Abstract
T cell immunoglobulin and mucin domain (TIM) proteins are cell-surface signaling receptors in T cells and scavenger receptors in antigen-presenting cells and kidney tubular epithelia. Here, we demonstrated a function for TIM proteins in mediating the degradation of NUR77, a nuclear receptor implicated in the cellular response to injury. TIM proteins interacted with and mediated the lysosomal degradation of NUR77 in a phosphoinositide 3-kinase–dependent pathway. We also showed dynamic cycling of TIM-1 to and from the cell surface through clathrin-dependent constitutive endocytosis. Blocking this process or mutating the phosphatidylserine-binding pocket in TIM-1 abrogated TIM-1–mediated degradation of NUR77. In an in vitro model of kidney injury, silencing TIM-1 increased NUR77 abundance and decreased epithelial cell survival. These results show that TIM proteins may affect immune cell function and the response of the kidney to injury.
Introduction
The members of the TIM (T cell immunoglobulin and mucin domain protein) family including TIM-1, TIM-3 and TIM-4 are conserved in mice and human and are associated with or implicated in several important immunological processes, including T cell proliferation (1), T cell survival (2), tissue inflammation (3) and atopy (4). TIM-1, the prototypical member of TIM family was initially cloned as a receptor for the hepatitis A virus (5). This protein is also called kidney injury molecule-1 (KIM-1) and shows increased abundance in renal tubular epithelial cells following injury (6) and serves as a biomarker of the severity of kidney injury (7). TIM-1 was also identified within the airway hypersensitivity loci by a positional cloning approach (4) and polymorphisms in human TIM-1 confer susceptibility to asthma and atopy (8–9). Members of the TIM family share common structural motifs namely extracellular IgV and mucin domains, a hydrophobic transmembrane domain, and a short cytoplasmic tail; however high identity of TIM family proteins at the amino acid level is found only in their extracellular IgV domain.
Although TIM proteins are best characterized as immune cell signaling receptors, identification of new ligands have offered unique insights into diverse biological functions of these proteins. TIM-4 is distributed widely on antigen presenting cells, and interacts with TIM-1 and fosters T cell activation (10). Murine TIM-2, which does not have a conserved homolog in humans, binds to H-ferritin and facilitates its uptake (11). Galactin-9, a protein present on antigen presenting cells and endothelial cells, binds to TIM-3 on activated Th1 cells. The resulting ligation of TIM-1 results in a pro-apoptotic signal, thereby restricting the number of activated Th1 cells, thus mediating T cell homeostasis (12). The IgV domain of TIM-1 and TIM-4 binds to phosphatidylserine (PS) present on the outer leaflet of cells that are undergoing apoptosis, resulting in their engulfment (13–14) and hence a major role of TIM proteins is to clear apoptotic cells during renal injury and immune surveillance (13, 15).
We set out to identify additional ligands for TIM family proteins as a means to further elucidate the biology of this family. We identified the nuclear orphan receptor NUR77 as a ligand of all three human TIM proteins (TIM-1, −3 and −4). NUR77 [also known as NGFI-B (Nerve growth factor inducible-B), TR3 (Thyroid hormone receptor 3) and NR4A1 (Nuclear receptor subfamily 4 group A member 1)], is an immediate early gene induced by serum, nerve growth factor and other stimuli and it regulates cell proliferation, differentiation, survival and death (16–17). Various reports have revealed the Janus face of NUR77, as an effector of cell survival in TNF pathway (18) and mitogenic effector in cancer cells (19) on one side and as a pro-apoptotic molecule mediating cell death during thymic selection (17) and in lung cancer cells on the other side (20).
We found that the interaction between TIM proteins and NUR77 resulted in the degradation of NUR77 through a lysosomal-dependent pathway. Furthermore, we showed that TIM-1 was constitutively endocytosed and dynamic cycling of TIM-1 through clathrin-dependent vesicles was essential for the targeting of NUR77 for degradation in lysosomes. Moreover, the interaction between TIM-1 and NUR77 in renal tubular epithelial cells confers protection against apoptosis in an in vitro epithelial cell injury model. TIM-mediated regulation of is likely to influence cell survival in various cell types, because the transcriptional activity of NUR77, as well as its translocation to mitochondria (21–22), promotes cellular apoptosis in multiple physiological systems including T cell clonal selection (23) and acute kidney injury (24).
Results
NUR77 is a binding partner of TIM proteins
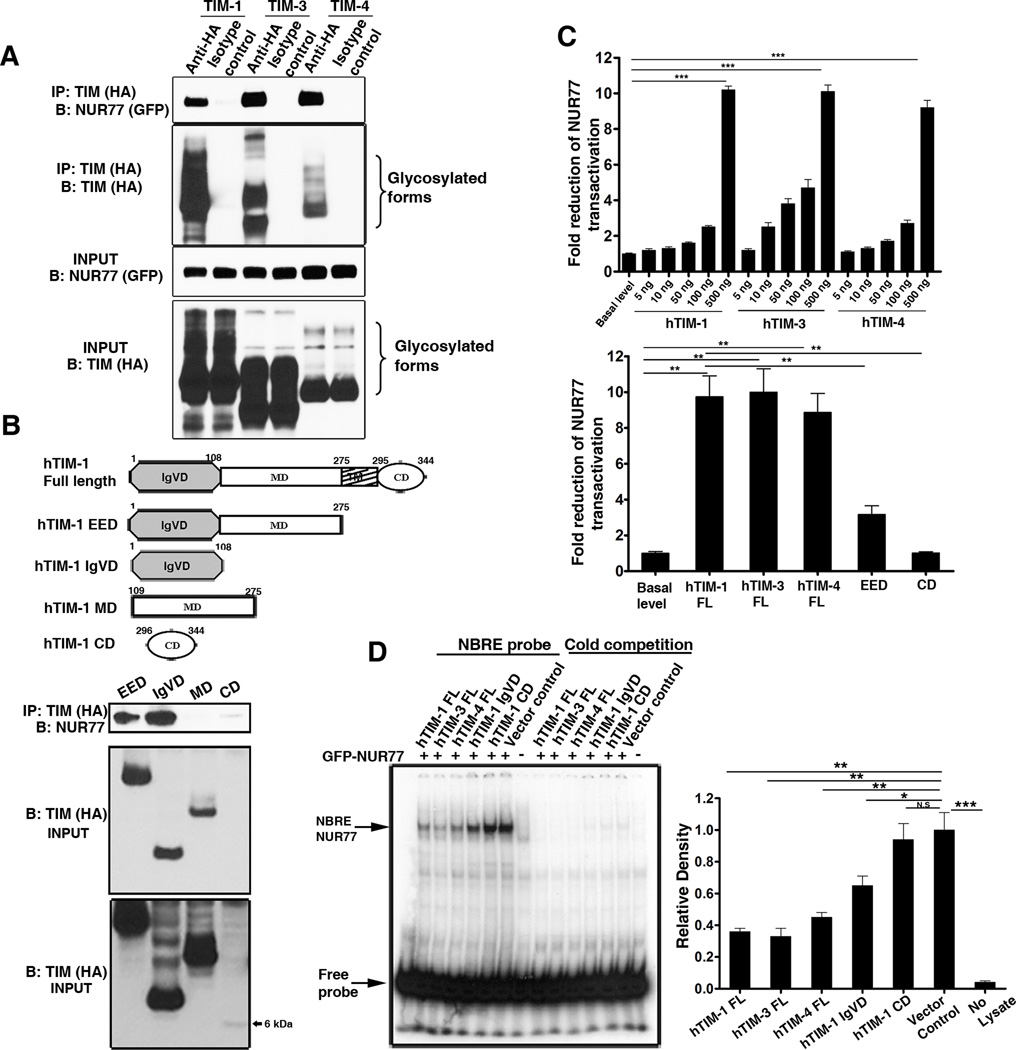
To identify TIM-1 ligands, we performed a yeast two hybrid screening of human spleen cDNA library using the IgV domain of human TIM-1 as bait and identified NUR77 and several other candidates as ligands for TIM-1 (Table S1). Because of the important role of NUR77 in altering the balance between cell survival and death, we selected this candidate for further evaluation. We validated the interaction between TIM-1 and NUR77 in coimmunoprecipitation experiments and determined that TIM-3 and TIM-4 also interacted with NUR77 (Figure 1A). Immunoprecipitation assays using deletion constructs of TIM-1 and NUR77 revealed that the IgV domain of TIM-1 and the ligand binding domain of NUR77 were necessary and sufficient to mediate this interaction (Figures 1B and S1).
Figure 1. TIM family proteins interact with NUR77 and repress its transactivation function.
