SUMMARY
MicroRNAs are frequently deregulated in cancer. Here we show that miR-22 is upregulated in myelodysplastic syndrome (MDS) and leukemia, and its aberrant expression correlates with poor survival. To explore its role in hematopoietic stem cell function and malignancy, we generated transgenic mice conditionally expressing miR-22 in the hematopoietic compartment. These mice displayed reduced levels of global 5-hydroxymethylcytosine (5-hmC) and increased hematopoietic stem cell self-renewal, accompanied by defective differentiation. Conversely, miR-22 inhibition blocked proliferation in both mouse and human leukemic cells. Over time, miR-22 transgenic mice developed MDS and hematological malignancies. We also identify TET2 as a key target of miR-22 in this context. Ectopic expression of TET2 suppressed the miR-22-incuced phenotypes. Downregulation of TET2 protein also correlated with poor clinical outcomes and miR-22 overexpression in MDS patients. Our results therefore identify miR-22 as a potent proto-oncogene, and suggest that aberrations in the miR-22-TET2 regulatory network are common in hematopoietic malignancies.
INTRODUCTION
Epigenetic mechanisms such as DNA methylation and histone modification drive stable and clonally propagated changes in gene expression, and have recently been shown to serve as molecular mediators of pathway dysfunction in neoplasia (Esteller, 2008; Rodriguez-Paredes and Esteller, 2011). Indeed, both myelodysplastic syndrome (MDS) and leukemias are characterized by frequent epigenetic abnormalities and aberrant DNA hypermethylation has been linked to poor prognosis in cases of MDS, as it is associated with more rapid progression to acute myeloid leukemia (AML) (Shen et al., 2010; Shih et al., 2012). Importantly, genome-wide discovery efforts in patients with myeloid malignancies have led to the identification of a number of causal genetic abnormalities that impact the epigenetic landscape. These genes include TET2, IDH1, IDH2, DNMT3a and EZH2, all of which affect DNA and/or histone lysine methylation (Shih et al., 2012).
Recent studies have examined in detail one such gene, TET2 (ten-eleven-translocation gene 2), which is located in 4q24, and whose mutation or deletion is extremely frequent in hematological malignancies, affecting 19% of patients with MDS, 12% of patients with myeloproliferative neoplasm (MPN), and 24% of patients with secondary AML (Abdel-Wahab et al., 2009; Delhommeau et al., 2009; Jankowska et al., 2009; Tefferi et al., 2009). TET2 mutations are observed in CD34+ progenitor cells and detected most frequently after MPN and MDS progression to AML (Abdel-Wahab et al., 2010).
TET2 catalyzes the oxidation of 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC) and is known to control hematopoiesis, presumably by regulating gene expression through its effect on DNA methylation (Branco et al., 2012; Ito et al., 2011; Ko et al., 2010; Shih et al., 2012). Tet2 deficiency in mice impairs 5-mC hydroxylation and leads to skewed differentiation, as well as enhancement of the self-renewal and repopulating capacity of hematopoietic stem cells (HSCs). This in turn promotes malignant transformation in mice, causing disorders resembling MDS and MPN, as well as chronic myelomonocytic leukemia (CMMoL) (Moran-Crusio et al., 2011; Quivoron et al., 2011). Recent studies have likewise shown that leukemia-associated Isocitrate Dehydrogenase (IDH) 1 or 2 mutant enzymes can induce carboxylation of glutamine-derived α-ketoglutarate (α-KG), with a concomitant increase in synthesis of 2-hydroxyglutarate (2-HG), an inhibitory metabolite of TET2 (Figueroa et al., 2010; Sasaki et al., 2012). Collectively, these studies implicate functional and mutational losses of TET2 as a key event underlying aberrant hematopoiesis and leukemogenesis, in turn suggesting that the inactivation of TET2 function by additional upstream cues could play a critical role in hematological malignancies.
MicroRNA (miRNA) deregulation is also known to contribute to hematological malignancies, including MDS and AML (Bartel, 2004; Chen et al., 2004; Gangaraju and Lin, 2009). A combined approach involving assessment of miRNA expression (using microarrays) and bioinformatic prediction of mRNA targets has revealed that distinct miRNA signatures fine-tune each step of hematopoiesis, including the reconstitution potential of HSCs (Arnold et al., 2011; Gangaraju and Lin, 2009). In turn, miRNAs that target critical leukemia suppressors could act as powerful proto-oncogenes in the hematopoietic compartment (Calin and Croce, 2006).
In this study, we demonstrate that miR-22 controls hematopoiesis and HSC maintenance by negatively regulating TET2 protein levels. Importantly, we identify miR-22 as an epigenetic modifier and the key oncogenic determinant for the pathogenesis of MDS and hematological malignancies in vivo, findings with novel prognostic and critical therapeutic implications for a large number of hematological disorders.
RESULTS
miR-22 is highly expressed in human hematological malignancies and increases the replating ability of hematopoietic stem cells
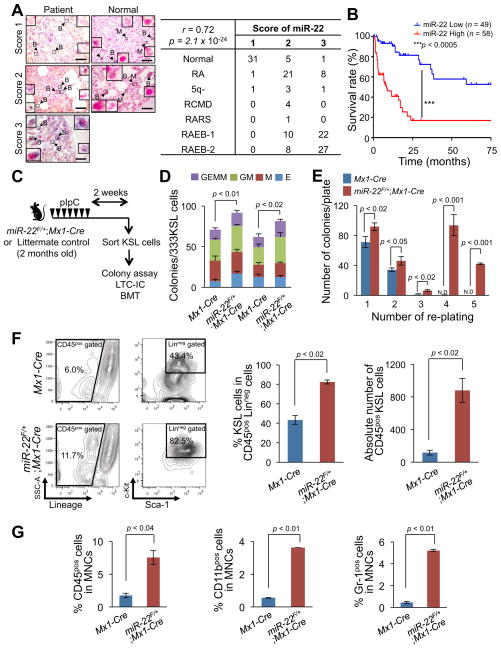
In order to build on the insights of microRNA biology into epigenetic alterations in myeloid malignancies, we attempted to identify those miRNAs playing a critical role in the pathogenesis of the stem cell disorder myelodysplastic syndrome (MDS), and its progression to overt leukemia. We first analyzed a previously reported miRNA microarray profiling database and found that miR-22 is markedly upregulated in patients with MDS (Figures S1A and S1B and Ref. (Dostalova Merkerova et al., 2011)). These results prompted us to perform expression analysis by in situ hybridization in a large set of patient samples, which included bone marrow with no sign of disease (n=37), early stage MDS (refractory anemia (RA), 5q syndrome (5q-), refractory cytopenia with multilineage dysplasia (RCMD) and RA with ringed sideroblasts (RARS)) (n=40) and RAEB (RA with excess blasts) (n=67). Our comprehensive in situ hybridization analysis revealed that miR-22 is highly expressed in patients with MDS (Figure 1A and Figure S1C). We next estimated the survival rates of the sets of MDS patients by using a Kaplan-Meier analysis, and applied a log-rank test to compare the survival rates of MDS patients on the basis of miR-22 expression levels. Remarkably, we found that aberrant expression of miR-22 directly correlates with poor survival rates in patients (Figure 1B). It is noteworthy that the distribution of blasts into each WHO (World Health Organization) category of MDS stages was not directly associated with the expression level of miR-22, but a significant correlation between miR-22 levels and poor survival rates was observed within each WHO classification, as well as among MDS patients harboring a normal karyotype (Figures S1D–S1F). This indicates that the poorer survival expectancy linked to miR-22 overexpression does not simply reflect confounding factors such as blast count and cytogenetic karyotype. Taken together, these findings led us to investigate the role of miR-22 in the hematopoietic system and the genesis of hematological malignancies.
Figure 1. miR-22 is upregulated in MDS patients and leads to increased replating capacity in vitro.
