SD.
(G and K) Cell lysates from MCF-10A cells expressing TET2 (G) or TET3 (K) shRNA were subjected to Western blot analysis for the indicated proteins.
(H and L) Real-time qPCR analysis of miR-200c with RNAs from MCF-10A cells expressing TET2 (H) or TET3 (L) shRNA. The data are represented as mean ±SD.
(I and M) GlucMS-qPCR analysis of CpG islands within the mir-200c promoter regions specifically enriched for 5hmC in MCF-10A cells expressing TET2 (I) or TET3 (M) shRNA. The data are represented as mean ±SD.
“see also Figures S4A–S4F”.
Next, we investigated whether repression of TET family members could account for the phenotypes caused by miR-22 overexpression in EMT, stemness and metastasis. To this end, we knocked down TET2 or TET3 in MCF-10A cells by using RNA interference (RNAi), and analyzed the resulting effect on EMT and stemness. Notably, knockdown of TET2 or TET3 by RNAi enhanced epithelial cell migration, EMT and mammosphere formation, suggesting that repression of the TET family creates phenotypes similar to those of miR-22 overexpression (Figures 5F, 5G, 5J and 5K and Figures S4C–S4E). Furthermore, downregulation of TET2 or TET3 resulted in a reduction in 5hmC levels at the mir-200 promoter, and caused a decline in miR-200 expression (Figures 5H, 5I, 5L and 5M).
To further understand the metastasis-suppression function of TET family members in vivo, we analyzed lung metastases in MCF-7 xenograft models with stable knockdown of TET2. Importantly, mice that received the orthotopic implantation of TET2-depleted MCF-7 cells displayed a suggestive lung metastasis (Figure S4F). Taken together, these data indicate that TET family members inhibit EMT, stemness and tumor metastasis, and positively regulate miR-200 expression.
miR-22-induced EMT, stemness and repression of miR-200 rely on its ability to silence TET family members
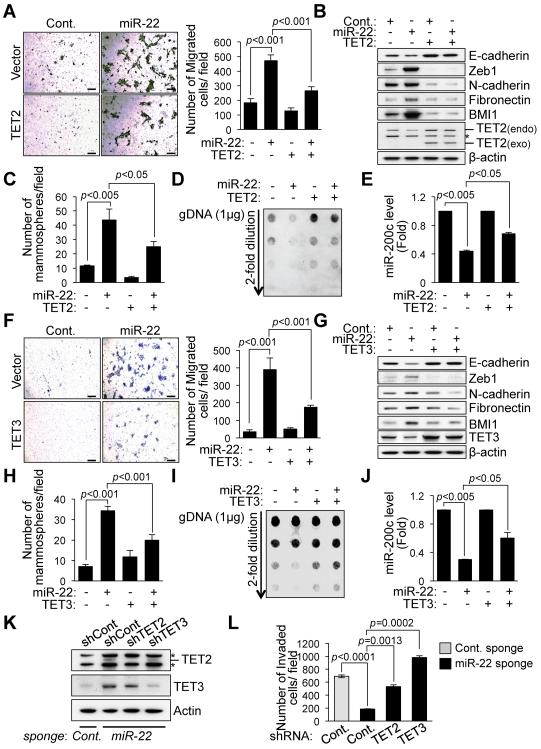
To finally determine whether TET family members directly contribute to miR-22 function, we ectopically expressed TET2 or TET3 in miR-22-overexpressing MCF-10A cells and then analyzed epithelial cell migration, EMT and stemness. Importantly, the exogenously expressed TET proteins significantly diminished the cell migration, EMT and mammosphere formation elicited by miR-22 overexpression (Figures 6A–6C and 6F–6H). Furthermore, silencing the expression of global 5hmC and miR-200 caused by miR-22 was rescued by the ectopic expression of TET2 or TET3 (Figures 6D, 6E, 6I and 6J and Figure S4G), suggesting that TET proteins are very likely to be major factors in reliable miR-22 function in suppression of demethylation of miR-200.