(A) All three members of human TIM family interact with NUR77 as shown by coimmunoprecipitation assay. N=3 independent experiments (B) Top panel: Domain architecture of TIM-1. Middle Panel: Coimmunoprecipitation experiment demonstrating that only the entire extracellular domain (EED) and IgV domain (IgVD) of TIM-1 interact with NUR77 and not the mucin (MD) or the cytoplasmic domain (CD). Bottom panel: long exposure of the input blot showing the CD protein band. N=2 independent experiments. (C) The transactivation function of NUR77 is repressed by all three human TIM proteins in a dose dependent manner (Top panel). Full length TIM-1 protein is required for repression of NUR77 transcriptional activity (Bottom panel). Data from at least three independent experiments is presented. (D) Binding of NUR77 to its response elements (NBRE) is abrogated by the full length TIM proteins in an electrophoretic mobility shift assay using p32 labeled NBRE oligo. Competition with unlabelled probe (cold competition) reveals the specificity of NUR77 binding. A representative autoradiograph is shown (left) and the relative amount of NUR77 protein binding to NBRE probe was quantified from three independent experiments (right). ***, P<0.001, **, P<0.01, *,P<0.05.
TIM proteins inhibit the transcriptional activity of NUR77
The transactivation function of NUR77 is essential for its pro-apoptotic and pro-survival functions (18, 25) and is mediated by its binding to the NGFI-B response element (NBRE) or the Nur response element (NuRE) in the promoters of its target genes (26). We used a NBRE-driven luciferase reporter assay to probe the effects of full length TIM proteins or individual domains of TIM-1 on NUR77-directed transactivation. The transactivation function of NUR77 was significantly impaired in cells co-expressing full length TIM proteins (Figure 1C). In the presence of full length TIM proteins, NUR77 binding to NBRE DNA elements was significantly decreased as indicated by electrophoretic mobility shift assays (EMSA) (Figure 1D). Although binding of the IgV domain and full length TIM-1 to NUR77 appeared equivalent in immunoprecipitation experiments, expression of the IgV or the entire extracellular domains of TIM-1 alone did not impede either the transactivation or DNA binding ability of NUR77. These results suggested that TIM-1 inhibits the transcriptional activity of NUR77 in a manner that requires both the transmembrane and cytoplasmic domains.
TIM proteins mediate NUR77 protein degradation and impact cell survival
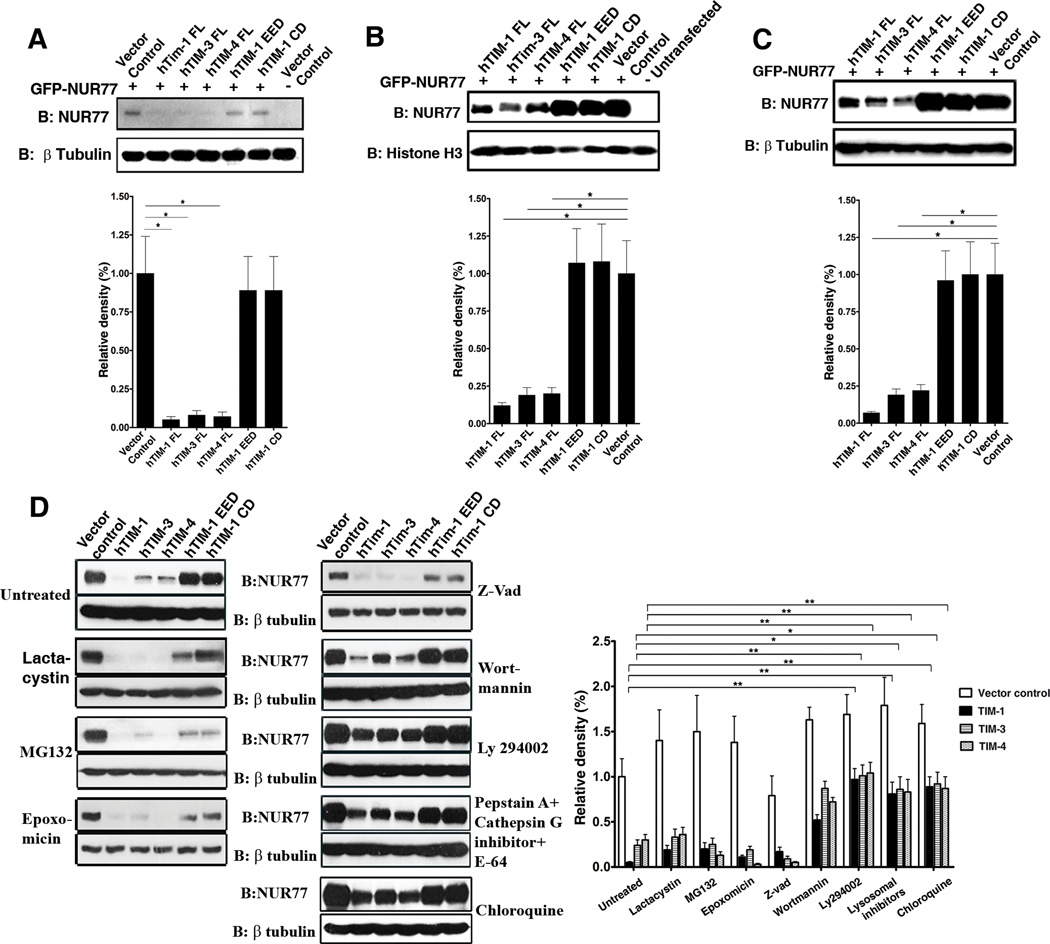
To further investigate the mechanism by which TIM proteins inhibit the transcriptional activity of NUR77, we analyzed the abundance of NUR77 protein in the lysates used for transactivation and EMSA assays. The abundance of NUR77 protein was decreased when full length, but not truncated TIM proteins, were coexpressed, both in the total cell lysates from the luciferase assay (Figure 2A) and the nuclear extracts from the EMSA assay (Figure 2B). The cellular abundance of NUR77 protein was decreased in the presence of TIM family proteins in several independent experiments using myc tagged or GFP tagged NUR77 (Figure 2C). Moreover, TIM-1-mediated modulation of cellular protein homeostasis was specific to NUR77 because the protein abundance of GFP was not altered by TIM proteins (Figure S2) and was not due to promoter interference or squelching because NUR77 mRNA abundance was equal among the individual samples (Figure S3). These results demonstrated modulation of NUR77 by TIM proteins at the protein level.
Figure 2. TIM family proteins mediate specific protein degradation of NUR77 in a lysosomal-dependent manner.
(A and B) Western blot analysis (top panels) of the protein lysates from the luciferase assay (A) and EMSA (B) showing the reduced protein abundance of NUR77 specifically in the presence of full length TIM proteins. Bottom panels: Quantitation of protein abundance from three independent experiments. (C) Representative western blot (top panel) showing that the protein abundance of NUR77 in total cell lysates is altered by the presence of full length TIM proteins. Bottom panel: Quantitation of protein abundance from three independent experiments. (D) TIM-mediated degradation of NUR77 is not affected by specific inhibitors of ubiquitin proteasome pathway (Lactacystin, MG132, or Epoxomycin) (Left panel) or by the pan-caspase inhibitor z-vad (right panel). It is abrogated by the PI3K inhibitor Ly294002, a lysosomal protease inhibitor cocktail (Pepstatin A, Cathepsin inhibitor I and E-64) and chloroquine (Right panel). A representative western blot is shown for each treatment and relative densities of the protein abundance were calculated from three independent experiments. **, P<0.01, *,P<0.05 and N.S, Not significant.
NUR77 protein abundance was not restored by various specific inhibitors of ubiquitin-proteasome machinery (27) (Figure 2D, left panel) or by the pan-caspase inhibitor z-vad. In contrast, specific inhibitors of the lysosomal protein degradation pathway partially restored NUR77 protein abundance (Figure 2D, right panel). In addition, TIM protein-mediated degradation of NUR77 was partially inhibited by the phosphoinositide 3-kinase (PI3K) inhibitor Ly294002. Taken together, these results suggest that TIM proteins bind NUR77 and target its lysosomal degradation through a PI3K–dependent pathway.
