Abstract
Recent progress in substrate nanofabrication has led to the development of Ag nanorod arrays as uniform, reproducible, large area SERS-active substrates with high signal enhancement. These novel nanostructures fabricated by oblique angle vapor deposition (OAD) offer a robust platform for the rapid detection of biological agents and open new perspectives for the development and integration of biomedical diagnostic for clinical and therapeutic applications. Ag nanorod arrays have been investigated as SERS-active substrates for the detection and identification of pathogens, including bacteria and viruses, as well as to evaluate the potential of this biosensing platform for bio-recognition of high affinity events using oligonucleotide-modified substrates. This review summarizes the various nanostructured substrates designed for SERS-based applications, highlights the nanofabrication methodology used to produce Ag nanorod arrays, outlines their morphological and physical properties, and provides a summary of the most recent uses of these substrates for clinical diagnostic and biomedical applications.
Introduction
Since its discovery in the 1970s, surface-enhanced Raman spectroscopy (SERS) has become a powerful analytical technique that provides identification and quantitative information about a variety of biomolecules with a high degree of sensitivity and specificity [1]. SERS is a nano-optical technique that has been applied to a wide array of bioanalytical problems, e.g. genetics and proteomics [2], medical diagnostics [3, 4], biochemistry [5], and the life sciences [6]. For example, over the last few years, SERS methods have been applied to the detection of infectious diseases [7–10] and therapeutic drugs [11–14].
SERS originates from molecules located in close proximity to a metallic (typically Au or Ag) nanostructured surface that is capable of generating a localized surface plasmon (LSP) [15, 16]. The LSP results from surface electrons that oscillate in resonance with the frequency of the incident radiation. For SERS, the optimum excitation frequency is provided by a visible/near-IR laser whose wavelength is in close proximity to the maximum in the extinction profile of the localized surface plasmon [17]. The SERS enhancement has been amply described in the literature and is primarily thought to be the result of two distinct mechanisms: a long-range classical electromagnetic (EM) and a short-range chemical enhancement (CE) [18]. An EM enhancement on the order of E2 results from an increase in the electromagnetic field due to the generation of LSPs at nanostructured metallic surfaces. An additional E2 EM enhancement arises from the Stokes-shifted Raman scattering due to the induced dipole of the surface-adsorbed analyte molecule, thus leading to a total SERS EM field enhancement at the nanoparticle surface that scales as E4 [19]. The CE mechanism originates from the analyte-specific interactions between the surface-adsorbed analyte molecule and the metal through the formation of localized adsorbate electronic resonances or metal-to-adsorbate charge-transfer complexes [20]. In this scenario, the metal is irradiated by the incoming radiation source to create an electron-hole pair that results in a transfer of energy to the analyte molecule through its bonds connecting it to the metal surface. The subsequent energy is transferred back into the metal, resulting in the emission of the scattered light from the analyte molecule [21]. It is commonly thought that the EM enhancement contributes the greater extent (~104-107) to the signal enhancement observed in SERS while the CE enhancement is thought to contribute to a smaller extent (~10-102) to the overall enhancement [22].
The inherent biochemical specificity of Raman spectroscopy offers a number of attractive features for detection and identification of biological samples. Raman provides a unique, chemically specific molecular fingerprint for each analyte, which eliminates the need for indirect detection via conjugated synthetic fluorophores. Raman has inherently narrow spectral lines that enable multi-component analysis without special experimental techniques or multi-fluorophore approaches [23]. For the SERS method, in particular, the phenomenon is due to the nanoparticle physics of the substrate and not the chemistry of a fluorophore; thus, photobleaching and quenching are of minor concern.
SERS has proven itself to be highly adaptable as a bioassay platform for biomedical sensing [24]. Routine sensitivity of SERS-based analytical methods rivals that of fluorescence, with pico/femto/attomolar detection limits, and the ability to detect single molecules or binding events [25]. The SERS approach also has advantages over other label-free biosensing methods such as surface plasmon resonance (SPR), in that SERS is molecule-specific, unlike SPR, which relies only on a general signal response for any captured analyte. In addition, detection, identification, classification and quantification of individual analytes are possible using well-validated multivariate statistical methods [26].
The unique analytical assets of SERS have enabled the development of a variety of diagnostic applications requiring low detection levels and a high degree of specificity. However, despite the remarkable advances in recent years, the overall sensitivity and reproducibility of SERS has been constrained by non-optimized nanofabrication methods that result in variability in substrate nano-morphology. Since the LSP is dependent on nanoparticle morphology, stringent control of feature size and inter-particle distance is vital to ensure high and reproducible enhancements [27]. The successful transition of SERS to a practical biosensing methodology requires the development of SERS substrates that can not only provide strong enhancement factors, but that can also be fabricated easily, reproducibly, inexpensively, and with potential for long-term storage.
Recently, a nanofabrication technique based on an oblique angle vapor deposition (OAD) has been developed that offers a versatile, simple and inexpensive way of producing Ag nanorod arrays for high sensitivity SERS applications [28]. OAD is a physical vapor deposition technique that offers a reliable means of fabricating uniform, reproducible, large area SERS-active substrates with high signal enhancement. Ag nanorod arrays fabricated by OAD have been investigated as SERS-active substrates for the detection of pathogens including viruses and bacteria, as well as to assess the potential of nucleotide-modified Ag nanorod arrays for a variety of bio-recognition and biosensing applications. This review summarizes the nanofabrication methodology used to produce Ag nanorod arrays for SERS, highlights their structural and physical properties, and provides a summary of the most recent uses of these substrates as applied to bioanalytical sensing.
Effective SERS-Active Substrates
Since the discovery of the SERS effect, a wide variety of fabrication techniques have been utilized to produce SERS-active substrates. In recent years, advances in nanofabrication have led to the emergence of a number of novel nanostructures of various composition, morphology, size and shape that have proven to be successful SERS substrates. SERS enhancement theory stipulates that the creation of a high electromagnetic field is dependent on the metallic surface morphology [15, 27]. For example, the electromagnetic (EM) field is at maximum for regions with high curvature [29]. As a result, experimental conditions that affect the geometry and size of the particles must be carefully defined and controlled for generation of optimum EM enhancement.
Original fabrication of SERS-active substrates was based on chemical and electrochemical methods such as electrochemical reduction cycles (ORC) [30], chemical etching [31], and metal island films [32]. ORC is an electrochemical method in which Ag is subjected to successive oxidation-reduction cycles that create nano-roughened, SERS-active substrates. Chemical etching has also been widely used to produce roughened metallic substrates. This method consists of immersing a metallic substrate into a highly concentrated acidic solution (typically HNO3) for a short period of time to chemically etch the metallic surface. The resulting roughened surface can then be used for SERS characterization. Metal island films are another form of specific nanostructures with high aspect ratios that have been developed as SERS substrates. They consist of vapor-deposited Ag film on a suitable surface to produce discontinuous island-like particles that can act as SERS substrates. Although these methods allow production of large surface area substrates, they provide relatively low SERS enhancements and suffer from poor reproducibility.
Other nanofabrication techniques have been investigated to produce substrates for SERS applications. High aspect ratio nanostructures have been fabricated using lithographic techniques such as electron beam lithography (EBL) [33], nanosphere lithography (NSL) [34], as well as other approaches such as template [35] and hybrid methods [36]. SERS-active substrates produced by EBL consist of an array of highly ordered structures produced by a focused beam of electrons capable of drawing patterns over the resist wafer in a serial manner with nanometer resolution. This mask-less semiconductor fabrication technique allows patterning of SERS-active nanostructures. Substrates fabricated by NSL consist of arrays of well-ordered nanospherical structures produced by vapor deposition of metal through a template usually consisting of a self-assembly of polystyrene nanoparticles. Lithographic techniques are reproducible, flexible and easily implemented to a large variety of templates and hence can produce numerous types of nano-features with different plasmonic properties, however, EBL methods suffer from high cost and the need for specialized infrastructure.
Similar in structure and morphology to the substrates produced by lithographic techniques, SERS-active substrates produced by template methods consist of an array of nanostructures onto which Ag or Au is deposited via an electrochemical patterning method. This procedure results in the formation of metallic nanorod arrays of high aspect ratios capable of supporting surface plasmons. Likewise, the hybrid method results in the formation of a variety of nanostructures capable of producing EM hotspots. These substrates are produced by depositing metal particles onto nanoporous scaffolds such as porous silicon, nanowire arrays, and highly ordered pillars or toroids that can sustain plasmonic behavior for SERS applications. These nanofabrication techniques allow stringent and rigorous control over the growth of the various nanostructures and hence have demonstrated enhancements up to 109. While these relatively simple methods provide inexpensive and flexible ways of producing highly sensitive SERS-active substrates, they have not proven to be scalable to large surface areas.
Perhaps the most widely used method of producing ordered nanostructures for SERS is by self-assembly of colloidal nanoparticles [37]. These SERS substrates are easily fabricated, have long-term stability and are relatively inexpensive to produce. Unlike flat substrates, metal nanoparticles are composed of suspended particles that are smaller and more homogeneous. The dispersive colloidal aggregates are readily prepared by reducing a dilute solution of metal salts. Despite the simplicity of nanoparticle preparation, undesired experimental effects remain as metal colloids tend to aggregate into macroscopic clusters and precipitate from solution following the addition of an adsorbate. The variation in aggregation of the particles results in discrepancy of the resulting SERS intensity.
