Abstract
Mesenchymal stem cells (MSC) are heterogeneous cell populations with promising therapeutic potentials in regenerative medicine. The therapeutic values of MSC in various clinical situations have been reported. Clonal assays (expansion of MSC from a single cell) demonstrated that multiple types of cells with different developmental potential exist in a MSC population. Due to the heterogeneous nature of MSC, molecular characterization of MSC in the absence of known biomarkers is a challenge for cell therapy with MSC. Here, we review potential therapeutic applications of MSC and discuss a systematic approach for molecular characterization of heterogeneous cell population using single-cell transcriptome analysis. Differentiation/maturation of cells is orchestrated by sequential expression of a series of genes within a cell. Therefore, single-cell mRNA expression (transcriptome) profiles from consecutive developmental stages are more similar than those from disparate stages. Bioinformatic analysis can cluster single-cell transcriptome profiles from consecutive developmental stages into a dendrogram based on the similarity matrix of these profiles.
Because a single-cell is an ultimately “pure” sample in expression profiling, these dendrograms can be used to classify individual cells into molecular subpopulations within a heterogeneous cell population without known biomarkers. This approach is especially powerful in studying cell populations with little molecular information and few known biomarkers, for example the MSC populations. The molecular understanding will provide novel targets for manipulating MSC differentiation with small molecules and other drugs to enable safer and more effective therapeutic applications of MSC.
Keywords: Single-cell transcriptome, Heterogeneity, Mesenchymal stem cells
1. Introduction
There is a long history of harnessing cellular plasticity in clinical situations. While hematopoietic stem cell is the best studied system, there is a growing interest in mesenchymal stem cells (MSC). Cellular plasticity of bone marrow cells has been utilized to rebuild the hematopoietic system since 1957 (Brecher and Cronkite, 1951). Recently, cells with differentiation plasticity such as MSC have been proposed for diverse applications in regenerative medicine. The focus of regenerative medicine has gradually shifted from whole bone marrow to specific plastic cells (e.g. mobilized stem cells, cord blood and MSC). However, most primary cell populations are heterogeneous due to isolation methods, culture conditions and potentially the need for interaction among different cell types for surviving and functioning. This naturally existing cellular heterogeneity presents a challenge for molecular characterization of these cells in order to improve their medical values. MSC is one of these cell populations. MSC studies using different isolation, expansion and characterization methods have raised concerns on the consistency of results among different studies due to the heterogeneous nature of MSC and lack of precise molecular characterization of MSC (Dominici et al., 2006; Wagner et al., 2006). Consequently, developing drugs to improve MSC therapeutic efficacy is limited by the lack of molecular knowledge of MSC. Here, we briefly review some medical applications of MSC, and discuss a potential methodology, single-cell transcriptome analysis, for MSC molecular characterization and the possibility of using small molecules to improve MSC therapeutic efficacy.
2. Mesenchymal stem cells
Mesenchymal stem cells (MSC) are a highly heterogeneous subset of stromal stem cells which are difficult for molecular characterization with traditional methods. Bone marrow and adipose tissue are two main sources of MSCs for clinical applications (Mendez-Ferrer et al., 2010; Morando et al., 2012; Rodriguez et al., 2005). The International Society for Cellular Therapy (ISCT) provided 3 minimal criteria to define human MSC with culture conditions, biomarkers and developmental potentials for facilitating comparison of different studies (Dominici et al., 2006). Expression of biomarkers is only one of the 3 criteria and is often different in different studies. Although various biomarkers are reported for association with MSC populations, the exhaustive list of markers is from various subpopulations within the MSC populations. Therefore, these biomarkers are not the unique characteristics of MSC, but rather a reflection of the heterogeneity of MSCs. It is still lack of consensus on a set of well defined biomarkers for MSC (Buhring et al., 2007; Mendez-Ferrer et al., 2010; Uccelli et al., 2008). In addition, because their phenotype may be affected by the culture medium, the plating density and the oxygen tension, the precise phenotype of cultured MSC is still debated, and their identification remains ambiguous (Gnecchi et al., 2012). Numerous studies including clonal assays followed by transcriptomic analyses have been carried out for molecular characterization of MSC and demonstrated the difficulty of molecular characterization of MSC (Jia et al., 2002; Kuznetsov et al., 1997; Muraglia et al., 2000; Panepucci et al., 2004; Russell et al., 2011; Silva et al., 2003; Tremain et al., 2001; Wagner et al., 2005, 2006; Wislet-Gendebien et al., 2012a). The limitations of clonal selection are that not all cells can be clonally expanded and cellular properties could be altered during clonal expansion (Wislet-Gendebien et al., 2012b). Clonal expansion itself may eliminate and alter cellular characteristics (Gnecchi et al., 2012). Single cell transcriptomes from freshly isolated MSC could overcome this limitation and reveal the natural characteristics of MSCs without the need for cell culture and expansion.