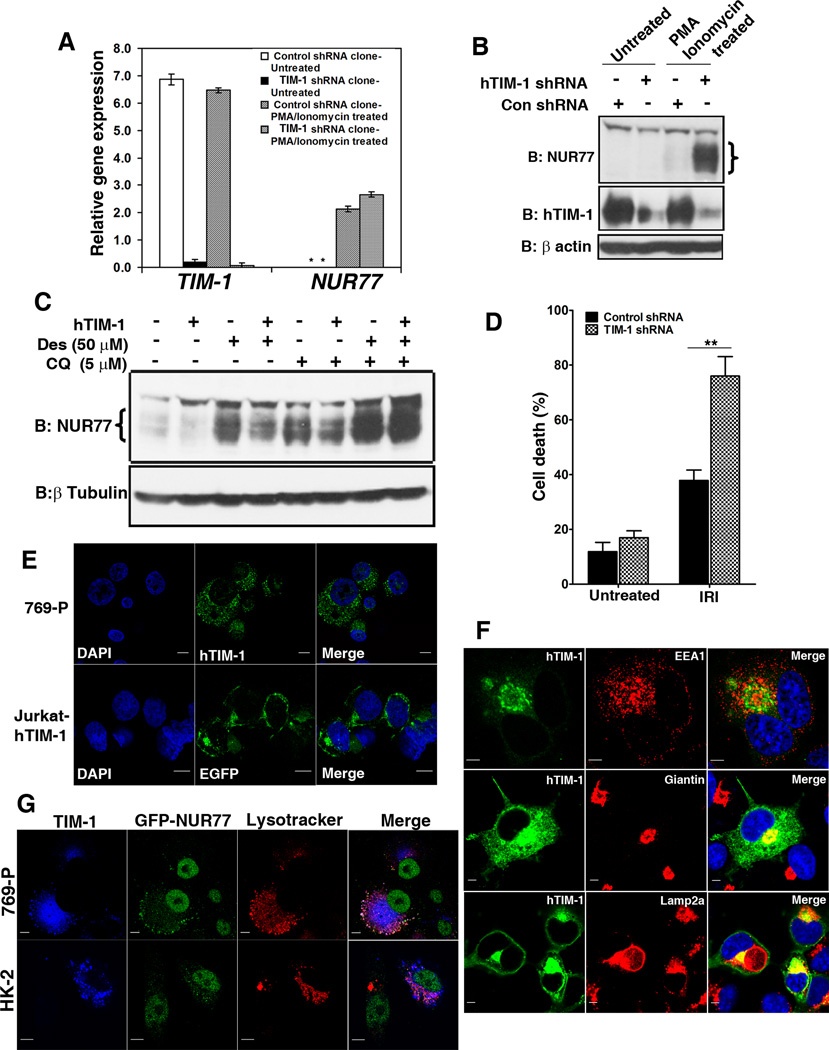
To investigate whether TIM mediated degradation of NUR77 occurs with endogenous TIM-1 and NUR77, we induced NUR77 expression using the protein kinase C activator PMA and the calcium ionophore ionomycin (28) in control and TIM-1 shRNA knockdown lines of 769-P renal cells. Although the induction of NUR77 mRNA was equal between the control and knockdown cell lines (Figure 3A), NUR77 protein abundance was increased in the absence of TIM-1 thus demonstrating TIM-1-dependent modulation of NUR77 protein abundance (Figure 3B). Similarly, overexpression of TIM-1 in the human kidney tubular epithelial cell line HK-2 resulted in a substantial decrease in NUR77 protein abundance induced by treatment of cells with desferrioxamine (Figure 3C, lanes 3 and 4), an iron chelator that induces expression of NUR77 (29). We restored the abundance of NUR77 protein by treating cells with chloroquine, a lysosomotrophic agent that perturbs lysosomal acidification (Figure 3C, lanes 7 and 8). These experiments proved that TIM-1-dependent degradation of NUR77 protein requires lysosomes and occurs endogenously in cells such as renal tubular epithelial cells.
Figure 3. Biology of the TIM-1-NUR77 interaction.
(A) Quantitative RT-PCR measurement of NUR77 transcript induced by PMA and ionomycin treatment in control and TIM-1 shRNA stable knockdown clones of 769-P cells (N=3 experiments). (B) Western analysis of the protein samples from (A) indicating the decrease in NUR77 abundance in the presence of TIM-1. Con, control. N= 3 independent experiments. (C) Overexpression of TIM-1 in HK-2 cells resulted in decreased NUR77 protein abundance induced by desferrioxamine (Des) (compare lanes: 3 and 4), which is perturbed by chloroquine treatment (CQ) (Lanes: 7 and 8). N=3 independent experiments. (D) Silencing of TIM-1 expression in HK-2 cells resulted in increased cell death in an in vitro epithelial cell injury model induced by a combination of ATP and glucose depletion and calcium overload. N= 3 independent experiments. IRI, Ischemia reperfusion injury (E) Confocal microscopy analysis of the localization pattern of endogenous hTIM-1 (green) in 769-P cells (Top panel) and GFP-tagged hTIM-1 stably expressed in Jurkat cells (Bottom panel). Scale bar, 10 µm. N=2 experiments. (F) Colocalization of ectopically expressed TIM-1 (green) in Cos-7 cells with markers of early endosomes (EEA1), Golgi complex (Giantin), and lysosomes (Lamp2a). Scale bar, 5 µm. Colocalization coefficients: hTIM-1/EEA1:0.34±0.06; hTIM-1/Giantin: 0.74±0.04; hTIM-1/Lamp2a: 0.55±0.08; n=5 cells each from two independent experiments (G). Colocalization of TIM-1 (blue) and ectopically expressed NUR77 (green) with lysosomes (Lysotracker red) in 769-P cells. Colocalization coefficients: hTim-1/NUR77: 0.59±0.12; hTIM-1/Lysotracker: 0.47±0.09; NUR77/Lysotracker: 0.51±0.08; and HK-2 cells; Colocalization coefficients: hTim-1/NUR77: 0.52+−.06; hTIM-1/Lysotracker: 0.56+0.08; NUR77/Lysotracker: 0.62+0.07; n=5 cells each from two independent experiments. Scale bar, 10 µm.
Further, we tested the biological importance of TIM-1 mediated depletion of NUR77, using an in vitro renal epithelial cell injury model through a combination of ATP and glucose depletion (antimycin A and 2-deoxyglucose) superimposed with calcium overload (A23187), an effect that has been previously reported to mimic the effects of ischemic renal injury (30). We have previously demonstrated that NUR77 protein abundance is rapidly and robustly increased in renal tubular epithelial cells in vivo during acute renal injury as well as in HK-2 cells after in vitro injury (24). Genetic ablation or pharmacological blockade of NUR77 induction results in reduced epithelial cell death and inflammation and confers protection against renal injury in mice, and increased NUR77 protein abundance correlates with increased renal epithelial cell apoptosis (24). TIM-1 protein abundance is also increased during renal injury (6). The biological function of TIM-1 in renal epithelial cells, beside enhancing their ability to take up dead cells (15), is not fully understood. In our in vitro epithelial cellular injury model, NUR77 transcript is induced in HK-2 cells, which also have TIM-1. Because sustained NUR77 protein abundance results in cell death, we tested the hypothesis that silencing TIM-1 would exacerbate cell death arising from increased NUR77 protein abundance. Silencing TIM-1 in HK-2 cells enhanced apoptosis as compared to control shRNA cells in this injury model (Fig. 3D). This increase in apoptosis was associated with increased abundance of NUR77 protein but not transcript (Figure S4), suggesting that TIM-1 mediated protection occurs at least in part by decreasing the abundance of NUR77. These data indicates that reduced TIM-1 abundance in injured HK-2 cells results in increased abundance of NUR77 protein and cell death.
TIM-1 exists in a large intracellular pool
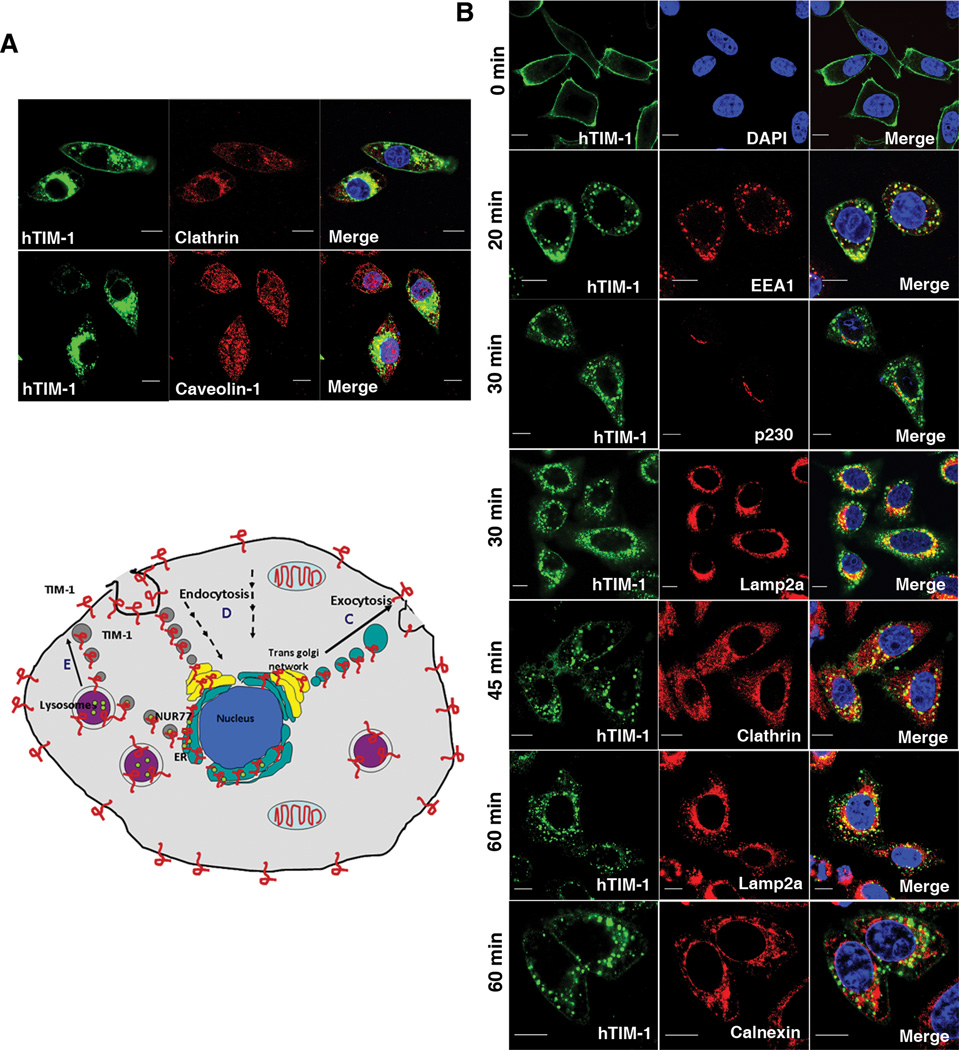
Because TIM-1 is a plasma membrane protein, its involvement in intracellular protein degradation was surprising. We analyzed the localization pattern of endogenous TIM-1 protein in 769-P cells by immunofluorescence microscopy using an antibody directed against the TIM-1 extracellular domain. In addition to its presence on the cell surface, substantial intracellular pools of endogenous TIM-1 proteins were evident within 769-P renal cancer cells and in Jurkat T cells stably transfected with full length TIM-1 (Figure 3E). Intracellular TIM-1 colocalized with markers of early endosomes and lysosomes but not with a marker for mitochondria (Figure S5). Ectopically expressed TIM-1 was also found in the Golgi complex, early endosome and lysosomal compartments in COS-7 cells (Figure 3F). TIM-1 colocalized with NUR77 in lysosomes in HK-2 cells (Fig 3G). These results reveal the presence of large intracellular pools of TIM-1 in various sub-cellular compartments which we reasoned may participate in the process leading to the degradation of specific target proteins.
