Abstract
Background & objectives:
Bacillus anthracis, Yersinia pestis, Burkholderia pseudomallei and Brucella species are potential biowarfare agents. Classical bacteriological methods for their identification are cumbersome, time consuming and of potential risk to the handler.
Methods:
We describe a sensitive and specific multiplex polymerase chain reaction (mPCR) assay involving novel primers sets for the simultaneous detection of B. anthracis, Y. pestis, B. pseudomallei and Brucella species. An additional non-competitive internal amplification control (IAC) was also included.
Results:
The mPCR was found to be specific when tested against closely related organisms. The sensitivity of the assay in spiked blood samples was 50 colony forming units (cfus)/25 μl reaction, for the detection of B. anthracis, Y. pestis and Brucella species; and 150 cfus/25 μl reaction, for B. pseudomallei. The assay proved useful in correctly and promptly identifing the clinical isolates of the targeted agents recovered from patients, compared to the gold standard culture methods.
Interpretation & conclusion:
The assay described in this study showed promise to be useful in application as a routine detection cum diagnostic method for these pathogens.
Keywords: Biowarfare, fever of unknown origin, internal amplification control, multiplex PCR, spiking
Bacillus anthracis, Yersinia pestis, Burkholderia pseudomallei and Brucella species are all listed by the Centers for Disease Control and Prevention (CDC) in ‘Category A’ and ‘Category B’, based on their ability to be used as bioterrorism agents and to cause disease in general. B. anthracis and Y. pestis are listed under Category A, which can be easily disseminated or transmitted from person to person causing high mortality rates and possessing the potential for major public health impact. B. pseudomallei and Brucella species are under Category B, easy to disseminate and result in moderate morbidity and low mortality rates1. Except for B. anthracis, the other three organisms also act as causative agents for infectious fevers of unknown origin (FUO), but are not routinely investigated. Despite recent advances in diagnostic techniques, FUO still remain a formidable diagnostic challenge2. Thus, rapid and reliable detection of these organisms is imperative for prompt and efficient patient management and disease control. For the identification of these organisms, generally direct methods based on bacteriological isolation are applied, which are cumbersome, time consuming and a potential threat to the handler. PCR is a useful approach for the diagnosis of bacterial infections, not detectable by classical bacteriology. Several PCR based molecular diagnostics in uniplex reactions have already been reported for these organisms3,4,5,6. Multiplex PCR (mPCR) first described by Chamberlain et al in 19887 is a useful tool for the identification of bacteria as well as other organisms8,9,10,11,12 with the possibility of detecting multiple organisms in a single tube reaction.
Here we describe a sensitive and specific mPCR involving novel primer sets for the simultaneous detection of B. anthracis, Y. pestis, B. pseudomallei and Brucella species. An additional 16S rRNA universal primer set was also added to the reaction as a non-competitive internal amplification control (IAC). The specificity of this mPCR was evaluated with closely related organisms and its sensitivity for its application as a routine diagnostic method, was evaluated on spiked blood samples.
Material & Methods
The study was carried out in Microbiology Division of Defence Research & Development Establishment (DRDE), Gwalior, India, during March 2010 to September 2010.
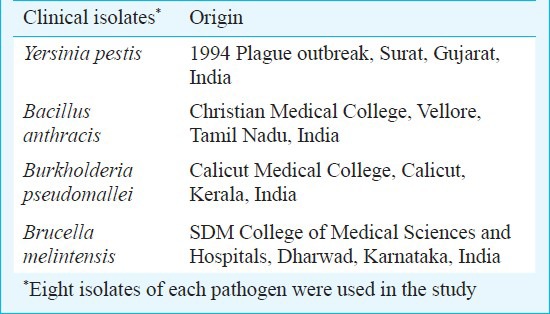
Standard bacterial strains, clinical isolates and culture conditions: Standard bacterial strains used in the study are shown in Table I and the clinical isolates in Table II. These organisms were grown overnight at 37°C on brain heart infusion (BHI) agar/broth (Difco Laboratories, USA).
Table I.