Commercial available Au substrates are also available for SERS sensing. Among several companies, Renishaw Diagnostics (Glasgow, UK) has commercialized the Klarite® (and more recently the Klarite Plus®) substrates designed from silicon surfaces made of regular inverse pyramidal pattern arrangements coated with Au [38, 39]. The plasmonic bands generated by this substrate permit a variety of wavelengths to be used for spectroscopic interrogations. Real Time Analyzers (Middeltown, CT) produces sol-gel based SERS substrates in solution, microplate, or capillary form.
SERS-active substrates produced by the aforementioned methods differ greatly in morphology and electromagnetic environments, and result in a great disparity in terms of performance and applicability. Although all the methods reviewed possess their respective benefits in terms of preparation, electromagnetic enhancement properties and SERS performance, they suffer from disadvantages such as reproducibility, performance, stability, cost, and inconsistent SERS enhancements. These limitations reinforce the need for practical fabrication methods for producing uniform, high enhancement, and larger area SERS substrates.
Fabrication, Characterization, and Morphology of SERS-Active Ag Nanorod Arrays
Recently, a nanofabrication technique called oblique angle vapor deposition (OAD) has been developed that allows for the growth of aligned Ag nanorod arrays for high sensitivity SERS applications [28]. OAD is a physical vapor deposition technique in which the substrate is rotated to a specific angle such that the vapor from the metal source is incident on the substrate close to the grazing angle. The two phenomena controlling the growth of the Ag nanorod arrays during OAD are the shadowing effect and surface diffusion. These conditions result in preferential growth of cylindrical, irregularly shaped rods that are randomly, yet uniformly, distributed on the surface. The growth of the nanorods arises from initial metal nucleation sites in the direction of vapor deposition. The nanorods produced by OAD are cylindrical in shape but encompass a variety of random protrusions and irregularities [40].
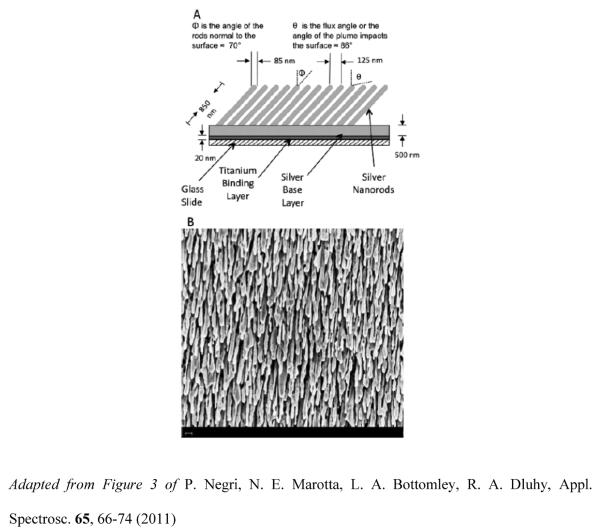
SERS-active Ag nanorod substrates are fabricated by OAD using an electron-beam/sputtering evaporation system. A 20 nm layer of Ti is first deposited onto cleaned glass substrates at a rate no greater than 1.0 Å/s for adhesion and stability of the subsequent silver layer. Next, a 500 nm layer of Ag is deposited on the substrates at a rate of 3.0–4.0 Å/s. The addition of this first silver layer prior to the growth of the Ag nanorods enhances the SERS signal, as compared to growing the nanorods directly on the underlying Ti layer. After deposition of the Ti:Ag underlayer, the substrate is rotated 86° relative to the incident Ag vapor plume. Ag nanorods are then grown at a constant rate of 2.5–3.0 Å/s to a final nominal thickness of 2000 nm. Deposition rate and thickness measurements are controlled by a quartz crystal microbalance (QCM) located inside the chamber and positioned at normal incidence to the vapor source. During the course of the deposition, the growth of the metallic layers is performed in a background deposition pressure maintained at less than 4 × 10−6 Torr. Following the growth of the Ag nanorods, the substrates are cooled down to room temperature in vacuum for a couple hours, then removed from the chamber and stored in a N2 purged glove box to minimize surface contamination.
SERS-active substrates prepared by the OAD method consist of aligned Ag nanorod arrays with: i) nanorod lengths of ~800–900 nm, ii) diameters ~80–90 nm, iii) tilt angles of ~71° with respect to the surface normal, iv) nanorod densities of 15–25 per μm2, and v) average spacing between two adjacent rods of approximately 150 nm. These nanofabrication parameters have been shown to yield SERS enhancement factors of ~108 [40]. Figure 1A depicts a schematic side-view of the Ag nanorod arrays and includes the approximate dimensions of the tilted parallel nanorods resulting from the shadowing effect during the OAD deposition procedure. Scanning electron microscopy (SEM) has been used to characterize Ag nanorod arrays. Figure 1B displays top-view SEM image of a Ag nanorod array substrate. The micrograph (40K) shows that the Ag nanorods are randomly, yet uniformly, distributed on the substrate and preferentially aligned along the vapor deposition direction. Minor variations in nanorod morphology, including length, diameter, and shape are caused by the polycrystalline nature of the Ag nanorods. The preferential growth of the Ag nanorods results in uniform, homogeneous and reproducible substrates. These attributes make OAD-fabricated Ag nanorod arrays excellent sources of SERS substrates.
Figure 1.
(A) Schematic of the Ag nanorod array with approximate dimensions. (B) Top-view micrograph (40K) of a Ag nanorod array depositied onto a glass slide using the OAD method. The scale bar represents 200nm.
Enhancements associated with high curvature regions have been deemed the “lightning rod effect” and contribute to the high enhancement factor observed for molecules at the tips of the nanorods or in pores [41–43]. It has been determined that oscillations of the localized surface plasmon take place along the width (transverse plasmon band) and the length (longitudinal plasmon band) of the nanorods [44]. The overlap of both the longitudinal and lateral plasmon bands is primarily responsible for the strong electromagnetic (EM) enhancement at the tips of the nanorods. As a result, the maximum SERS intensity observed for these Ag nanorod arrays is in the polarization direction perpendicular to the long axis of the nanorods [45]. The high aspect ratio of the Ag nanorods results in a blue shift to below 400 nm of the transverse mode (TM) of the plasmon resonance in the nanorods and a red shift to above 1000 nm of the longitudinal mode of the resonant wavelength. However, the two waves can be tuned accordingly based on the aspect ratio of the nanorods to produce the enhanced signal observed in SERS. Based on these effects, the large SERS enhancement observed on Ag nanorod substrates can be attributed to either a plasmon mode in the gap between two adjacent nanorods or a multimode plasmon resonance. Theoretical calculations on these localized plasmon modes and previous experiments are in agreement with these observations [46].
Presence of surface contamination has been a major limitation to the sensitivity of measurements and utility of SERS as an analytical technique. Graphitization of SERS-active substrates results in an intense background that tends to interfere with the desired spectrum. A number of methods have been developed to chemically or physically remove absorbed organic and carbonaceous contamination from SERS-active metallic surfaces [47–51]. Unfortunately, none of the methods investigated to remove surface contamination have proven fully effective at completely displacing contaminants from SERS-active surfaces.
A mild and efficient method has been reported for removing carbonaceous and organic contaminants from Ag nanorod arrays [52]. The results from this investigation demonstrated that under controlled plasma conditions and exposure times (<4 minutes), the Ar+ plasma cleaning procedure essentially eliminated any detectable background organic and carbonaceous contamination from Ag nanorod substrates without substantially changing their morphology. Plasma cleaning also proved efficient at removing self-assembled monolayers (SAMs) from SERS-active Ag substrates, using 1-propanethiol as the model SAM. The findings from this study showed that Ar+ plasma can effectively remove carbonaceous and organic contaminants from Ag nanorod arrays before their use in sensitive analytical applications.
The thermal stability of the Ag nanorod arrays produced by OAD was investigated [53]. In this study, morphological changes in the arrays were studied as a function of thermal cycling and method of preparation. The findings showed that the method of fabrication heavily influenced the resulting thermal stability. Substrates in which the underlying glass/Ti/Ag surface was exposed to air prior to deposition of the Ag nanorods experienced a higher level of surface restructuring than those substrates held in vacuum for the entire deposition process. These observations were consistent with previously published observations showing that oxygen exposure activates low-temperature coarsening, ultimately resulting in variation of the crystal structure of the polycrystalline Ag. The structural changes seen in this study are consistent with the results found when Ag nanorod arrays were over-exposed to the Ar+ plasma. The results indicated that the Ag nanorods are subject to kinetically-controlled Ag diffusion and coalescence; however, storage in inert conditions reduces the effect and resulting loss of SERS enhancement.
A patterning method for Ag nanorod substrates has been developed in which patterned wells are formed by contact printing of a polymer [54]. The patterning protocol produced an array of three rows containing twelve circular wells each with 1 mm spacing between adjacent wells. Contact with the stamp by contact printing applied a UV-curable epoxy onto the Ag nanorod substrates, which after curing produced the patterned nanorod substrate. The performance of the patterned substrate showed spectral reproducibility and minimal variability in peak intensity across a single well, suggesting uniformity in both analyte concentration and SERS enhancement within the array. The performance repeatability of these patterned Ag nanorod substrates from batch to batch was found to be acceptable for spectroscopic investigations. The benefits of this patterned Ag nanorod array substrate include multiplexing detection, spectral uniformity, physical isolation of samples which minimizes cross contamination and small sample volumes.