(A) miR-22 is highly expressed in MDS patient samples. Expression levels of miR-22 in blasts in subtypes of MDS were measured by in situ hybridization analysis. Normal (n=37), RA (n=30), 5q- (n=5), RCMD (n=4), RARS (n=1), RAEB-1 (n=32) and RAEB-2 (n=35). Representative images of in situ hybridization for miR-22 in MDS patients and normal bone marrow are shown (left). Insets of panels show miR-22 staining in blasts (arrow with B) and in differentiated myeloid cells (e.g. neutrophil, arrow with M) with a high magnification. The expression levels of miR-22 were also scored as described in Extended Experimental Procedures (right). The data were analyzed by Chi-square test. r, Pearson’s r. Scale bars, 60 μm.
(B) miR-22 overexpression correlates with poor survival rates of human MDS patients. MDS patients were divided into two groups; low miR-22 expressing patients group (miR-22 Score 1 or 2, n=49, blue) and high miR-22 expressing patients group (miR-22 Score 3, n=58, red). Overall survival of these patients is demonstrated. P-value was generated by log-rank test.
(C) Overview of the experimental design for the conditional miR-22 expression in hematopoietic compartment. 2 months-old miR-22F/+;Mx1-Cre mice or Mx1-Cre littermate controls were treated with pIpC for 2 weeks. 2 weeks after pIpC administration, KSL (c-KitposSca-1posLinneg) cells were sorted to evaluate the characteristics of miR-22 expressing hematopoietic stem/progenitor cells. LTC-IC, long-term culture initiating cells, BMT, bone marrow transplantation.
(D) Colony forming capacity of miR-22 expressing progenitor cells. 2 weeks after pIpC administration, sorted KSL cells from miR-22F/+;Mx1-Cre mice or littermate controls were cultured in semi-solid medium. Counting and classification of colonies were performed in independent littermate pairs (n=3). GEMM, Colony-forming unit-granulocyte, erythroid, macrophage, megakaryocyte, GM, Colony-forming unit-granulocyte, macrophage, M, Colony-forming unit-macrophage, E, Burst-forming unit-erythroid.
(E) miR-22 expression retains the ability to serially replate and generate colonies. In vitro colony replating assay was performed as shown in Figure S2D and colony counts in the indicated replatings were scored (n=3). N.D., non-detectable.
(F) Ectopic expression of miR-22 results in an increase of HSPCs in in vitro long-term culture with stromal cells. KSL cells isolated as described in (C) were co-cultured with OP-9 stromal cells for 2 weeks. Maintained Linneg cells and KSL cells were evaluated (n=3). Representative flow cytometry data of Linneg and the KSL cells (left) are shown. Mean percentage ± S.D. of KSL cells in Linneg cells (middle) and mean absolute numbers ± S.D. of CD45posKSL cells (right) are also shown.
(G) miR-22 increases the myeloid compartment. The percentages of CD45pos cells (left), CD11bpos cells (middle), or Gr-1pos cells (right) in total mononuclear cells (MNCs) were investigated 2 weeks after the co-culture with stromal cells from (F) (n=3). All error bars represent ± S.D.
“see also Figures S1, S2A–S2D and S7”.
To explore the function of miR-22 in hematopoiesis and leukemia, we first transduced mouse hematopoietic stem/progenitor cells (HSPCs; c-KitposSca-1posLinneg hereafter referred to as KSL cells) isolated from wild-type donor mice with either a retroviral vector encoding miR-22 and a green fluorescent protein (GFP), or a control vector encoding GFP only. The GFP-expressing KSL cells were then re-sorted by flow cytometry and plated in methylcellulose for an in vitro CFU (colony-forming unit) assay (Figure S1G). We observed in the first plating that the cells expressing miR-22 produced fewer colonies compared to control cells; however, when GFP+KSL cells were re-sorted and re-plated, miR-22–transduced cells produced more colonies in the second plating (Figure S1H). Strikingly, these cells also maintained this ability throughout serial replatings – whereas control cells only formed colonies in the first two platings, miR-22–expressing KSL cells were able to produce colonies after being re-plated at least seven times (Figure S1H). Microscopic analysis of the cells from these colonies revealed a homogenous blast-like morphology (Figure S1I, top). Consistent with these results, miR-22–expressing KSL cells maintained their characteristics and c-Kit expression (a marker of undifferentiated cells) during their replatings (Figure S1I, bottom and Figures S1J and S1K). To ensure that miR-22–transduced and control cells were subjected to the same experimental treatment, we also performed an in vitro competitive colony assay by mixing GFP+KSL cells expressing miR-22 and GFP− control KSL cells (Figure S1L, top). This analysis confirmed that miR-22–expressing GFP+, but not GFP−, KSL cells remained mostly in colonies through consecutive replatings (Figure S1L, bottom). Taken together, these data suggest that miR-22 can drive aberrant replating capacity of HSPCs and may contribute to malignant transformation.
miR-22 affects hematopoietic stem cell biology in vivo
We next sought to examine the biological effects of miR-22 in vivo and generated a conditional transgenic mouse model overexpressing miR-22 in the hematopoietic compartment (Figure S2A). miR-22 expression was induced by Mx1-Cre–mediated excision of a LoxP flanked transcriptional STOP cassette by administrating seven doses of polyinosine-polycytidine (pIpC) over 14 days (Figures S2B and S2C). Two weeks after pIpC administration, we isolated KSL cells from miR-22F/+;Mx1-Cre mice and Mx1-Cre littermate controls and analyzed their properties by using CFU and LTC-IC (long-term culture-initiating cell) assays (Figures 1C–1G and Figure S2D). In agreement with our previous retroviral transduction experiments, we found that KSL cells from miR-22F/+;Mx1-Cre transgenic mice displayed an enhanced replating potential (Figures 1D and 1E). Furthermore, ectopic expression of miR-22 caused a robust increase in the number of both KSL and mature myeloid cells, as determined by LTC-IC assay, indicating that miR-22 enhances HSPC maintenance and predisposes those cells to differentiate into the myeloid lineage (Figures 1F and 1G).
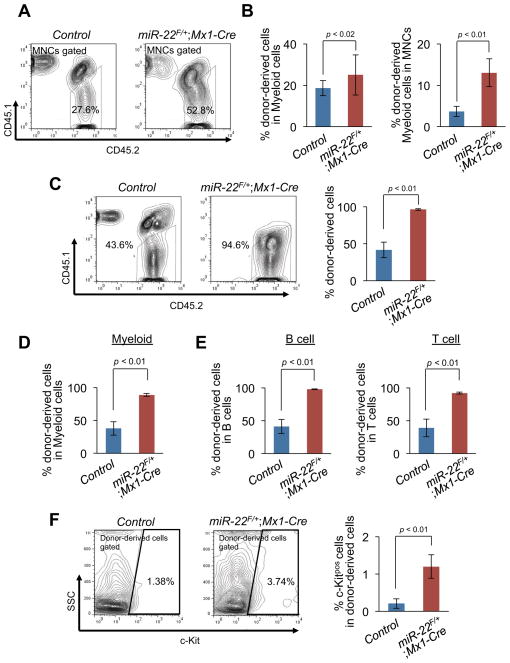
Next, we examined the ability of miR-22–overexpressing HSPCs to compete with wild-type counterparts in vivo by utilizing an in vivo competitive transplantation assay. KSL cells from miR-22F/+;Mx1-Cre and Mx1-Cre mice (Ly45.2) were purified 2 weeks after pIpC administration, mixed with Ly45.1/Ly45.2 competitor bone marrow mononuclear cells (BM MNCs) and transplanted into lethally irradiated Ly45.1 congenic recipient mice. The ability of donor cells to contribute to the reconstitution was then determined by flow cytometric analysis of peripheral blood stained for Ly45.1 and Ly45.2. Within 3 weeks of transplantation, donor cells from miR-22F/+;Mx1-Cre mice exhibited a slight increase in chimerism compared to those from control animals (Figure S2E). Over time this difference became more pronounced as miR-22F/+;Mx1-Cre KSL cells outcompeted their wild-type counterparts (Figures 2A–2C and Figure S2F). At 6 weeks of post-transplantation, an increased chimerism was observed in all BM MNCs as well as within the myeloid fraction, and by 9 weeks this effect was also evident in the c-Kitpos fraction (Figures 2A–2F). Taken together, these data strongly suggest that miR-22 enhances the self-renewal capacity of HSPCs in vivo.