Bacterial strains used to evaluate the mPCR specificity
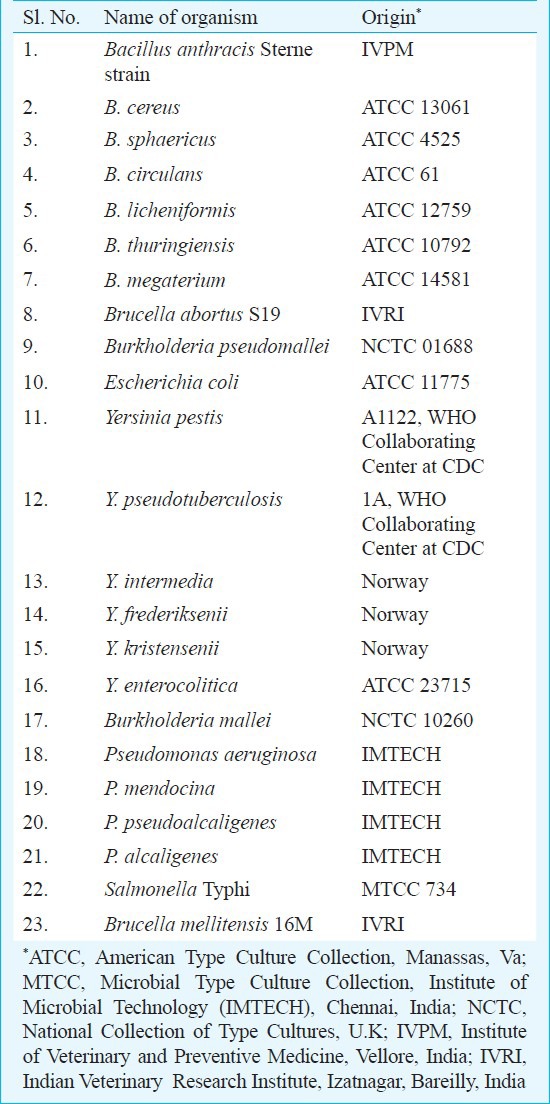
Table II.
List of clinical isolates used in the study
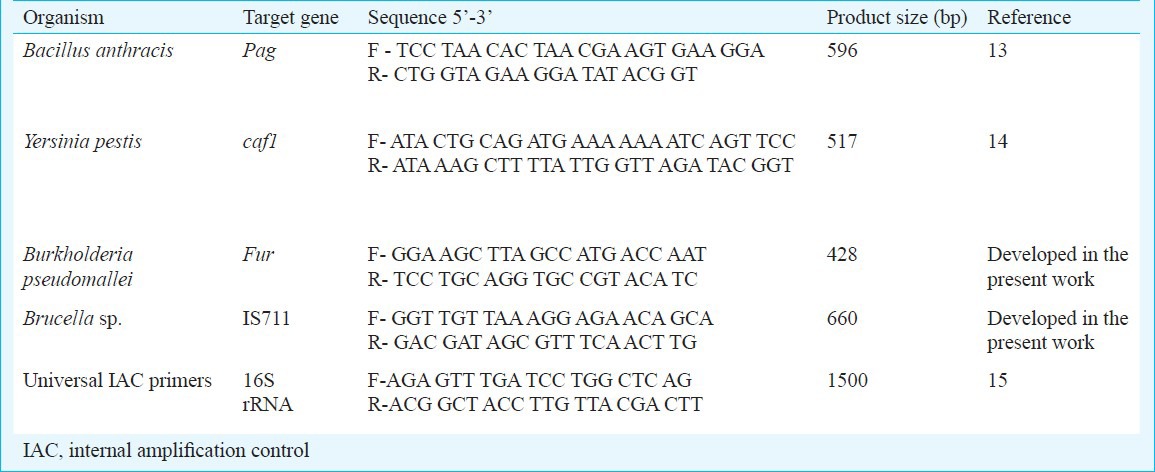
Oligonucleotide primers: The sequences of primers used in the study are listed in Table III. Specific primers for the pag gene in B. anthracis13, caf1 gene in Y. pestis16 and 16s rRNA gene for Universal IAC15 were obtained from published literature, while two sets of primers targeting the IS711 gene in Brucella species and the fur gene in B. pseudomallei, respectively were self-designed. All the primer sets were designed using the Gene Runner software (www.generunner.com) and obtained from Operon Biotechnologies, Cologne, Germany.
Table III.
