Abstract
Topoisomerases are expressed throughout the developing and adult brain and are mutated in some individuals with autism spectrum disorder (ASD). However, how topoisomerases are mechanistically connected to ASD is unknown. Here we found that topotecan, a Topoisomerase 1 (TOP1) inhibitor, dose-dependently reduced the expression of extremely long genes in mouse and human neurons, including nearly all genes >200 kb. Expression of long genes was also reduced following knockdown of Top1 or Top2b in neurons, highlighting that each enzyme was required for full expression of long genes. By mapping RNA polymerase II density genome-wide in neurons, we found that this length-dependent effect on gene expression was due to impaired transcription elongation. Interestingly, many high confidence ASD candidate genes are exceptionally long and were reduced in expression following TOP1 inhibition. Our findings suggest that chemicals and genetic mutations that impair topoisomerases could commonly contribute to ASD and other neurodevelopmental disorders.
Introduction
Autism is a neurodevelopmental disorder with symptoms that include repetitive behaviors and deficits in social interactions. Hundreds of genes are now associated with ASD1,2, suggesting there are diverse genetic risk factors for autism. Environmental factors, including chemicals that are ingested during critical periods of brain development3, can also increase autism risk. Many ASD candidate genes regulate synapse function4-6; however, whether there are additional mechanisms that unite ASD patients or expression of ASD genes is unclear.
Recently, we found that topoisomerase inhibitors can transcriptionally unsilence the paternal allele of Ube3a in neurons7. Ube3a is located adjacent to a cluster of imprinted genes, is normally expressed only from the maternal allele in neurons and regulates synaptic function8. In addition, Ube3a is associated with two distinct neurodevelopmental disorders. Specifically, deletion or mutation of maternal Ube3a causes Angelman syndrome while duplication of the chromosomal region containing maternal Ube3a is frequently detected in individuals with autism9,10.
Intriguingly, mutations in topoisomerases were recently identified in some individuals with ASD11,12. However, precisely how topoisomerases regulate expression of Ube3a and possibly other genes associated with autism is unknown. Topoisomerases, including Top1 and Top2b, are expressed throughout the developing and adult brain13,14. Topoisomerases are integral to gene expression, as they resolve DNA supercoiling that is generated during transcription15-18. Here, we sought to determine if topoisomerases preferentially regulate expression of additional imprinted genes in neurons, or if topoisomerases have broader effects on gene expression. Using genome-wide approaches, we unexpectedly found that topoisomerases facilitate expression of long genes, including numerous long genes associated with synaptic function and ASD. In addition, our study uncovers a transcriptional mechanism that is particularly important for maintaining expression of numerous ASD genes at normal levels.
Gene length effects
To determine if topotecan, a TOP1 inhibitor, altered expression of imprinted genes, we treated cultured cortical neurons from C57BL/6J (B6) × CASTEi/J (CAST) F1 hybrid mice with vehicle or 300 nM topotecan, then used high-throughput transcriptome sequencing (RNA-seq) to survey changes in gene expression genome-wide. Single nucleotide polymorphisms (SNPs) were used to determine the parent-of-origin of sequence reads for autosomal genes19. We defined imprinted genes as those displaying statistically significant parent-of-origin expression bias in reciprocal C57/CAST crosses (Fisher’s exact test, P<0.05 after adjustment for multiple comparisons). We found that cortical neurons expressed 49 known autosomal imprinted genes (Extended Data Table 1), yet Ube3a was the only imprinted gene that showed a significant change in parental allele bias in reciprocal crosses upon topotecan treatment (Fisher’s exact test, P<0.05 after correction; Extended Data Table 1). Indeed, topotecan increased expression of the paternal allele of Ube3a, driving Ube3a levels significantly above wild-type levels (Extended Data Fig. 1a,b).
As we previously found7, topotecan reduced expression of Ube3a-ATS (Extended Data Fig.1a,b). Ube3a-ATS is an extremely long (>1 Mb), paternally-expressed antisense-transcript that overlaps Ube3a and is required for paternal Ube3a silencing20,21. Other imprinted genes in the same genomic region as Ube3a did not show changes in allelic expression following topotecan treatment (Extended Data Fig. 1b, Extended Data Table 1). Importantly, topotecan also reduced expression of UBE3A-ATS and increased expression of UBE3A in induced pluripotent stem cell (iPSC)-derived neurons from an Angelman syndrome patient (Extended Data Fig. 1c). Topotecan thus had similar transcriptional effects at the Ube3a locus in mouse and human neurons.
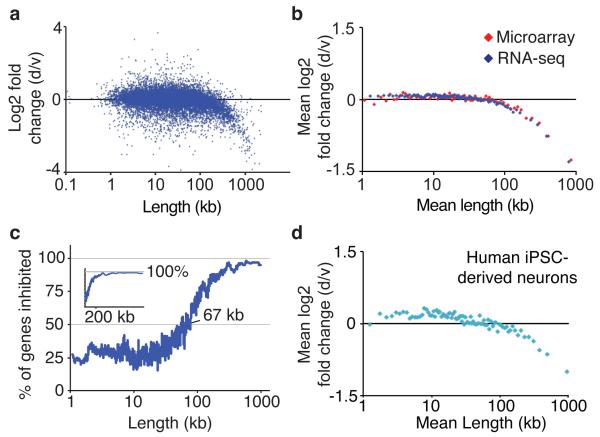
Since Ube3a-ATS is extremely long and was strongly downregulated, we hypothesized that topotecan might reduce expression of other long genes. Remarkably, using RNA-seq and Affymetrix microarrays to quantify gene expression, we found that topotecan reduced expression of nearly all extremely long genes in mouse cortical neurons (Fig. 1a-c), with a strong correlation between gene length and reduced expression (for genes longer than 67 kb; Pearson’s R = −0.69). Topotecan also reduced expression of long genes in iPSC-derived human neurons (Fig. 1d). Topotecan did not exclusively reduce expression of extremely long genes, but instead acted over a continuum of gene lengths (Fig. 1c). Specifically, the percentage of genes that were inhibited (to any extent) by 300 nM topotecan increased from 50% for genes 67 kb in length to nearly 100% for genes ~200 kb and longer. And, inhibition of long genes by topotecan was highly dose-dependent (Extended Data Fig. 2).
Figure 1. TOP1 inhibition reduces expression of long genes in neurons.
a, Mouse cortical neurons treated with vehicle (v) or 300 nM topotecan (drug; d) for 3 days (n=5 biological replicates). RNA-seq gene expression versus gene length. b, Mean expression change in bins of 200 genes by length. c, Percentage of genes that were reduced in expression by topotecan; plotted as a sliding window of 100 genes by length, RNA-seq data (log scale). Inset, same data on linear scale. d, iPSC-derived human neurons treated with 1 μM topotecan for 6 days relative to vehicle, RNA-seq data in bins of 200 genes by length (n=2 biological replicates).
In contrast, topotecan increased expression of a majority of genes that were <67 kb in length (Fig. 1c), although the magnitude of this increase was very small for most genes (Fig. 1a,b). For some genes, this increase may reflect regulation by longer overlapping transcripts, like for Ube3a, or it might reflect other stimulatory effects of topoisomerase inhibitors22,23.
The length-dependent effects on gene expression were not due to cell death or persistent DNA damage, as topotecan (300 nM for 3 days) did not kill neurons or damage DNA (Extended Data Fig. 3a,b). Moreover, agents that damage DNA in neurons (paraquat and H2O2) did not reduce expression of long genes (Extended Data Fig. 3b-d). Notably, all length-dependent effects were fully reversible upon drug washout (Extended Data Fig. 3e), ruling out the possibility that gene expression changes were due to permanent effects (such as irreversible DNA damage and/or killing neurons).