Figure 2. miR-22 leads to an enhanced repopulating capacity of hematopoietic stem progenitor cells in vivo.
(A and B) Ly45.1 recipient mice were transplanted with the 1500 KSL cells from miR-22F/+;Mx1-Cre mice (n=15) or littermate controls (n=10) after pIpC administration together with 4×105 Ly45.1/Ly45.2 competitor BM MNCs. Donor-derived chimerism in peripheral blood was analyzed 6 weeks after the transplantation. Representative flow data of the CD45.1/CD45.2 positivity (A) and the percentages of donor-derived cells in myeloid (CD11bpos and/or Gr-1pos) (B, left) and donor-derived myeloid cells in MNCs (B, right) of recipient mice are shown.
(C–E) Donor contribution in hematopoiesis of recipient mice 9 weeks after the transplantation. Representative flow data of the CD45.1/CD45.2 positivity with the percentages of donor-derived CD45.1negCD45.2pos cells (C, left) are shown. Mean percentages ± S.D. of donor-derived cells in MNCs (C, right), myeloid cells (D), B cells (E, left) and T cells (E, right) are shown (n=10 for littermate controls and n=15 for miR-22F/+;Mx1-Cre).
(F) c-Kitpos cells are observed in peripheral blood of recipient mice transplanted with KSL cells from miR-22F/+;Mx1-Cre mice. Representative flow cytometry data of c-Kit positivity in donor-derived cells (left) and the percentages of c-Kitpos cells in donor-derived cells (right) are shown. (n=10 for littermate controls and n=15 for miR-22F/+;Mx1-Cre). All error bars represent ± S.D.
“see also Figures S2E, S2F and S7”.
miR-22 triggers MDS-like syndromes and hematological malignancies in vivo
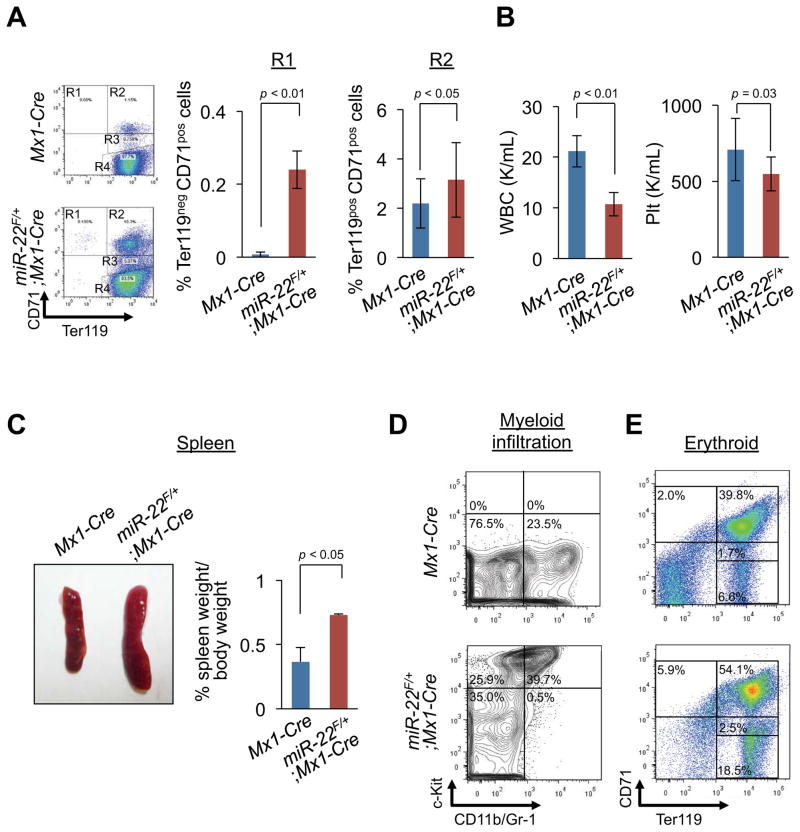
To further test whether increased aberrant hematopoiesis caused by miR-22 overexpression ultimately progresses to malignant hematological disease, we continued to follow up primarily transplanted recipients. Notably, at 12 weeks of post-transplantation, recipient mice transplanted with KSL cells from miR-22F/+;Mx1-Cre donors, but not from Mx1-Cre donors, developed an illness which closely resembled human MDS, as characterized by defective erythroid maturation and anemia, splenomegaly with myeloid infiltration, low white blood cell (WBC) counts and dysplastic myeloid cells in the peripheral circulation (Figure 3 and Figures S2G and S2H). To determine whether this disorder is transplantable, the donor-derived KSL cells re-sorted from primary recipient mice were subjected to a secondary transplantation using fresh competitor cells. In agreement with our earlier results, all secondary recipients developed a lethal hematological disease within four weeks after secondary transplantation (Figure S2I).
Figure 3. miR-22 overexpression develops hematological syndromes.
(A and B) miR-22 overexpression leads to human MDS-like phenotypes characterized by defective erythropoiesis (A) and cytopenia (B) in miR-22 transgenic mice. After pIpC administration, 1500 KSL cells from miR-22F/+;Mx1-Cre mice or littermate controls were transplanted into lethally irradiated Ly45.1 recipient mice with 4×105 Ly45.1/Ly45.2 competitor BM MNCs (n=10 for littermate controls and n=15 for miR-22F/+;Mx1-Cre). 12 weeks after transplantation, peripheral blood of recipient mice was evaluated. Representative flow data of the Ter119/CD71 positivity (A, left), the percentages of R1 (Ter119negCD71pos) (A, middle) and R2 (Ter119posCD71pos) (A, right) are shown. White blood cell (WBC) (B, left) and platelet (Plt) counts (B, right) are also shown.
(C and D) SplenomegaIy and myeloid infiltration into spleen were observed in recipient mice transplanted with KSL cells from miR-22F/+;Mx1-Cre mice. Representative images of spleens (C, left), spleen weight (n=3) (C, right), and representative flow data of the positivity of c-Kit and CD11b and/or Gr-1 in spleen (D) are shown.
(E) miR-22 expression leads to a differentiation defect in erythroid compartment. Representative flow data of the CD71/Ter119 positivity in spleen are shown. All error bars indicate ± S.D.
“see also Figures S2G–S2I”.
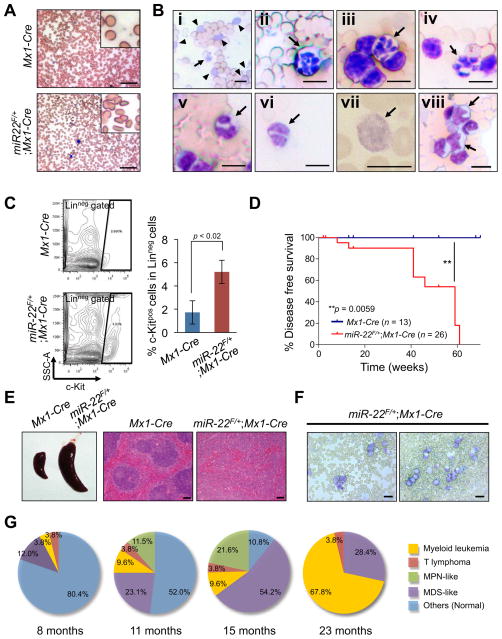
Furthermore, we also monitored disease onset and progression to malignancy in miR-22 transgenic mice. The development of disease similar to that found in our chimeric transplantation model was observed in miR-22F/+;Mx1-Cre mice but not Mx1-Cre littermate controls at 8 months after pIpC administration: the hallmarks of this distress included dysplasia in multi-lineage including erythroid, myeloid and platelet, splenomegaly and an increased number of c-Kit-positive undifferentiated blastic cells in the peripheral blood (Figures 4A–4C). By 16 months after pIpC administration, all miR-22 transgenic mice had developed MDS, affecting multi-lineage hematopoiesis. Importantly, ~13% of mice succumbed to myeloid leukemia as well as T cell lymphoma by 11 months, and all miR-22 transgenic mice suffered from hematological abnormality by 16 months. Finally, ~70% of miR-22 transgenic mice developed myeloid leukemia within two years (Figures 4D–4G).