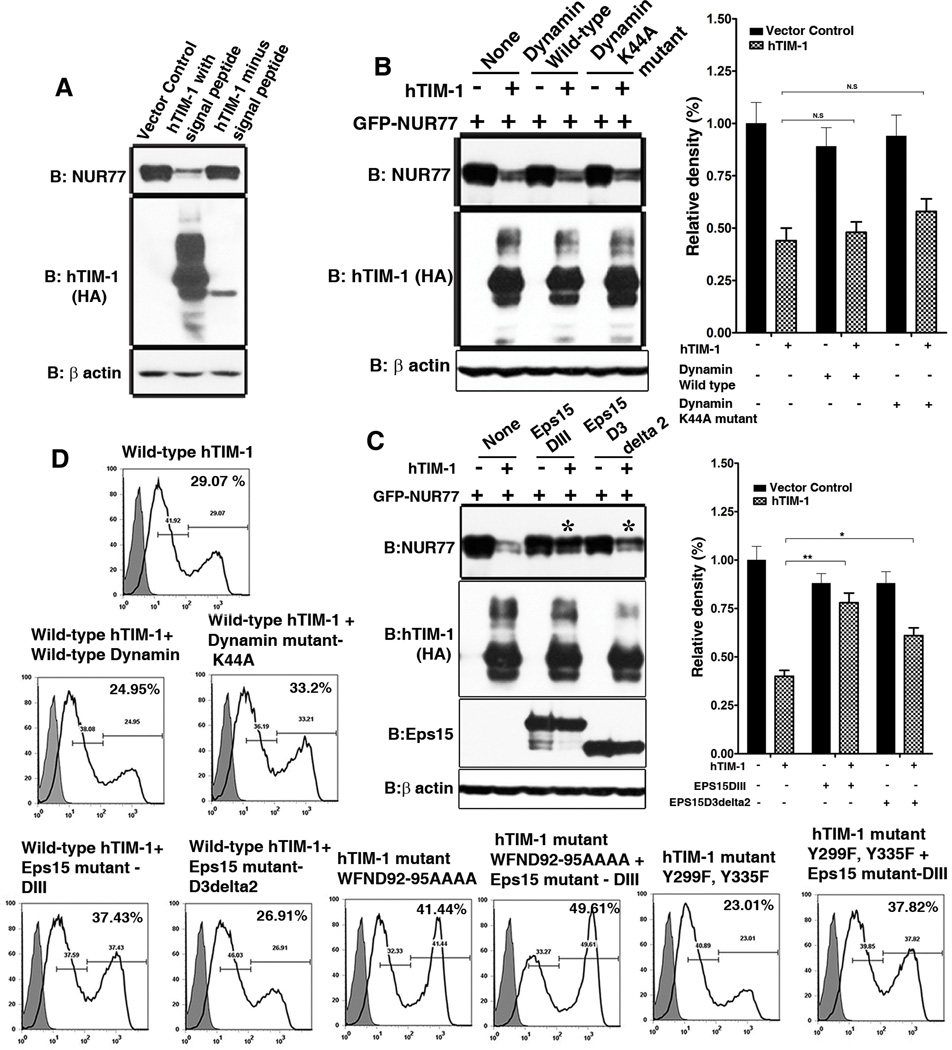
The Metal-ion-dependent Ligand Binding Site of TIM-1 is essential for TIM-1-mediated degradation of NUR77
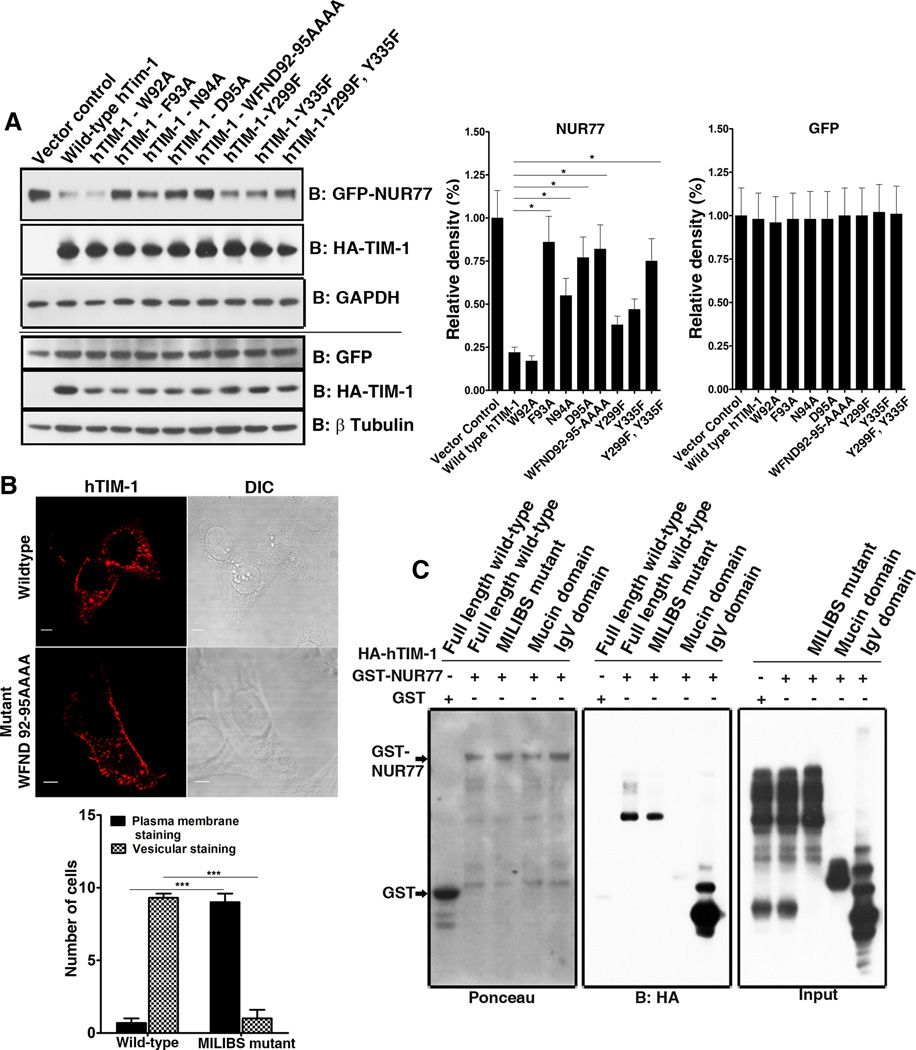
Depending on the strength of their binding to PS, certain membrane proteins are localized in intracellular endosome and lysosomal compartments through binding of PS in the organellar membranes (31). Both TIM-1 and TIM-4 bind to PS through the metal ion dependent ligand binding site (MILIBS) in the conserved CC’FG loop in their IgV domains (32), a sequence that is also conserved in human TIM-3 (Figure S6) and that mediates the uptake of apoptotic bodies by TIM proteins. We reasoned that the MILIBS might determine the cellular distribution of TIM proteins and therefore the ability to mediate intracellular protein degradation. To determine whether the MILIBS contributes to TIM-1-mediated degradation of NUR77, four amino acids (Trp92 to Asp95) lining this pocket were substituted with alanine, both individually and collectively. Substituion of all four alanine substitutions (WFND92–95AAAA) compromises the binding of TIM-4 to PS on the surface of apoptotic bodies (13, 32). Except for the W92A substitution, individual or collective substitutions in the MILIBS pocket attenuated the ability of TIM-1 to mediate NUR77 degradation to varying degrees (Figure 4A). These results suggest that TIM proteins could bind PS in intracellular membranes to facilitate increased retention of TIM proteins in subcellular compartments which in turn could enable TIM proteins to specifically target binding partners for degradation. We observed partial loss of NUR77 degradation in cells expressing forms of TIM-1 with mutations in cytoplasmic tyrosine residues Y299F and Y335F of TIM-1 (Figure 4A). This is in agreement with the loss-of-function of cytoplasmic domain deletion constructs of TIM-1 in mediating NUR77 degradation (Figures 1 and 2) and point to the general requirement of cytoplasmic domain in this phenomenon.