A different TOP1 inhibitor, irinotecan, had a highly correlated length-dependent effect on gene expression in cortical neurons (Extended Data Fig. 4). Additionally, we re-analyzed published data from other labs and found that irinotecan and camptothecin (a TOP1 inhibitor) strongly reduced expression of long genes and moderately increased expression of shorter genes in several human cell lines (Extended Data Fig. 5a-e). Thus, the length-dependent effects we observed were not unique to postmitotic neurons and could be reproducibly detected in expression data acquired by other labs.
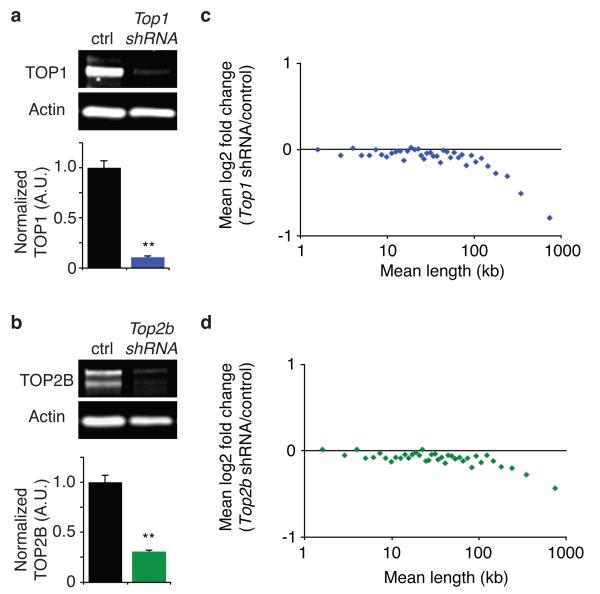
We also found that lentiviral shRNA knockdown of TOP1 reduced expression of long genes in neurons (Fig. 2a,c), providing independent genetic support that TOP1 facilitates expression of long genes. These gene knockdown results also rule out the possibility that TOP1-DNA covalent complexes, which form only in the presence of TOP1 inhibitors18, block expression of long genes. Unlike TOP1 inhibitors (Fig. 1a-c, Extended Data Fig. 4a, Extended Data Fig. 5a-e), TOP1 knockdown did not globally increase expression of shorter genes (Fig. 2c). Thus TOP1 inhibitors likely increase expression of shorter genes via a drug-specific effect that is unrelated to TOP1 depletion.
Figure 2. Lentiviral shRNA knockdown of Top1 or Top2b in mouse cortical neurons reduces expression of long genes.
a, b, Representative western blots and quantification of TOP1 and TOP2B seven days post infection. Normalized to β-actin in arbitrary units (A.U.). **P<0.01 relative to scrambled (Scr) control, Student’s t-test. Error bars, s.e.m. n=3 biological replicates. c, d, Gene expression from Affymetrix microarrays, relative to scrambled control shRNA. Plotted as mean expression change in bins of 200 genes by length.
TOP2 enzymes (particularly TOP2B) also participate in gene transcription15,16,24. We next tested whether genetic or pharmacological inhibition of TOP2 enzymes could reduce the expression of long genes. Indeed, with new experiments and by re-analyzing data from others14,25, we found that the TOP2A/TOP2B inhibitor ICRF-193 reduced gene expression in a length-dependent manner in cultured mouse cortical neurons, embryonic stem (ES) cells, and ES cell-derived neurons (Extended Data Fig. 6a, Extended Data Fig. 7a,b). There was extensive overlap between genes affected by ICRF-193 and topotecan in cortical neurons, particularly for long genes, and the magnitudes of these effects were highly correlated (Extended Data Fig. 6b-e). Thus, TOP1 and TOP2 enzymes regulate expression of many of the same genes.
Since Top2b is the predominant TOP2 expressed in neurons25, we next knocked down Top2b with shRNA (Fig. 2b,d). We found that Top2b knockdown reduced expression of long genes (Fig. 2d). Moreover, re-analysis of published datasets showed that expression of long genes was reduced in embryonic brain and ES cell-derived neurons from Top2b−/− mice14 (Extended Data Fig. 7c,f,g). In contrast, long genes were expressed normally in Top2b−/− ES cells and neuronal progenitors25 (Extended Data Fig. 7d,e), presumably because these cell types express Top2a in addition to Top2b25. Lastly, two additional TOP2 inhibitors (doxorubicin and etoposide) reduced expression of long genes in a human cancer cell line26 (Extended Data Fig. 5f,g). Together, our data show that TOP1 and TOP2 enzymes are both required for proper expression of long genes in mammalian cells. This stands in contrast to yeast, in which length-dependent transcriptional effects specific to TOP2 were observed27.
Length-dependent effect on elongation
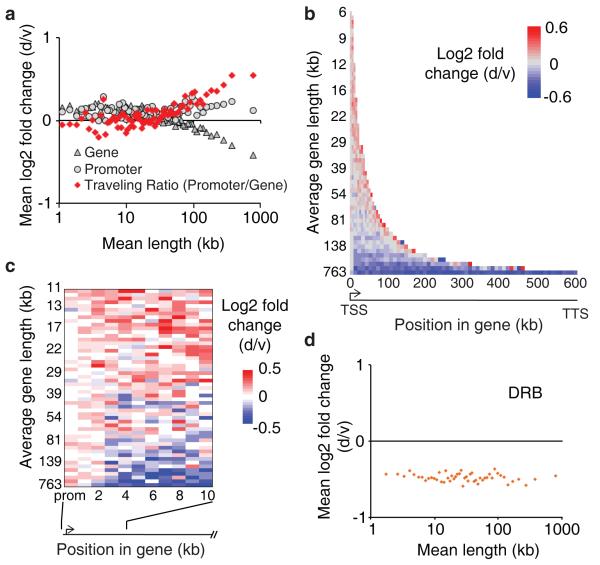
Topoisomerases interact directly with RNA polymerase II (Pol II) and are required for transcription elongation17,28,29. To study genome-wide effects of topotecan on transcription in neurons, we measured Pol II distribution by chromatin immunoprecipitation followed by high-throughput DNA sequencing (ChIP-seq). We calculated the Traveling Ratio (TR) for all genes bound by Pol II, defined as the ratio of Pol II density (read counts per million mapped reads; RPM) near transcription start sites to Pol II density in the body of the gene, a parameter that is increased when transcription elongation is inhibited30-32. We found that topotecan produced a distinct length-dependent increase in TR for very long genes, consistent with an impairment of transcription elongation (Fig. 3a, Extended Data Fig. 8).
Figure 3. Topotecan impairs transcription elongation of long genes.
a, Mouse cortical neurons treated with vehicle or topotecan (300 nM for 3 days). Pol II density in gene bodies and promoter regions, averaged for bins of 200 genes by length. b, c, Change in Pol II density across genes and (c) across the first 10 kb, averaged for groups of 200 genes by length. Gene bins aligned relative to the transcription start site (TSS). d, Mouse cortical neurons treated with 100 μM DRB for 3 days. Affymetrix microarray expression compared to controls in bins of 200 genes by length.