An alternative procedure uses a polymer-molding assembly to pattern SERS-active Ag nanorod array substrates deposited on standard glass microscope slides [55]. This well-array patterning technique consists of pouring liquid polydimethylsiloxane (PDMS) into a molding case and curing the polymer at low temperature to produce a uniform 4 × 10 PDMS-patterned microwell substrate for high throughput biosensing and multiplexing. Less spectral variations were observed on patterned Ag nanorod arrays when compared to un-patterned substrates. The patterning procedure facilitated reproducible spotting and drying of the sample in a well-defined area on the substrate, which resulted in more spectral homogeneity and more reproducible SERS results.
Ag Nanorod SERS Substrates For Bioanalytical Sensing
Rapid and sensitive detection of pathogens is an essential tool in disease outbreak intervention strategies. Current diagnostic methods available for the detection of pathogens rely on immunofluorescence tests and antigen-capture immunoassays such as enzyme-linked immunosorbent assay (ELISA) [56–58], hemi-nested multiplex RT-PCR [59–61], or hemadsorption [62, 63]. Many of these assay techniques suffer from lack of sensitivity and reproducibility or require the use of synthetic labels, species-specific reagents (genotyping primers), or amplification of nucleic acid coupled with polymerase chain reaction (PCR). In addition, most of the currently used biochemical assays are limited by the time required to grow cell or bacterial cultures. Consequently, new diagnostic approaches for rapid and sensitive means of detecting pathogens both in the laboratory and in the field are urgently needed to successfully curb potential disease outbreaks.
The unique ability of SERS to provide an analyte-specific response has allowed the development of this method for rapid bio-medical diagnosis of infectious diseases [64–66]. The ultrasensitive, rapid, and label-free attributes of SERS seem to satisfy all criteria needed for routine analysis of different species and strains of pathogens. As a label-free, molecularly specific detection method, SERS allows detection of pathogens with a high degree of sensitivity and specificity. In contrast with the other diagnostic methods previously noted, SERS offers several advantages, including the ability to provide molecular vibrational information for both in-vivo and in-vitro applications, hence allowing discrimination of subtle structural differences and classification of pathogen types and species [26, 67–69]. Another advantage of SERS over fluorescent-based assays includes the multiplexed detection capabilities due to the narrower SERS bands relative to broad fluorescent bands. In addition, the intrinsic detection mode of SERS eliminates the need for synthetic labels and enables the direct detection of the spectral fingerprint of the whole organism [70].
Over the last several years, Ag nanorod arrays have been evaluated as SERS-active substrates for a variety of biosensing applications. The unique signal enhancement offered by Ag nanorod substrates allows nondestructive detection and discrimination of pathogens at the strain level with minimal sample or culture preparation. Ag nanorod arrays have been employed for the SERS-based detection of a variety of bacterial and viral species to allow classification of bacterial and viral types as well as discrimination between pathogen strains and pathogens having single gene mutations. In addition, oligonucleotide-modified Ag nanorod arrays have been used to directly probe sequence information and detect binding of complementary targets. The following sections summarize recent applications of the use of these SERS substrates for pathogen detection and provide an outlook on the future of Ag nanorod arrays for bioanalytical and diagnostic applications.
Detection Of Bacteria
Bacteria are single-celled prokaryote microorganisms that contain neither a nucleus nor other membrane-enclosed organelles like mitochondria and chloroplasts [71]. Rapid means of detecting and identifying pathogenic bacteria play a pivotal role in medical diagnostics, food safety, water quality, pharmaceutical production and biological warfare. Conventional detection methods of bacteria range from immunological approaches [72] to nucleic acid-based assays [73]. These techniques can be cumbersome, time-consuming, and suffer from other shortcomings such as lack of specific chemical information, and a high false positive rate. There is an urgent need for rapid, sensitive and specific detection of pathogenic bacteria. As a vibrational spectroscopic method, SERS provides specific chemical information about the bioanalyte and presents real-time diagnostic capabilities for rapid species identification.
SERS substrates fabricated by OAD have been assessed as a potential analytical sensor for rapid pathogenic bacteria detection. The first report on the bio-detection and identification of bacteria by SERS reported the use of Ag nanorod arrays to detect and identify foodborne pathogenic bacteria [74]. PCA was used to analyze and identify the SERS spectra of generic Escherichia coli (E. coli), E. coli O157:H7, Staphylococcus aureus, Staphylococcus epidermidis, Salmonella typhimurium 1925-1 poultry isolate, E. coli DH 5α and bacteria mixtures. This multivariate statistical method demonstrated the ability to distinguish different bacterial species and allowed differentiation between viable and nonviable cells. The results of this study also highlighted the potential of this technique to classify different bacterial strains as well as the ability to differentiate pure cell samples from mixed cell samples of high spectral similarity.
A separate study compared the SERS spectra of several bacterial species with commonly used bacterial growth media [75]. The findings demonstrated that Raman spectra of various bacterial growth media shared similarities with those of the bacteria themselves, suggesting that previously published reports on Raman detection of bacteria may have misattributed the spectral features of the organisms with their background matrix. The findings of this study called for a re-investigation of the current protocols used for SERS-based detection and identification of bacteria and suggested development of new protocols for acquisition of SERS spectra on bacterial samples so that the spectral bands specific to the bacteria's structure and biomolecular composition can be identified and differentiated from those of the growth medium.
Mycoplasma is a genus of small bacteria lacking a cell wall, which are unaffected by many common antibiotics, and are a major cause of respiratory disease with an increasing threat to human and animal health [76]. Detection of mycoplasma is typically achieved using serologic tests including serum plate agglutination, hemagglutination inhibition, and enzyme-linked immunosorbent assay (ELISA) [77]. These assays are usually time-consuming, cumbersome, expensive, lack sensitivity, and often yield false positives. Therefore, there is a critical need for a new diagnostic platform to detect mycoplasmas with high sensitivity, specificity, and expediency.
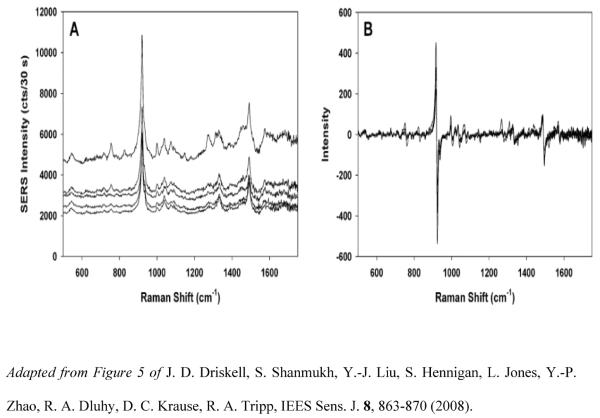
Ag nanorod arrays have been evaluated as a diagnostic platform for the SERS-based detection and identification of different species of mycoplasmas. One of the first studies on the bio-detection of Mycoplasma focused on the detection and differentiation of different strains of Mycoplasma pneumoniae in culture and in both spiked and true clinical throat swab samples by SERS [78]. Figure 2 shows five spectra of M. pneumoniae collected from various locations on the Ag nanorod substrate, illustrating the high degree of reproducibility of the data. Principal Components Analysis (PCA), Hierarchical Cluster Analysis (HCA), and Partial Least Squares Discriminant Analysis (PLS-DA) were used to analyze and identify SERS spectra of three M. pneumonia strains (FH, II-3, and M129). These multivariate statistical techniques allowed robust differentiation of all three mycoplasma strains. In particular, the PLS-DA model generated from 90 spectra (30 of each strain) classified FH, II-3, and M129 with 93–100% sensitivity and specificity. The limit of detection of M. pneumoniae was also assessed using spectra from the serial 10-fold dilution of strain II-3, starting as a concentration of 1.8 × 109 CFU/mL. PLS regression analysis revealed a lower limit of detection of 0.02 CFU/sample, a concentration exceeding the sensitivity of standard PCR.
Figure 2.
(a) Five SERS spectra for M. pneumonia collected from different locations on the substrate. (B) The first derivative spectra of the data plotted in (A).
Simulated and true clinical throat samples were also evaluated using PLS-DA to assess the performance of the assay in a biologically complex background. Spectra from individual and pooled throat swab samples were spiked with M. pneumonia and analyzed against control throat swab samples. A PLS-DA model was built with all dilutions and classification as positive or negative for M. pneumoniae indicated >90% accuracy in cross-validated analysis, showing the ability of Ag nanorod array SERS substrates to detect mycoplasma in a biologically complex, clinically relevant background with 98.1% specificity and 95.2% sensitivity.
In a more recent study, Raman spectral signatures of multiple strains of avian mycoplasma species including Acholeplasma laidlawii, Mycoplasma gallinarum, Mycoplasma gallinaceum, Mycoplasma synoviae, and M. gallisepticum, and vaccine strains 6/85, F, and ts-11 were collected and processed by multivariate analysis to differentiate laboratory cultures of mycoplasma species [79]. A PLS-DA model classified the spectra for all species with 93 to 100% sensitivity and with similar specificity for all species except Mycoplasma synoviae, which was distinguishable with only 80% specificity. Table 1 summarizes the results from the PLS-DA model for the discrimination of the avian mycoplasma species. The M. gallisepticum strains Rlow, S6, and A5969 were examined under the same conditions and were able to be correctly classified with 69 to 100% sensitivity and specificity using PLS-DA. The findings of this study also revealed that when the vaccine strains 6/85, F, and ts-11 were included in the model, PLS-DA was unable to correctly distinguish the strains with statistical significance. When the same data set was examined according to virulence, differentiation of all members of each class occurred with >96% accuracy.
Table 1.