Figure 6. TET family members are required for miR-22-induced EMT, stemness and miR-200 repression.
(A and F) MCF-10A cells infected with a combination of the miR-22, TET2b (A) and TET3 (F) expressing vector were subjected to the cell migration assay. Representative fields of the migrated cells are shown (left). Scale bars, 100 μm. The migrated cells were also quantified (right). The data are represented as mean ±SD from three independent experiments.
(B and G) Cell lysates from MCF-10A cells expressing a combination of the miR-22, TET2b (B) and TET3 (G) were subjected to Western blot analysis for the indicated proteins. Asterisk indicates non-specific band.
(C and H) Mammospheres derived from MCF-10A cells expressing a combination of the miR-22, TET2b (C) and TET3 (H) were measured. The number of mammospheres per 1000-plated cells in each culture was then quantified. The data are represented as mean ±SD.
(D and I) Genomic DNA purified from MCF-10A cells expressing a combination of the miR-22, TET2b (D) and TET3 (I) was denatured and neutralized. Global 5hmC levels were then measured by a dot blot assay using anti-5hmC antibody.
(E and J) Real-time qPCR analysis of miR-200c with RNAs from MCF-10A cells expressing a combination of the miR-22, TET2b (E) and TET3 (J). The data are represented as mean ±SD.
(K and L) LM2 cells infected with a combination of the control and miR-22 sponge and the TET2 or TET3 shRNA expressing vector were subjected to Western blot analysis for the indicated proteins (K) and the cell invasion assay (L). Asterisk indicates non-specific band. The data are represented as mean ±SD.
“see also Figures S4G and S4H”.
To further evaluate whether the invasion-suppression capacity of miR-22 inhibition directly relates to the TET family, we knocked down TET2 or TET3 by RNAi in LM2 metastatic breast cancer cell line after applying the miR-22 sponge (Figure 6K). Noticeably, the dramatic reduction in cell invasion caused by miR-22 inhibition was overcome by knockdown of TET proteins (Figure 6L and Figure S4H). Taken together, these data suggest that TET family members are the major players responsible for miR-22 function in EMT, stemness and repression of miR-200.
miR-22 overexpression correlates with poor clinical outcomes and silencing of the TET-miR-200 axis in patients
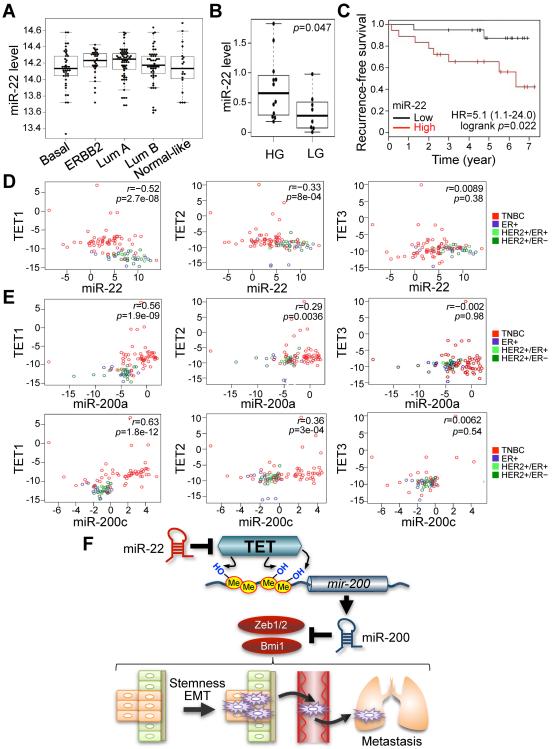
Finally, we sought to determine whether our observations could be verified in human breast cancer patients. Indeed, we observed a high expression rate of miR-22 in non-triple-negative breast cancer (non-TNBC) subtypes of breast cancer when compared to normal breast tissue (Figure 7A and Figure S5A and Refs. (Buffa et al., 2011; Enerly et al., 2011)).
Figure 7. miR-22 overexpression correlates with poor clinical outcomes and silencing of the TET-miR-200 axis in patients.