3. Treating disease with MSC
In recent years, MSCs have been considered a promising therapeutic resource to treat many central nervous system diseases caused by acute trauma and progressive degeneration, such as Parkinson's disease (PD) (Danielyan et al., 2011), amyotrophic lateralizing sclerosis (ALS) (Choi et al., 2010), multiple system atrophy (MSA) (Lee and Park, 2009), experimental allergic encephalomyelitis (EAE) (Matysiak et al., 2011) and spinal cord injury(Nakajima et al., 2012; J.H. Park et al., 2012; S.-S. Park et al., 2012). A study by Seung et al. suggested that MSCs derived from human fat had the potential to treat brainstem gliomas (Choi et al., 2012). It has also been shown that MSCs could suppress middle cerebral artery occlusion (MCAO) focal ischemia-induced inflammation by expressing fractalkine and IL-5 (Sheikh et al., 2011). Transplanted MSCs derived from bone marrow promoted the behavioral recovery, endogenous neurogenesis as well as protected newly generated cells in rats with cerebral ischemia (Bao et al., 2011). By enhancing the expression of neurotrophic factors, MSCs derived from human cord blood can alleviate cerebellar atrophy in mice with dyskinesia and ataxia (Zhang et al., 2011). MSCs have the potential to reduce the injury and infarction area of myocardium, restore its mechanical energy, improve local and global ventricular functions, and increase blood vessel density and myocardial perfusion (Madonna and De Caterina, 2011; Ohnishi et al., 2007; Psaltis et al., 2008). It is now clear that MSCs not only contribute to reconstituting the capillary network and increasing the vessel density, and differentiating into cardiomyocyte-like cells, but also serve as a sponge cell source which can release multiple paracrine factors with protective function, and also in turn prevent advert remodeling while promoting neovascularization and activation of endogenous stem cells and proliferation of cardiomyocytes(Gnecchi et al., 2008; Li et al., 2012). Toghraie et al. directly injected MSCs derived from the infrapatellar fat pad to the damaged knee of a rabbit osteoarthritis model and found that after 20 weeks, animals injected with MSCs showed lower cartilage degeneration rate, and less degree of osteophyte formation and cartilage ossification compared to control group (Toghraie et al., 2011). Autologous MSCs effectively alleviated symptoms of type 1 diabetes for as long as 16 weeks in beagles (Zhu et al., 2011).
4. Single-cell transcriptome analysis
The molecular foundation of cellular plasticity that has led to all the therapeutic possibilities described above is largely unknown. Previous clonal assays indicated that there are many different types of pluripotent cells within a MSC population (Digirolamo et al., 1999; Kuznetsov et al., 1997; Muraglia et al., 2000; Russell et al., 2011; Wislet-Gendebien et al., 2012a). The heterogeneity of cell populations makes investigating the molecular foundation of MSC cellular plasticity very challenging. In order to obtain sufficient material for measurement, traditional molecular methods typically measure gene expression or protein levels in cell lysates which is a physical-average of all cells in the sample. Therefore, cellular heterogeneity becomes an inherent noise in the measurement of gene expressions (Evsikov et al., 2003; Ivanova et al., 2002; Ramalho-Santos et al., 2002). The cell lysate provides limited molecular knowledge. One approach for molecular characterization of heterogeneous cell populations is single-cell transcriptome analysis.
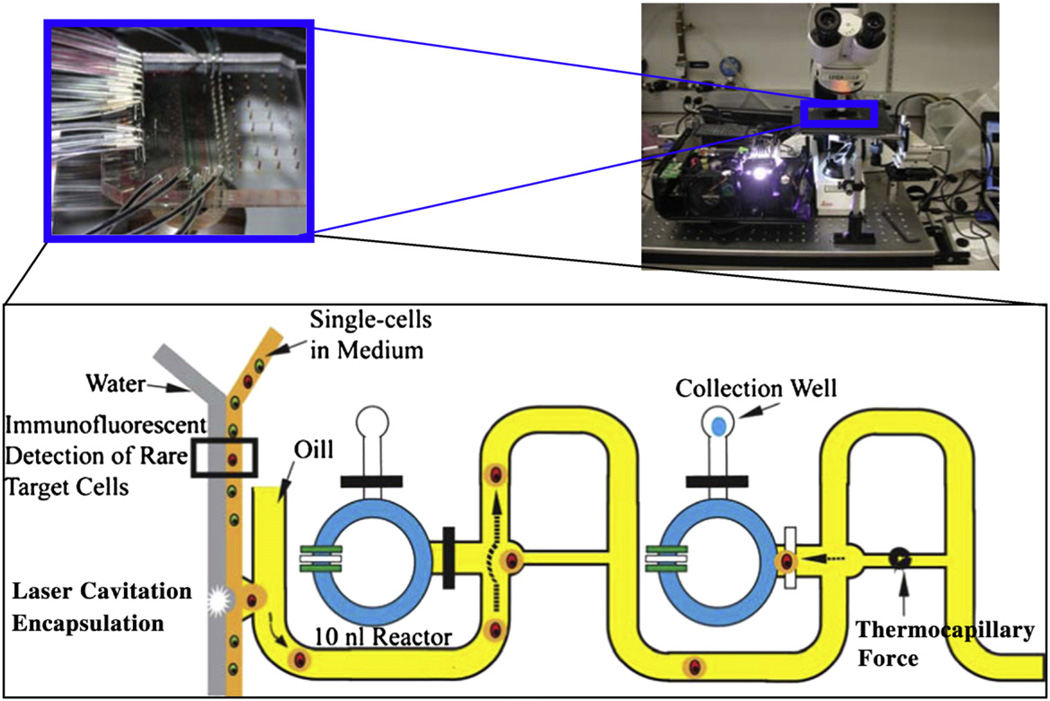