Oligonucleotide primers used in the study
DNA isolation: Genomic DNA from Gram-negative and Gram-positive bacterial cultures was isolated using Dneasy Tissue kit (Qiagen, Germany) according to the manufacturer's instructions. The concentration of the extracted DNA was measured using Smartspec Plus spectrophotometer (Bio-Rad Lab, USA). The purity of the genomic DNA used in the study was within the acceptable range of 1.8-2.0 A260/A280.
DNA amplification: The primers chosen for developing mPCR were initially tested in uniplex PCRs. For each of the uniplex analysis, PCR was performed17 using MBI (Fermentas, Germany) reagents. Each 25 μl reaction mixture consisted of 10 pmol of the primers, 1x Dream Taq buffer (2mM MgCl2), 0.2 mM dNTP mix, Template DNA (50-70 ng/μl) and 1.2 units of Dream Taq Polymerase enzyme. PCRs were run using the following steps: initial denaturation for four min at 95°C, and 30 cycles of 95°C denaturation temperature for one min, annealing gradient temperature ranging from 50 to 60°C for one min and two min of 72°C extension temperature followed by another five min of final extension temperature at 72°C.
mPCR was performed employing Multiplex PCR Kit (Qiagen). Appropriate adjustments were made in the concentration of critical reagents and thermocycling conditions including reaction volume, template DNA concentration, primer concentration and annealing temperature to obtain optimal amplification of targeted genes. A 20 μl template DNA mix containing 2 μl (100 ng) DNA from each of the chosen organisms was prepared. Similarly, a primer mixture containing all the five primer pairs was made by mixing 10 μl (10 pmol/μl) of each of the forward and reverse primers. A 25 μl mPCR reaction volume contained 4 μl of template DNA mix (20ng of each organism DNA), 0.5-3 μl of primer mix (1 pmol), mPCR master mix and RNAse-Free Water (Qiagen Multiplex PCR Kit). The mPCR consisted of: initial denaturation at 940C for four min, and 30 amplification cycles constituting denaturation for one min at 94°C, annealing at 50-55°C for one min and extension at 72°C for two min followed by 10 min of final extension at 72°C for incompletely synthesized DNA. The mPCR products were visualized in 2 per cent agarose (Amresco, USA) gels stained with ethidium bromide (0.5 μg/ml), after electrophoresis in a 0.5 X TBE buffer. DNA ladder of 100 bp (Fermentas) was used as size standard.
Evaluation of mPCR: The specificity of the multiplex PCR was evaluated for its cross-reactivity with closely related organisms (Table I). In order to determine the sensitivity, based on colony forming units (cfu) was determined18 on BHI agar from cultures of B. anthracis, Y. pestis, B. melitensis and B. pseudomallei and was spiked into one ml anti-coagulant human blood of healthy volunteers drawn intravenously at concentration starting from 106 to 101 cfu/ml. DNA was then extracted from the spiked blood using the Dneasy Tissue kit (Qiagen, Hiden, Germany) following manufacturer's instructions for use in mPCR. Following optimization of the mPCR assay, its efficiency was evaluated on eight clinical isolates of each of the pathogens (Table II) and compared with gold standard culture methods.
Results
Multiplex PCR: The concentration of primers was optimized after standardizing on individual PCRs with their respective genes. All the chosen primers gave optimum amplification at concentrations ranging from 10-15 pmol. Gradient annealing temperature studies revealed satisfactory amplification for all the primers at 50, 52 and 55°C. Primers targeting specific genes in B. anthracis and Y. pestis gave optimum amplification up to 60°C, while in B. pseudomallei and Y. pestis the band intensity began to diminish after 55°C. Hence, for the mPCR gradient temperatures ranging from 50 to 55°C were tested and amplification at 52°C was found to be optimum. The primer concentrations tested in the mPCR yielded optimum amplification at 10-fold lower concentrations than the uniplex setups. The 16S rRNA universal non-competitive IAC co-amplified with the target DNA and had the amplicon size of 1.5 kb. The standardized mPCR revealed the presence of five amplicons after agarose gel electrophoresis (428 bp for B. pseudomallei, 517 bp for Y. pestis, 596 bp for B. anthracis, 660 bp for Brucella species and 1.5 kb for IAC) (Figure).
Fig.