Figure 4. Determinants of TIM-1 mediated NUR77 degradation: Contribution of MILIBS and cytoplasmic domain of TIM-1 and clathrin-mediated endocytic pathway.
(A) Left panel: Mutations in the MILIBS of TIM-1 abrogated TIM-1-mediated degradation of NUR77 protein (top). The double mutant of cytoplasmic tyrosine residues Y299 and Y335 partially restored the protein abundance of NUR77, but did not affect that of GFP (bottom). Right panel: Quantitation of NUR77 and GFP abundance in the presence of wild type and mutant TIM-1 based on three independent experiments. (B) Confocal microscopy analysis of the localization pattern of wild type, MILIBS, cytoplasmic tyrosine residue mutants of TIM-1, revealing increased cell surface localization of the MILIBS TIM-1 mutants. Scale bar, 5 µm (Left). Quantification of the localization of wild-type TIM-1 and the MILIBS mutant. N=10 cells each from three independent experiments (Right). (C) Inability of MILIBS mutants to mediate NUR77 degradation does not arise from lack of interaction with NUR77: Wild type and MILIBS mutant TIM-1 interact with NUR77 with equal efficiency demonstrated by GST pull down assay (Middle panel). IgV and mucin domains served as positive and negative controls of the interaction between TIM-1 and NUR77. Shown is a representative of three independent experiments.
Furthermore, when expressed in HeLa cells, the TIM-1 MILIBS mutant exhibited increased surface membrane staining compared to wild type TIM-1 protein (Figure 4B), which agrees with an earlier study that described the cell surface localization of MILIBS domain mutants of TIM-1 in pre-B cells (32). This reinforces the notion that binding of TIM proteins to PS in the membranes of subcellular organelles might facilitate their intracellular localization, as has been previously reported for other PS binding proteins (31). This intracellular localization appear essential for the intracellular function of TIM-1 in mediating NUR77 protein degradation, which is abrogated in the PS-binding mutants of TIM-1 that also exhibits decreased intracellular retention and increased cell surface localization. In addition, the inability of MILIBS mutant TIM-1 to mediate NUR77 degradation arose largely from altered cellular localization but not loss of interaction with NUR77 (Figure 4C). In a GST pull down assay, both wild-type TIM-1 and the MILIBS mutant interacted with GST tagged NUR77 with equal efficiency (Figure 4C, middle panel).
TIM-1 protein trafficking through clathrin coated vesicles is crucial for TIM-1-mediated protein degradation
We wanted to determine whether TIM-1 trafficked between various subcellular compartments or remained in distinct static pools such as cell surface or intracellular pools. We stalled the transit of either the newly synthesized TIM-1 from the Golgi complex to the cell surface by removing the signal peptide or the endocytic routing of cell surface TIM-1 to cytoplasm by specifically perturbing receptor endocytosis. The signal peptide deleted TIM-1 mutant protein failed to undergo glycosylation; showed reduced ability to mediate NUR77 degradation (Figure 5A); and did not appear to enter the secretory system or the vesicles en route to the plasma membrane (Figure S7). These results raised the possibility that glycosylation or a prior targeting of TIM-1 to the cell surface through secretory pathway might be required to mediate the specific degradation of NUR77. This prompted us to examine whether TIM-1 dynamically cycles to and from the cell surface and identify the cellular components of the protein trafficking system required for the TIM-1–dependent degradation of NUR77. To this end, we specifically perturbed the functions of dynamin-2, a GTPase that mediates vesicle scission in both caveolae- and clathrin-mediated endocytic pathways (33), and Eps15, which dictates clathrin coated pit formation (34). The ability of TIM-1 to mediate degradation of NUR77 was significantly impaired by expression of the dominant negative construct of Eps15, DIII, but not of the dominant negative dynamin construct Dyn2K44A (Figure 5, B and C). Hence, following synthesis and glycosylation, TIM-1 is targeted to the cell surface and constitutively traffics to and from the cell surface to the sub-cellular compartments through clathrin-mediated vesicles, which is essential for TIM-1 mediated specific protein degradation.
Figure 5. Role of constitutive TIM-1 trafficking in NUR77 degradation.
(A) A form of TIM-1 lacking the signal peptide deleted TIM-1 is not glycosylated and unable to mediate degradation of NUR77. N= 2 independent experiments. (B) Perturbation of endocytic pathway using a dominant negative Dynamin construct (Dyn2K44A) did not affect TIM-1 mediated degradation of NUR77. N= 3 independent experiments (Left panel) and quantitation of protein abundance based on three independent experiments (Right panel). (C) Dominant negative constructs of Eps15 (DIII and D3delta2) substantially abrogated TIM-1 mediated degradation of NUR77. N=3 independent experiments (left panel). NUR77 protein abundance was quantified based on three independent experiments (right panel). (D) TIM-1 is constitutively endocytosed as revealed by the increased cell surface localization of transiently transfected TIM-1 in 293T cells, upon blockade of endocytosis using dominant negative constructs of dynamin-2 and Eps15 as assayed by flow cytometry. Perturbation of clathrin vesicle formation further enhanced the increased cell surface localization of the MILIBS mutant. Shaded histogram represents the control antibody staining and the open histogram represents hTIM-1 staining. Two peaks of TIM-1 staining reveal the dynamic cycling of TIM-1. N= 2 independent experiments.
The constitutive endocytosis of TIM-1 may also explain the ability of the PI3K inhibitor Ly294002 to rescue the protein abundance of NUR77 (Figure 2D) and the inability of the cytoplasmic domain deletion construct (hTIM-1-EED) (Figure 2A, B and C) and the cytoplasmic tyrosine residue mutant (hTIM-1 Y299F, Y335F) TIM-1 (Figure 4A) to mediate NUR77 degradation. The potency of PI3K inhibitors in blocking PI3K associated receptor endocytosis as well as sorting events has been previously documented (35–36). However, the differential effects of the PI3K inhibitors Wortmannin and Ly294002 (Figure 2D) on TIM-1-dependent NUR77 degradation required further exploration. As reported earlier (37–38), the p85 subunit of PI3K binds to the cytoplasmic domain of human TIM-1. Along these lines, the inability of cytoplasmic tyrosine TIM-1 mutants to mediate NUR77 degradation (Figure 4A) further suggests that physical association of TIM-1 and p85 subunit of PI3K might be essential for endocytosis of TIM-1. In the ensuing process, PI3K apparently influences the endocytic and sorting events and signaling downstream of PI3K seemed to not be involved because stable knockdowns of Akt1 and Akt 2 isoforms did not appear to block TIM-1-mediated degradation of NUR77 (Figure S8).
Constitutive endocytosis and retrograde translocation of TIM-1
We confirmed the constitutive intracellular trafficking of TIM-1 by showing increased surface localization of the MILIBS TIM-1 mutant (Figure 5D: compare wild-type and WFND92–95AAA mutant hTIM-1) and wild-type TIM-1 in the cells in which clathrin-mediated endocytosis was stalled using dominant negative Eps15 constructs (Figure 5D: compare wild type hTIM-1 with and without Eps15 mutants DIII. Compared to the Eps15 mutant DIII, D3delta2 is apparently not potent in altering the endocytosis of TIM-1 in the settings that we studied. Although the perturbation of endocytosis using dominant negative dynamin construct (Dyn2K44A) did not diminish TIM-1 mediated degradation of NUR77 (Figure 5B), the surface localization of TIM-1 appeared to be enhanced (Figure 5D: compare wild type hTIM-1 with and without wild type or mutant dynamin K44A). This differential effect of the Dyn2K44A mutant agrees with the findings that dynamin mutants affect receptor endocytosis from the plasma membrane, but not the vesicle budding from trans-Golgi network (39). Also, although the TIM-1 tyrosine mutant did not by itself exhibit higher cell surface localization, increased cell surface retention was observed upon perturbation of clathrin-dependent endocytosis (Figure 5D, compare wild type and Y299F–Y33F mutant hTIM-1 with and without EPS15 mutant-DIII). Taken together, these data reveal that TIM-1 mediated degradation of NUR77 is a complex process that might be regulated at multiple levels including receptor endocytosis, intracellular vesicle traffic, endosomal sorting and possibly traffic through trans-Golgi. The predominant role of clathrin-mediated trafficking of TIM-1 was further confirmed by colocalization of TIM-1 with clathrin coat protein (Figure 6A).