Specificity and sensitivity of NA-SERS discrimination of avian mycoplasma species by PLS-DA.
| Modeled classa | Sensitivity (CVb) | Specificity (CV) | Class error (CV) | RMSECVc |
|---|---|---|---|---|
| M. gallisepticum | 0.944 | 0.951 | 0.053 | 0.246 |
| M. gallinaceum | 0.933 | 0.992 | 0.038 | 0.201 |
| A. laidlawii | 0.933 | 0.950 | 0.058 | 0.236 |
| M. synoviae | 0.933 | 0.800 | 0.133 | 0.2B7 |
| M. gallinarum | 1.000 | 1.000 | 0 | 0.033 |
| Methanol | 1.000 | 0.930 | 0.035 | 0.238 |
Six latent variables (n = 135), accounting for 77% of the captured x variance, were used to generate the model.
CV, cross validation by Venetian Blinds with 10% of the data.
RMSECV, root mean squared error in cross validation.
Adapted from Table 1 of S. L. Hennigan, J. D. Driskell, N. Ferguson-Noel, R. A. Dluhy, Y. P. Zhao, R. A. Tripp, D. C. Krause, Appl. Environ. Microbiol. 78, 1930–1935 (2012).
M. gallisepticum samples were also analyzed based on their virulence in a biochemically complex clinical background as part of a vaccine challenge study involving ts-11. Samples were classified as positive or negative for M. gallisepticum based on serologic testing. PLS-DA showed that samples from the positive field isolate and negative background/vaccine strain classes could be accurately distinguished with 88% cross-validated sensitivity and 85% cross-validated sensitivity. Moreover, the ts-11 vaccine strain and the ts-11-like field isolate K6216D were discriminated with 90% sensitivity and 82% specificity with a 14% error rate using the same multivariate statistical technique. Using the Ag nanorod arrays as the diagnostic platform, the limit of detection for M. pneumonia was assessed on 10 serial 10-fold dilutions with a starting concentration of 4.67 × 105 CFU/μL and was able to be detected down to 1 CFU.
The results suggest that Ag nanorod arrays for SERS-based detection constitute a promising biosensing platform for rapid and sensitive detection of mycoplasma in various complex, biological backgrounds with excellent sensitivity and specificity, and show similar, if not improved, limits of detection when compared to standard molecular strategies. The results demonstrate the potential of this technology for practical diagnosis of pathogens for clinical and point-of-care applications.
Detection of Viruses
Viruses are microscopic infectious agents that can infect the cells of a wide variety of organisms [80]. The current state-of-the-art viral diagnosis methods rely on antibody-based methods such as immunofluorescent testing of isolated viral preparations [81], enzyme-linked immunosorbent (ELISA) [82], or PCR [60], a method that amplifies genetic material fragments for detection. Methods relying on antigen detection and serology are unable to discriminate between virus species or strains of the same virus. Therefore, development of novel strategies to overcome limitations of current viral detection methods is in the forefront of infectious disease detection strategies.
Ag nanorod arrays produced by OAD provide a flexible and sensitive method for SERS-based detection of viruses. An early study demonstrated that SERS using Ag nanorod arrays could detect and differentiate the molecular fingerprints of several important human pathogenic viruses [83]. The spectral variations observed result from the differences in molecular composition of the genomes and proteins of these pathogens as well as adsorption of these components to the Ag surface. PCA and HCA were used to classify the four sample types successfully based solely on their intrinsic spectra. These initial results demonstrated that SERS could be used in combination with multivariate statistical methods for rapid identification and classification of various infectious agents.
Ag nanorod arrays were also used to establish molecular fingerprints of several human viruses [84]. SERS spectra of adenovirus, rhinovirus, and HIV virus were collected; the Raman bands of the viral nucleic acid bases and protein amino acids were identified. The biological media did not confound the viral spectra or prevent band assignments. The results demonstrate that Ag nanorod arrays can readily detect viruses in a variety of biological media.
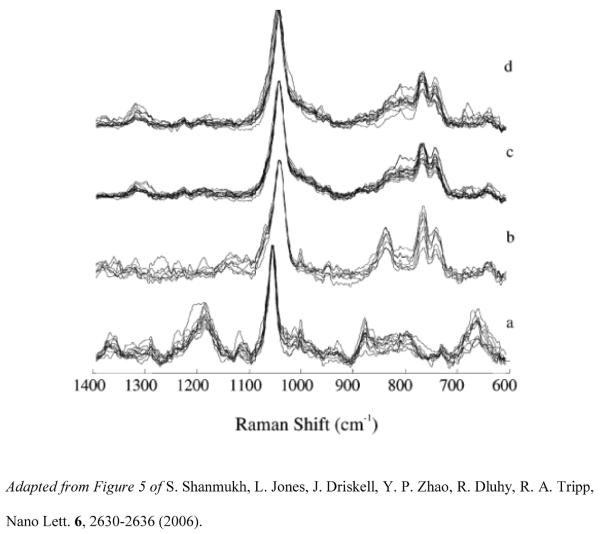
SERS spectra of different virus strains of influenza and RSV were collected to determine if Ag nanorod arrays provide enough sensitivity to distinguish different strains from a single pathogen. Analysis of the SERS spectra collected on the three strains of influenza A (A/HKx31, A/WSN33, and A/PR/8/34) and the four strains of RSV (A/Long, B1, A2, and A2 with gene deletion (ΔG)) suggests that although the spectra of each strain from a single pathogen are very similar, distinct spectral differences between the spectra of the strains of each of the two virus species exist and can be detected via SERS. Figure 3 shows the SERS spectra of the four RSV strains examined in this study. The results suggested that SERS offers a high level of sensitivity for the detection of viruses at extremely low concentrations and permits their rapid and accurate identification, including differentiation of a single pathogen at the strain level.
Figure 3.
SERS spectra of the RSV strains (a) strain A/Long (A/Long), (b) strain B1 (B1), (c) strain A2 with a G gene deletion (ΔG), and (d) strain A2 (A2), collected from several spots on multiple substrates and normalized to the peak intensity of the most intense band (1045 cm−1) and overlaid to illustrate the reproducibility on the Ag nanorod substrate.
In a follow up to that study, PCA and HCA were used to classify four different strains of respiratory syncytial virus (RSV) [85]. The Raman spectra of RSV strains A/Long, B1, A2, and ΔG were acquired. The reproducibility of measurements in this study allowed distinction of the SERS spectra based on the slight nucleic acid and protein composition differences between the four strains. PCA clustered the three main virus strains (A/Long, A2, and B1) together into individual groups. RSV strains A2 and ΔG clustered together due to their extreme similarity in biochemical composition. HCA allowed classification of the RSV strains into their respective classes. SERS spectra of RSV strains A/Long and B1 were identified with 100% accuracy, whereas 88% of the ΔG spectra and 63% of the A2 were identified correctly using HCA. The mismatches accounting for these percentages are due to their close molecular similarity resulting from G gene deletion differences. Table 2 shows the results of the RSV virus strains classification based on HCA.
Table 2.
Virus strain classification based on hierarchical cluster analysis (HCA).
| Viral strain | Correctly classified | Falsely classified | Also classified as | Sensitivitya | Specificityb |
|---|---|---|---|---|---|
| RSV A/Long | 17 | 0 | - | 1.0 | 1.0 |
| RSV B1 | 17 | 0 | - | 1.0 | 0.92 |
| RSV ΔG | 15 | 2 | A2(2) | 0.88 | 0.94 |
| RSV A2 | 12 | 7 | ΔG(3), B1(4) | 0.63 | 0.96 |
Probability of correctly classifing a SERS virus spectrum as belonging to the virus strain class (i.e., a true positive)
Probability of correctly classifing a SERS virus spectrum as not belonging to the virus strain class (i.e., a true negative)
Adapted from Table 1 of S. Shanmukh, L. Jones, Y. P. Zhao, J. D. Driskell, R. A. Tripp, R. A. Dluhy, Anal Bioanal Chem 390, 1551–1555 (2008).
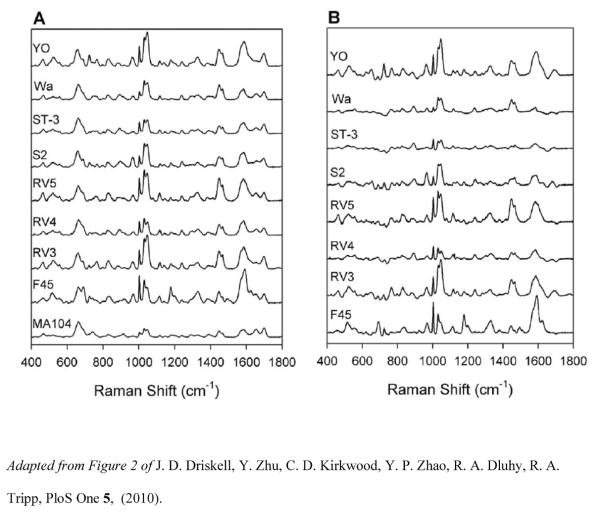
A SERS-based detection scheme has been developed to qualitatively identify eight rotavirus strains and classify them according to genotype [86]. SERS spectra of each of the eight rotavirus strains were collected on three different OAD-fabricated substrates to assess the reproducibility of measurements and the variation in terms of substrate and sampling homogeneity. SERS spectra of rotavirus strains F45, RV3, RV4, RV5, S2, ST-3, Wa, and YO corresponding to either G or P genotypes were analyzed in cell lysate, applied directly to the Ag nanorod array substrates and allowed to dry at room temperature prior to SERS measurements. Figure 4 shows the SERS spectra of the eight strains of rotavirus and the negative control MA104 cell lysate (left panel), as well as the difference SERS spectra of the eight strains after subtraction of MA104 spectrum (right panel). The spectra collected showed a high degree of similarity among each strain. Subtle structural differences inherent to antigenic variations between the virus strains permitted discrimination of the strains, and differentiation of rotavirus positive and negative samples was accomplishedusing PLS-DA with 100% accuracy. Overall, the model resulted in >98% sensitivity and 100% specificity for G and P genotypes, as seen in Table 3. Partial least squares (PLS) regression analysis was also used to quantitatively assess the detection limits of these rotavirus strains. The PLS regression model predicted concentration based on intrinsic SERS spectra to be accurate for concentrations ≥ 104 pfu/mL.