Cross-reactivity testing of the mPCR with closely related organisms. Lane 1: 100 bp DNA ladder, Lane 2: Burkholderia pseudomallei, Lane 3: Burkholderia mallei, Lane 4: Y. pestis, Lane 5: Y. pseudotuberculosis, Lane 6: Bacillus anthracis, Lane 7: Bacillus cereus, Lane 8: Brucella mellitus, Lane 9: Multiplex positive control. Lane 10: Y. enterocolitica.
Evaluation of mPCR assay: The mPCR was checked for its cross-reactivity with DNA of closely related organisms (Table I). No amplified product except for IAC was observed with DNA of other organisms, even on repeated tests (Figure). This showed that the developed mPCR assay had 100 per cent specificity for the chosen organisms. Spiking studies revealed a sensitivity of 50 cfus/25 μl reaction, for the detection of B. anthracis, Y. pestis and Brucella species; and 150 cfus/25 μl reaction, for the detection of B. pseudomallei. The mPCR assay when performed on the clinical isolates could selectively amplify the targeted genes and thus rapidly detect the chosen pathogens.
Discussion
Appearance of clinical symptoms with any of the acute systemic bacterial disease is inadvertently associated with bacteremia. Thus, at the initial appearance of clinical symptoms, the presence of bacteria in the blood can be effectively detected by PCRs. However, the use of uniplex PCR in the diagnostic laboratories is limited in view of cost and the availability of adequate test sample which can be obviated by the mPCR approach19,20,21,22. Considerable sensitivity has been reported in uniplex PCRs targeting fur gene in B. pseudomallei, pag gene in B. anthracis, IS711 element in Brucella species and caf1 in Y. pestis14,23,24,25. Using reported primers (for pag, caf1, 16S rRNA genes) and by designing new primers (for IS711 and fur genes), a novel mPCR was developed in the present study. It was optimized to a common annealing temperature, maintaining a minimum 80 bp difference between the amplified products for better readability and resolution in the agarose gel. In order to make this mPCR assay acceptable to the present norms of a diagnostic PCR26, a non-competitive IAC primer targeting the 16S rRNA universal bacterial gene was also added as a positive control. The mPCR system besides conferring enhanced applicability requires additional careful optimizations. The inclusion of additional primer sets in the reaction invariably results in a higher probability of mis-priming and primer dimerization. In addition to effective primer design and selection, annealing temperature standardization, Mg2+ ion optimization and maintenance of adequate primer-template DNA ratio is necessary. Further, the use of Hot start PCR eliminates non-specific reactions caused by primer annealing at low temperatures (4-25°C) before the commencement of thermocycling27. Hot start PCR was applied in the present study to eliminate non-specific reactions after fulfilling the other optimization requirements.
All the genes targeted in this reaction are established virulence associated factors in each of the organisms. The protective antigen (part of the tripartite exotoxin complex) gene in B. anthracis has earlier been used in the diagnosis of anthrax13. In the current study, primers targeting the pag gene were optimized to mPCR setup and applied as reliable markers for the detection of B. anthracis. The IS711 insertion sequence element is exclusively present in the genome of all described species of the genus Brucella28 and the gene sequence was thus used to design specific primers. The fraction 1 (caf1) antigen, a major component of the surface of Y. pestis29, encoded by f1 (110) kb plasmid has been classically used for diagnosis of Y. pestis infections16. The gene is highly conserved and was tried in the present study to provide the characteristic signature of the organism. The ferric uptake regulator (FUR) transcription factor plays an important role in the pathogenesis of B. pseudomallei30. Specific primers targeting this transcription factor were designed and evaluated for cross-reactivity with other closely related organisms namely, B. mallei and Pseudomonas aeruginosa in uniplex setup (data not shown). However, the primer sequences utilized in the present study showed homology with other sequences in blast search. In PCR/mPCR, these yielded specific amplification product with the DNA of targeted organism. The specificity of the designed mPCR was evaluated by testing the primer sets against DNA from closely related organisms.