Figure 6. Retrograde translocation of TIM-1.
(A) Confocal immunofluorescence analysis of TIM-1 (green) colocalization with Clathrin (Top panel) and Caveolin-1 (Bottom panel) where yellow indicates colocalization. Scale bar, 10 µm. Colocalization coefficients: hTIM-1/Clathrin: 0.68±0.05; hTIM-1/Caveoli: 0.12±0.04; n=5 cells each from two independent experiments. (B). Retrograde translocation of TIM-1(green) from cell surface to lysosomes and endoplasmic reticulum through the trans-Golgi network in a clathrin-mediated pathway demonstrated by endocytosis assay; subcellular organelle markers are in red. Shown is a representative image. Scale bar, 10 µm. Colocalization coefficients: hTIM-1/EEA1: 0.57±0.05; hTIM-1/p230: 0.35±0.08; hTIM-1/Lamp2a (30 min): 0.46±0.09; hTIM-1/Clathrin: 0.38±0.06; hTIM-1/Lamp2a (60 min): 0.55±0.03; hTIM-1/Calnexin: 0.31±0.03; n=5 cells each from two independent experiments. (C to E) Model depicting the clathrin-mediated constitutive trafficking of TIM-1 and the phenomenon of TIM-1 mediated NUR77 degradation. TIM-1 and other TIM family members (red) are targeted to cell surface (C), prior to constitutive endocytosis through clathrin-coated vesicles (D). Following fusion with the trans-golgi network, TIM-1 undergoes retrograde translocation to endoplasmic reticulum (ER) and lysosomes (purple circles). TIM-1 might come in contact with NUR77 possibly in ER or vesicles leading to recruitment of this protein complex to PS-rich endosomes and lysosomes depending on the TIM-1-PS interaction, facilitating sorting of NUR77 to lysosomes and ultimately resulting in the degradation of NUR77 by lysosomal enzymes. TIM-1 is presumably retained in the vesicles (E).
Based on the above observations, we hypothesized that TIM-1 might translocate in a retrograde fashion from the Golgi or endosomes to ER as well as lysosomes. Indeed, an interaction between TIM-1 and NUR77 at this site might be possible and result in the degradation of NUR77 through lysosomal sorting. We performed endocytosis assays in which we selectively labeled only cell surface TIM-1 with an antibody directed against the extracellular domain of TIM-1 and subsequently chased the trafficking of the antibody labeled TIM-1 at different time points. Immunofluorescence assays performed at distinct time points revealed trafficking of TIM-1 – antibody complex to various subcellular organelles including clathrin-coated vesicles, lysosomes (as identified by Lamp2a immunoreactivity), endoplasmic reticulum (as identified by calnexin immunoreactivity) and trans-Golgi network (as identified by p230 immunoreactivity) (Figure 6B). These data indicate that TIM-1 undergoes retrograde trafficking through clathrin-dependent endocytosis followed by fusion with the trans-Golgi network and offer a mechanistic explanation for the ability of cell surface TIM proteins to participate in the lysosomal degradation of an intracellular protein. There was no apparent change in the abundance of either wild type or mutant TIM proteins in the presence and absence of NUR77 during the process of mediating NUR77 degradation (Figure S9). This suggests that TIM proteins bound to NUR77 are not necessarily subjected to lysosomal degradation, but rather TIM proteins specifically deliver target protein cargo to lysosomes for degradation. Therefore a model in which TIM proteins are retained in the vesicles could be possible (Figure 6, C to E). Further we tested the possibility of incorporation of NUR77 into vesicular compartment such as autophagosomes. Under steady state conditions, NUR77 and TIM-1 colocalized with the autophagosome marker LC3 (Figure S10), thus indicating the possibility of intracellular vesicles as a site of interaction for TIM-1 and NUR77.
Discussion
TIM proteins function as cell surface signaling receptors at the plasma membrane (1–3) or phagocytic receptors (13–14). The ligands previously identified to bind IgV domain of TIM proteins so far have been extracellular or membrane bound (10–12, 40). In the present study, we identified a role of TIM family proteins in targeting the intracellular protein NUR77 for degradation and NUR77 as a ligand for the IgV domain of TIM proteins.
The ability of TIM proteins to mediate degradation of intracellular NUR77 protein requires the structural and functional integrity of the IgV domain, which harbors the MILIBS domain which binds PS. Mutations in the MILIBS domain of TIM-1 rescued NUR77 from degradation, an effect we attribute to the predominant cell surface localization of the MILIBS mutant, as opposed to largely intracellularly localized wild type TIM-1. The protein degradative function of TIM-1 utilizes a constitutive endocytosis pathway, which is principally clathrin vesicle dependent, followed by lysosomal sorting of the target protein. The routing of NUR77 protein to lysosomes does not result in the degradation of TIM-1, which suggests that TIM-1 may continue to participate in this process.
We report the functional role of a large cytoplasmic pool of TIM-1, a protein with established cell surface signaling functions. The ability of residues in the MILIBS domain to bind PS in organellar membranes might contribute to the retention of TIM-1 in endosomal- and lysosomal- compartments and might facilitate its interaction with target proteins. Our postulate regarding the contribution of PS binding site in the cellular distribution and therefore the protein degradatory function of TIM-1 was based on two previous studies. First, Santiago et al. (32) described the crystal structure of TIM-1 and TIM-4 and determined the MILIBS domain that binds PS, providing the structural basis for the cellular distribution of TIM proteins. They proposed a flip-flop model in which the extracellular domain of TIM proteins reside in the cytoplasmic side of the cell, with the MILIBS domain interacting with PS on the cytoplasmic side of the cell (32). Consequently, the extracellular domain of TIM proteins would be exposed to the extracellular milieu under certain conditions. Second, Yeung et al. (31) documented the cellular distribution of PS including vesicles and endocytic compartments and the contribution of intracellular PS in determining the cellular distribution of PS binding proteins. These two studies as well as our imaging data led us to test the role of MILIBS domain in the protein degradatory function of TIM-1. In addition, intracellular PS is essential for retrograde membrane traffic through endosomes (41). Besides supporting the intracellular trafficking and localization of TIM-1 that we observed, these studies (31–32, 41) provide further evidence for the possibility of physical proximity and therefore interaction between TIM-1, a cell surface receptor and NUR77, a nucleo-cytoplasmic protein.
Indeed the intracellular retention and constitutive endocytosis of TIM proteins could potentially regulate crucial signaling intermediates in the endosomal vesicular compartments, and therefore the outcome of cellular phenotype and function. Further, this mechanism might also regulate TIM protein localization on the cell surface and the outcome of receptor signaling upon binding to its extracellular ligands. The intracellular protein degradative function of TIM family proteins demonstrated using the candidate protein NUR77 may play fundamental roles in human diseases such as inflammation and kidney injury, conditions that are characterized by increases in the abundance of TIM family proteins and NUR77.
Materials and Methods
Yeast two hybrid screen
Yeast two hybrid screen was performed using the IgV domain of human TIM-1 (21–128 aa) cloned in pPC97 as bait and human spleen cDNA library (Kind gift from Prof. Marc Vidal) constructed in the yeast vector pPC86 as prey following the procedure as described previously (42). The interactants were selected on 3AT containing synthetic dropout medium lacking histidine. The positive clones were further checked for the expression of the nutritional markers Ura and LacZ.
Constructs
Full length human TIM-1 with an N-terminal HA tag and full length TIM-3 and TIM-4 with a C terminal HA tag were cloned in pKH3. TIM-1 deletion constructs, EED, the IgVD domain, the mucin domain, the cytoplasmic domain were cloned as HA tag fusion proteins in pKH3. The MILIBS TIM-1 mutants (W92A, F93A, N94A, D95A, and WFND92–95AAAA) and the cytoplasmic tyrosine TIM-1 mutants (Y229A, Y335A and double mutant Y229A Y335A) were constructed by overlapping PCR. Signal peptide deleted full length TIM-1 was cloned with an N-terminal HA tag. GFP-tagged full length hTIM-1 full length was cloned in pEGFPN1. Full length and deletion constructs of NUR77 were cloned in pcDNA3.1 with a C-terminal Myc tag. GFP-tagged full length wild type and NBRE luciferase reporter constructs were kind gift from Dr. Yuri Pekarasky (43). Wild type and dominant negative (Dyn2K44A) constructs of Dynamin were generous gifts of Dr. Sean Egan and GFP-tagged dominant negative constructs of Eps15: DIII and D3delta2 were kindly provided by Dr. Alexandre Benmerah. RFP-LC3 was provided by Dr. Walter Beron (Universidad Nacional de Cuyo Conicet, Argentina).