(A) miR-22 expression profiling was analyzed from a previously published Illumina Human RefSeq-8 and miRNAv1 array dataset (superSeries GSE22220). Breast tumors were classified by molecular subtypes, estrogen receptor (ER), progesterone receptor (PR), ERBB2 (HER-2) and epithelial clusters.
(B) miR-22 expression was analyzed by using a real-time qPCR with RNAs from human breast cancer patient samples. Tumor grade was determined by the parameters, ER, PR, ERBB2 (HER-2) and epithelial clusters. HG, ER-positive high-grade tumors; LG, ER-positive low-grade tumors.
(C) Non triple-negative breast cancer (TNBC) patient samples were sub-divided into two groups according to low and high expression of miR-22 with median split of all samples. A Kaplan-Meier plot representing the disease-free survival of patients was stratified.
(D) Anti-correlation between miR-22 and TET family expressions was analyzed using a real-time qPCR with RNAs from breast cancer patient samples.
(E) Co-expression analysis of TET family and miR-200a/miR-200c was analyzed by using a real-time qPCR with RNAs from breast cancer patient samples.
(F) Proposed model of the role of miR-22 for EMT, stemness and metastasis through epigenetic inactivation of miR-200 by directly targeting the TET family. miR-22 decreases the level of 5hmC by negatively regulating TET family, followed by epigenetic inactivation of miR-200 due to the reduced 5hmC levels. Ultimately, dysfunction of miR-200 triggers EMT and stemness, which in turn increases mammary tumorigenesis and metastasis.
To further evaluate the relationship between miR-22 overexpression and tumor aggressiveness in non-TNBC patients, we analyzed miR-22 levels in breast cancer tissue samples positive for ERα (estrogen receptor α) by real-time qPCR, using RNU6B as an internal control. Critically, the breast cancer patients found to have high expression levels of miR-22 had high-grade tumors of an advanced stage (p=0.047; Figure 7B), and poor survival rates as compared to the patients with low miR-22 expression levels (p=0.022, Figure 7C).
Interestingly, in this analysis miR-22 was also found to be significantly co-expressed with genes involved in breast cancer metastasis, including those that encode matrix metalloproteases 11 and 1 (Pearson r=0.536 and 0.392, respectively), cathepsin B (0.500), integrin α11 (0.425), TGF-βR-associated protein (0.416), collagen Xα1 (0.411), TGF-β1 (0.407), fibronectin 1 (0.403) and laminin α5 (0.399) (Bierie and Moses, 2006; McCarthy et al., 1985; Melisi et al., 2008; Radisky and Radisky, 2010; Vuoristo et al., 2007; Withana et al., 2012) (Table S1). This observation also supports the notion that in breast cancer miR-22 is pro-metastatic and contributes to aggressive disease.
More importantly, and consistent with our functional studies, miR-22 expression was directly anti-correlated with the expression of TET1 and TET2 in human breast cancer patients; in addition, a statistically significant anti-correlation between miR-22 and TET3 was also observed, but only in the non-TNBC breast cancer subset (Figure 7D and p=0.014, Figure S5B).
Lastly, we examined whether downregulation of TET family members by miR-22 can explain its effects on miR-200s expression in human breast cancer tissues. In support of this possibility, we found a clear correlation between the expression of TET1 and TET2 and miR-200s in human breast cancer patients; while a correlation was also found between TET3 and miR-200s, but this was not statistically significant (Figure 7E and Figures S5C). Taken together, these results reveal miR-22 as a crucial and unexpected switch for a metastatic phenotype of breast cancer via repression of TET family members/miR-200s.