Figure 4. miR-22 transgenic mice develop primary hematological diseases.
(A and B) miR-22 transgenic mice develop MDS-like hematological syndromes. Representative smears of peripheral blood of 8 months-old mice after pIpC administration are shown (A). Representative images of dysplastic erythroid cells (poikilocytosis, A; polychromasia, B-i arrowhead), dysplastic platelets (giant platelet, B-i arrow, B-vii), dysplastic neutrophils (hypersegmented neutrophils, B-ii-iv arrows; a pseudo-Pelger-Huet anomaly, B-v and vi) and dysplastic blasts (B-viii arrows) in miR-22 transgenic mice are also shown. Scale bars, 50 μm (A) and 10 μm (B).
(C) c-Kitpos immature blasts are increased in miR-22 transgenic mice. Representative flow cytometry data (left) and mean percentages ± S.D. of c-Kitpos cells in Linneg compartment (right) are shown (n=3).
(D) Disease free survival of miR-22F/+;Mx1-Cre mice (n=26) and littermate controls (n=13).
(E and F) Representative lethal hematological syndromes observed in miR-22 transgenic mice. Representative images of spleens (E, left) and H&E staining (E, right) are shown. Scale bars, 100 μm. Representative smears of peripheral blood of miR-22 transgenic mice (6 months old) with increased myeloid blasts (F) are also shown. Scale bars, 20 μm.
(G) Pie charts representing the disease spectrum in miR-22F/+;Mx1-Cre mice at the indicated ages (n=26). MPN, myeloproliferative neoplasm. All error bars indicate ± S.D.
“see also Figure S3”.
We also confirmed that the disease was triggered by miR-22 from KSL cells sorted from miR-22–expressing colonies. To this end, miR-22–expressing cells were subjected to consecutive colony replating and at the 6th replating, the residual KL (c-KitposLinneg) cells were transplanted into lethally irradiated recipient mice (Figure S3A). Consistent with our in vivo miR-22 transgenic mice data, by 14 weeks after transplantation, recipient mice transplanted with miR-22–expressing cells developed myeloid leukemia, as characterized by increased myeloid blasts in the peripheral blood, defective erythroid maturation and anemia, splenomegaly with the infiltration of myeloid blasts, and increased c-Kit-positive blasts in bone marrow (Figures S3B–S3H). Taken together, these data strongly suggest that miR-22 can enhance HSPC function and contribute to leukemia development in vivo.
miR-22 directly targets TET2 and affects the epigenetic landscape of hematopoietic compartment in vivo
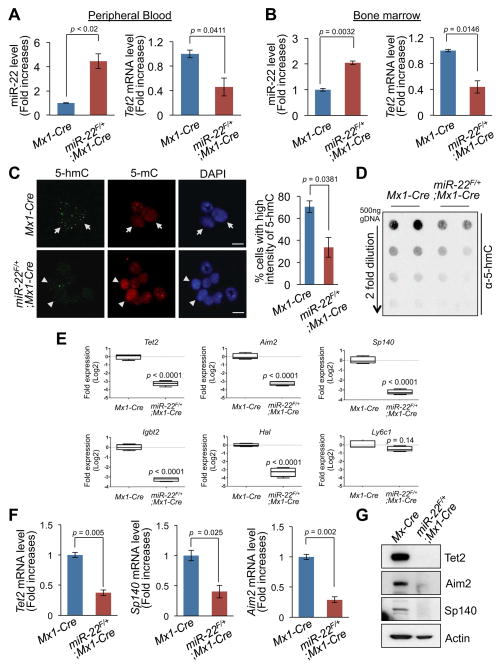
To identify relevant molecular targets of miR-22, we next undertook a bioinformatic analysis using TargetScan 6.0 (http://targetscan.org) (Lewis et al., 2005), microRNA.org (http://www.microrna.org) (Betel et al., 2008) and miRBase (http://www.mirbase.org) (Griffiths-Jones et al., 2006). Among various potential targets, we focused on TET2 (ten eleven translocation 2) as it possesses critically conserved nucleotides indicative of a legitimate target (Figure S4A). We then confirmed a direct interaction of miR-22 with the 3′UTR region of the TET2 gene by luciferase reporter assay (Figure S4B and Song et al., in press). TET2 is a member of the TET methylcytosine dioxygenase family, which catalyzes the conversion of 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC). TET proteins have been implicated in epigenetic re-programming, embryonic stem cell maintenance and early development (Ficz et al., 2011; Ito et al., 2010). Importantly, recurrent TET2 mutations have been identified in a variety of hematological malignancies including MDS, MPN (myeloproliferative neoplasm), CMMoL and AML (Abdel-Wahab et al., 2009; Delhommeau et al., 2009; Jankowska et al., 2009; Tefferi et al., 2009). It is noteworthy that the phenotypes elicited by miR-22 overexpression closely phenocopy the inactivation of Tet2 in the hematopoietic system both in vitro and in vivo (Li et al., 2011; Moran-Crusio et al., 2011; Quivoron et al., 2011). It has also been documented that Tet2 expression is increased during hematopoietic differentiation, and conversely, that miR-22 is highly expressed in HSPCs while its expression declines in mature cells (Figure S4C and Ref. (Moran-Crusio et al., 2011)). We therefore tested whether miR-22 could act as an authentic TET2-targeting microRNA in the hematopoietic compartment by utilizing our conditional transgenic mouse model. To this end, we measured Tet2 expression in the peripheral blood and bone marrow of miR-22F/+;Mx1-Cre or Mx1-Cre littermate mice at 2~3 weeks after pIpC administration. Importantly, real-time qPCR and Western blot analyses revealed a marked reduction in levels of Tet2 mRNA and protein, respectively, in miR-22F/+;Mx1-Cre mice compared to littermate controls (Figures 5A, 5B, 5E–5G and Figure S4D).
Figure 5. miR-22 acts as an epigenetic modifier by directly targeting TET2.
(A and B) A reduction in the levels of Tet2 mRNA in miR-22 transgenic mice. Expression levels of miR-22 (left) and Tet2 mRNA (right) in mononuclear cells from peripheral blood (A) and bone marrow (BM) (B) of Mx1-Cre control or miR-22F/+;Mx1-Cre mice 2–3 weeks after pIpC administration were determined by real-time qPCR (n=6).
(C) miR-22 expression results in a significant reduction in the levels of 5-hmC in hematopoietic compartment. 5-hmC and 5-mC were evaluated by immunofluorescence analysis in BM cells isolated from miR-22F/+;Mx1-Cre mice and littermate controls 3 weeks after pIpC administration (left). Cells expressing high levels of 5-hmC show low levels of 5-mC (arrow) and conversely, cells expressing low levels of 5-hmC exhibit high levels of 5-mC (arrowhead). Bar graph depicts mean percentages ± S.D. of cells expressing high levels of 5-hmC (n=3) (right). Scale bars, 10 μm.
(D) miR-22 expression leads to a significant reduction in global 5-hmC expression levels in the genome in BM cells. 5-hmC levels were analyzed by quantitative dot blot assay with genomic DNA purified from BM cells of miR-22F/+;Mx1-Cre or littermate control mice.
(E) Plots depict relative expression levels of putative Tet2 target genes (Aim2, Sp140, Igbt2 and Hal) in CD150posCD48negFlt-3negCD34negKSL cells of miR-22F/+;Mx1-Cre mice and littermate controls 2~3 weeks after pIpC administration (n=4). Ly6c1 and Actb were used as a Tet2 unrelated gene control and an internal control, respectively.
(F and G) Repression of putative downstream genes of Tet2 in miR-22 transgenic mice. Expression levels of Tet2, Sp140 and Aim2 mRNAs (F) or proteins (G) in BM of miR-22F/+;Mx1-Cre mice or littermate controls 2–3 weeks after pIpC administration were evaluated (n=3). All error bars indicate ± S.D.