Fever of unknown origin have always been a diagnostic challenge. Routine diagnosis of a suspected FUO based infection evaluates the occurrence of common pathogens causing malaria, typhoid, dengue fever, leptospirosis etc. In an earlier study31, brucellosis was found to be the second most common causative agent of infectious FUO. Human brucellosis is a significant public health problem in India15. The occurrences of the other organisms detected in this study as causative agents of FUO are not routinely inspected in diagnostic laboratories. These organisms although rare, have the ability to cause significant damage to public health. The designed mPCR would also find utility in finding the source of the outbreak from among other available environmental samples in such conditions. This would render response teams in mitigating the spread of the disease and consequently aide management and control strategies. The reaction is easy to perform and requires basic PCR apparatus and the mPCR reagents kit. In conclusion, the mPCR described in this study appears to have potential to be used for diagnosis of these potentially harmful pathogens in routine practice.
Acknowledgment
The authors thank Drs K. Thavachelvam and Vanlalhmuaka of Microbiology Division, Defence Research and Development Establishment (DRDE), Jhansi, for their constant help, and acknowledge Dr R. Vijayaraghavan, Director, DRDE, Ministry of Defence, Government of India, and Prof. J. Sashidhar Prasad, Vice Chancellor, Sri Sathya Sai Institute of Higher Learning, for providing the requisite facilities to undertake this study.
References
- 1. [accessed on December 1, 2011]. http://www.bt.cdc.gov/agent/agentlist-catagory.asp .
- 2.Bleeker-Rovers CP, van der Meer JWM. Pyrexia of unknown origin. Medicine. 2005;33:33–6. [Google Scholar]
- 3.Haase A, Brennan M, Barrett S, Wood Y, Huffam S, O’Brien D, et al. Evaluation of PCR for diagnosis of melioidosis. J Clin Microbiol. 1998;36:1039–41. doi: 10.1128/jcm.36.4.1039-1041.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rahalison L, Vololonirina E, Ratsitorahina M, Chanteau S. Diagnosis of bubonic plague by PCR in Madagascar under field conditions. J Clin Microbiol. 2000;38:260–3. doi: 10.1128/jcm.38.1.260-263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamdy ME, Amin AS. Detection of Brucella species in the milk of infected cattle, sheep, goats and camels by PCR. Vet J. 2002;163:299–305. doi: 10.1053/tvjl.2001.0681. [DOI] [PubMed] [Google Scholar]
- 6.Berg T, Suddes H, Morrice G, Hornitzky M. Comparison of PCR, culture and microscopy of blood smears for the diagnosis of anthrax in sheep and cattle. Lett Appl Microbiol. 2006;43:181–6. doi: 10.1111/j.1472-765X.2006.01931.x. [DOI] [PubMed] [Google Scholar]
- 7.Chamberlain JS, Gibbs RA, Ranier JE, Nguyen PN, Caskey CT. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988;16:11141–56. doi: 10.1093/nar/16.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elnifro EM, Ashshi AM, Cooper RJ, Klapper PE. Multiplex PCR: optimization and application in diagnostic virology. Clin Microbiol Rev. 2000;13:559–70. doi: 10.1128/cmr.13.4.559-570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mason WJ, Blevins JS, Beenken K, Wibowo N, Ojha N, Smeltzer MS. Multiplex PCR protocol for the diagnosis of staphylococcal infection. J Clin Microbiol. 2001;39:3332–8. doi: 10.1128/JCM.39.9.3332-3338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo G, Mitchell TG. Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR. J Clin Microbiol. 2002;40:2860–5. doi: 10.1128/JCM.40.8.2860-2865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strålin K, Törnqvist E, Kaltoft MS, Olcén P, Holmberg H. Etiologic diagnosis of adult bacterial pneumonia by culture and PCR applied to respiratory tract samples. J Clin Microbiol. 2006;44:643–5. doi: 10.1128/JCM.44.2.643-645.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehmann LE, Hunfeld KP, Emrich T, Haberhausen G, Wissing H, Hoeft A, et al. A multiplex real-time PCR assay for rapid detection and differentiation of 25 bacterial and fungal pathogens from whole blood samples. J Clin Microbiol. 2008;197:313–24. doi: 10.1007/s00430-007-0063-0. [DOI] [PubMed] [Google Scholar]