Cell lines
769-P, 293T, HeLa, Cos-7 and HK-2 cell lines were obtained from ATCC and were maintained in DMEM supplemented with 10% FBS. Stable knockdown 293T lines of Akt1 and Akt2 were kindly provided by Prof. Joan Brugge (Harvard Medical School, Boston, USA).
Coimmunoprecipitation
293T cells were cotransfected with either pEGFP-N1 or pEGFP-NUR77 along with full length or deletion constructs of TIM-1, full length TIM-3, or TIM-4. 36 h post transfection, the cells were harvested and cell lysates were prepared (10mM Tris pH 7.6, 150 mM NaCl, 0.5% NP-40, 0.5% Triton X-100, Protease inhibitor cocktail) and protein concentrations in the total cell lysate were analyzed in places indicated. For Coimmunoprecipitation, 500 µg of total cell lysate was incubated overnight with 2 µg of respective antibody; further incubated with protein A sepharose for 1h and the complex was washed three times with cell lysis buffer and the beads were processed for western blot analysis.
Western blotting and quantification of proteins
Protein concentrations were quantified using Bio-rad DC kit and equal amounts of protein samples were separated on either 8% or 4–12% polyacrylamide gel and blotted onto PVDF membrane and probed with different antibodies as indicated (Anti-human NUR77 (Pharmingen), Anti-mouse NUR77 (BD), Anti-human TIM-1 (R&D), Anti-HA (Upstate/Millipore corp), Anti-β actin (Abcam), Anti-GFP (Abcam). In most cases, the blots were stripped and probed with antibodies against house keeping proteins. Protein band intensities were quantified using the Image J software.
Inhibitor treatment
293T cells were cotransfected with TIM (full length or deletion constructs) and NUR77 constructs and 24 h post transfection, the cells were treated for 24 h with various chemical inhibitors including 50 µM Ly294002 (Biosource, Invitrogen), 200 nM Wortmannin (Biosource, Invitrogen), mixture of E64 (10 µM) (Calbiochem), Pepstatin A (10 µg/ml) (Sigma) and Cathepsin inhibitor I (12.5 µM) (Calbiochem), 10 µM Lactacystin (BioMol), 5µM MG132 (Biomol), 50 µM Z-vad-fmk (Sigma), 150 nM Epoxomycin (BioMol) or 5 µM Chloroquine (Sigma). The cell lysate was then subjected to immunoblot analysis.
Luciferase assay
293T cells were cotransfected with the plasmids: pNBRE-Luc (which contained four copies of NBRE elements (TTTTAAAAGGTCATGC) cloned downstream of the minimal thymidine kinase promoter), pCMV Renilla, pKH3 or pKH3-TIM-1, TIM-3, TIM-4 or deletion constructs of TIM-1 along with pEGFP-NUR77 and cell lysates were prepared 36 h post transfection and analyzed for luciferase activity using a luminometer (Analytical Luminescence Laboratory Monolight 2010). The relative light units were normalized against the renilla luciferase activity and the results were plotted as fold reduction of transactivation function of NUR77.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay was performed using p32 labeled NBRE probe (prepared by annealing the oligos: TCGAGTTTTAAAAGGTCATGCTCAATTTG; CAAATTGAGCATGACCTTTTAAAACTCGA). Nuclear extracts from 293T cells transfected with pGFP-NUR77 and full length or deletion constructs of TIM proteins were incubated with the labeled NBRE probe and separated on a 5% polyacrylamide gel; the gel was dried and exposed to × ray film.
Immunofluroscence assay
For immunofluorescence assay, cell lines were grown on cover slips; transfected with respective plasmids if necessary and at 36 h post transfection, the cells were fixed with either 4% formaldehyde or ice cold methanol and permeabilised using 0.2% Triton X-100. The cover slips were blocked with 5% normal serum corresponding to secondary antibody species, incubated overnight with primary antibody at 4°C and then with appropriate secondary antibody at room temperature for 45 min; the cover slips were then mounted using Vectashield. Images were obtained using Zeiss LSM 510 confocal scanning microscope (Carl Zeiss, Germany). Colocalization was quantified by calculating Pearson’s correlation coefficients using Image J software. Data are represented as Ave±SEM and n= number of cells analyzed. Following are the sources of the primary antibodies: Anti-hTIM-1 (R&D), Anti-HA (Upstate), Anti-Giantin (Abcam), Anti-EEA1, Anti-Lamp2a, Anti-HSP60 (Santa Cruz), Anti-Caveolin-1, Anti-Clathrin, Anti-Calnexin, Anti-p230 (BD transduction laboratories).
Realtime PCR assay
RNA was prepared using RNeasy kit from Qiagen. DNA contamination was eliminated by DNase treatment. Equal amounts of RNA were reverse transcribed using Taqman reverse transcription kit (Applied Biosystem) using random hexamers. Real time quantification of messages was performed either by Taqman or Sybr green assay, using the gene-specific primer pairs (hTIM-1: sense 5’ CAGGGAGCAATAAGGAGAGA; anti-sense 5’ AAGGCCATCTGAAGACTCTG; hNUR77: sense 5’TCCCATATTGGGCTTGGA TA; anti-sense 5’ ATC TTG GGA TTC TCC CTT CG) and normalized to the expression of GAPDH in 7900HT sequence detection system (Applied Biosystems). Transcript quantities were calculated according to the 2−ddCt method.
GST pull down assay
Glutathione S-transferase (GST) was expressed in the E. coli BL21 (DE3) cells by induction with 1 mM IPTG at 30°C for 2.5 – 4 hours. Bacterial cells were resuspended in lysis buffer (50 mM Tris.Cl pH 8.0, 300 mM NaCl, 1 mM DTT, 1 mM EDTA, PMSF, Protease inhibitor cocktail) and were lysed by sonication at 4°C (3 times with 30 second pulses at medium setting) using a probe sonicator (LABSONIC, B. Braun, USA). Following sonication, bacterial lysates were cleared by centrifugation at 13,000 rpm for 20 minutes at 4°C. Cleared bacterial lysates were incubated with pre-equilibrated Glutathione- Sepharose 4B at 4°C for 2 hours with continuous gentle rotation. The beads were then washed 3 times at 4°C with lysis buffer.
5µg of GST or GST-NUR77 fusion protein (Invitrogen) were immobilised on Glutathione- sepharose 4B beads. Total cell lysates prepared from cells transfected with pKH3, pKH3-hTIM-1 wild type, pKH3-hTIM-1 WFND92–95AAAA or hTIM-1 mucin domain constructs were pre-cleared with glutathione-sepharose 4B. The pre-cleared cell lysates were then incubated with GST or GST-NUR77 bound glutathione-sepharose 4B beads for 4h at 4°C and subsequently washed five times with the lysis buffer (10mM Tris pH 7.6, 150 mM NaCl, 0.5% NP-40, Protease inhibitor cocktail). The beads were then subjected to western blot analysis.
Generation of stable knockdown of TIM-1 in 769-P cells
shRNA constructs targeting human TIM-1 were cloned into pLKO.1 puro (Kind gift from Dr. William C Hahn, DFCI, Harvard Medical School, Boston) and the silencing efficiency was determined using the 293T–hTIM-1 stable cell line. The strongest silencing construct (hairpin sequence: sense: 5’-CCG GTG GTC ACG ACT ACT CCA ATT GTC TTC CTG TCA ACA ATT GGA GTA GTC GTG ACC TTT TTG-3’; anti-sense: 5’-AAT TCA AAA AGG TCA CGA CTA CTC CAA TTG TTG ACA GGA AGA CAA TTG GAG TAG TCG TGA CCA-3’) was transfected into 769-P cells, selected using Puromycin (5 µg/ml) and a single cell clone was isolated. TIM-1 stable knockdown was confirmed at the RNA and protein level in the single cell knockdown clone.
Desferrioxamine treatment
HK-2 cells, which are human proximal kidney tubular cells, were transiently transfected with either vector (pKH3) or full length hTIM-1. 24 h post transfection, the cells were treated with 50 µM Desferrioxamine for 16 h and the cells lysates were subjected to immunoblot analysis with anti-NUR77 and anti-hTIM-1 antibodies. Chloroquine treated cells received 5 µM final concentration for a period of 16 h.
In vitro renal epithelial cell injury model
HK-2 cells were maintained in K-SFM serum free medium. The cells were subjected to in vitro injury that mimics the effects of ischemia (30), through depletion of ATP and glucose and overloading of calcium. Briefly cells were washed with Hank’s Buffered Salt Solution (HBSS) and incubated with HBSS containing antimycin A (10 µM), 2-deoxyglucose (10 mM) and A23187 (2 µM) for 1 hour at 37°C, 5% CO2. Subsequently the cells were washed with HBSS and cultured in complete culture medium for 24h. After 24h, both the floating as well as adherent cells were collected and stained with Propidium iodide as described elsewhere (44). The sub-G1 population is expressed as percentage dead cells.