The change in TR observed with topotecan treatment could reflect either a progressive inhibition of Pol II as it transcribes long genes, or it could reflect a block in the transition to productive elongation. To distinguish between these possibilities, we examined the change in Pol II density across the entire length of genes at high resolution (Fig. 3b). We found that Pol II density was slightly increased throughout the gene bodies of smaller genes, consistent with the modest increase in expression seen for smaller genes and with studies showing that TOP1 inhibitors can stimulate the transition to elongation of two genes (both shorter than 67 kb)5,6. However, for longer genes, topotecan strongly reduced Pol II density across the entire length of the gene body (Fig. 3b). These results are consistent with topotecan affecting the transition to productive elongation at long genes. On average, all genes showed a slight increase in Pol II density in the promoter proximal region (Fig. 3c) and stronger increase in Pol II density near the transcription termination site (TTS; Fig. 3b). However, these changes were independent of length, making it unlikely that they contributed to differential expression of short versus long genes.
TOP1 inhibitors can stimulate the release of the positive elongation factor pTEF-B17. Thus, we next tested whether pTEF-B inhibition would affect expression of longer genes differently from shorter genes by treating neurons with 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB; a pTEF-B inhibitor that can block the transition to elongation17,30). We found that DRB (100 μM) reduced gene expression equally across all gene lengths (Fig. 3d), suggesting that the requirement for pTEF-B is not influenced by length. Note that 1 μM and 10 μM DRB had no length-dependent effects on expression (data not shown), ruling out the possibility that lower DRB concentrations preferentially impair expression of longer genes.
Numerous long ASD genes affected
To further investigate the biological consequences of TOP1 inhibition in neurons, we defined a list of genes that were differentially expressed with high confidence. From our RNA-seq expression data, we found that topotecan significantly downregulated 155 genes and significantly upregulated 28 genes (Benjamini-Hochtberg method, 5% false discovery rate) (Supplementary Data 1). The topotecan-downregulated genes were significantly longer (mean 591 kb, median 548 kb) than all expressed genes in cortical neurons (mean 59.3 kb, median 23.5 kb) and were significantly longer than topotecan upregulated genes (mean 29.3 kb, median 16.4 kb) (One-way ANOVA vs. all expressed genes, P=2.2×10−16, vs. upregulated genes P=3.7×10−14), further indicating that topotecan has pronounced effects on long genes.
Based on Gene Ontology and functional annotation terms, we found that many topotecan-downregulated genes were involved in neuronal development and synaptic function (Supplementary Data 2). Since ASD is thought to be a neurodevelopmental disorder that affects synapses, we cross-referenced our list of downregulated genes with known ASD candidate genes, combining genes in the SFARI Gene database with candidates identified in recent exome sequencing studies11,12,33-35 (Supplementary Data 3). Remarkably, 27% (n=49) of the 183 differentially expressed genes are known ASD candidate genes (Table 1, Supplementary Data 3), a proportion that is highly significant compared to chance (P=4.4×10−8, Fisher’s Exact Test). Independent microarray experiments showed that these ASD genes were dose-dependently downregulated by topotecan (Extended Data Fig. 9). Notably, ASD candidate genes are exceptionally long as a group; genes in the SFARI Gene database (as of June 20, 2013) are 3.7-fold longer on average than all genes expressed in cortical neurons (means of 217.3 kb versus 59.3 kb). Thus, mutations that alter topoisomerase activity might reduce expression of numerous long ASD genes and might contribute significantly to ASD. Consistent with this hypothesis, recent sequencing studies of autism patient cohorts uncovered rare de novo missense mutations in TOP1, TOP3B, TOPORS (a TOP1-SUMO ligase36) and several other genes that directly connect to TOP111,12.
Table 1. Topotecan reduces expression of numerous ASD candidate genes in neurons.
| Gene | Length in Mouse (kb) |
Length in Human (kb) |
Log2 fold change (mouse) |
p (adjusted) | SFARI Gene |
Mutation |
|---|---|---|---|---|---|---|
| Cntnap2 * | 2241.3 | 2304.6 | −4.5718 | 0.0055 | Yes | H 275 R34 |
| Csmdl | 1642.8 | 2059.4 | −3.4896 | <0.0001 | No | Q 2254 R33 |
| Nrxn3 | 1612.1 | 1460.7 | −2.9494 | <0.0001 | Yes | |
| Fhit | 1611.9 | 1502.1 | −2.1229 | 0.0490 | Yes | |
| Rbfoxl | 1527.7 | 1694.2 | −1.3178 | 0.0441 | Yes | |
| Grid2 | 1409.4 | 1468.1 | −2.4100 | 0.0204 | Yes | |
| Illrapll | 1368.4 | 1368.3 | −3.0444 | 0.0213 | Yes | |
| Il1rapl2 | 1275.4 | 1200.8 | −2.6076 | 0.0034 | Yes | |
| Park2 | 1223.0 | 1380.2 | −2.8055 | 0.0003 | Yes | |
| Cntn5 | 1243.9 | 1337.9 | −2.3360 | 0.0009 | Yes | |
| Ptprt | 1133.5 | 1117.2 | −2.0706 | <0.0001 | Yes | |
| Frmpd4 | 1105.9 | 586.1 | −1.5332 | 0.0028 | Yes | |
| Erbb4 | 1068.1 | 1162.9 | −3.3890 | <0.0001 | Yes | |
| Nrxn1 | 1059.2 | 1114.0 | −2.1914 | <0.0001 | Yes | Y 587 STOP12 |
| Gpc6 | 1054.2 | 1181.2 | −2.3530 | <0.0001 | Yes | |
| Astn2 | 1023.7 | 989.8 | −2.7763 | 0.0001 | Yes | |
| Cntn4 | 1021.6 | 959.1 | −2.0717 | 0.0092 | Yes | |
| Pard3b | 1003.5 | 1074.4 | −2.9457 | <0.0001 | Yes | |
| Epha6 | 952.0 | 934.3 | −1.8366 | 0.0301 | Yes | |
| Grm7 | 921.6 | 880.4 | −2.2647 | <0.0001 | No | R 622 Q33 |
| Nckap5 | 917.0 | 896.7 | −1.3453 | 0.0089 | Yes | |
| Nlgnl | 900.1 | 884.9 | −1.8339 | <0.0001 | Yes | H 795 Y34 |
| Pcdh9 | 875.2 | 927.5 | −1.8314 | 0.0002 | Yes | |
| Pcdh15 | 827.9 | 998.5 | −3.2033 | <0.0001 | Yes | |
| Grid1 | 761.0 | 766.9 | −1.4922 | 0.0448 | Yes | |
| Mdga2 | 756.5 | 835.3 | −1.6942 | 0.0012 | Yes | |
| Dpp10 | 713.4 | 1402.4 | −1.7914 | 0.0007 | Yes | |
| Kcnmal | 705.4 | 768.2 | −1.9021 | 0.0004 | Yes | F 372 V11 |
| Grik2 | 689.3 | 669.2 | −1.5500 | 0.0028 | Yes | |
| Plcb1 | 689.1 | 752.3 | −1.4067 | 0.0075 | Yes | |
| Ptprm | 687.6 | 839.5 | −1.7610 | 0.0057 | No | A 535 T12 |
| Rbms3 | 679.7 | 729.1 | −1.6432 | 0.0191 | Yes | |
| Vps13b | 559.7 | 864.3 | −1.0465 | 0.0317 | Yes | |
| Nbea | 558.5 | 730.5 | −1.3664 | 0.0147 | Yes | |
| Grm5 | 550.9 | 559.1 | −1.3240 | 0.0343 | Yes | Aaa67912 |
| Kalrn | 545.0 | 626.5 | −1.3646 | 0.0186 | Yes | |
| Ptprk | 522.6 | 551.9 | −1.6458 | 0.0005 | No | R 784 H34 |
| Atrnll | 522.3 | 855.3 | −1.2194 | 0.0211 | Yes | |
| Robo2 | 518.6 | 1712.5 | −1.4921 | 0.0017 | Yes | |
| Cacna1c | 515.9 | 644.7 | −1.1665 | 0.0236 | Yes | |