DISCUSSION
Collectively, our findings have allowed us to reach a number of important conclusions:
-
i)
miR-22 promotes EMT and tumor invasion and metastasis. Using in vivo miR-22 transgenic mouse models we further show miR-22 can promote lobuloalveolar structures and MSCs stemness, and impart mammary tumor development and invasive and metastatic properties. Other studies have demonstrated that miR-22 could play a role in the pathogenesis of breast cancer by interacting with the 3'UTR region of ERα (estrogen receptor α) and regulating ERα expression (Pandey and Picard, 2009; Xiong et al., 2010). However, we and others found no effect of miR-22 on expression of ERα in either clinical samples or miR-22F/+;MMTV-Cre transgenic animals (Figure 1H and Figure S1I and Refs. (Pandey and Picard, 2009; Yoshimoto et al., 2011)).
The findings presented here are coherent with a proto-oncogenic role for miR-22, and with previous findings implicating miR-22 in the regulation of PTEN (phosphatase and tensin homolog deleted on chromosome 10) tumor suppressor (Poliseno et al., 2010). While inactivation of PTEN may contribute to the role of miR-22 in breast cancer initiation, however, it has little effect on EMT in the various tissues including breast (our unpublished observation and Refs. (Mulholland et al., 2012; Shorning et al., 2011; Wang et al., 2012)). Moreover, unlike miR-22F/+;MMTV-Cre transgenic mice, the 5hmC levels in precancerous mammary glands of Pten mutant mice were not appreciably different from littermate controls, indicating that miR-22 functions as an epigenetic modifier and EMT promoter independently of its ability to target PTEN (Figure S3H).
-
ii)
We propose a new mechanism and a new model for antagonistic miRNA crosstalks whereby one miRNA (e.g. miR-22) can suppress the expression of another miRNA (e.g. miR-200) via direct targeting of chromatin remodeling enzymes such as TET family members, which in this specific example in turn leads to the hypermethylation of the mir-200 promoter and induction of EMT and stemness (Figure 7F). Indeed, aberrant DNA methylation of CpG islands from mir-200 family appears to be closely linked to their silencing during EMT and metastasis, while epigenetic modifier drugs such as 5-aza-2'-deoxycytidine can reactivate miR-200 family expression, indicating that epigenetic mechanisms play a functional role for controlling the expression of this family in cancer (Davalos et al., 2011; Li et al., 2010; Vrba et al., 2010).
-
iii)
TET2 inactivating deletion and mutations have been observed in hematopoietic malignancies (Cimmino et al., 2011; Shih et al., 2012), and yet to date, whether TET family members play a role in solid tumors and metastasis remained an open question. Our results, i.e., that inhibition of TET proteins triggers a similar phenotype as miR-22 overexpression in EMT and stemness, and that the phenotypes caused by miR-22 are abolished by ectopic expression of TET proteins, demonstrate that the TET family acts as a major target in mediating the function of miR-22 in breast cancer and metastasis. In turn, these data suggest that the functional and concomitant loss of TET family members may be extremely relevant to the pathogenesis of solid tumors.
It has also been recently reported that mutations of the isocitrate dehydrogenase genes IDH1 and IDH2 can lead to the aberrant production of 2-hydroxyglutarate (2-HG), a metabolite that inhibits TET2 enzymatic activity, resulting in a hypermethylated promoter phenotype in acute myeloid leukemia (AML) tumors carrying IDH1/2 mutations (Figueroa et al., 2010), further highlighting a possible critical role for TET family members in the epigenetic deregulation observed in many cancers. Importantly, beside mutations specifically targeting TET2 or IDH1/2 genes, our results now reveal an additional and global silencing mechanism of the whole TET family by miR-22, which can block the 5mC-to-5hmC conversion and thus DNA demethylation. The fact that miR-22 can target multiple TET family members at once explains on the one hand its powerful effects on the global epigenetic landscape, but also its potent proto-oncogenic role in multiple tissues (Song and Ito et al. Cell Stem Cell, in press).
-
iv)
Our findings not only help at further defining the molecular basis of tumor metastasis, but also have important implications for the diagnosis, prognosis and treatment of cancer. As miR-22 triggers EMT and a metastatic phenotype and its overexpression correlates with poor clinical outcomes in patients, it might be utilized as a useful biomarker to identify metastatic forms of breast cancer. Critically, our results that inhibiting miR-22 by the miR-22 sponge in highly metastatic cells leads to a reduction of metastatic phenotypes as well as an elevation of the expression of TET and miR-200, provide a rationale for the therapeutic targeting of miR-22 (e.g. by using the LNA (locked nucleic acid) miR-22 inhibitors) in order to prevent EMT and metastatic phenotypes.