Single-cell transcriptome analysis has the potential to turn cellular heterogeneity from noise into news. Since gene–gene interaction typically happens inside a cell, analysis of the single-cell transcriptome is the most efficient method to reveal the regulatory relationship among genes. The small amount of RNA within a mammalian cell (~20–40 pg of total RNA (Uemura, 1980)) is a challenge for transcriptome analysis. Our group has overcome technical hurdles for single-cell molecular analysis by using microfluidic technology (Chen and Zhong, 2008; Fan et al., 2012; Zhong et al., 2008a). Traditional molecular analysis methods are performed in microliter scale while a mammalian cell typically has a volume of ~0.065 pl. Therefore, the total RNA is often diluted for more than 106 folds in classic molecular assay(Chen and Zhong, 2008; Zhong et al., 2008a, 2008b). Unlike PCR which is a repetitive reaction, reverse transcription (RT) for obtaining cDNA from mRNA is a single biochemical event. The concentration of RNA in RT is critical for an effective RT reaction. We have developed a microfluidic device to reduce the volume of RT reactions to nanoliter level for increasing the RNA concentration to a level that is compatible with bulk assay for effective single-cell cDNA synthesis (Chen and Zhong, 2008; Fan et al., 2012; Zhong et al., 2008a, 2008b). These microfluidic devices allow us to obtain reliable single-cell transcriptomes for investigating gene regulation. More recently, we developed phase-switch microfluidic devices with various designs to encapsulate individual cells into droplets for nanoliter RT reaction (Fig. 1). Using these microfluidic devices, we have successfully obtained whole genome transcriptomes with microarrays from individual cells (Fig. 2). The quality of single-cell data is similar to that from the population cell experiment in which 1 ng cDNA inputs was used (Fan et al., 2012).
Fig. 1.
Phase-switch microfluidic system for single-cell transcriptome analysis. The system consists of microfluidic structures and encapsulates a live single-cell into a 500-pl droplet for biochemical reactions in a 10nl reactor. Inside the 10-nl reactor, single-cells are lysed and reverse transcription is carried out as described previously (Zhong et al., 2008a, 2008b). The resulting cDNAs are then flushed out with oil for gene expression profiling with qPCR or microarrays (one of the designs is illustrated).
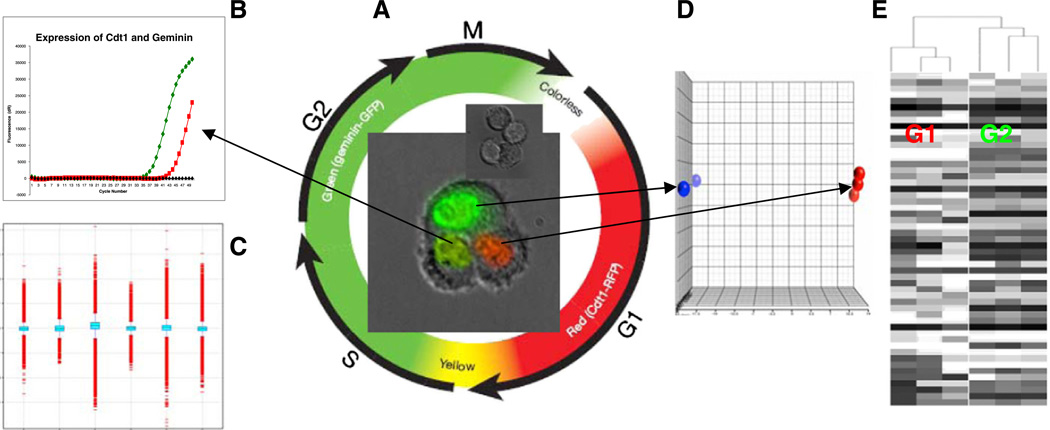
Fig. 2.
Whole genome single-cell transcriptome analysis of cell cycle regulation. A. Images of cells at different stages are shown in the center of the color chart. The cells were genetically modified to have a red (RFP) and a green (GFP) fluorescent protein fused to two cell cycle regulators Cdt1 and geminin respectively. In G1 phase, geminin is broken down and only Cdt1 tagged with RFP may be visualized in the red nuclei. In the S, G2, and M phases, Cdt1 is degraded and only geminin tagged with GFP remains (green nuclei). During the G1/S transition, as Cdt1 levels decrease and geminin levels increase, both proteins are present in the cells (yellow nuclei) B. Single-cell multiplex qRT-PCR of a G1/S (yellow) cell confirms the expression of both Cdt1 and geminin. C. All 6 Affymetrix chips passed data QC. D. PCA analysis of the 6 chips shows that red and green cells have different expression patterns. E. Unsupervised clustering shows that the 3 red cells and the 3 green cells form distinguishable branches. Adding more profiles from continuous stages and repeating unsupervised clustering, the dendrogram will expand into a continuous spectrum of cell cycles. The completed dendrogram will have expression profiles of all cell cycle stages and these profiles will be arranged in sequential order as a molecular map of cell cycles.