| Lrrc7 | 477.8 | 363.3 | −1.6330 | 0.0340 | Yes | |
| Reln | 460.3 | 517.7 | −2.0687 | <0.0001 | Yes | Q 417 STOP11 R 2290 C12 |
| Exoc6b | 451.0 | 646.7 | −0.9691 | 0.0285 | Yes | |
| Gabrb1 | 437.2 | 395.2 | −1.4909 | 0.0041 | Yes | |
| Nfia | 341.3 | 385.5 | −1.8218 | 0.0186 | Yes | R 30 STOP12 |
| Nxph1 | 298.6 | 319.0 | −1.8182 | 0.0181 | Yes | R 45 Q33 |
| Myb | 36.1 | 37.9 | −3.8779 | 0.0011 | No | |
| C3 | 24.1 | 42.8 | −2.2892 | <0.0001 | No | I 1569 M11 |
| C4b | 15.5 | 1.7 | −1.5240 | 0.0121 | Yes |
isoform a (uc009bst.2)
Discussion
Our study shows that topoisomerases facilitate expression of a large number of ASD candidate genes, including many that are long and that are thought to have large effects on ASD pathology in isolation1,2,37. Pharmacological inhibition of topoisomerases also reduced expression of long genes in other cell types, suggesting this length-dependent transcriptional effect is fundamental to all mammalian cells. Our data rule out numerous possibilities for why topoisomerase inhibitors reduce expression of long genes (e.g., cell death, DNA damage, formation of covalent complexes), and instead implicate a gene length-dependent block in transcription elongation. Pol II and topoisomerases dynamically form and remodel large supercoiling domains38, and the effects of topoisomerases on gene expression are strongly influenced by genomic structure and context23,39. Thus, we speculate that higher order structure differentially constrains shorter and longer genes, and that this creates distinct length-dependent requirements for topoisomerases in transcription elongation.
Some long genes were not strongly reduced in expression following topotecan treatment (Fig. 1a). In many cases this reflected ambiguity in gene annotation (data not shown). For example, a number of long genes also express shorter transcripts, making it difficult to distinguish expression of short isoforms from long isoforms. Alternatively, some long genes might be located within genomic regions that are more permissive for expression when TOP1 is inhibited.
Intriguingly, numerous genes associated with transcription are mutated in autism patients11,40,41, although how these diverse transcriptional regulators contribute to autism is unclear. Our study highlights a mechanistic link between a critical step in transcription elongation and expression of numerous long ASD candidate genes. Our data suggest that chemicals or genetic mutations that impair topoisomerases, and possibly other components of the transcription elongation machinery that interface with topoisomerases, have the potential to profoundly affect expression of long ASD candidate genes. Length-dependent impairment of gene transcription, particularly in neurons and during critical periods of brain development, may thus represent a unifying cause of pathology in many individuals with ASD and other neurodevelopmental disorders.
Methods
Mouse cell culture
Cortical neurons were cultured from E13.5-E15.5 mouse embryos as described7. For RNA-seq and ChIP-seq, neurons were seeded on 10 cm diameter poly-D-lysine treated culture plates at a density of 5-10×106/plate. Microarray experiments used 6-well plates seeded at 1×106 cells per well. After 7 days in culture, drugs or an equivalent amount of vehicle were added and left in the culture medium for 24 or 72 hours. For topotecan, irinotecan, DRB and paraquat; vehicle was 0.1% DMSO. For ICRF-193, vehicle was 0.02% DMSO.
Lentiviral shRNA experiments used viruses from The RNAi Consortium (acquired from Sigma-Aldrich and from the UNC Lenti-shRNA core): Top1 knockdown used clone TRCN0000011884 (CCGGCCAGCGAAGATTCTATCTTATCTCGAGATAAGATAGAATCTTCGCTGG TTTTT) and Top2b knockdown used clone TRCN0000070988 (CCGGCCTTGTGTTGTCCTTTGTCTTCTCGAGAAGACAAAGGACAACACAAGG TTTTTG). Virus expressing non-targeting hairpin RNA (SHC002, Sigma; CCGGCGTGATCTTCACCGACAAGATCTCGAGATCTTGTCGGTGAAGATCACG TTTTT) was used as a control. Neurons were seeded on 24-well plates at 2×105 per well. After 3 days in culture, cells were treated with lentivirus at a multiplicity of infection of at least 1. Virus was removed after 24 hours, and RNA harvested after a further 6 days in culture. Western blotting to assess knockdown was performed with anti-TOP1 (Santa Cruz Biotechnology, H5) or anti-TOP2B (Santa Cruz, H286) antibodies, with signal normalized to β-actin (Millipore, C4).
Cell death was assayed using Sytox Green (Invitrogen Molecular Probes). γH2AX foci were measured by immunohistochemistry. Primary antibodies were anti-γH2AX (Millipore, 1:500 dilution) and anti-NeuN (Millipore, 1:500), used to mark neurons.
iPSC culture and neuronal differentiation
Human iPSC work was approved by the University of Connecticut Stem Cell Research Oversight Committee. iPSCs that carry a large deletion of maternal 15q11-q13 (AGdel1-0; see also (ref. 43), this cell line deemed exempt from IRB approval at the University of Connecticut due to its establishment in 1995 and lack of identifying information), were cultured on irradiated mouse embryonic fibroblasts and manually passaged as described43. iPSCs were differentiated into forebrain cortical neurons as described43 with the following modifications: neural progenitors were generated by culturing iPSCs on feeders in N2B27 medium supplemented with Noggin (500 ng/mL) for 8 days and then manually picking neural rosettes for two additional passages using trypsin and standard cell culture protocols. Topotecan was applied to mature neurons and RNA was collected by standard protocols 6 days following the addition of drug or vehicle.
qPCR
qRT-PCR was carried out as described43 using Taqman (Life Technologies) gene expression assays for UBE3A (Hs00166580_m1) and UBE3A-ATS (Hs03454279_m1) according to the manufacturer’s instructions. The Taqman assay for GAPDH was used as a control.
RNA-seq
Total RNA was collected using Trizol reagent (Invitrogen). Mouse polyA-selected mRNA libraries were then prepared using the Illumina True-Seq kit for RNA. For RNA-seq on human iPSC-derived neuronal samples, stranded multiplexed mRNA libraries were prepared using Illumina kits. Cluster generation and sequencing were performed using the Illumina HiSeq 2000 platform. For allele-specific expression analysis, equal amounts of total RNA from 3-6 biological replicates were pooled before polyA mRNA purification and library preparation.
For non-allelic expression analysis, data from an additional three biological replicates were included. mRNA was isolated and libraries prepared independently for each replicate sample. Library preparation incorporated barcoded adapters and all samples were sequenced in one lane, using 50 bp paired-end reads.