“see also Figures S4A–S4F”.
On the basis of these observations, we then sought to determine whether miR-22–mediated TET2 inactivation is sufficient to impact the global epigenetic landscape in blood cells. We therefore analyzed 5-hmC and 5-mC levels in the genomic DNA of miR-22F/+;Mx1-Cre mice. Immunofluorescence and dot blot analyses revealed a drastic (more than two-fold) reduction in 5-hmC levels and a concomitant increase in 5-mC levels in bone marrow of these mice (Figures 5C and 5D), suggesting that changes in an important epigenetic mark (e.g. 5-hmC) may contribute to the ability of miR-22 to affect hematopoiesis and leukemogenesis.
To further understand the consequences of repression of TET2 by miR-22 in hematopoiesis, we next examined the effects of miR-22 overexpression on putative targets of TET2 (Kallin et al., 2012; Ko et al., 2010; Yamazaki et al., 2012). A comprehensive real-time qPCR analysis revealed that the expression levels of a substantial number of putative Tet2 target genes, including Aim2, Hal, Igbt2 and Sp140, were markedly decreased in the hematopoietic stem cell compartment (CD150posCD48negFlt3negCD34negKSL cells) purified from miR-22F/+;Mx1-Cre mice (Figure 5E and Figure S4E). Interestingly, AIM2 (absence in myeloma 2) has been reported to have a role in the reduction of cell proliferation by cell cycle arrest, and to also be highly methylated in MDS, especially in CMMoL patients with TET2 mutations (Figueroa et al., 2010; Patsos et al., 2010; Yamazaki et al., 2012). Therefore we hypothesized that its silencing via methylation of the promoter might provide a growth advantage to cancer cells. Additionally, polymorphisms of SP140 (SP140 nuclear body protein) have been found to correlate with chronic lymphoid leukemia, and its hypermethylation has been also demonstrated in CMMoL patients with TET2 mutations, and hence its hypermethylation may influence the risk of leukemogenesis (Di Bernardo et al., 2008; Ko et al., 2010; Yamazaki et al., 2012). Indeed, we confirmed that the expression of Aim2 and Sp140 was drastically reduced at the levels of mRNA and protein in the bone marrow of miR-22F/+;Mx1-Cre mice compared to littermate controls, and this reduction was associated with an increased promoter methylation (Figures 5F and 5G and Figure S4F). Taken together, these results suggest that miR-22 represses TET2 expression and thereby can remodel the epigenetic landscape, with global changes in 5-hmC levels and alteration in the expression of putative TET2 target genes including AIM2 and SP140.
miR-22 enhances stem cell function and triggers hematological transformation through repression of TET2
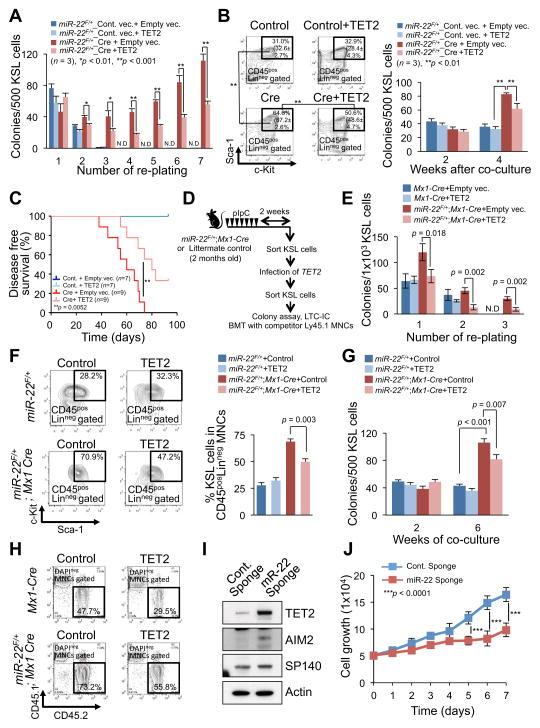
Next, we explored the relevance of the cross talk between miR-22 and TET2 in hematopoiesis and hematological malignancies. To test whether TET2 directly contributes to the function of miR-22 in hematopoietic compartment, we first infected KSL cells isolated from wild-type mice with a retroviral vector encoding miR-22 and GFP. After the re-sorting, GFP+KSL cells were additionally infected with a TET2-expressing lentiviral particle. In vitro colony replating and LTC-IC assays demonstrated that the phenotypes caused by miR-22 overexpression in hematopoietic compartment are potently suppressed by ectopic expression of TET2 (Figures S4G–S4J). Notably, an in vivo transplantation assay revealed that ectopic expression of TET2 leads to survival advantages for recipient mice transplanted with miR-22–expressing KSL cells (Figure S4K).
To further examine this interaction, we purified KSL cells from miR-22 transgenic mice and infected them with a retroviral vector encoding Cre recombinase and GFP. After the re-sorting of GFP+KSL cells, cells were additionally infected with a TET2-expressing lentiviral particle and the resulting phenotypes were analyzed. In vitro colony replating and LTC-IC analyses demonstrated that the oncogenic function of miR-22 was antagonized by ectopic expression of TET2 (Figures 6A and 6B). Notably, when KSL cells of miR-22F/+ mice were simultaneously co-infected with a Cre recombinase construct and a TET2-expressing construct, ectopic expression of TET2 more strikingly impaired the replating potential of GFP+KSL cells (Figures S5A and S5B). In line with these observations, an in vivo transplantation assay revealed significant survival advantages in recipient mice transplanted with miR-22F/+ KSL cells co-infected with Cre and TET2 compared to those with miR-22F/+ KSL cells co-infected with Cre and control vector (Figure 6C). Importantly, the positive survival effect drawn by ectopic expression of TET2 was more pronounced in secondary transplantation (Figure S5C).
Figure 6. miR-22–TET2 regulatory network affects hematopoietic stem cell function and hematological transformation.
(A and B) Ectopic expression of TET2 reduces the colony forming capacity of miR-22 expressing hematopoietic stem/progenitor cells. The sorted KSL cells from miR-22F/+ mice were infected with Cre-GFP or GFP control vector. GFP+KSL cells were resorted and infected with empty vector or TET2 expressing lentiviral particles. Cells were then incubated in semi-solid medium. Colony replating assay was performed and the resulting colonies were counted at the indicated replatings (n=3) (A). Some of TET2 infected cells were also subjected to LTC-IC assay. Representative flow cytometry data of the positivity of c-Kit/Sca-1 in LinnegCD45pos gated cells 2 weeks after co-culture with stromal cells and mean percentages ± S.D. of KSL cells in Linneg CD45pos (in brackets) are shown (n=3) (B, left). Colony forming capacities were also determined at the indicated weeks after co-culture with stromal cells (n=3) (B, right).
(C) TET2 attenuates the hematopoietic malignancies induced by miR-22 overexpression. KSL cells sorted from miR-22F/+ mice were infected with Cre-GFP or GFP control vector followed by TET2 infection, and 1500 GFP+KSL cells were transplanted into recipient mice with 2.0×105 Ly45.1 competitor bone marrow mononuclear cells (BM MNCs). Disease free survival of recipient mice was examined by Kaplan-Meier survival curves. Log-rank test was used to generate P-value.
(D) Overview of the experimental design for introduction of TET2 into KSL cells from miR-22 transgenic mice. KSL cells purified from miR-22 transgenic mice were subjected to infection with TET2 expressing vector, and their characteristics were evaluated in vivo and in vitro.
(E) Colony forming capacity of miR-22 transgenic progenitor cells is attenuated by ectopic expression of TET2. 2 weeks after pIpC administration, sorted KSL cells from miR-22F/+;Mx1-Cre mice were infected with TET2 expressing vector and resorted KSL cells were then subjected to the incubation in semi-solid medium. The resulting colony numbers were scored in three independent littermate pairs in the indicated platings (n=3). N.D., non-detectable.