This single-cell technology has been demonstrated on cultured cells (Chen and Zhong, 2008; Fan et al., 2012; Zhong et al., 2008a). It can also be applied to molecular characterization of freshly sorted MSCs and other primary cell populations, although there are challenges associated with such applications. Freshly isolated MSC populations are much more heterogeneous than cultured cells. Our microfluidic devices can isolate thousands of single-cells, but profiling a single-cell will cost as much as profiling a cell lysate. Therefore, there is a limitation on how many single-cells can be analyzed due to the profiling cost. At the same time, a sufficient number of single-cells must be profiled for statistically sound results. The more heterogeneous a population is, the more single-cells are needed for molecular profiling. Enriching MSCs with various methods prior to single-cell analysis can minimize the profiling cost while ensuring sufficient pluripotent MSCs are profiled. Multi-color flow cytometry has been used to isolate MSCs freshly from mouse and from human bone marrows with empirical biomarkers (Morikawa et al., 2009; Qian et al., 2012; Tormin et al., 2011). These freshly isolated MSCs are not homogenous and their purities are dependent on the isolation biomarkers. Combining advanced flow cytometry enrichment (enriching MSCs to more than 10% of the isolated population) with our single-cell technique, only one or two hundred single-cells will need to be profiled to have sufficient MSCs (with >20 profiles) to form a distinct molecular cluster. The clusters of cells with similar expression profiles will not only reveal how many subpopulations (cells with similar expression profiles) there are within a MSC-enriched population, but will also provide new biomarkers (surface biomarkers from their expression profiles) to isolate each molecular subpopulation for functional assays. The functional assay is the gold standard to determine and validate which subpopulation is the pluripotent MSCs. The quality of single-cell expression profiles can also be validated with known biomarkers during single-cell isolation such as DNA and KI67 staining for validation of cell cycle status.
Heterogeneity is a problem for traditional transcriptome analysis because the expressions of genes are physically averaged in the cell lysate. Although large amounts of materials (e.g. cell lysate) warrant repeatable measurements, the unknown heterogeneity (the composition of the cell population) allows measurement only of highly expressed genes. On the other hand, single-cell transcriptome has more measurement noise due to the small amount of RNA from one cell. However, the sample is homogenous (from a single cell) and cellular heterogeneity becomes a beneficial element by providing samples (single cells) at various developmental stages. Each single-cell transcriptome is a snapshot of a molecular event (i.e. blood cell differentiation, somatic cell reprogramming, and apoptosis). Because transcriptome from consecutive developmental stages are more similar than those from disparate stages, single-cell transcriptome profiles can be arranged with similarity to reconstructing a stepwise developmental event at the molecular level. These stepwise molecular maps reveal the sequential perturbation of the gene network. That is similar to organizing a collection of still pictures (transcriptome) into a “video” describing a cellular event at the molecular level. We are currently in the progress of building and distributing these “molecular videos” for the molecular biology community. Such molecular maps can reveal signature genes involved in cell differentiation, which serve as potential targets of genetic or pharmacological manipulation in the ongoing efforts to direct cell fate.
5. Improving MSC efficacy with small molecules
Molecular maps of single-cell transcriptome can predict possible target genes and facilitate chemical optimization of lead compounds for MSC therapeutic applications. Small molecules are critically important tools for stem cell therapeutic application (Lukaszewicz et al., 2010; Xu et al., 2008). Unlike viral vectors that typically have low efficiency delivering target genes to a cell, small molecules can enter and affect almost all cells. More importantly, small molecules do not alter DNA sequences in target cells and can be readily removed post-differentiation. Various studies including ours also demonstrated that transient perturbation of transcriptome can change cell fate permanently(Li et al., 2011; Takahashi et al., 2007; Wernig et al., 2008; Yu et al., 2007). These features make small molecules ideal tools for controlling the fate of stem cells in vitro or in vivo for therapeutic interventions. The identification of small molecules that can efficiently control the fate of stem cells poses a major challenge in the field (Xu et al., 2008). Intensive efforts have been made to meet this challenge. Based on the methodology, these efforts can be broadly divided into two categories — forward chemical genetics and reverse chemical genetics. Forward chemical genetics typically involves screening of a small-molecule library to search for hits that cause a phenotypic change to target cells (Specht and Shokat, 2002). Reverse chemical genetics, on the other hand, entails identification of small-molecule modulators of particular genes in a pathway followed by characterization of phenotypic effects of such modulators on cells (Specht and Shokat, 2002). The application of reverse chemical genetics requires the generation of a list of candidate genes which can be provided by single-cell transcriptomes from MSCs.