EXPERIMENTAL PROCEDURES
Mice
The details of generation of miR-22 transgenic mice and the sources of primers used for genotyping are described in the Extended Experimental Procedures.
Plasmids
The details of generation of plasmids used are described in the Extended Experimental Procedures.
Cell culture and Transfection
HMEC, MCF-10A, MCF-7, HEK293 and 293T cell lines were maintained as described in the Extended Experimental Procedures. The details of transfection of siRNA/miRNAs mimic or microRNA inhibitors are also described in the Extended Experimental Procedures.
Immunohistochemistry
The details of immunohistochemical analysis are described in the Extended Experimental Procedures.
Immunocytochemistry
For immunofluorescence analysis of 5hmC and 5mC, cells were fixed and stained as de scribed (Ko et al., 2010). The details of staining of cells with antibodies against E-cadher in, Vimentin or Fibronectin are also described in the Extended Experimental procedures.
Orthotopic injection
4-weeks old female Nu/Nu immunodeficient mice (Jackson Laboratory) were used for surgery. The details of mammary gland orthotopic injection are described in the Extended Experimental Procedures.
Whole mount staining
The fourth inguinal glands were dissected at the indicated ages and were spread on a glass slide. After fixation with acidic alcohol for 2 hrs, the tissues were hydrated and stained in Carmine alum for overnight as described (Jackson-Fisher et al., 2004). Samples were then dehydrated, cleared with xylene and mounted.
Mouse mammary stem cell analysis
Mammary glands were dissected from 7-week old mice. After mechanical dissociation, mammary stem cells were analyzed as described in detail in the Extended Experimental Procedures.
Human patient samples
Human patient samples were analyzed as described in detail in the Extended Experimental Procedures. This study was approved by the institutional review boards of Dana Farber/Harvard Cancer Center.
Real-time quantitative PCR
The details of RNA isolation and real-time qPCR analysis are described in the Extended Experimental Procedures.
Glucosylation of genomic 5hmC followed by methylation sensitive qPCR (glucMS-qPCR)
The details of glucMS-qPCR analysis and all the sources of primers used are described in the Extended Experimental Procedures.
Quantitative analysis of 5hmC and 5mC levels using a dot-blot analysis
Genomic DNA was denatured, neutralized and analyzed for 5hmC and 5mC levels as described in detail in the Extended Experimental Procedures.
Methylation specific PCR
DNA methylation status was examined by the methylation-specific PCR with genomic DNA treated with sodium bisulfite using the EZ DNA Methylation-Direct kit (Zymo Research). Primers as described were used (Davalos et al., 2011).
Luciferase reporter assay
The details of luciferase reporter assay are described in the Extended Experimental Procedures.
Western blot analysis
The details of Western blot analysis are described in the Extended Experimental Procedures.
Migration and invasion assay
The details of cell migration and invasion assay are described in the Extended Experimental Procedures.
Statistical analysis
Student's t test was utilized to determine statistical significance unless otherwise specified. p values lower than 0.05 were considered statistically significant.
Supplementary Material
Highlights
miR-22 triggers epithelial-mesenchymal transition, invasion and metastasis.
miR-22 enhances murine mammary gland side-branching, stemness and tumor development.
miR-22 antagonizes miR-200 by directly targeting TET family members.
miR-22 is linked to poor clinical outcomes and silencing of the TET-miR-200 axis in patients.