RNA-seq expression analysis
For allelic expression analysis, informative CAST/B6 SNPs were downloaded from (http://www.sanger.ac.uk/resources/mouse/genomes/). CAST alleles were then substituted into their corresponding mm9 positions and sequence reads were aligned to mm9 and the CAST version of mm9 using Bowtie, selecting for unique matches. Filtered read counts for autosomal genes were tested for allelic bias using Fisher’s Exact Test against a background model derived from autosomes, and P-values adjusted for multiple comparisons using the Benjamini-Hochberg procedure. Statistical analysis was performed using R. Genomic intervals were derived from UCSC known genes or created manually where annotation was absent, namely for Ube3a-ATS.
For non-allelic analysis, reads were aligned to the reference genome (mm9) using Bowtie. Read counts were obtained using DEGseq, and normalization and analysis of differential gene expression was performed using the R package, edgeR, employing a negative binomial model.
ChIP-seq
ChIP-seq against RNA Pol II was performed as described previously30. Cultures (n=4/condition) totaling approximately 2×107 neurons were treated with vehicle or 300 nM topotecan as described above. Nuclear lysates were sheared to an average fragment size of approximately 200 bp. 2 μg anti-RNA Pol II N20 (Santa Cruz Biotechnology) was added, and the sample incubated at 4°C for 16 hours. Chromatin immunoprecipitation was performed as described previously30,42
ChIP-seq libraries were prepared from immunoprecipitated samples and their corresponding inputs using the Illumina Tru-Seq kit for ChIP-seq. Ligation products were size-selected by purification on 2% PippinPrep gels (Sage Science). Samples from vehicle and drug treated cells and their inputs were sequenced using the HiSeq 2000 platform with single-end reads of 50 bp.
ChIP-seq analysis
Short read sequences were aligned to the mouse reference genome (mm9) with Bowtie. Duplicate reads were removed. The quality of the experiment and false discovery rate for enriched peaks was assessed using MACS 1.4.2. CoverageBED was used to obtain read counts covering the promoter region (from −30 to +300 bp, relative to TSS and gene bodies (from +300 after TSS to 3000 bp after the annotated TTS, and to count reads in intervals across genes. Read counts were normalized to the number of mapped unique reads per sample (RPM) per base.
Affymetrix microarrays
For single-dose microarray experiments, cultured cortical neurons were treated with 300 nM or 1 μM topotecan, 10 μM irinotecan (Sigma) or 3 μM ICRF-193 (Santa Cruz Biotechnology), 100 μM DRB (Sigma), 100 μM H2O2 (Fisher Scientific), or 10 μM paraquat (Sigma) for 24 or 72 hours. For topotecan dose response, cells were treated with 3 nM, 30 nM, 150 nM, 300 nM, 500 nM and 1000 nM topotecan or vehicle for 72 hours. Total RNA was used for all Affymetrix microarray experiments. Comparative expression with topotecan and ICRF-193 was measured with Affymetrix mouse genome 430 2.0 arrays. All other microarray experiments used Affymetrix Mouse Gene 1.0 ST 24-array plates. Linear RMA background correction and normalization was used for all microarray data.
Supplementary Material
Extended Data Figure 2. Topotecan dose-response. Mouse cortical neurons were treated with 3 nM, 30 nM, 150 nM, 300 nM and 1000 nM topotecan for 3 days (n=3 for 300 nM topotecan, all other doses n=1). Gene expression was analyzed by Affymetrix microarrays, plotted as mean expression change in bins of 200 genes.
Extended Data Figure 3. DNA damage does not inhibit gene expression in a length-dependent manner. a, Cultured mouse cortical neuron viability, assayed by Sytox Green staining after 72 hours treatment with 300 nM topotecan, and after 24 hours with 100 μM H2O2 or 10 μM paraquat, compared to vehicle-treated controls. Error bars represent s.e.m. n=4. b, Average number of γH2AX foci per nucleus for cultured cortical neurons treated with 300 nM topotecan for 72 hours, 100 μM H2O2 for 24 hours and 10 μM paraquat for 24 hours, compared to vehicle-treated controls. **P<0.01, Student’s t-test. Number of cells counted is indicated for each sample. c, Gene expression compared to vehicle controls in bins of 200 genes by length, for cultured cortical neurons treated with 100 μM H2O2 for 24 hours. d, Gene expression compared to vehicle controls in bins of 200 genes by length, for cultured cortical neurons treated with 10 μM paraquat for 24 hours. e, Gene expression in cultured cortical neurons treated with 300 nM topotecan for 24 hours, or treated for 24 hours followed by 48 hours without drug (washout). Average change in expression for bins of 200 genes by length.
Extended Data Figure 4. Topotecan and irinotecan have highly similar effects on gene expression. a, Affymetrix microarray analysis of gene expression in cultured mouse cortical neurons treated with vehicle or 10 μM irinotecan (n=3 biological replicates), an inhibitor of TOP1, for 3 days. Mean expression fold change in bins of 200 genes, plotted by average gene length. b, Scatterplot of fold change with 1 μM topotecan (n=6 biological replicates) versus fold change with 10 μM irinotecan for all expressed genes, measured by Affymetrix microarray. Pearson’s R = 0.860. c, Overlap between genes showing positive or negative fold change of log2 = 0.5 or greater with topotecan and irinotecan treatment. d, Overlap between genes reduced or increased in expression. e, Overlap between differentially expressed genes that are greater or less than 67 kb.
Extended Data Figure 5. TOP1 and TOP2 inhibitors reduce expression of long genes in human cell lines. Re-analysis of microarray gene expression datasets from other labs. All plots are mean fold change in expression compared to vehicle controls in bins of 200 genes, plotted by average gene length. a, MCF7 cells treated with 10 μM irinotecan for 24 hours, from the CMAP2 project44. b, MCF7 cells treated with 165 nM SN38, the active metabolite of irinotecan, for 6 hours45. c-e, Gene expression in three human cell lines treated with camptothecin. c, MCF7 cells treated for 24 hours with 10 μM camptothecin, from CMAP2. d, 293T cells treated with 2 μM camptothecin for 48 hours46. e, HeLa cells treated with 10 μM camptothecin for 8 hours47. f, g, Re-analysis of microarray data from Troester et al.26 comparing gene expression in TOP2 inhibitor and vehicle treated ME16C cells. f, ME16C cells treated with 0.5 μM doxorubicin for 36 hours. g, ME16C cells treated with 50 μM etoposide for 36 hours.
Extended Data Figure 6. Topotecan and the TOP2 inhibitor ICRF-193 have similar effects on gene expression. a, Affymetrix microarray analysis of gene expression in cultured mouse cortical neurons treated with vehicle or 3 μM ICRF-193 (n=3 biological replicates), an inhibitor of TOP2 enzymes, for 3 days. Mean expression fold change in bins of 200 genes, plotted by average gene length. b, Scatterplot of fold change with 300 nM topotecan (n=3 biological replicates) versus fold change with 3 μM ICRF-193 for all expressed genes, measured by Affymetrix microarray. Pearson’s R=0.588. c, Overlap between genes showing positive or negative fold change of log2 = 0.5 or greater with topotecan and ICRF-193 treatment. d, Overlap between genes reduced or increased in expression. e, Overlap between differentially expressed genes that are greater or less than 67 kb in length.