Figure 4.
Rotavirus SERS spectra. (A) Average SERS spectra for eight strains of rotavirus and the negative control (MA104 cell lysate). Spectra were baseline corrected, normalized to the band at 633cm−1, and offset for visualization. (B) Difference SERS spectra for eight strains after subtraction of MA104 spectrum.
Table 3.
Summary of the PLS-DA cross-validation results for classification according to three different models based on the strain, G genotype and P genotype.
| P genotype Classification | |||||||||
| P8 | P4 | P6 | Neg ctrl | ||||||
| sensitivity | 0983 | 1.000 | 1.000 | 1.000 | |||||
| specificity | 1.000 | 1.000 | 1.000 | 1.000 | |||||
| G genotype Classification | |||||||||
| G9 | G3 | G1 | G2 | G4 | neg ctrl | ||||
| sensitivity | 1.000 | 1.000 | 0.967 | 1.000 | 1.000 | 1.000 | |||
| specificity | 0.992 | 1.000 | 0.990 | 1.000 | 1.000 | 1.000 | |||
| Strain Classification | |||||||||
| F45 | RV3 | RV4 | RV5 | S2 | ST-3 | Wa | YO | neg Ctrl | |
| sensitivity | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| specificity | 0.992 | 1.000 | 1.000 | 1.000 | 1.000 | 0.992 | 1.000 | 1.000 | 1.000 |
doi:10.1371/journal.pone0010222.t002
Adapted from Table 2 of J. D. Driskell, Y. Zhu, C. D. Kirkwood, Y. P. Zhao, R. A. Dluhy, R. A. Tripp, Plos One 5, (2010).
Ag nanorod arrays have also been used for rapid genotyping of four separate measles virus (MeV) genotypes (Mev A, D4, D9, and H1) [87]. Analysis of the genetic sequences of the MeV genotypes examined in this study determined that the four MeV genotypes exhibited >96% genetic similarity. PCA analysis based on the SERS spectra of each genotype showed close similarity between the genotypes, but effective identification of MeV genotypes and differentiation of MeV from controls was possible. The dendrogram generated by HCA correctly distinguished the spectra of the four MeV samples, the media controls, and the solvent background, but neither PCA nor HCA could differentiate the SERS spectra of the closely related MeV-D4 and Mev-D9 genotypes. However, PLS-DA was able to correctly differentiate between each of the four MeV genotypes and the negative controls with high accuracy. The PLS-DA model demonstrated cross-validated sensitivities of >90% and specificities of >96% for each MeV genotype. A summary table of the PLS-DA cross-validated results is shown in Table 4. The results of this study demonstrated the accuracy these multivariate statistical methods to effectively differentiate between four MeV genotypes, two media controls, and a background with high sensitivity and specificity.
Table 4.
Summary of the PLS-DA cross-validation results for classification of the 4 separate measles genotypes along with 2 negative controls and the blanks.
| Measles Genotype Classification | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Aa | D4 | D9 | H1 | C1 | C2 | Blank | |
| sensitivityb | 1.000 | 0.900 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| specificityc | 1.000 | 0.982 | 0.964 | 1.000 | 1.000 | 1.000 | 1.000 |
A, D4, D9 and H1 denote the individual measles genotype used in this study (see Table 1). C1 denotes control sample 1 (i.e. the Vero cell medium). C2 denotes control sample 2 (i.e. Vero/hSLAM cell medium). Blank denotes the bare Ag nanorod substrate with only solvent added.
sensitivity = probability of classifying a SERS spectrum as belonging to the correct measles virus class (i.e. a true positive).
specificity = probability of classifying a SERS spectrum as not belonging to the correct measles virus class (i.e. a true negative).
Adapted from Table 2 of V. Hoang, R. A. Tripp, P. Rota, R. A. Dluhy, Analyst 135, 3103–3109 (2010).
The results of these studies highlight the potential of Ag nanorod arrays for SERS-based detection of important pathogenic viruses. This platform allows rapid and accurate identification and classification of viruses at the species, strain, and genotype levels. SERS-based virus detection appears to be a highly promising alternative to current diagnostic assays and has the potential to be broadly applicable to a variety of viral targets.
Oligonucleotide-Modified Ag Nanorod Substrates for Detection of Pathogenic Targets
The studies on SERS-based detection of bacteria and viruses established the proof of principle that Ag nanorod SERS has the ability to directly probe biochemical information in a label-free fashion. In addition to direct detection of whole organisms, chemical modification of Ag nanorod substrates with oligonucleotides allows for specific detection of various high affinity genomic targets.
MicroRNAs (miRNA) have been studied using Ag nanorod substrates. miRNAs are non-coding RNAs that play a key role in the regulation of gene expression and have been extensively studied as candidates for diagnostic and/or prognostic biomarkers [88, 89]. However, miRNA detection involves a number of analytical challenges. First, miRNA are small (19–25 nucleotides), which makes it difficult for traditional DNA-based methods to sensitively detect these sequences with any reliability. Second, current technologies for detection and profiling miRNA expression may have limited specificity when closely related miRNA family members exhibiting single base-pair polymorphism are analyzed. These drawbacks emphasize the need for novel methods with high specificity that can discriminate between single nucleotide mismatches.
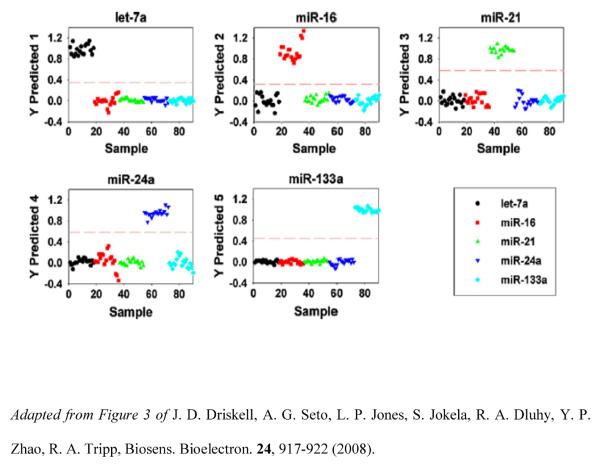
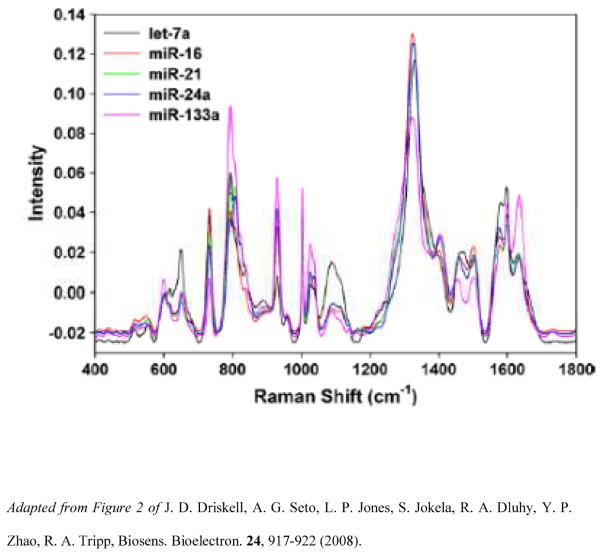
miRNA profiling has been demonstrated using Ag nanorod of SERS-based array substrates [90]. Five unrelated miRNAs having dissimilar sequences were examined, as well as eight members of the hsa-let-7 family containing similar miRNA sequences, including sequences differing by a single base mismatch. Analysis of the SERS spectra of let-7a reveals reproducible spectral features that can be attributed to nucleic acid vibrations, with similar band position and shape and with only slight variations in relative intensities. Figure 6 shows the average SERS spectra for five unrelated miRNA sequences. PLS-DA classified the five unrelated miRNAs based on unique SERS molecular signatures and correctly assigned each let-7a sequence in its correct class, with 100% sensitivity and specificity. Figure 6 shows the PLS-DA Y predicted plots for the 5 miRNA classes. The SERS spectra of the seven has-let-7a family members (having 71–95% sequence homology) were classified with 100% sensitivity and 99–100% specificity, proving that SERS can be used as a rapid, specific and sensitive miRNA detection technology without target amplification.
Figure 6.
PLS-DA Y predicted plots. Each plot predicts a sample as belonging to or not belonging to the specified miRNA class. Let-7a (●), miR-16( ), miR-21 (
), miR-21 ( ), miR-24a (
), miR-24a ( ), and miR-133a (◇).
), and miR-133a (◇).