ACKNOWLEDGEMENTS
We are grateful to all members of the Pandolfi lab and in particular to Keisuke Ito and Leonardo Salmena for advice and discussion. We thank Thomas Garvey for critical editing of the manuscript as well as Yvonne Tay, Luo Man-Lee, April Greene-Colozzi and Soon Jung Kim for experimental and technical support. This work was supported by NIH grant CA 082328-14 awarded to P.P.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin. Cancer Res. 2009;15:5060–5072. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bendoraite A, Knouf EC, Garg KS, Parkin RK, Kroh EM, O'Briant KC, Ventura AP, Godwin AK, Karlan BY, Drescher CW, et al. Regulation of miR-200 family microRNAs and ZEB transcription factors in ovarian cancer: evidence supporting a mesothelial-to-epithelial transition. Gynecol. Oncol. 2010;116:117–125. doi: 10.1016/j.ygyno.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–153. doi: 10.1093/nar/gkm995. microRNA.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierie B, Moses HL. Tumour microenvironment: TGFbeta: the molecular Jekyll and Hyde of cancer. Nat. Rev. Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- Breving K, Esquela-Kerscher A. The complexities of microRNA regulation: mirandering around the rules. Int. J. Biochem. Cell Biol. 2010;42:1316–1329. doi: 10.1016/j.biocel.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Buffa FM, Camps C, Winchester L, Snell CE, Gee HE, Sheldon H, Taylor M, Harris AL, Ragoussis J. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011;71:5635–5645. doi: 10.1158/0008-5472.CAN-11-0489. [DOI] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- Cimmino L, Abdel-Wahab O, Levine RL, Aifantis I. TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell. 2011;9:193–204. doi: 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos V, Moutinho C, Villanueva A, Boque R, Silva P, Carneiro F, Esteller M. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene. 2011;31:2062–2074. doi: 10.1038/onc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enerly E, Steinfeld I, Kleivi K, Leivonen SK, Aure MR, Russnes HG, Ronneberg JA, Johnsen H, Navon R, Rodland E, et al. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One. 2011;6:e16915. doi: 10.1371/journal.pone.0016915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat. Rev. Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 1992a;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc. Natl. Acad. Sci. USA. 1992b;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, Toubaji A, Argani P, Iacobuzio-Donahue C, Nelson WG, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2:627–637. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 2007;21:3238–3243. doi: 10.1101/gad.1616307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell. 2010;39:761–772. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Bernardi R, Pandolfi PP. A novel signaling network as a critical rheostat for the biology and maintenance of the normal stem cell and the cancer-initiating cell. Curr. Opin. Genet. Dev. 2009;19:51–59. doi: 10.1016/j.gde.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Fisher AJ, Bellinger G, Ramabhadran R, Morris JK, Lee KF, Stern DF. ErbB2 is required for ductal morphogenesis of the mammary gland. Proc. Natl. Acad. Sci. USA. 2004;101:17138–17143. doi: 10.1073/pnas.0407057101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat. Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li A, Omura N, Hong SM, Vincent A, Walter K, Griffith M, Borges M, Goggins M. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer Res. 2010;70:5226–5237. doi: 10.1158/0008-5472.CAN-09-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling B, Wang GX, Long G, Qiu JH, Hu ZL. Tumor suppressor miR-22 suppresses lung cancer cell progression through post-transcriptional regulation of ErbB3. J. Cancer Res. Clin. Oncol. 2012;138:1355–1361. doi: 10.1007/s00432-012-1194-2. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Git A, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA, Delaloye JF, Huelsken J. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2012;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- McCarthy JB, Basara ML, Palm SL, Sas DF, Furcht LT. The role of cell adhesion proteins--laminin and fibronectin--in the movement of malignant and metastatic cells. Cancer Metastasis Rev. 1985;4:125–152. doi: 10.1007/BF00050692. [DOI] [PubMed] [Google Scholar]