(F and G) Ectopic expression of TET2 causes a reduction in LTC-IC capacity of miR-22 transgenic progenitor cells. TET2 infected KSL cells from miR-22F/+;Mx1-Cre mice or littermate controls, as shown in Figure 6D, were co-cultured with stromal cells for the indicated weeks. Maintained KSL cells were evaluated 2 weeks after co-culture (n=3) (F). At the indicated times after co-culture, the capacity of colony formation was also determined in semi-solid medium (n=3) (G).
(H) Increased reconstitution capacity of KSL cells of miR-22 transgenic mice is reduced by ectopic expression of TET2. The sorted KSL cells from miR-22F/+;Mx1-Cre mice or littermate controls were infected with TET2 expressing vector. 1×104 KSL cells were resorted and subjected to the bone marrow transplantation with 5×105 competitor BM MNCs. Representative flow cytometry data of the CD45.1/CD45.2 positivity at 6 weeks after transplantation are shown.
(I) Inhibition of miR-22 increases the expression of TET2 and its putative target genes in human leukemic cells. K562 cells were infected with the vector encoding miR-22 sponge or control sponge, and 48 hrs after the infection cell lysates were subjected to immunoblot analysis for the indicated proteins.
(J) Inhibition of miR-22 suppresses the proliferation of human leukemic cells. K562 cells were infected with the vector encoding miR-22 sponge or control sponge. 48 hrs after puromycin selection, 5×104 cells were incubated for 7 days. Optical densities of the cells were determined at the indicated times (n=3). All error bars indicate ± S.D. “see also Figures S4G–S4K and S5”.
Finally, we purified KSL cells from miR-22 transgenic mice after pIpC administration, infected them with a TET2-expressing lentiviral particle and evaluated the resulting phenotypes (Figure 6D). We found that all the phenotypes elicited by an elevation of miR-22 in vitro and in vivo in hematopoietic compartment, including repression of the expression of Aim2 and Sp140 in bone marrow, were also significantly suppressed by ectopic expression of TET2 (Figures 6E–6H and Figure S5D).
Importantly, when we purified KSL cells from primary miR-22 transgenic mice and infected them with TET2 followed by in vivo transplantation together with competitor cells, ectopic expression of TET2 delayed the onset of hematological disorder, accompanied with significant advantages in disease-free survival (Figure S5E). Taken together, these data suggest that TET2 is one of the major targets responsible for the miR-22 proto-oncogenic function in hematopoiesis and development of hematological malignancies.
miR-22 decoying offers novel therapeutic opportunities and its overexpression correlates with the silencing of TET2 protein in MDS and AML patients
Next, we investigated whether the miR-22/TET2 interaction may offer a therapeutic opportunity for the treatment of hematological malignancies. We therefore tested whether inhibition of miR-22 affects TET2 expression and activity, as well as the biology of leukemias. To this end, we inhibited miR-22 in human leukemia cell lines, K562 and U937, both of which express relatively high levels of miR-22 (Figure S5F and Ref. (Wang et al., 2012)), by using a miR-22 retroviral sponge or a custom-made LNA (locked nucleic acid (Petersen and Wengel, 2003)) miR-22 decoy as the miR-22 sponge. Critically, inhibition of miR-22 in human leukemic cells resulted in a significant reduction in cell proliferation as well as an elevation of the expression of TET2 and its targets, AIM2 and SP140 (Figures 6I and 6J and Figures S5G–S5I). It is worth noting that U937 cell line harbors PTEN (phosphatase and tensin homologue) mutant with premature termination of coding sequence (Liu et al., 2000). Hence in this setting the effects observed upon miR-22 decoying are not mediated by the ability of miR-22 to downregulate PTEN expression.
In full agreement with these observations, an in vitro colony replating assay revealed the inhibition of miR-22 by a miR-22 retroviral sponge in KSL cells purified from miR-22 transgenic mice led to a reduction in their colony forming capability (Figure S5J). Taken together, these data provide a rationale for the therapeutic potential of targeting miR-22 with LNA miR-22 inhibitors in the treatment of hematological malignancies.
The pathological roles of TET2 in myeloid malignancies have been previously highlighted as its mutations are widely observed in myeloid malignancies and MDS patients. Most of these earlier studies, however, were limited to mutation analysis, and found little relationship between the progression of MDS to AML and TET2 expression levels, although the phenotypes of Tet2 knockout mice gave indications to the contrary (Moran-Crusio et al., 2011; Quivoron et al., 2011). Likewise, emerging evidence demonstrated no significant prognostic association between TET2 mutations in MDS patients and WHO subtypes, the IPSS (International Prognostic Scoring System) score, cytogenetic status, or transformation to AML (Kosmider et al., 2009; Smith et al., 2010).
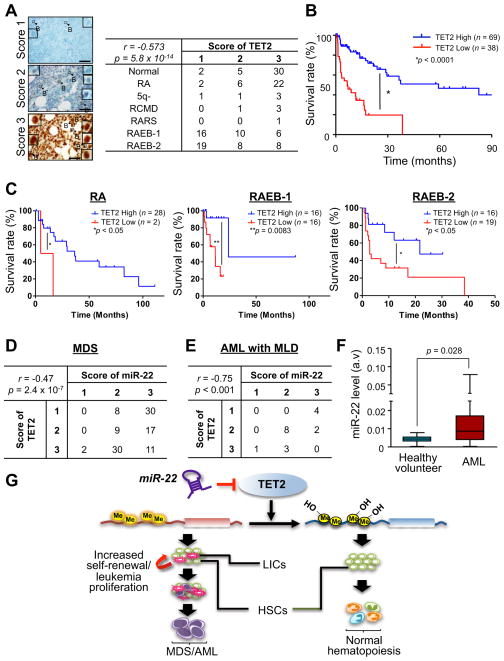
However, on the basis of our data we hypothesized that a miR-22–dependent reduction in the levels of TET2 might have been overlooked, and could play a key role in the development of myeloid malignancies. To test this hypothesis, we investigated a large cohort of patients with MDS by an immunohistochemical analysis. Strikingly, this analysis revealed that ~60% of 107 MDS patients analyzed displayed a reduction in the levels of TET2 (36% of 107 MDS patients show Score 1 and 24% show Score 2), and that this reduction was directly correlated with poor patient survival rates of patients (Figures 7A and 7B and Figures S6A–S6D). Importantly, the percentage of blasts distributed in each WHO category of MDS stage was not directly associated with expression level of TET2, but a significant correlation was found between TET2 expression and poor survival rates within each WHO classification, as well as among MDS patients harboring a normal karyotype, suggesting that the poorer survival expectancy linked to TET2 downregulation does not simply reflect confounding factors, such as blast count and cytogenetic karyotype (Figure 7C and Figures S6E and S6F). Together, these results suggest that the contribution of TET2 to the pathogenesis of hematological malignancies extends far beyond the initial impact of its mutations.
Figure 7. miR-22 overexpression directly correlates with the silencing of TET2 in human MDS and AML patients.
(A) TET2 expression is downregulated in human MDS patients. Expression levels of TET2 protein in CD34pos blasts in subtypes of MDS were evaluated by immunohistochemical analysis. Representative images of immunohistochemical analysis for TET2 protein in patients with subtypes of MDS are shown (left). Insets represent TET2 staining in blasts (arrow with B) with a high magnification. Scoring the expression levels of TET2 in blasts was performed as described in Extended Experimental Procedures (right). The data were analyzed by Chi-square test. r, Pearson’s r. Scale bars, 60 μm.
(B) Downregulation of TET2 levels correlates with poor survival rates of human MDS patients. Overall survival of patients that express low TET2 (TET2 Score 1, n=38, red) or high TET2 (TET2 Score 2 or 3, n=69, blue) is shown. P-value is generated by log-rank test.
(C) Survival rates of low TET2 (Score 1) and high TET2 (Score 2 or 3) patients with RA (left), RAEB-1 (middle) and RAEB-2 (right). Survival rate of P-value is generated by log-rank test.