Extended Data Figure 7. Pharmacological inhibition of TOP2 or genetic deletion of Top2b reduces expression of long genes in embryonic stem (ES) cell-derived neurons. a, b, Re-analysis of Tiwari et al. microarray expression data25. Mean fold change in expression in bins of 200 genes, plotted by average gene length. a, Gene expression in ES cell-derived neurons treated with vehicle or 50 μM ICRF-193 for 3 days. b, Gene expression in ES cells treated with vehicle or 50 μM ICRF-193 for 3 days. c, Re-analysis of Lyu et al. microarray expression data14, comparing gene expression in whole brain from wild-type (wt) and Top2b-/- embryonic mice. Expression data from three developmental time points (E16.5, E17.5, E18.5) were averaged for each gene then plotted as mean fold change in expression between wt and Top2b-/- mice (in bins of 100 genes, by average gene length). d, e, Re-analysis of Tiwari et al. microarray expression data25. Mean fold change in gene expression between wt and Top2b-/-, for bins of 200 genes, plotted by average gene length. d, Expression data from ES cells, e, neuronal progenitors, f, ES cell-derived neurons, two days after plating of neuronal progenitors, and g, ES cell-derived neurons, six days after plating of neuronal progenitors. ES cells and neuronal progenitors express Top2a and Top2b (indicated within parentheses) and do not show reduced expression of long genes when Top2b is knocked out, suggesting that Top2a and Top2b redundantly regulate expression of long genes.
Extended Data Figure 8. Topotecan increases Travelling Ratio (TR) in genes larger than 67 kb. TR was calculated for all genes bound by Pol II in vehicle and topotecan-treated (300 nM for 3 days) cultured cortical neurons. 92.1% of all genes had TR values greater than 2, consistent with a previous report using mouse ES cells30. a, Frequency of TRs for genes less than 67 kb in length, and b, for genes greater than 67 kb in length. Mean TR of vehicle and topotecan treated samples was significantly different for genes greater than 67 kb (P=2.4×10−21, t-test) but not for genes less than 67 kb (P=0.648, t-test).
Extended Data Figure 9. Topotecan dose-dependently reduces expression of ASD candidate genes in cortical neurons. a, Topotecan dose-response for high confidence autism candidate genes (n=3 for 300 nM topotecan, all other doses n=1). b-e, Topotecan dose-response for other ASD candidate genes organized by length from longest to shortest. Mouse cortical neurons were treated with vehicle or the indicated doses of topotecan for 3 days. Gene expression was quantified using Affymetrix microarrays. Dose-responses are from all topotecan-downregulated ASD candidate genes that were identified by RNA-seq (Table 1) and that were present on the Affymetrix microarrays.
Extended Data Table 1. Topotecan does not alter parent-of-origin bias of imprinted genes. Genes with significant parent-of-origin expression bias in cultured cortical neurons. Shown are P values for allelic bias in baseline expression, and P values for change in expression bias with topotecan treatment (Fisher’s Exact Test, adjusted for multiple comparisons). ns = P>0.05. Embryos from CAST male × B6 female crosses (n=3) were pooled and B6 male × CAST female crosses (n=6) were pooled for sequencing. *Genes in the Ube3a imprinted cluster. †AF217545 is contained within Copg2, which has also been reported to be maternally expressed48.
Extended Data Figure 1. Topotecan affects allelic expression of Ube3a and Ube3a-ATS but not expression of nearby genes. a, Parent-of-origin specific RNA-seq reads for Ube3a and Ube3a-ATS in vehicle and topotecan-treated (300 nM for 3 days) mouse cortical neurons (n=5 biological replicates). Shown are SNP filtered read counts per million mapped reads (RPM) from the maternally (mat) and paternally (pat) inherited chromosomes. Reads from exons and 3′-untranslated region of Ube3a are indicated by arrows. b, Expression of Ube3a and imprinted genes near Ube3a in mouse cortical neurons ± 300 nM topotecan for 3 days. * P<0.05, t-test. Error bars represent s.e.m. c, Expression of UBE3A and UBE3A-ATS in iPSC-derived neurons from an Angelman syndrome patient carrying a maternal deletion of the 15q11-q13 region. Differentiated neuronal cultures were treated with 10 nM-10 μM topotecan or vehicle for 6 days. Expression quantified by qPCR. **P<0.01, one-way ANOVA with Dunnet’s Post-hoc Test. n=4. Error bars represent s.e.m.
Acknowledgements
We thank Mike Vernon at the UNC Expression Profiling Core for assistance with microarray experiments and with data analysis, Hemant Kelkar for pilot bioinformatics support, Piotr Mieczkowski and Alicia Brandt at the UNC High Throughput Sequencing Facility for advice and assistance with Illumina library preparation and sequencing and Tal Kafri and Ping Zhang at the UNC Lenti-shRNA Core for assistance with preparation of lentiviral vectors. This work was supported by grants to M.J.Z. and B.D.P. from The Angelman Syndrome Foundation, The Simons Foundation (SFARI 10-3625) and The National Institute of Mental Health (R01MH093372). I.F.K. and A.M.M. were supported by Joseph E. Wagstaff Postdoctoral Research Fellowships from the Angelman Syndrome Foundation. J.M.C. was supported by a grant from the American Cancer Society (117571-PF-09-124-01-DDC). J.M.C., J.S. and T.M. were supported by a grant from the NIH (R01GM101974). B.L.P. was supported by a NIH postdoctoral training grant (T32HD040127). S.J.C. was supported by a grant from NICHD (R01HD068730). The expression profiling and bioinformatics cores were funded by grants from NINDS (P30NS045892) and NICHD (P30HD03110).
Footnotes
Author Contributions. I.F.K., H.-S.H., A.M.M., J.S.H., S.J.C., B.D.P. and M.J.Z. conceived and designed experiments. I.F.K. performed RNA-seq and ChIP-seq experiments with mouse neurons. I.F.K., C.N.Y., J.M.C., J.S. and J.S.P. analyzed data from genome-wide experiments and from published datasets. J.M.C. performed SNP filtering of RNA-seq data and J.S. performed statistical analysis of RNA-seq data. A.M.M. performed lentiviral shRNA knockdown experiments. J.S.H. and S.J.C. performed all experiments with iPSC-derived human neurons. B.L.P. assessed propensity of compounds to kill neurons and damage DNA. T.M. provided bioinformatics support. I.F.K., H.-S.H. and A.M.M. performed microarray experiments. I.F.K. and M.J.Z. wrote the manuscript.
Author Information. Data from Microarray, RNA-seq and ChIP-seq experiments has been deposited in Gene Expression Omnibus (GSE43900). Reprints and permissions information is available at www.nature.com.
The authors declare no competing financial interests.