Forward chemical genetics is a powerful approach for identifying molecules that affect cell differentiation. Peter Schultz and coworkers were among the first to employ forward chemical genetics to identify a number of different small molecules that affect various cell fate, including stem cell self-renewal, differentiation, and lineage-specific reprogramming (Chen et al., 2004; Ding et al., 2003; Wu et al., 2002). Using the same strategy, Sheng Ding and co-workers went on to identify a series of small molecules and conditions that can replace reprogramming transcription factors in generating iPS cells from somatic cells (Li et al., 2009; Shi et al., 2008). While forward chemical genetics proves quite powerful in identifying novel small molecules that regulate the fate of stem cells, the mechanism of action of these molecules can be elusive. Different methods such as gene deletion, siRNA and affinity chromatography have been applied toward target identification of small molecules (Wu et al., 2004; Zhang et al., 2007). However they suffer from various drawbacks such as distinct phenotypes from pharmacological inhibition versus gene ablation and poor sensitivity to low-abundance proteins (Knight and Shokat, 2007). This results in a lack of mechanistic understanding of small-molecule actions in many cases, which hampers chemical optimization of lead compounds for therapeutic application.
Reverse chemical genetics yields small molecules with known mechanism of action, because these molecules have been characterized toward target proteins in vitro. The better mechanistic understanding of small-molecule actions offered by reverse chemical genetics allows for efficient chemical optimization of lead compounds, especially when the 3-dimensional structures of the target proteins are available. For example, a study identified multiple compounds that affect MSC differentiation by screening a small library of known inhibitors for different protein kinases (Song et al., 2012). These results revealed the relevance of several kinases such as PKA and ROCK in MSC differentiation and provide excellent starting points for medicinal chemistry efforts to generate compounds having improved control of MSC differentiation. Despite the advantage of reverse chemical genetics, there is a bottleneck for its application to MSC differentiation, namely the generation of a list of druggable genes that have validated roles in MSC differentiation. Multiple genes in diverse pathways including Wnt and Notch pathways were found to play important roles in MSC differentiation (Satija et al., 2007). However, the druggable targets in these established pathways are limited to a small number of proteins such as Wnt and GSK3 (Ding et al., 2003; Liu et al., 2005). Accelerated understanding of MSC biology is required to expand this list, which serves as potential targets for small-molecule intervention.
While conventional genetic studies connect individual genes to phenotypes, transcriptome analysis simultaneously associates hundreds of genes to particular biological processes. This makes transcriptome analysis a highly powerful means for generating lists of candidate genes that regulate or affect particular biological processes. As described in the previous sections, single cell transcriptome analysis is ideally suited to dissect the heterogeneity of MSC populations and elucidate the cell differentiation path.
6. Conclusion and prospects
The potential of therapeutic application of MSC under various disease settings has been amply demonstrated by multiple research groups as discussed above. However, the modest efficacy of MSC transplantation limits its clinical application currently. One of the major hurdles for improving MSC efficacy is lack of molecular characterization of MSC which is highly heterogeneous without biomarkers for classification. Single-cell transcriptome analysis is a potential systematic approach to address this issue. With the technologies developed in our group and other laboratories, single-cell transcriptomes can be as reliable as those from standard inputs (Fan et al., 2012). Using reliable single-cell transcriptomes, cells in a heterogeneous population such as MSCs can be classified molecularly based on transcriptome similarity without known biomarkers. The molecular classification in turn will provide targets for controlling MSC differentiation with small molecules for improving MSC therapeutic efficacy. The single-cell transcriptome analysis approach provides a promising solution for molecular characterization of heterogeneous MSC population for therapeutic applications and medicinal research.
Acknowledgments
This work was supported by grant R01CA164509 from the National Institutes of Health, USA (JFZ), and CHE1213161 from the National Science Foundation (JFZ), USA, and an internal grant from the University of Southern California (CZ).
References
- Bao X, Wei J, Feng M, Lu S, Li G, Dou W, et al. Transplantation of human bone marrow-derived mesenchymal stem cells promotes behavioral recovery and endogenous neurogenesis after cerebral ischemia in rats. Brain Res. 2011;1367:103–113. doi: 10.1016/j.brainres.2010.10.063. [DOI] [PubMed] [Google Scholar]