(D) miR-22 overexpression correlates with downregulation of TET2 in MDS patients. 107 MDS patient specimen were subjected to in situ hybridization and immunohistochemical analyses, and the expression levels of miR-22 and TET2 were evaluated. The data were analyzed by Chi-square test. r, Pearson’s r.
(E) The expression levels of miR-22 and TET2 in 18 AML with MLD patient samples were evaluated. The data were analyzed by Chi-square test. r, Pearson’s r. AML, acute myeloid leukemia; MLD, multiple lineage dysplasia.
(F) miR-22 is highly expressed in human AML patients. Expression levels of miR-22 in CD34pos bone marrow cells from healthy donors (n=9) and AML patients (n=215) are shown.
(G) Proposed model of oncogenic role of miR-22 in hematopoiesis. miR-22 negatively regulates TET2 tumor suppressor, leading to a reduction of 5-hmC and a potentiation of global gene methylation. This genetic remodeling in turn enhances hematopoietic stem cell function and promotes hematological transformation. HSCs, hematopoietic stem cells, LIC, leukemia-initiating cells.
“see also Figure S6”.
Importantly, combined in situ hybridization and immunohistochemical analyses revealed that miR-22 expression was directly anti-correlated with the levels of TET2 in a large-cohort data set of MDS patients (n=107) (Pearson’s r = −0.47, P = 2.4e-07): 28% of MDS patients that showed the reduced levels of TET2 (Score 1) also exhibited high miR-22 levels (Score 3) (Figure 7D and Figure S6G and see also Figure 1A).
Furthermore, as our in vivo analysis demonstrated that miR-22 triggers overt leukemia (Figure 4G), we investigated the miR-22/TET2 interactive relationship in AML with MLD (n=18). Our combined analyses of in situ hybridization and immunohistochemistry revealed a clear anti-correlation between miR-22 expression and the levels of TET2 (Pearson’s r = −0.75, P = 2.9e-04); in addition, 22.2% of AML with MLD patients showing the reduced levels of TET2 (Score 1) displayed a high expression of miR-22 (Score 3) (Figure 7E). Western blot and real-time qPCR analyses also corroborated a relationship between miR-22 and TET2 in primary AML patients (Figure S6H). Finally, we confirmed these findings by analyzing the expression levels of miR-22 in a very large cohort of AML patients from a previously reported real-time qPCR database (Jongen-Lavrencic et al., 2008). Here miR-22 was found highly expressed in 58.1% of 214 AML patient samples compared to normal bone marrow from healthy donors (P = 0.028, Figure 7F). Taken together, these data strongly suggest that aberration of the miR-22-TET2 regulatory network is one of the common events in hematopoietic malignancies.
DISCUSSION
The findings presented in this study point to a number of important conclusions:
The first of these is that miR-22 is a powerful regulator of HSPC maintenance and self-renewal. These effects are so pronounced that miR-22–expressing HSPCs out-competed their wild-type counterparts in all the assays that we performed in vivo as well as in vitro. Importantly, in so doing miR-22 also skews hematopoietic differentiation, mainly toward myeloid compartment.
We have also identified miR-22 as a key determinant of the pathogenesis of both MDS and hematological malignancies. Remarkably, miR-22 plays a role in the initiation of MDS as well as full-blown leukemia; hence it exerts a bona fide proto-oncogenic activity. MDS is a genetically heterogeneous disease and we are only now beginning to fully appreciate its diversity. This study will therefore help in the reclassification of subtypes of MDS, and possibly AML, on the basis of miR-22 expression.
Furthermore, our comprehensive analysis of MDS patients has identified TET2 protein downregulation as a common event in hemopoietic malignancy (35.5% of total MDS patient samples, and 22.2% of AML samples with MLD, with most of those patients exhibiting high expression of miR-22) as well as a novel factor in poor prognosis, findings which directly impact the management and therapeutic decisions for patients with myeloid malignancies. Our findings thus imply that the contribution of TET2 to the pathogenesis of myeloid malignancies could be related to more than its mutations, and identify the deregulation of the miR-22–TET2 pathway as one of the most frequent events in hematological malignancies. Importantly, they also provide a compelling rationale for targeted therapies that impact the mechanisms of regulation responsible for the control of TET2 levels (e.g. through miR-22 decoying).
We show that miR-22 negatively regulates the expression of TET2 and that its overexpression closely phenocopies, both in vitro and in vivo, many of the characteristics observed upon inactivation of TET2 (Figure 7G). Extensive mouse modeling efforts support these similarities in our current study as well as in other Tet2 related studies (Li et al., 2011; Moran-Crusio et al., 2011; Quivoron et al., 2011). Furthermore, our comprehensive analyses of a large cohort of MDS and AML patients likewise have demonstrated this antagonistic cross talk.
miR-22 also appears to exert a tumor-promoting role in other tissues, including mammary glands, through its ability to suppress the expression of other TET family members such as TET1 and TET3 (Figure S4D and Song et al., in press). Thus, miR-22 represents an alternative mechanism contributing to inactivation of TET2 in hematological malignancies, and more generally of TET family members, in addition to their mutations and deletion.
We previously demonstrated that miR-22 also targets the PTEN tumor suppressor (Poliseno et al., 2010). Pten loss has also been implicated in the pathogenesis of hematological malignancies (Yilmaz et al., 2006; Zhang et al., 2006). However, miR-22 through TET2 regulation can bestow to hemopoietic cells further competitive advantage as also supported by the fact that miR-22 decoying suppresses proliferation even in a PTEN mutant setting (Figure S5I). This may in turn explain the potent proto-oncogenic activity of miR-22. Indeed, there are several intriguing phenotypical differences between PtenF/F;Mx1-Cre and miR-22F/+;Mx1-Cre mouse models that could illuminate a unique and powerful role exerted by miR-22 in leukemogenesis. First, miR-22 increased the ability of HSCs to repopulate, which was then followed by leukemia development, whereas loss of Pten resulted in the exhaustion of normal HSCs. In addition, while Pten-loss induces short-term expansion in HSCs through excessive cell cycle entry followed by exhaustion, no significant alteration in stem cell compartment was observed at 2~3 weeks after the introduction of miR-22 (Figure S7). Additionally, while loss of Pten in HSCs is known to lead to MPN-like disease followed by leukemia development, hematological dysplasia with ineffective hematopoiesis is one of the main phenotypes caused by miR-22 expression, with bi- or pan-cytopenia observed in miR-22 transgenic mice. Furthermore, although HSCs from PtenF/+;Mx1-Cre mice are known to be transplantable, the recipients do not develop hematopoietic syndromes (Yilmaz et al., 2006). In contrast, recipient mice transplanted with HSCs purified from miR-22F/+;Mx1-Cre mice developed aberrant hematopoiesis followed by MDS and leukemia. Hence, the ability of miR-22 to concomitantly downregulate TET2 and PTEN provides it a unique mode of action, and confers on it the ability to overcome PTEN-loss driven stem cell exhaustion while favoring MDS development and leukemogenesis. This in turn raises the possibility that concomitant silencing of TET2 and PTEN by miR-22 could represent a powerful event underlying the pathogenesis of human cancer, as it may overcome the fail-safe mechanisms elicited by loss of PTEN (Berger et al., 2011).
Finally, since the technology to inhibit microRNAs for therapy is rapidly evolving (Elmen et al., 2008; Stenvang et al., 2008), this will impact on treatment options for a range of diseases and disorders in the years to come. Indeed, LNA-based miRNA decoying has already come of age for the treatment of hepatitis-C (Lanford et al., 2010), and may prove very effective for the treatment of blood-borne diseases (Zhang et al., 2012). The data presented in this study thus support the notion that an LNA-based therapeutic targeting of miR-22 may represent an effective strategy for TET2 reactivation as a treatment modality for incurable diseases such as MDS and leukemia.
EXPERIMENTAL PROCEDURES
Mice
The details of generation of miR-22 transgenic mice are described in the Extended Experimental Procedures. C57BL/6 mice (B6-CD45.2) and C57BL/6 mice congenic for the CD45 locus (B6-CD45.1) were purchased from The Jackson Laboratory and used as recipients in transplantation assay.