Full methods accompany this paper as supplementary material.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nature Reviews Genetics. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.State MW, Levitt P. The conundrums of understanding genetic risks for autism spectrum disorders. Nature Neuroscience. 2011;14:1499–1506. doi: 10.1038/nn.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen J GT. Prenatal valproate exposure and risk of autism spectrum disorders and childhood autism. JAMA. 2013;309:1696–1703. doi: 10.1001/jama.2013.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delorme R, et al. Progress toward treatments for synaptic defects in autism. Nat Med. 2013;19:685–694. doi: 10.1038/nm.3193. [DOI] [PubMed] [Google Scholar]
- 5.Betancur C, Sakurai T, Buxbaum JD. The emerging role of synaptic cell-adhesion pathways in the pathogenesis of autism spectrum disorders. Trends in Neurosciences. 2009;32:402–412. doi: 10.1016/j.tins.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peça J, Feng G. Cellular and synaptic network defects in autism. Current Opinion in Neurobiology. 2012;22:866–872. doi: 10.1016/j.conb.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang H-S, et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2012;481:185–189. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mabb AM, Judson MC, Zylka MJ, Philpot BD. Angelman syndrome: insights into genomic imprinting and neurodevelopmental phenotypes. Trends in Neurosciences. 2011;34:293–303. doi: 10.1016/j.tins.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook EH, et al. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet. 1997;60:928–934. [PMC free article] [PubMed] [Google Scholar]
- 10.Moreno-De-Luca D, et al. Using large clinical data sets to infer pathogenicity for rare copy number variants in autism cohorts. Molecular Psychiatry. 2012 doi: 10.1038/mp.2012.138. doi:10.1038/mp.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iossifov I, et al. De Novo Gene Disruptions in Children on the Autistic Spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plaschkes I, Silverman FW, Priel E. DNA topoisomerase I in the mouse central nervous system: Age and sex dependence. The Journal of Comparative Neurology. 2005;493:357–369. doi: 10.1002/cne.20793. [DOI] [PubMed] [Google Scholar]
- 14.Lyu YL, et al. Role of Topoisomerase IIβ in the Expression of Developmentally Regulated Genes. Mol. Cell. Biol. 2006;26:7929–7941. doi: 10.1128/MCB.00617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol. 2011;12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baranello L, Levens D, Gupta A, Kouzine F. The importance of being supercoiled: How DNA mechanics regulate dynamic processes. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2012;1819:632–638. doi: 10.1016/j.bbagrm.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Capranico G, Marinello J, Baranello L. Dissecting the transcriptional functions of human DNA topoisomerase I by selective inhibitors: Implications for physiological and therapeutic modulation of enzyme activity. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2010;1806:240–250. doi: 10.1016/j.bbcan.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem. Biol. 2013;8:82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Babak T, et al. Global Survey of Genomic Imprinting by Transcriptome Sequencing. Current Biology. 2008;18:1735–1741. doi: 10.1016/j.cub.2008.09.044. [DOI] [PubMed] [Google Scholar]
- 20.Meng L, Person RE, Beaudet AL. Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Hum. Mol. Genet. 2012;21:3001–3012. doi: 10.1093/hmg/dds130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landers M, et al. Regulation of the large (1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf/Snrpn. Nucl. Acids Res. 2004;32:3480–3492. doi: 10.1093/nar/gkh670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ljungman M, Hanawalt PC. The anti-cancer drug camptothecin inhibits elongation but stimulates initiation of RNA polymerase II transcription. Carcinogenesis. 1996;17:31–36. doi: 10.1093/carcin/17.1.31. [DOI] [PubMed] [Google Scholar]
- 23.Collins I, Weber A, Levens D. Transcriptional Consequences of Topoisomerase Inhibition. Mol. Cell. Biol. 2001;21:8437–8451. doi: 10.1128/MCB.21.24.8437-8451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. PNAS. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tiwari VK, et al. Target genes of Topoisomerase IIβ regulate neuronal survival and are defined by their chromatin state. PNAS. 2012;109:E934–E943. doi: 10.1073/pnas.1119798109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Troester MA, Hoadley KA, Parker JS, Perou CM. Prediction of Toxicant-Specific Gene Expression Signatures after Chemotherapeutic Treatment of Breast Cell Lines. Environ Health Perspect. 2004;112:1607–1613. doi: 10.1289/txg.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joshi RS, Piña B, Roca J. Topoisomerase II is required for the production of long Pol II gene transcripts in yeast. Nucl. Acids Res. 2012 doi: 10.1093/nar/gks626. doi:10.1093/nar/gks626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu H-Y, Shyy S, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 29.Francis Stewart A, Herrera RE, Nordheimt A. Rapid induction of c-fos transcription reveals quantitative linkage of RNA polymerase II and DNA topoisomerase I enzyme activities. Cell. 1990;60:141–149. doi: 10.1016/0092-8674(90)90724-s. [DOI] [PubMed] [Google Scholar]
- 30.Rahl PB, et al. c-Myc Regulates Transcriptional Pause Release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muse GW, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeitlinger J, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basu SN, Kollu R, Banerjee-Basu S. AutDB: a gene reference resource for autism research. Nucleic Acids Research. 2009;37:D832–D836. doi: 10.1093/nar/gkn835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammer E, Heilbronn R, Weger S. The E3 ligase Topors induces the accumulation of polysumoylated forms of DNA topoisomerase I in vitro and in vivo. FEBS Letters. 2007;581:5418–5424. doi: 10.1016/j.febslet.2007.10.040. [DOI] [PubMed] [Google Scholar]
- 37.Zhao X, et al. A unified genetic theory for sporadic and inherited autism. PNAS. 2007;104:12831–12836. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naughton C, et al. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nature Structural & Molecular Biology. 2013 doi: 10.1038/nsmb.2509. doi:10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sano K, Miyaji-Yamaguchi M, Tsutsui KM, Tsutsui K. Topoisomerase IIβ Activates a Subset of Neuronal Genes that Are Repressed in AT-Rich Genomic Environment. PLoS ONE. 2008;3:e4103. doi: 10.1371/journal.pone.0004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ben-David E, Shifman S. Combined analysis of exome sequencing points toward a major role for transcription regulation during brain development in autism. Molecular Psychiatry. 2012 doi: 10.1038/mp.2012.148. doi:10.1038/mp.2012.148. [DOI] [PubMed] [Google Scholar]
- 41.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forsberg EC, Downs KM, Bresnick EH. Direct interaction of NF-E2 with hypersensitive site 2 of the β-globin locus control region in living cells. Blood. 2000;96:334–339. [PubMed] [Google Scholar]
- 43.Chamberlain SJ, et al. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. PNAS. 2010;107:17668–17673. doi: 10.1073/pnas.1004487107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamb J, et al. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 45.Iorio F, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. PNAS. 2010;107:14621–14626. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Groschel B, Bushman F. Cell Cycle Arrest in G2/M Promotes Early Steps of Infection by Human Immunodeficiency Virus. J. Virol. 2005;79:5695–5704. doi: 10.1128/JVI.79.9.5695-5704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carson JP, et al. Pharmacogenomic Identification of Targets for Adjuvant Therapy with the Topoisomerase Poison Camptothecin. Cancer Res. 2004;64:2096–2104. doi: 10.1158/0008-5472.can-03-2029. [DOI] [PubMed] [Google Scholar]
- 48.MacIsaac JL, Bogutz AB, Morrissy AS, Lefebvre L. Tissue-specific alternative polyadenylation at the imprinted gene Mest regulates allelic usage at Copg2. Nucl. Acids Res. 2012;40:1523–1535. doi: 10.1093/nar/gkr871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extended Data Figure 2. Topotecan dose-response. Mouse cortical neurons were treated with 3 nM, 30 nM, 150 nM, 300 nM and 1000 nM topotecan for 3 days (n=3 for 300 nM topotecan, all other doses n=1). Gene expression was analyzed by Affymetrix microarrays, plotted as mean expression change in bins of 200 genes.
Extended Data Figure 3. DNA damage does not inhibit gene expression in a length-dependent manner. a, Cultured mouse cortical neuron viability, assayed by Sytox Green staining after 72 hours treatment with 300 nM topotecan, and after 24 hours with 100 μM H2O2 or 10 μM paraquat, compared to vehicle-treated controls. Error bars represent s.e.m. n=4. b, Average number of γH2AX foci per nucleus for cultured cortical neurons treated with 300 nM topotecan for 72 hours, 100 μM H2O2 for 24 hours and 10 μM paraquat for 24 hours, compared to vehicle-treated controls. **P<0.01, Student’s t-test. Number of cells counted is indicated for each sample. c, Gene expression compared to vehicle controls in bins of 200 genes by length, for cultured cortical neurons treated with 100 μM H2O2 for 24 hours. d, Gene expression compared to vehicle controls in bins of 200 genes by length, for cultured cortical neurons treated with 10 μM paraquat for 24 hours. e, Gene expression in cultured cortical neurons treated with 300 nM topotecan for 24 hours, or treated for 24 hours followed by 48 hours without drug (washout). Average change in expression for bins of 200 genes by length.