- 13.Hutson RA, Duggleby CJ, Lowe JR, Manchee RJ, Turnbull PC. The development and assessment of DNA and oligonucleotide probes for the specific detection of Bacillus anthracis. J Appl Bacteriol. 1993;75:463–72. doi: 10.1111/j.1365-2672.1993.tb02803.x. [DOI] [PubMed] [Google Scholar]
- 14.Engelthaler DM, Gage KL, Montenieri JA, Chu M, Carter LG. PCR detection of Yersinia pestis in fleas: comparison with mouse inoculation. J Clin Microbiol. 1999;37:1980–4. doi: 10.1128/jcm.37.6.1980-1984.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panda SK, Nanda SK, Ghosh A, Sharma C, Shivaji S, Kumar GS, et al. The 1994 plague epidemic of India: molecular diagnosis and characterization of Yersinia pestis isolates from Surat and Beed. Curr Sci. 1996;71:794–9. [Google Scholar]
- 17.Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–50. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 18.Koch AC. Growth measurement. In: Gerhardt R, Murray GE, Wood WA, Krieg NR, editors. Methods for general and molecular bacteriology. Washington, D.C: ASM Press; 1994. pp. 254–7. [Google Scholar]
- 19.Mehrotra M, Wang G, Johnson WM. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J Clin Microbiol. 2000;38:1032–5. doi: 10.1128/jcm.38.3.1032-1035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neogi SB, Chowdhury N, Asakura M, Hinenoya A, Haldar S, Saidi S, et al. A highly sensitive and specific multiplex PCR assay for simultaneous detection of Vibrio cholerae, Vibrio parahaemolyticus and Vibrio vulnificus. Lett Appl Microbiol. 2010;51:293–300. doi: 10.1111/j.1472-765X.2010.02895.x. [DOI] [PubMed] [Google Scholar]
- 21.Yin Ngan GJ, Ng LM, Lin RTP, Teo JWP. Development of a novel multiplex PCR for the detection and differentiation of Salmonella enterica serovars Typhi and Paratyphi A. Res Microbiol. 2010;161:243–8. doi: 10.1016/j.resmic.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Kawasaki S, Fratamico P, Horikoshi N, Okada Y, Takeshita K, Sameshima T, et al. Development of the multiplex PCR detection kit for Salmonella spp., Listeria monocytogenes, and Escherichia coli O157: H7. Jpn Agri. 2011;45:77–81. [Google Scholar]
- 23.Lynch KH, Dennis JJ. Development of a species-specific fur gene-based method for identification of the Burkholderia cepacia complex. J Clin Microbiol. 2008;46:447–55. doi: 10.1128/JCM.01460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryu C, Lee K, Yoo C, Seong WK, Oh HB. Sensitive and rapid quantitative detection of anthrax spores isolated from soil samples by real-time PCR. Microbiol Immunol. 2003;47:693–9. doi: 10.1111/j.1348-0421.2003.tb03434.x. [DOI] [PubMed] [Google Scholar]
- 25.Leyla G, Kadri G, Umran O. Comparison of polymerase chain reaction and bacteriological culture for the diagnosis of sheep brucellosis using aborted fetus samples. Vet Microbiol. 2003;93:53–61. doi: 10.1016/s0378-1135(02)00442-x. [DOI] [PubMed] [Google Scholar]
- 26.Hoorfar J, Cook N, Malorny B, Wagner M, De Medici D, Abdulmawjood A, et al. Making internal amplification control mandatory for diagnostic PCR. J Clin Microbiol. 2003;41:5835. doi: 10.1128/JCM.41.12.5835.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou Q, Russell M, Birch DE, Raymond J, Bloch W. Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications. Nucleic Acids Res. 1992;20:1717–23. doi: 10.1093/nar/20.7.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ocampo Sosa AA, Garcia Labo JM. Demonstration of IS 711 transposition in Brucella ovis and Brucella pinnipedialis. BMC Microbiol. 2008;8:17. doi: 10.1186/1471-2180-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry RD, Fetherston JD. Yersinia pestis - etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loprasert S, Sallabhan R, Whangsuk W, Mongkolsuk S. Characterization and mutagenesis of fur gen from Burkholderia pseudomallei. Gene. 2000;254:129–37. doi: 10.1016/s0378-1119(00)00279-1. [DOI] [PubMed] [Google Scholar]
- 31.Ergönül Ö, Willke A, Azap A, Tekeli E. Revised definition of fever of unknown origin: limitations and opportunities. J Infect. 2005;50:1–5. doi: 10.1016/j.jinf.2004.06.007. [DOI] [PubMed] [Google Scholar]