- Melisi D, Ishiyama S, Sclabas GM, Fleming JB, Xia Q, Tortora G, Abbruzzese JL, Chiao PJ. LY2109761, a novel transforming growth factor beta receptor type I and type II dual inhibitor, as a therapeutic approach to suppressing pancreatic cancer metastasis. Mol. Cancer Ther. 2008;7:829–840. doi: 10.1158/1535-7163.MCT-07-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland DJ, Kobayashi N, Ruscetti M, Zhi A, Tran LM, Huang J, Gleave M, Wu H. Pten Loss and RAS/MAPK Activation Cooperate to Promote EMT and Metastasis Initiated from Prostate Cancer Stem/Progenitor Cells. Cancer Res. 2012;72:1878–1889. doi: 10.1158/0008-5472.CAN-11-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor CE, Ottaviano R, Reddington J, Sproul D, Reinhardt D, Dunican D, Katz E, Dixon JM, Harrison DJ, Meehan RR. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res. 2012;22:467–477. doi: 10.1101/gr.126417.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves R, Scheel C, Weinhold S, Honisch E, Iwaniuk KM, Trompeter HI, Niederacher D, Wernet P, Santourlidis S, Uhrberg M. Role of DNA methylation in miR-200c/141 cluster silencing in invasive breast cancer cells. BMC Res. 2010;3:219. doi: 10.1186/1756-0500-3-219. Notes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson P, Lu J, Zhang H, Shai A, Chun MG, Wang Y, Libutti SK, Nakakura EK, Golub TR, Hanahan D. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–2165. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey DP, Picard D. miR-22 inhibits estrogen signaling by directly targeting the estrogen receptor alpha mRNA. Mol. Cell. Biol. 2009;29:3783–3790. doi: 10.1128/MCB.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci. Signal. 2010;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radisky ES, Radisky DC. Matrix metalloproteinase-induced epithelialmesenchymal transition in breast cancer. J. Mammary Gland Biol. Neoplasia. 2010;15:201–212. doi: 10.1007/s10911-010-9177-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the protooncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat. Rev. Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorning BY, Griffiths D, Clarke AR. Lkb1 and Pten synergise to suppress mTOR-mediated tumorigenesis and epithelial-mesenchymal transition in the mouse bladder. PLoS One. 2011;6:e16209. doi: 10.1371/journal.pone.0016209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP. Metastasis: alone or together? Curr. Biol. 2009;19:R1121–1123. doi: 10.1016/j.cub.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat. Rev. Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, Stampfer MR, Futscher BW. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One. 2010;5:e8697. doi: 10.1371/journal.pone.0008697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuoristo M, Vihinen P, Vlaykova T, Nylund C, Heino J, Pyrhonen S. Increased gene expression levels of collagen receptor integrins are associated with decreased survival parameters in patients with advanced melanoma. Melanoma Res. 2007;17:215–223. doi: 10.1097/CMR.0b013e328270b935. [DOI] [PubMed] [Google Scholar]
- Wang T, Zhang L, Shi C, Sun H, Wang J, Li R, Zou Z, Ran X, Su Y. TGF-beta-induced miR-21 negatively regulates the antiproliferative activity but has no effect on EMT of TGF-beta in HaCaT cells. Int. J. Biochem. Cell Biol. 2012;44:366–376. doi: 10.1016/j.biocel.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Withana NP, Blum G, Sameni M, Slaney C, Anbalagan A, Olive MB, Bidwell BN, Edgington L, Wang L, Moin K, et al. Cathepsin B Inhibition Limits Bone Metastasis in Breast Cancer. Cancer Res. 2012;72:1199–1209. doi: 10.1158/0008-5472.CAN-11-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Yu D, Wei N, Fu H, Cai T, Huang Y, Wu C, Zheng X, Du Q, Lin D, et al. An estrogen receptor alpha suppressor, microRNA-22, is downregulated in estrogen receptor alpha-positive human breast cancer cell lines and clinical samples. FEBS J. 2010;277:1684–1694. doi: 10.1111/j.1742-4658.2010.07594.x. [DOI] [PubMed] [Google Scholar]
- Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013;32:663–669. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto N, Toyama T, Takahashi S, Sugiura H, Endo Y, Iwasa M, Fujii Y, Yamashita H. Distinct expressions of microRNAs that directly target estrogen receptor alpha in human breast cancer. Breast Cancer Res. Treat. 2011;130:331–339. doi: 10.1007/s10549-011-1672-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.