Extended Data Figure 4. Topotecan and irinotecan have highly similar effects on gene expression. a, Affymetrix microarray analysis of gene expression in cultured mouse cortical neurons treated with vehicle or 10 μM irinotecan (n=3 biological replicates), an inhibitor of TOP1, for 3 days. Mean expression fold change in bins of 200 genes, plotted by average gene length. b, Scatterplot of fold change with 1 μM topotecan (n=6 biological replicates) versus fold change with 10 μM irinotecan for all expressed genes, measured by Affymetrix microarray. Pearson’s R = 0.860. c, Overlap between genes showing positive or negative fold change of log2 = 0.5 or greater with topotecan and irinotecan treatment. d, Overlap between genes reduced or increased in expression. e, Overlap between differentially expressed genes that are greater or less than 67 kb.
Extended Data Figure 5. TOP1 and TOP2 inhibitors reduce expression of long genes in human cell lines. Re-analysis of microarray gene expression datasets from other labs. All plots are mean fold change in expression compared to vehicle controls in bins of 200 genes, plotted by average gene length. a, MCF7 cells treated with 10 μM irinotecan for 24 hours, from the CMAP2 project44. b, MCF7 cells treated with 165 nM SN38, the active metabolite of irinotecan, for 6 hours45. c-e, Gene expression in three human cell lines treated with camptothecin. c, MCF7 cells treated for 24 hours with 10 μM camptothecin, from CMAP2. d, 293T cells treated with 2 μM camptothecin for 48 hours46. e, HeLa cells treated with 10 μM camptothecin for 8 hours47. f, g, Re-analysis of microarray data from Troester et al.26 comparing gene expression in TOP2 inhibitor and vehicle treated ME16C cells. f, ME16C cells treated with 0.5 μM doxorubicin for 36 hours. g, ME16C cells treated with 50 μM etoposide for 36 hours.
Extended Data Figure 6. Topotecan and the TOP2 inhibitor ICRF-193 have similar effects on gene expression. a, Affymetrix microarray analysis of gene expression in cultured mouse cortical neurons treated with vehicle or 3 μM ICRF-193 (n=3 biological replicates), an inhibitor of TOP2 enzymes, for 3 days. Mean expression fold change in bins of 200 genes, plotted by average gene length. b, Scatterplot of fold change with 300 nM topotecan (n=3 biological replicates) versus fold change with 3 μM ICRF-193 for all expressed genes, measured by Affymetrix microarray. Pearson’s R=0.588. c, Overlap between genes showing positive or negative fold change of log2 = 0.5 or greater with topotecan and ICRF-193 treatment. d, Overlap between genes reduced or increased in expression. e, Overlap between differentially expressed genes that are greater or less than 67 kb in length.
Extended Data Figure 7. Pharmacological inhibition of TOP2 or genetic deletion of Top2b reduces expression of long genes in embryonic stem (ES) cell-derived neurons. a, b, Re-analysis of Tiwari et al. microarray expression data25. Mean fold change in expression in bins of 200 genes, plotted by average gene length. a, Gene expression in ES cell-derived neurons treated with vehicle or 50 μM ICRF-193 for 3 days. b, Gene expression in ES cells treated with vehicle or 50 μM ICRF-193 for 3 days. c, Re-analysis of Lyu et al. microarray expression data14, comparing gene expression in whole brain from wild-type (wt) and Top2b-/- embryonic mice. Expression data from three developmental time points (E16.5, E17.5, E18.5) were averaged for each gene then plotted as mean fold change in expression between wt and Top2b-/- mice (in bins of 100 genes, by average gene length). d, e, Re-analysis of Tiwari et al. microarray expression data25. Mean fold change in gene expression between wt and Top2b-/-, for bins of 200 genes, plotted by average gene length. d, Expression data from ES cells, e, neuronal progenitors, f, ES cell-derived neurons, two days after plating of neuronal progenitors, and g, ES cell-derived neurons, six days after plating of neuronal progenitors. ES cells and neuronal progenitors express Top2a and Top2b (indicated within parentheses) and do not show reduced expression of long genes when Top2b is knocked out, suggesting that Top2a and Top2b redundantly regulate expression of long genes.
Extended Data Figure 8. Topotecan increases Travelling Ratio (TR) in genes larger than 67 kb. TR was calculated for all genes bound by Pol II in vehicle and topotecan-treated (300 nM for 3 days) cultured cortical neurons. 92.1% of all genes had TR values greater than 2, consistent with a previous report using mouse ES cells30. a, Frequency of TRs for genes less than 67 kb in length, and b, for genes greater than 67 kb in length. Mean TR of vehicle and topotecan treated samples was significantly different for genes greater than 67 kb (P=2.4×10−21, t-test) but not for genes less than 67 kb (P=0.648, t-test).
Extended Data Figure 9. Topotecan dose-dependently reduces expression of ASD candidate genes in cortical neurons. a, Topotecan dose-response for high confidence autism candidate genes (n=3 for 300 nM topotecan, all other doses n=1). b-e, Topotecan dose-response for other ASD candidate genes organized by length from longest to shortest. Mouse cortical neurons were treated with vehicle or the indicated doses of topotecan for 3 days. Gene expression was quantified using Affymetrix microarrays. Dose-responses are from all topotecan-downregulated ASD candidate genes that were identified by RNA-seq (Table 1) and that were present on the Affymetrix microarrays.
Extended Data Table 1. Topotecan does not alter parent-of-origin bias of imprinted genes. Genes with significant parent-of-origin expression bias in cultured cortical neurons. Shown are P values for allelic bias in baseline expression, and P values for change in expression bias with topotecan treatment (Fisher’s Exact Test, adjusted for multiple comparisons). ns = P>0.05. Embryos from CAST male × B6 female crosses (n=3) were pooled and B6 male × CAST female crosses (n=6) were pooled for sequencing. *Genes in the Ube3a imprinted cluster. †AF217545 is contained within Copg2, which has also been reported to be maternally expressed48.
Extended Data Figure 1. Topotecan affects allelic expression of Ube3a and Ube3a-ATS but not expression of nearby genes. a, Parent-of-origin specific RNA-seq reads for Ube3a and Ube3a-ATS in vehicle and topotecan-treated (300 nM for 3 days) mouse cortical neurons (n=5 biological replicates). Shown are SNP filtered read counts per million mapped reads (RPM) from the maternally (mat) and paternally (pat) inherited chromosomes. Reads from exons and 3′-untranslated region of Ube3a are indicated by arrows. b, Expression of Ube3a and imprinted genes near Ube3a in mouse cortical neurons ± 300 nM topotecan for 3 days. * P<0.05, t-test. Error bars represent s.e.m. c, Expression of UBE3A and UBE3A-ATS in iPSC-derived neurons from an Angelman syndrome patient carrying a maternal deletion of the 15q11-q13 region. Differentiated neuronal cultures were treated with 10 nM-10 μM topotecan or vehicle for 6 days. Expression quantified by qPCR. **P<0.01, one-way ANOVA with Dunnet’s Post-hoc Test. n=4. Error bars represent s.e.m.