Quantitative detection of miRNA sequences in multicomponent mixtures has also been accomplished [91]. In this study, two-, three-, and five-component mixtures of miRNA were prepared with the total miRNA concentration in the component mixtures held constant at 1.00 μg/mL while the relative ratios of each component varied from 6 to 150 μM. Plots of PLS cross-validated predictions for each of the miRNA sequences in the mixtures for the calibration model were generated for all three sample mixtures. The results indicated that the PLS regression model was able to accurately detect and quantify individual miRNA sequences in multicomponent mixtures in a label-free fashion.
The binding affinities of miRNAs in ssRNA and dsRNA:DNA duplexes have been evaluated, both with and without thiol modification [92]. The findings from this study showed that thiolated probes adsorb better than non-thiolated probes on the surface of Ag nanorod arrays. Additionally, it was shown that a thiolated RNA:DNA solution complex does not bind to the Ag nanorod substrate due to the rigid structure of the duplex. Following these observations, an assay was designed for miRNA detection in which a constant concentration (500 nM) of let-7f probe was mixed with various concentrations of let-7f and allowed to hybridize prior to incubation of the mixture on the Ag nanorod substrate. As expected, the SERS intensity linearly decreased as the concentration of the let-7f increased from 125 to 500 nM. Incubation of the let-7a probe with a mismatched miRNA indicated no detectable hybridization. These results show that miRNA sequences can be detected and quantified based on affinity of the probe to its complementary target. Intrinsic SERS detection allowed detection of target miRNA by providing a molecularly specific signal, thus offering the double advantage of multiplexed label-free detection.
Other oligonucleotide sequences, and in particular aptamers, have been studied with Ag nanorod SERS. Aptamers are single-stranded DNA or RNA oligonucleotides synthesized in-vitro that are capable of binding with high affinity and specificity to specific target molecules associated with a number of disease states [93, 94]. Aptamers are promising candidates for the development of therapeutic and diagnostic-based assays due to their robustness, lack of immunogenicity and stability against biodegradation and denaturation [95, 96].
The high affinity and specificity of aptamers for their molecular targets leads to binding of the target molecule to the aptamer-modified surface, which results in a change of the oligonucleotide secondary structure [93, 94]. Therefore, the use of aptamers in biomedical vibrational spectroscopy can provide a unique Raman spectral signature [97–99]. Ag nanorod SERS was shown to detect the binding of a commercial diagnostic aptamer to the nucleoproteins of three virus strains contained in the Fluarix™ Influsplit SSW® 2009/2010 split-virion inactivated influenza vaccine [100]. In this study, the ssDNA aptamer was first immobilized on the surface of the Ag nanorod arrays as the 5' C6 disulfide. Binding of the three separate virus nucleoproteins to the aptamer-functionalized surface was accomplished by incubating the respective influenza cell lysate. As expected, distinct spectral changes were observed in the SERS spectra of the anti-influenza aptamer after incubation with the cell lysates from the three monovalent influenza virus strains when compared to the SERS spectrum of the anti-influenza aptamer alone.
A negative control experiment immobilized a random 22-mer DNA sequence to the Ag nanorod arrays, followed by binding of the three separate virus strains in the same fashion as previously noted. As expected, no measurable changes in the SERS spectrum of the immobilized 22-mer DNA were observed following incubation with any of the three influenza virus samples, reflecting a high degree of specificity for the binding of the influenza nucleoproteins only to the anti-influenza aptamer.
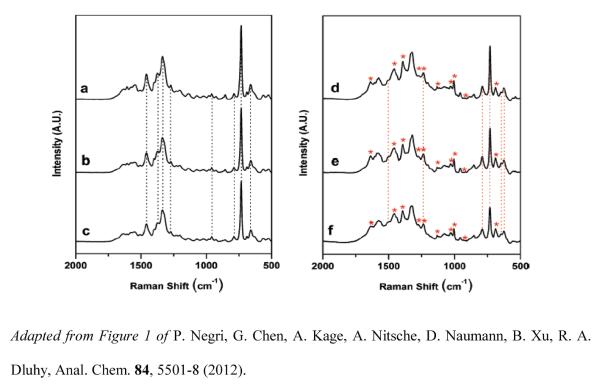
A further study extended this investigation and presented an additional negative control experiment, a detailed chemometric spectral analysis of the aptamer-target complexes, and AFM images of the aptamer-target complex [101]. Figure 7 shows the SERS spectra of the anti-influenza aptamer- spacer complex, blank control (binding buffer), negative control (RSV) (left panel) and the spectra of the aptamer-spacer complex incubated with the three strains contained in the split-virion influenza vaccine (right panel). Spectral features attributed to addition of nucleoproteins to the aptamer-coated surface as well as changes in oligonucleotide bands in the SERS spectra resulting from conformational changes upon binding were identified. The binding specificity of the influenza viral nucleoproteins to the anti-influenza aptamer was investigated using a 5'-C6-thiolated anti-vasopressin aptamer as an additional negative control.
Figure 7.
SERS spectrum of the anti-influenza aptamer (1000 nM) - PEG spacer (100 nM) complex on a Ag nanorod substrate, (b) anti-influenza aptamer complex and binding buffer blank control, (c) anti-influenza aptamer complex and RSV (105 PFU/mL) negative control, (d) anti-influenza aptamer complex and nucleoproteins from A/Uruguay, (e) anti-influenza aptamer complex and nucleoproteins from A/Brisbane, and (f) anti-influenza aptamer complex and nucleoproteins from B/Brisbane. Virus concentrations in (d) – (f) were adjusted to 1μg/mL (relative to HA content). Each spectrum shown is an average of 10 individual spectra for each particular sample. The dashed vertical lines in (a), (b), and (c) indicate the characteristic oligonucleotide bands for the influenza aptamer and its controls. The dashed vertical lines in (d), (e), and (f) indicate the oligonucleotide bands that changed after binding of the nucleoproteins. Asterisks indicate the presence of new bands in the aptamer complex corresponding to binding of the protein target.
The chemometric methods HCA, PCA, and PLS-DA were used to differentiate the SERS spectra of the unbound aptamers and their controls from the aptamer-target complexes. In the case of the anti-influenza aptamer, the SERS spectra of the blanks and negative controls were differentiated from those of the influenza nucleoprotein complexes with 100% accuracy. Likewise, perfect statistical differentiation was achieved between the aptamer controls and the vasopressin targets. The three multivariate statistical methods used confirmed selectivity of binding of the anti-influenza aptamer to its nucleoprotein target, and showed lack of binding of the influenza nucleoproteins to a non-influenza aptamer, thus providing a measure of diagnostic sensitivity and specificity of the assay. These results demonstrated the use of a novel aptamer-modified SERS substrate as a diagnostic tool for virus detection in a complex biological matrix and provide support for the use of SERS-based arrays for high-throughput sensing and multiplexing.
Summary
The ability of SERS to provide a unique, chemically specific molecular fingerprint with high sensitivity and specificity offers a powerful diagnostic platform that is well-suited for a variety of bioanalytical problems. Aligned Ag nanorod arrays fabricated by OAD have been investigated as potential SERS-active substrates for the detection of a variety of pathogens and oligonucleotide-based bio-recognition and biosensing applications. This novel biosensing platform has the advantage to rapidly, accurately, and cost-effectively detect extremely low levels of virus and bacteria, thus fulfilling a critical need for sensitive and reliable means of pathogens detection not currently existing. The level of spectral reproducibility offered by the Ag nanorod arrays allows acquisition of molecular fingerprints of pathogens and permits differentiation between microorganisms or viral pathogens based on their inherent structural and biochemical differences. Of the current nano-based bioanalytical methods under investigation, SERS shows potential for multiplexed detection and identification of the biochemical components of pathogenic microorganisms with minimum sample preparation. The inherent attributes of SERS overcome many of the limitations of current immunologic- or PCR-based diagnostic tests for pathogenic microorganisms, most notably, the elimination of amplification and labeling steps. The results of investigations conducted using Ag nanorod arrays as SERS-active substrates demonstrate the use of spectral fingerprinting in biomedical vibrational spectroscopy can potentially: i) provide a unique spectral signature specific for a pathogen of interest, ii) facilitate identification through comparison with spectral libraries, iii) eliminate the use of species-specific monoclonal antibodies, and iv) minimize false positives common to indirect methods. Additionally, the high sensitivity of the SERS data coupled with the statistical method of data opens new perspectives for the development and integration of ultrasensitive detection of biorecognition events for early clinical diagnostic and biomedical applications. Ag nanorod arrays also have the potential to be integrated into biomedical diagnostic platforms accommodating a wide variety of biospecific recognition events in a label-free fashion. The use of these substrates for SERS-based detection of pathogenic targets has potential use as a diagnostic tool for virus detection in a complex biological matrix. The advantages of this biosensing platform open new perspectives for the development and integration of ultrasensitive detection of pathogens for early clinical diagnostic and biomedical applications central to human health. The attributes of Ag nanorod arrays as SERS-active substrates open the door to various sensor applications and demonstrate the applicability of this biosensing platform to a wide range of infectious agents for rapid medical diagnostics of infectious diseases.
Figure 5.
Overlaid average SERS spectra (n=18) for each unrelated miRNA. The spectra have been baseline corrected and unit-vector normalized for visualization of spectral differences.
Acknowledgements
Support for the work described here was provided by the UGA Office of the Vice President for Research through a UGA Faculty of Infectious Disease Seed Grant, by the U.S. Army Research Laboratory (Cooperative Agreement # W911NF-07-R-001-04), as well as the National Institutes of Health (GM102546).