- Brecher G, Cronkite EP. Post-radiation parabiosis and survival in rats. Proc Soc Exp Biol Med. 1951;77:292–294. doi: 10.3181/00379727-77-18754. [DOI] [PubMed] [Google Scholar]
- Buhring H-J, Battula VL, Treml S, Schewe B, Kanz L, Vogel W. Novel markers for the prospective isolation of human MSC. Ann N Y Acad Sci. 2007;1106:262–271. doi: 10.1196/annals.1392.000. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhong JF. Microfluidic devices for high-throughput gene expression profiling of single hESC-derived neural stem cells. Methods Mol Biol. 2008;438:293–303. doi: 10.1007/978-1-59745-133-8_22. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhang Q, Wu X, Schultz PG, Ding S. Dedifferentiation of lineage-committed cells by a small molecule. J Am Chem Soc. 2004;126:410–411. doi: 10.1021/ja037390k. [DOI] [PubMed] [Google Scholar]
- Choi M, Kim H, Park J, Lee T, Baik C, Chai Y, et al. Selection of optimal passage of bone marrow-derived mesenchymal stem cells for stem cell therapy in patients with amyotrophic lateral sclerosis. Neurosci Lett. 2010;472:94–98. doi: 10.1016/j.neulet.2010.01.054. [DOI] [PubMed] [Google Scholar]
- Choi SA, Lee JY, Wang KC, Phi JH, Song SH, Song J, et al. Human adipose tissue-derived mesenchymal stem cells: characteristics and therapeutic potential as cellular vehicles for prodrug gene therapy against brainstem gliomas. Eur J Cancer. 2012;48:129–137. doi: 10.1016/j.ejca.2011.04.033. [DOI] [PubMed] [Google Scholar]
- Danielyan L, Schafer R, von Ameln-Mayerhofer A, Bernhard F, Verleysdonk S, Buadze M, et al. Therapeutic efficacy of intranasally delivered mesenchymal stem cells in a rat model of Parkinson disease. Rejuvenation Res. 2011;14:3–16. doi: 10.1089/rej.2010.1130. [DOI] [PubMed] [Google Scholar]
- Digirolamo CM, Stokes D, Colter D, Phinney DG, Class R, Prockop DJ. Propagation and senescence of human marrow stromal cells in culture: a simple colony-forming assay identifies samples with the greatest potential to propagate and differentiate. Br J Haematol. 1999;107:275–281. doi: 10.1046/j.1365-2141.1999.01715.x. [DOI] [PubMed] [Google Scholar]
- Ding S, Wu TY, Brinker A, Peters EC, Hur W, Gray NS, et al. Synthetic small molecules that control stem cell fate. Proc Natl Acad Sci U S A. 2003;100:7632–7637. doi: 10.1073/pnas.0732087100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- Evsikov AV, Solter D, Evsikov AV, Solter D. Comment on " ‘Stemness’: transcriptional profiling of embryonic and adult stem cells" and "a stem cell molecular signature". Science. 2003;302:393. doi: 10.1126/science.1082380. [author reply] [DOI] [PubMed] [Google Scholar]
- Fan J-B, Chen J, April CS, Fisher JS, Klotzle B, Bibikova M, et al. Highly parallel genome-wide expression analysis of single mammalian cells. PLoS One. 2012;7:e30794. doi: 10.1371/journal.pone.0030794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnecchi M, Zhang Z, Ni A, Dzau VJ. Paracrine mechanisms in adult stem cell signaling and therapy. Circ Res. 2008;103:1204–1219. doi: 10.1161/CIRCRESAHA.108.176826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnecchi M, Danieli P, Cervio E. Mesenchymal stem cell therapy for heart disease. Vascul Pharmacol. 2012;57:48–55. doi: 10.1016/j.vph.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR, et al. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- Jia L, Young MF, Powell J, Yang L, Ho NC, Hotchkiss R, et al. Gene expression profile of human bone marrow stromal cells: high-throughput expressed sequence tag sequencing analysis. Genomics. 2002;79:7–17. doi: 10.1006/geno.2001.6683. [DOI] [PubMed] [Google Scholar]
- Knight ZA, Shokat KM. Chemical genetics: where genetics and pharmacology meet. Cell. 2007;128:425–430. doi: 10.1016/j.cell.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Kuznetsov SA, Krebsbach PH, Satomura K, Kerr J, Riminucci M, Benayahu D, et al. Single-colony derived strains of human marrow stromal fibroblasts form bone after transplantation in vivo. J Bone Miner Res. 1997;12:1335–1347. doi: 10.1359/jbmr.1997.12.9.1335. [DOI] [PubMed] [Google Scholar]
- Lee PH, Park HJ. Bone marrow-derived mesenchymal stem cell therapy as a candidate disease-modifying strategy in Parkinson's disease and multiple system atrophy. J Clin Neurol. 2009;5:1–10. doi: 10.3988/jcn.2009.5.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, et al. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell Stem Cell. 2009;4:16–19. doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Li SC, Jin Y, Loudon WG, Song Y, Ma Z, Weiner LP, et al. Increase developmental plasticity of human keratinocytes with gene suppression. Proc Natl Acad Sci U S A. 2011;108:12793–12798. doi: 10.1073/pnas.1100509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T-S, Cheng K, Malliaras K, Smith RR, Zhang Y, Sun B, et al. Direct comparison of different stem cell types and subpopulations reveals superior paracrine potency and myocardial repair efficacy with cardiosphere-derived cells. J Am Coll Cardiol. 2012;59:942–953. doi: 10.1016/j.jacc.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wu X, Mitchell B, Kintner C, Ding S, Schultz PG. A small-molecule agonist of the Wnt signaling pathway. Angew Chem Int Ed Engl. 2005;44:1987–1990. doi: 10.1002/anie.200462552. [DOI] [PubMed] [Google Scholar]