Real-time quantitative PCR
The details of RNA isolation and real-time qPCR analysis are described in the Extended Experimental Procedures.
Immunofluorescence
Cells were fixed and stained as described (Ko et al., 2010). The details of staining of cells and the sources of antibodies used are also described in the Extended Experimental procedures.
Flow cytometry
The sources of antibodies used for flow cytometric analysis are described in the Extended Experimental procedures.
Long-term cultures and in vitro colony-forming assays
The details of colony replating assays are described in the Extended Experimental Procedures.
Competitive reconstitution assay
The details of competitive reconstitution assay are described in the Extended Experimental Procedures
Human sample analysis
Human patient samples were analyzed as described in detail in the Extended Experimental Procedures. This study was approved by the Institutional Review Board waiver and Human Tissue Utilization Committee, and clinical and hematological data were recorded after patients provided their informed consent in accordance with the Declaration of Helsinki.
microRNA in situ hybridization
The details of in situ hybridization analysis for miR-22 are described in the Extended Experimental Procedures.
Immunohistochemistry
The details of immunohistochemical analysis are described in the Extended Experimental Procedures.
Dynamic transcriptome analysis
The details of dynamic transcriptome analysis are described in the Extended Experimental Procedures.
Methylation specific PCR
DNA methylation status of Sp140 in purified KSL cells (or Linneg cells) was examined by the methylation-specific PCR with genomic DNA treated with sodium bisulfite using the EZ DNA Methylation-Direct kit (Zymo Research). Primer sequences were previously described (Yamazaki et al., 2012). β-actin was used as internal control of genomic DNA.
Luciferase reporter assay
The details of luciferase reporter assay are described in the Extended Experimental Procedures.
Statistical Analysis
Student’s t test was utilized to determine statistical significance unless otherwise specified. P-values lower than 0.05 were considered statistically significant.
Supplementary Material
Highlights.
miR-22–TET2 regulatory network controls hematopoietic stem cell biology.
miR-22 triggers myelodysplastic syndrome and hematological malignancies.
Levels of miR-22 and TET2 are reliable prognostic factors in MDS patients.
miR-22 decoying offers novel therapeutic opportunities for MDS and leukemia patients.
Acknowledgments
We thank all the members of the Pandolfi laboratory for their comments and discussion. We are grateful to Thomas Garvey and Min Sup Song for critical editing of the manuscript. We are also especially thankful to Akinobu Matsumoto, Dina Stroopinsky and Mesruh Turkekul for technical support and help on in situ hybridization, to Lourdes Mendez for comments and help on hematological analysis, to Aviv Bergman for comments and support on statistical analysis of human patients data and to Miriam Fayad for support on cohort study of patients with hematological malignancies. K.I. was supported by NIH grants. LK was supported by an NHMRC Overseas Biomedical Fellowship. This work was supported by CA141457-03 NIH grants to P.P.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdel-Wahab O, Manshouri T, Patel J, Harris K, Yao J, Hedvat C, Heguy A, Bueso-Ramos C, Kantarjian H, Levine RL, et al. Genetic analysis of transforming events that convert chronic myeloproliferative neoplasms to leukemias. Cancer Res. 2010;70:447–452. doi: 10.1158/0008-5472.CAN-09-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara O, Bhat R, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold CP, Tan R, Zhou B, Yue SB, Schaffert S, Biggs JR, Doyonnas R, Lo MC, Perry JM, Renault VM, et al. MicroRNA programs in normal and aberrant stem and progenitor cells. Genome Res. 2011;21:798–810. doi: 10.1101/gr.111385.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2012;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- Di Bernardo MC, Crowther-Swanepoel D, Broderick P, Webb E, Sellick G, Wild R, Sullivan K, Vijayakrishnan J, Wang Y, Pittman AM, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008;40:1204–1210. doi: 10.1038/ng.219. [DOI] [PubMed] [Google Scholar]
- Dostalova Merkerova M, Krejcik Z, Votavova H, Belickova M, Vasikova A, Cermak J. Distinctive microRNA expression profiles in CD34+ bone marrow cells from patients with myelodysplastic syndrome. Eur J Hum Genet. 2011;19:313–319. doi: 10.1038/ejhg.2010.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowska AM, Szpurka H, Tiu RV, Makishima H, Afable M, Huh J, O’Keefe CL, Ganetzky R, McDevitt MA, Maciejewski JP. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009;113:6403–6410. doi: 10.1182/blood-2009-02-205690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Lowenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111:5078–5085. doi: 10.1182/blood-2008-01-133355. [DOI] [PubMed] [Google Scholar]
- Kallin EM, Rodriguez-Ubreva J, Christensen J, Cimmino L, Aifantis I, Helin K, Ballestar E, Graf T. Tet2 facilitates the derepression of myeloid target genes during CEBPalpha-induced transdifferentiation of pre-B cells. Mol Cell. 2012;48:266–276. doi: 10.1016/j.molcel.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmider O, Gelsi-Boyer V, Cheok M, Grabar S, Della-Valle V, Picard F, Viguie F, Quesnel B, Beyne-Rauzy O, Solary E, et al. TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs) Blood. 2009;114:3285–3291. doi: 10.1182/blood-2009-04-215814. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, Yang FC, Xu M. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TC, Lin PM, Chang JG, Lee JP, Chen TP, Lin SF. Mutation analysis of PTEN/MMAC1 in acute myeloid leukemia. Am J Hematol. 2000;63:170–175. doi: 10.1002/(sici)1096-8652(200004)63:4<170::aid-ajh2>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsos G, Germann A, Gebert J, Dihlmann S. Restoration of absent in melanoma 2 (AIM2) induces G2/M cell cycle arrest and promotes invasion of colorectal cancer cells. Int J Cancer. 2010;126:1838–1849. doi: 10.1002/ijc.24905. [DOI] [PubMed] [Google Scholar]
- Petersen M, Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 2003;21:74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brustle A, Harris IS, Holmes R, Wakeham A, Haight J, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Kantarjian H, Guo Y, Lin E, Shan J, Huang X, Berry D, Ahmed S, Zhu W, Pierce S, et al. DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. J Clin Oncol. 2010;28:605–613. doi: 10.1200/JCO.2009.23.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- Smith AE, Mohamedali AM, Kulasekararaj A, Lim Z, Gaken J, Lea NC, Przychodzen B, Mian SA, Nasser EE, Shooter C, et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood. 2010;116:3923–3932. doi: 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- Stenvang J, Silahtaroglu AN, Lindow M, Elmen J, Kauppinen S. The utility of LNA in microRNA-based cancer diagnostics and therapeutics. Semin Cancer Biol. 2008;18:89–102. doi: 10.1016/j.semcancer.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Lim KH, Abdel-Wahab O, Lasho TL, Patel J, Patnaik MM, Hanson CA, Pardanani A, Gilliland DG, Levine RL. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia. 2009;23:1343–1345. doi: 10.1038/leu.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xiang G, Zhang K, Zhou Y. Expression signatures of intragenic miRNAs and their corresponding host genes in myeloid leukemia cells. Biotechnol Lett. 2012;34:2007–2015. doi: 10.1007/s10529-012-1018-0. [DOI] [PubMed] [Google Scholar]
- Yamazaki J, Taby R, Vasanthakumar A, Macrae T, Ostler KR, Shen L, Kantarjian HM, Estecio MR, Jelinek J, Godley LA, et al. Effects of TET2 mutations on DNA methylation in chronic myelomonocytic leukemia. Epigenetics. 2012;7:201–207. doi: 10.4161/epi.7.2.19015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, Haug JS, Rupp D, Porter-Westpfahl KS, Wiedemann LM, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Roccaro AM, Rombaoa C, Flores L, Obad S, Fernandes SM, Sacco A, Liu Y, Ngo H, Quang P, et al. LNA-mediated anti-miR-155 silencing in low-grade B-cell lymphomas. Blood. 2012;120:1678–1686. doi: 10.1182/blood-2012-02-410647. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.