References
- [1].Hering K, Cialla D, Ackermann K, Dorfer T, Moller R, Schneidewind H, Mattheis R, Fritzsche W, Rosch P, Popp J. Anal Bioanal Chem. 2008;390:113–124. doi: 10.1007/s00216-007-1667-3. [DOI] [PubMed] [Google Scholar]
- [2].Chou IH, Benford M, Beier HT, Cote GL, Wang M, Jing N, Kameoka J, Good TA. Nano Lett. 2008;8:1729–1735. doi: 10.1021/nl0808132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kneipp K, Kneipp H, Itzkan I, Dasari RR, Feld MS. Curr. Sci. 1999;77:915–924. [Google Scholar]
- [4].Vo-Dinh T, Yan F, Wabuyele MB. J Raman Spectrosc. 2005;36:640–647. [Google Scholar]
- [5].Kneipp K, Kneipp H, Itzkan I, Dasari RR, Feld MS. J. Phys.-Condens. Mat. 2002;14:R597–R624. [Google Scholar]
- [6].Carey PR. Biochemical Applications of Raman and Resonance Raman Spectroscopies. Academic Press; New York: 1982. p. xi.p. 262. [Google Scholar]
- [7].Kalasinsky KS, Hadfield T, Shea AA, Kalasinsky VF, Nelson MP, Neiss J, Drauch AJ, Vanni GS, Treado PJ. Anal. Chem. 2007;79:2658–2673. doi: 10.1021/ac0700575. [DOI] [PubMed] [Google Scholar]
- [8].Vo-Dinh T, Allain LR, Stokes DL. J Raman Spectrosc. 2002;33:511–516. [Google Scholar]
- [9].Zhang XY, Young MA, Lyandres O, Van Duyne RP. J. Am. Chem. Soc. 2005;127:4484–4489. doi: 10.1021/ja043623b. [DOI] [PubMed] [Google Scholar]
- [10].Monaghan PB, McCarney KM, Ricketts A, Littleford RE, Docherty F, Smith WE, Graham D, Cooper JM. Anal. Chem. 2007;79:2844–2849. doi: 10.1021/ac061769i. [DOI] [PubMed] [Google Scholar]
- [11].Sutherland WS, Laserna JJ, Angebranndt MJ, Winefordner JD. Anal. Chem. 1990;62:689–693. doi: 10.1021/ac00206a008. [DOI] [PubMed] [Google Scholar]
- [12].Ruperez A, Montes R, Laserna JJ. Vib Spectrosc. 1991;2:145–154. [Google Scholar]
- [13].Farquharson S, Shende C, Inscore FE, Maksymiuk P, Gift A. J Raman Spectrosc. 2005;36:208–212. [Google Scholar]
- [14].Pinzaru SC, Pavel I, Leopold N, Kiefer W. J Raman Spectrosc. 2004;35:338–346. [Google Scholar]
- [15].Moskovits M. Rev Mod Phys. 1985;57:783–826. [Google Scholar]
- [16].Kneipp K, Kneipp H, Kneipp J. Acc. Chem. Res. 2006;39:443–450. doi: 10.1021/ar050107x. [DOI] [PubMed] [Google Scholar]
- [17].Dieringer JA, McFarland AD, Shah NC, Stuart DA, Whitney AV, Yonzon CR, Young MA, Zhang XY, Van Duyne RP. Faraday Discuss. 2006;132:9–26. doi: 10.1039/b513431p. [DOI] [PubMed] [Google Scholar]
- [18].Kneipp K, Kneipp H, I I, Dasari RR, Feld MS. Chem. Rev. 1999;99:2957–+. doi: 10.1021/cr980133r. [DOI] [PubMed] [Google Scholar]
- [19].Willets KA, Van Duyne RP. Annu Rev Phys Chem. 2007;58:267–297. doi: 10.1146/annurev.physchem.58.032806.104607. [DOI] [PubMed] [Google Scholar]
- [20].Guerrero AR, Aroca RF. Angew Chem Int Edit. 2011;50:665–668. doi: 10.1002/anie.201004806. [DOI] [PubMed] [Google Scholar]
- [21].Zhang YX, Aslan K, Previte MJR, Geddes CD. Appl Phys Lett. 2007;90 doi: 10.1063/1.2829798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Stiles PL, Dieringer JA, Shah NC, Van Duyne RR. Annu Rev Anal Chem. 2008;1:601–626. doi: 10.1146/annurev.anchem.1.031207.112814. [DOI] [PubMed] [Google Scholar]
- [23].Mulvaney SP, Musick MD, Keating CD, Natan MJ. Langmuir. 2003;19:4784–4790. [Google Scholar]
- [24].Bantz KC, Meyer AF, Wittenberg NJ, Im H, Kurtulus O, Lee SH, Lindquist NC, Oh SH, Haynes CL. Phys Chem Chem Phys. 2011;13:11551–11567. doi: 10.1039/c0cp01841d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kneipp K, Wang Y, Kneipp H, Perelman LT, Itzkan I, Dasari R, Feld MS. Phys. Rev. Lett. 1997;78:1667–1670. [Google Scholar]
- [26].Jarvis RM, Goodacre R. Chem Soc Rev. 2008;37:931–936. doi: 10.1039/b705973f. [DOI] [PubMed] [Google Scholar]
- [27].Moskovits M. J Raman Spectrosc. 2005;36:485–496. [Google Scholar]
- [28].Chaney SB, Shanmukh S, Zhao Y-P, Dluhy RA. Appl Phys Lett. 2005;87:31908–31910. [Google Scholar]
- [29].Fusco M. IEEE Trans Antennas Propag. 1990;38:76–89. [Google Scholar]
- [30].Fleischm M, Hendra PJ, Mcquilla Aj. Chem Phys Lett. 1974;26:163–166. [Google Scholar]
- [31].Xue G, Dong J, Zhang MS. Appl Spectrosc. 1991;45:756–759. [Google Scholar]
- [32].Royer P, Goudonnet JP, Warmack RJ, Ferrell TL. Phys. Rev. B. 1987;35:3753–3759. doi: 10.1103/physrevb.35.3753. [DOI] [PubMed] [Google Scholar]
- [33].Abu Hatab NA, Oran JM, Sepaniak MJ. ACS Nano. 2008;2:377–385. doi: 10.1021/nn7003487. [DOI] [PubMed] [Google Scholar]
- [34].Haynes CL, Van Duyne RP. J. Phys. Chem. B. 2001;105:5599–5611. [Google Scholar]
- [35].Yao JL, Pan GP, Xue KH, Wu DY, Ren B, Sun DM, Tang J, Xu X, Tian ZQ. Pure Appl. Chem. 2000;72:221–228. [Google Scholar]
- [36].Walsh RJ, Chumanov G. Appl Spectrosc. 2001;55:1695–1700. [Google Scholar]
- [37].Freeman RG, Grabar KC, Allison KJ, Bright RM, Davis JA, Guthrie AP, Hommer MB, Jackson MA, Smith PC, Walter DG, Natan MJ. Science. 1995;267:1629–1632. doi: 10.1126/science.267.5204.1629. [DOI] [PubMed] [Google Scholar]
- [38].Perney NMB, Baumberg JJ, Zoorob ME, Charlton MDB, Mahnkopf S, Netti CM. Opt Express. 2006;14:847–857. doi: 10.1364/opex.14.000847. [DOI] [PubMed] [Google Scholar]
- [39].Perney NMB, de Abajo FJG, Baumberg JJ, Tang A, Netti MC, Charlton MDB, Zoorob ME. Phys Rev B. 2007;76 [Google Scholar]
- [40].Driskell JD, Shanmukh S, Liu Y, Chaney SB, Tang XJ, Zhao YP, Dluhy RA. J. Phys. Chem. C. 2008;112:895–901. [Google Scholar]
- [41].Schatz GC. Acc. Chem. Res. 1984;17:370–376. [Google Scholar]
- [42].Gersten JI. J Chem Phys. 1980;72:5779–5780. [Google Scholar]
- [43].Gersten JI. J Chem Phys. 1980;72:5780–5781. [Google Scholar]
- [44].Zhao YP, Chaney SB, Shanmukh S, Dluhy RA. J. Phys. Chem. B. 2006;110:3153–3157. doi: 10.1021/jp057406o. [DOI] [PubMed] [Google Scholar]
- [45].Leverette CL, Jacobs SA, Shanmukh S, Chaney SB, Dluhy RA, Zhao YP. Appl Spectrosc. 2006;60:906–913. doi: 10.1366/000370206778062084. [DOI] [PubMed] [Google Scholar]
- [46].GarciaVidal FJ, Pendry JB. Physical Review Letters. 1996;77:1163–1166. doi: 10.1103/PhysRevLett.77.1163. [DOI] [PubMed] [Google Scholar]
- [47].Taylor CE, Garvey SD, Pemberton JE. Anal. Chem. 1996;68:2401–2408. [Google Scholar]
- [48].Li MD, Cui Y, Gao MX, Luo J, Ren B, Tian ZQ. Anal. Chem. 2008;80:5118–5125. doi: 10.1021/ac8003083. [DOI] [PubMed] [Google Scholar]
- [49].Cooney RP, Mahoney MR, Howard MW. Chem Phys Lett. 1980;76:448–452. [Google Scholar]
- [50].Schoenfisch MH, Ross AM, Pemberton JE. Langmuir. 2000;16:2907–2914. [Google Scholar]
- [51].Taylor CE, Schoenfisch MH, Pemberton JE. Langmuir. 2000;16:2902–2906. [Google Scholar]
- [52].Negri P, Marotta NE, Bottomley LA, Dluhy RA. Appl Spectrosc. 2011;65:66–74. doi: 10.1366/10-06037. [DOI] [PubMed] [Google Scholar]
- [53].Beavers KR, Marotta NE, Bottomley LA. Chem Mater. 2010;22:2184–2189. [Google Scholar]