- Lukaszewicz AI, McMillan MK, Kahn M. Small molecules and stem cells. Potency and lineage commitment: the new quest for the fountain of youth. J Med Chem. 2010;53:3439–3453. doi: 10.1021/jm901361d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madonna R, De Caterina R. Stem cells and growth factor delivery systems for cardiovascular disease. J Biotechnol. 2011;154:291–297. doi: 10.1016/j.jbiotec.2011.05.014. [DOI] [PubMed] [Google Scholar]
- Matysiak M, Orlowski W, Fortak-Michalska M, Jurewicz A, Selmaj K. Immunoregulatory function of bone marrow mesenchymal stem cells in EAE depends on their differentiation state and secretion of PGE2. J Neuroimmunol. 2011;233:106–111. doi: 10.1016/j.jneuroim.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morando S, Vigo T, Esposito M, Casazza S, Novi G, Principato MC, et al. The therapeutic effect of mesenchymal stem cell transplantation in experimental autoimmune encephalomyelitis is mediated by peripheral and central mechanisms. Stem Cell Res Ther. 2012;3:3. doi: 10.1186/scrt94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa S, Mabuchi Y, Kubota Y, Nagai Y, Niibe K, Hiratsu E, et al. Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. J Exp Med. 2009;206:2483–2496. doi: 10.1084/jem.20091046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraglia A, Cancedda R, Quarto R. Clonal mesenchymal progenitors from human bone marrow differentiate in vitro according to a hierarchical model. J Cell Sci. 2000;113:1161–1166. doi: 10.1242/jcs.113.7.1161. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Uchida K, Guerrero AR, Watanabe S, Sugita D, Takeura N, et al. Transplantation of mesenchymal stem cells promotes an alternative pathway of macrophage activation and functional recovery after spinal cord injury. J Neurotrauma. 2012;29:1614–1625. doi: 10.1089/neu.2011.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi S, Yanagawa B, Tanaka K, Miyahara Y, Obata H, Kataoka M, et al. Transplantation of mesenchymal stem cells attenuates myocardial injury and dysfunction in a rat model of acute myocarditis. J Mol Cell Cardiol. 2007;42:88–97. doi: 10.1016/j.yjmcc.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Panepucci RA, Siufi JLC, Silva WA, Jr, Proto-Siquiera R, Neder L, Orellana M, et al. Comparison of gene expression of umbilical cord vein and bone marrow-derived mesenchymal stem cells. Stem Cells. 2004;22:1263–1278. doi: 10.1634/stemcells.2004-0024. [DOI] [PubMed] [Google Scholar]
- Park JH, Kim DY, Sung IY, Choi GH, Jeon MH, Kim KK, et al. Long-term results of spinal cord injury therapy using mesenchymal stem cells derived from bone marrow in humans. Neurosurgery. 2012a;70:1238–1247. doi: 10.1227/NEU.0b013e31824387f9. [discussion 47] [DOI] [PubMed] [Google Scholar]
- Park S-S, Lee YJ, Lee SH, Lee D, Choi K, Kim W-H, et al. Functional recovery after spinal cord injury in dogs treated with a combination of Matrigel and neural-induced adipose-derived mesenchymal stem cells. Cytotherapy. 2012b;14:584–597. doi: 10.3109/14653249.2012.658913. [DOI] [PubMed] [Google Scholar]
- Psaltis PJ, Zannettino AC, Worthley SG, Gronthos S. Concise review: mesenchymal stromal cells: potential for cardiovascular repair. Stem Cells. 2008;26:2201–2210. doi: 10.1634/stemcells.2008-0428. [DOI] [PubMed] [Google Scholar]
- Qian H, Le Blanc K, Sigvardsson M. Primary mesenchymal stem and progenitor cells from bone marrow lack expression of CD44 protein. J Biol Chem. 2012;287:25795–25807. doi: 10.1074/jbc.M112.339622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA, Ramalho-Santos M, et al. "Stemness": transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- Rodriguez AM, Elabd C, Amri E-Z, Ailhaud G, Dani C. The human adipose tissue is a source of multipotent stem cells. Biochimie. 2005;87:125–128. doi: 10.1016/j.biochi.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Russell KC, Phinney DG, Lacey MR, Barrilleaux BL, Meyertholen KE, O'Connor KC. In vitro high-capacity assay to quantify the clonal heterogeneity in trilineage potential of mesenchymal stem cells reveals a complex hierarchy of lineage commitment. Stem Cells. 2011;28:788–798. doi: 10.1002/stem.312. [DOI] [PubMed] [Google Scholar]
- Satija NK, Gurudutta GU, Sharma S, Afrin F, Gupta P, Verma YK, et al. Mesenchymal stem cells: molecular targets for tissue engineering. Stem Cells Dev. 2007;16:7–23. doi: 10.1089/scd.2006.9998. [DOI] [PubMed] [Google Scholar]
- Sheikh AM, Nagai A, Wakabayashi K, Narantuya D, Kobayashi S, Yamaguchi S, et al. Mesenchymal stem cell transplantation modulates neuroinflammation in focal cerebral ischemia: contribution of fractalkine and IL-5. Neurobiol Dis. 2011;41:717–724. doi: 10.1016/j.nbd.2010.12.009. [DOI] [PubMed] [Google Scholar]
- Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Silva WA, Jr, Covas DT, Panepucci RA, Proto-Siqueira R, Siufi JLC, Zanette DL, et al. The profile of gene expression of human marrow mesenchymal stem cells. Stem Cells. 2003;21:661–669. doi: 10.1634/stemcells.21-6-661. [DOI] [PubMed] [Google Scholar]
- Song H, Chang W, Song BW, Hwang KC. Specific differentiation of mesenchymal stem cells by small molecules. Am J Stem Cell. 2012;1:22–30. [PMC free article] [PubMed] [Google Scholar]
- Specht KM, Shokat KM. The emerging power of chemical genetics. Curr Opin Cell Biol. 2002;14:155–159. doi: 10.1016/s0955-0674(02)00317-4. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Toghraie FS, Chenari N, Gholipour MA, Faghih Z, Torabinejad S, Dehghani S, et al. Treatment of osteoarthritis with infrapatellar fat pad derived mesenchymal stem cells in rabbit. Knee. 2011;18:71–75. doi: 10.1016/j.knee.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Tormin A, Li O, Brune JC, Walsh S, Schutz B, Ehinger M, et al. CD146 expression on primary nonhematopoietic bone marrow stem cells is correlated with in situ localization. Blood. 2011;117:5067–5077. doi: 10.1182/blood-2010-08-304287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremain N, Korkko J, Ibberson D, Kopen GC, DiGirolamo C, Phinney DG. MicroSAGE analysis of 2,353 expressed genes in a single cell-derived colony of undifferentiated human mesenchymal stem cells reveals mRNAs of multiple cell lineages. Stem Cells. 2001;19:408–418. doi: 10.1634/stemcells.19-5-408. [DOI] [PubMed] [Google Scholar]
- Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol. 2008;8:726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- Uemura E. Age-related changes in neuronal RNA content in rhesus monkeys (Macaca mulatta) Brain Res Bull. 1980;5:117–119. doi: 10.1016/0361-9230(80)90182-3. [DOI] [PubMed] [Google Scholar]
- Wagner W, Wein F, Seckinger A, Frankhauser M, Wirkner U, Krause U, et al. Comparative characteristics of mesenchymal stem cells from human bone marrow, adipose tissue, and umbilical cord blood. Exp Hematol. 2005;33:1402–1416. doi: 10.1016/j.exphem.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Wagner W, Feldmann RE, Jr, Seckinger A, Maurer MH, Wein F, Blake J, et al. The heterogeneity of human mesenchymal stem cell preparations—evidence from simultaneous analysis of proteomes and transcriptomes. Exp Hematol. 2006;34:536–548. doi: 10.1016/j.exphem.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Wernig M, Lengner CJ, Hanna J, Lodato MA, Steine E, Foreman R, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wislet-Gendebien S, Laudet E, Neirinckx V, Alix P, Leprince P, Glejzer A, et al. Mesenchymal stem cells and neural crest stem cells from adult bone marrow: characterization of their surprising similarities and differences. Cell Mol Life Sci. 2012a;69:2593–2608. doi: 10.1007/s00018-012-0937-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wislet-Gendebien S, Poulet C, Neirinckx V, Hennuy B, Swingland JT, Laudet E, et al. In vivo tumorigenesis was observed after injection of in vitro expanded neural crest stem cells isolated from adult bone marrow. PLoS One. 2012b;7:e46425. doi: 10.1371/journal.pone.0046425. [Electronic Resource] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Ding S, Ding Q, Gray NS, Schultz PG. A small molecule with osteogenesis-inducing activity in multipotent mesenchymal progenitor cells. J Am Chem Soc. 2002;124:14520–14521. doi: 10.1021/ja0283908. [DOI] [PubMed] [Google Scholar]
- Wu X, Walker J, Zhang J, Ding S, Schultz PG. Purmorphamine induces osteogenesis by activation of the hedgehog signaling pathway. Chem Biol. 2004;11:1229–1238. doi: 10.1016/j.chembiol.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Xu Y, Shi Y, Ding S. A chemical approach to stem-cell biology and regenerative medicine. Nature. 2008;453:338–344. doi: 10.1038/nature07042. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Major MB, Takanashi S, Camp ND, Nishiya N, Peters EC, et al. Small-molecule synergist of the Wnt/beta-catenin signaling pathway. Proc Natl Acad Sci U S A. 2007;104:7444–7448. doi: 10.1073/pnas.0702136104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MJ, Sun JJ, Qian L, Liu Z, Zhang Z, Cao W, et al. Human umbilical mesenchymal stem cells enhance the expression of neurotrophic factors and protect ataxic mice. Brain Res. 2011;1402:122–131. doi: 10.1016/j.brainres.2011.05.055. [DOI] [PubMed] [Google Scholar]
- Zhong JF, Chen Y, Marcus JS, Scherer A, Quake SR, Taylor CR, et al. A microfluidic processor for gene expression profiling of single human embryonic stem cells. Lab Chip. 2008a;8:68–74. doi: 10.1039/b712116d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong JF, Feng Y, Taylor CR. Microfluidic devices for investigating stem cell gene regulation via single-cell analysis. Curr Med Chem. 2008b;15:2897–2900. doi: 10.2174/092986708786848721. [DOI] [PubMed] [Google Scholar]
- Zhu S, Lu Y, Zhu J, Xu J, Huang H, Zhu M, et al. Effects of intrahepatic bone-derived mesenchymal stem cells autotransplantation on the diabetic beagle dogs. J Surg Res. 2011;168:213–223. doi: 10.1016/j.jss.2009.10.008. [DOI] [PubMed] [Google Scholar]