- [54].Marotta NE, Barber JR, Dluhy PR, Bottomley LA. Appl Spectrosc. 2009;63:1101–1106. doi: 10.1366/000370209789553174. [DOI] [PubMed] [Google Scholar]
- [55].Abell JL, Driskell JD, Dluhy RA, Tripp RA, Zhao YP. Biosens Bioelectron. 2009;24:3663–3670. doi: 10.1016/j.bios.2009.05.039. [DOI] [PubMed] [Google Scholar]
- [56].Chao RK, Fishaut M, Schwartzman JD, Mcintosh K, Infect J. Dis. 1979;139:483–486. doi: 10.1093/infdis/139.4.483. [DOI] [PubMed] [Google Scholar]
- [57].Pedneault L, Robillard L, Turgeon JP. J. Clin. Microbiol. Infect. Dis. 1994;32:2861–2864. doi: 10.1128/jcm.32.11.2861-2864.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Petitjean-Lecherbonnier J, Vabret A, Gouarin S, Dina J, Legrand L, Freymuth F. Pathol Biol. 2006;54:603–611. doi: 10.1016/j.patbio.2006.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Freymuth F, Eugene G, Vabret A, Petitjean J, Gennetay E, Brouard J, Duhamel JF, Guillois B. J. Clin. Microbiol. 1995;33:3352–3355. doi: 10.1128/jcm.33.12.3352-3355.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Henkel JH, Aberle SW, Kundi M, PopowKraupp T. J. Med. Virol. 1997;53:366–371. [PubMed] [Google Scholar]
- [61].Pitcher D, Chalker VJ, Sheppard C, George RC, Harrison TG. J. Med. Microbiol. 2006;55:149–155. doi: 10.1099/jmm.0.46281-0. [DOI] [PubMed] [Google Scholar]
- [62].Connor EM, Loeb MR. J Infect Dis. 1983;148:855–860. doi: 10.1093/infdis/148.5.855. [DOI] [PubMed] [Google Scholar]
- [63].Uhlendorff J, Matrosovich T, Klenk HD, Matrosovich M. Arch. Virol. 2009;154:945–957. doi: 10.1007/s00705-009-0393-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Patel IS, Premasiri WR, Moir DT, Ziegler LD. J Raman Spectrosc. 2008;39:1660–1672. doi: 10.1002/jrs.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Bao PD, Huang TQ, Liu XM, Wu TQ. J Raman Spectrosc. 2001;32:227–230. [Google Scholar]
- [66].Premasiri WR, Moir DT, Klempner MS, Krieger N, Jones G, Ziegler LD. J. Phys. Chem. B. 2005;109:312–320. doi: 10.1021/jp040442n. [DOI] [PubMed] [Google Scholar]
- [67].Jarvis RM, Brooker A, Goodacre R. Anal. Chem. 2004;76:5198–5202. doi: 10.1021/ac049663f. [DOI] [PubMed] [Google Scholar]
- [68].Jarvis RM, Brooker A, Goodacre R. Faraday Discuss. 2006;132:281–292. doi: 10.1039/b506413a. [DOI] [PubMed] [Google Scholar]
- [69].Jarvis RM, Goodacre R. Anal. Chem. 2004;76:40–47. doi: 10.1021/ac034689c. [DOI] [PubMed] [Google Scholar]
- [70].Goodacre R, Timmins EM, Burton R, Kaderbhai N, Woodward AM, Kell DB, Rooney PJ. Microbiol-UK. 1998;144:1157–1170. doi: 10.1099/00221287-144-5-1157. [DOI] [PubMed] [Google Scholar]
- [71].Mandell GLD, R. G., Bennett JE. Principles and practice of infectious diseases. Volumes 1 and 2. John Wiley & Sons; New York: 1979. [Google Scholar]
- [72].Mazenko RS, Rieders F, Brewster JD. J. Microbiol. Methods. 1999;36:157–165. doi: 10.1016/s0167-7012(99)00027-5. [DOI] [PubMed] [Google Scholar]
- [73].Graham C. R. N. a. A. PCR. Bios Scientific; Oxford: 1997. [Google Scholar]
- [74].Chu HY, Huang YW, Zhao YP. Appl Spectrosc. 2008;62:922–931. doi: 10.1366/000370208785284330. [DOI] [PubMed] [Google Scholar]
- [75].Marotta NE, Bottomley LA. Appl Spectrosc. 2010;64:601–606. doi: 10.1366/000370210791414326. [DOI] [PubMed] [Google Scholar]
- [76].Levisohn S, Kleven SH. Rev. Sci. Tech. OIE. 2000;19:425–442. [PubMed] [Google Scholar]
- [77].Avakian AP, Kleven SH, Glisson JR. Avian Dis. 1988;32:262–272. [PubMed] [Google Scholar]
- [78].Hennigan SL, Driskell JD, Dluhy RA, Zhao YP, Tripp RA, Waites KB, Krause DC. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0013633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hennigan SL, Driskell JD, Ferguson-Noel N, Dluhy RA, Zhao YP, Tripp RA, Krause DC. Appl. Environ. Microbiol. 2012;78:1930–1935. doi: 10.1128/AEM.07419-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Henle W. J. Immunol. 1950;64:203–236. [PubMed] [Google Scholar]
- [81].Barenfanger J, Drake C, Leon N, Mueller T, Troutt T. J. Clin. Microbiol. 2000;38:2824–2828. doi: 10.1128/jcm.38.8.2824-2828.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chao RK, Fishaut M, Schwartzman JD, Mcintosh K. J Infect Dis. 1979;139:483–486. doi: 10.1093/infdis/139.4.483. [DOI] [PubMed] [Google Scholar]
- [83].Driskell JD, Shanmukh S, Liu Y-J, Hennigan S, Jones L, Zhao Y-P, Dluhy RA, Krause DC, Tripp RA. IEES Sens. J. 2008;8:863–870. [Google Scholar]
- [84].Shanmukh S, Jones L, Driskell J, Zhao YP, Dluhy R, Tripp RA. Nano Lett. 2006;6:2630–2636. doi: 10.1021/nl061666f. [DOI] [PubMed] [Google Scholar]
- [85].Shanmukh S, Jones L, Zhao YP, Driskell JD, Tripp RA, Dluhy RA. Anal Bioanal Chem. 2008;390:1551–1555. doi: 10.1007/s00216-008-1851-0. [DOI] [PubMed] [Google Scholar]
- [86].Driskell JD, Zhu Y, Kirkwood CD, Zhao YP, Dluhy RA, Tripp RA. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0010222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Hoang V, Tripp RA, Rota P, Dluhy RA. Analyst. 2010;135:3103–3109. doi: 10.1039/c0an00453g. [DOI] [PubMed] [Google Scholar]
- [88].Bartel B. Nat. Struct. Mol. Biol. 2005;12:569–571. doi: 10.1038/nsmb0705-569. [DOI] [PubMed] [Google Scholar]
- [89].Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- [90].Driskell JD, Seto AG, Jones LP, Jokela S, Dluhy RA, Zhao YP, Tripp RA. Biosens Bioelectron. 2008;24:917–922. doi: 10.1016/j.bios.2008.07.060. [DOI] [PubMed] [Google Scholar]
- [91].Driskell JD, Primera-Pedrozo OM, Dluhy RA, Zhao YP, Tripp RA. Appl Spectrosc. 2009;63:1107–1114. doi: 10.1366/000370209789553183. [DOI] [PubMed] [Google Scholar]
- [92].Driskell JD, Tripp RA. Chem. Commun. 2010;46:3298–3300. doi: 10.1039/c002059a. [DOI] [PubMed] [Google Scholar]
- [93].Ellington AD, Szostak JW. Nature. 1990;346:818–22. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- [94].Hermann T, Patel DJ. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- [95].Wang GQ, Wang YQ, Chen LX, Choo J. Biosens Bioelectron. 2010;25:1859–1868. doi: 10.1016/j.bios.2009.11.012. [DOI] [PubMed] [Google Scholar]
- [96].Tombelli S, Minunni A, Mascini A. Biosens Bioelectron. 2005;20:2424–2434. doi: 10.1016/j.bios.2004.11.006. [DOI] [PubMed] [Google Scholar]
- [97].Ochsenkuhn MA, Campbell CJ. Chem. Commun. 2010;46:2799–2801. doi: 10.1039/b920941g. [DOI] [PubMed] [Google Scholar]
- [98].Pagba CV, Lane SM, Cho H, Wachsmann-Hogiu S. J Biomed Opt. 2010;15:047006. doi: 10.1117/1.3465594. [DOI] [PubMed] [Google Scholar]
- [99].Pagba CV, Lane SM, Wachsmann-Hogiu S. J Raman Spectrosc. 2010;41:241–247. [Google Scholar]
- [100].Negri P, Kage A, Nitsche A, Naumann D, Dluhy RA. Chem. Commun. 2011;47:8635–8637. doi: 10.1039/c0cc05433j. [DOI] [PubMed] [Google Scholar]
- [101].Negri P, Chen G, Kage A, Nitsche A, Naumann D, Xu B, Dluhy RA. Anal. Chem. 2012;84:5501–8. doi: 10.1021/ac202427e. [DOI] [PMC free article] [PubMed] [Google Scholar]