FACS analysis of surface expression of TIM-1
293T cells were transiently transfected with full length wild type, MILIBS, cytoplasmic domain mutants with and without wild type dynamin, Dyn2K44A, DIII, D3delta2. 24 h post-transfection, live cells were stained with anti-hTIM-1 (R&D) for 20 min on ice, subsequently fixed and analysed by flow cytometry.
Perturbation of endocytic pathway
293T cells were transiently transfected with TIM-1 and NUR77 together with dominant negative constructs of dynamin or Eps15. 24 h post-transfection, cells were again transfected with the corresponding dominant negative construct. 48 h post-first transfection, cell lysates were analysed for the abundance of NUR77 protein.
Endocytosis assay
HeLa cells were transiently transfected with full length hTIM-1. 24 h post-transfection, live cells were coated with anti-hTIM-1 antibody (R&D) for 1h in isotonic buffer (25 mM Hepes, 140 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 15 mM Glucose, pH 7.4) at 4°C, washed extensively with cold PBS and shifted to medium at 37°C and chased for different time points. The cells were then fixed, permeabilised and stained with primary and secondary antibodies against hTIM-1 and various subcellular organelle markers and analyzed by confocal microscopy.
Statistical analysis
All comparisons were performed using a two-tailed unpaired Student’s t test. Wherever applicable, statistical significance is denoted by * for p<0.05, ** for p<0.01 and *** for p <0.001. The data are expressed as means ±SEM. For data that are normalized, data analyses were performed following the method described in Valcu et al. (45). Briefly, values for control and experimental samples were divided by the mean of the control sample, which conserves the distribution and the relative variance of the samples, allowing the subsequent use of a t-test: mean (k*X) = k * mean(X); std error(k * X) = k * std error(X); where X=sample, k = 1/mean of the control and is a normalization factor.
Supplementary Material
Acknowledgements
This work is supported by funds from NIH to TBS (1P01 AI073748-01A1). We thank Prof Marc Vidal (DFCI, Harvard Medical School, Boston) who kindly provided the human spleen yeast two hybrid cDNA library. We thank Dr. Yuri Pekarsky (Human Cancer Genetics Program, Ohio State University) for the kind gift of pGL3- NBRE and GFP-NUR77 constructs; Dr. Sean Egan (The Hospital for Sick children, Toronto, Canada) for the wild type and K44A mutant Dynamin constructs, Dr. Alexandre Benmerah (INSERM U567, Institut Cochin, Paris, France) for the kind gift of GFP tagged dominant negative mutants of Eps 15- DIII, D3delta3, Prof. Joan Brugge (Harvard Medical School, Boston, USA) for the stable knockdown lines of Akt1 and Akt2 isoforms and Dr. Walter Beron (Universidad Nacional de Cuyo Conicet, Argentina for RFP-LC3 construct. We thank Dr. Xian C Li for his valuable comments on the manuscript.
Footnotes
“This manuscript has been accepted for publication in Science Signaling. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencesignaling.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.”
Supplementary Materials
Fig S1. NUR77 domains required for the interaction with TIM-1.
Fig S2. TIM proteins do not affect the abundance of GFP.
Fig S3. Reduction of NUR77 protein abundance is not through squelching.
Fig S4. Silencing of TIM-1 increases NUR77 protein abundance in HK-2 cells in an in vitro epithelial cell injury model.
Fig S5. Cellular localization of endogenous TIM-1.
Fig S6. The IgV domain of TIM proteins is highly conserved.
Fig S7. Localization pattern of wild-type and signal peptide deleted TIM-1 proteins.
Fig S8. The requirement for PI3K in TIM-1-mediated degradation of NUR77 is independent of Akt signaling.
Fig S9. TIM-1 is not subjected to lysosomal degradation.
Fig S10. Colocalization of NUR77 and TIM-1 with the autophagosome marker LC3.
Table S1. Interacting partners of hTIM-1 identified in the yeast two-hybrid screen
Author contributions
SB and TBS conceived, designed all the experiments and wrote the manuscript. SB carried out all the experiments. SK, VKK and BDH contributed in experimental design and helped through discussions.
Competing interests
The authors declare that they have no competing interests.
References and Notes
- 1.Mariat C, Degauque N, Balasubramanian S, Kenny J, DeKruyff RH, Umetsu DT, Kuchroo V, Zheng XX, Strom TB. Tim-1 signaling substitutes for conventional signal 1 and requires costimulation to induce T cell proliferation. J Immunol. 2009;182:1379–1385. doi: 10.4049/jimmunol.182.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodriguez-Manzanet R, Meyers JH, Balasubramanian S, Slavik J, Kassam N, Dardalhon V, Greenfield EA, Anderson AC, Sobel RA, Hafler DA, Strom TB, Kuchroo VK. TIM-4 expressed on APCs induces T cell expansion and survival. J Immunol. 2008;180:4706–4713. doi: 10.4049/jimmunol.180.7.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson AC, Anderson DE, Bregoli L, Hastings WD, Kassam N, Lei C, Chandwaskar R, Karman J, Su EW, Hirashima M, Bruce JN, Kane LP, Kuchroo VK, Hafler DA. Promotion of tissue inflammation by the immune receptor Tim-3 expressed on innate immune cells. Science. 2007;318:1141–1143. doi: 10.1126/science.1148536. [DOI] [PubMed] [Google Scholar]
- 4.McIntire JJ, Umetsu SE, Akbari O, Potter M, Kuchroo VK, Barsh GS, Freeman GJ, Umetsu DT, DeKruyff RH. Identification of Tapr (an airway hyperreactivity regulatory locus) and the linked Tim gene family. Nat Immunol. 2001;2:1109–1116. doi: 10.1038/ni739. [DOI] [PubMed] [Google Scholar]
- 5.Kaplan G, Totsuka A, Thompson P, Akatsuka T, Moritsugu Y, Feinstone SM. Identification of a surface glycoprotein on African green monkey kidney cells as a receptor for hepatitis A virus. Embo J. 1996;15:4282–4296. [PMC free article] [PubMed] [Google Scholar]
- 6.Ichimura T, Bonventre JV, Bailly V, Wei H, Hession CA, Cate RL, Sanicola M. Kidney injury molecule-1 (KIM-1), a putative epithelial cell adhesion molecule containing a novel immunoglobulin domain, is up-regulated in renal cells after injury. J Biol Chem. 1998;273:4135–4142. doi: 10.1074/jbc.273.7.4135. [DOI] [PubMed] [Google Scholar]
- 7.Han WK, Bailly V, Abichandani R, Thadhani R, Bonventre JV. Kidney Injury Molecule-1 (KIM-1): a novel biomarker for human renal proximal tubule injury. Kidney Int. 2002;62:237–244. doi: 10.1046/j.1523-1755.2002.00433.x. [DOI] [PubMed] [Google Scholar]
- 8.Chae SC, Song JH, Heo JC, Lee YC, Kim JW, Chung HT. Molecular variations in the promoter and coding regions of human Tim-1 gene and their association in Koreans with asthma. Hum Immunol. 2003;64:1177–1182. doi: 10.1016/j.humimm.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 9.McIntire JJ, Umetsu SE, Macaubas C, Hoyte EG, Cinnioglu C, Cavalli-Sforza LL, Barsh GS, Hallmayer JF, Underhill PA, Risch NJ, Freeman GJ, DeKruyff RH, Umetsu DT. Immunology: hepatitis A virus link to atopic disease. Nature. 2003;425:576. doi: 10.1038/425576a. [DOI] [PubMed] [Google Scholar]
- 10.Meyers JH, Chakravarti S, Schlesinger D, Illes Z, Waldner H, Umetsu SE, Kenny J, Zheng XX, Umetsu DT, DeKruyff RH, Strom TB, Kuchroo VK. TIM-4 is the ligand for TIM-1, and the TIM-1-TIM-4 interaction regulates T cell proliferation. Nat Immunol. 2005;6:455–464. doi: 10.1038/ni1185. [DOI] [PubMed] [Google Scholar]
- 11.Chen TT, Li L, Chung DH, Allen CD, Torti SV, Torti FM, Cyster JG, Chen CY, Brodsky FM, Niemi EC, Nakamura MC, Seaman WE, Daws MR. TIM-2 is expressed on B cells and in liver and kidney and is a receptor for H-ferritin endocytosis. J Exp Med. 2005;202:955–965. doi: 10.1084/jem.20042433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu C, Anderson AC, Schubart A, Xiong H, Imitola J, Khoury SJ, Zheng XX, Strom TB, Kuchroo VK. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol. 2005;6:1245–1252. doi: 10.1038/ni1271. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi N, Karisola P, Pena-Cruz V, Dorfman DM, Jinushi M, Umetsu SE, Butte MJ, Nagumo H, Chernova I, Zhu B, Sharpe AH, Ito S, Dranoff G, Kaplan GG, Casasnovas JM, Umetsu DT, Dekruyff RH, Freeman GJ. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity. 2007;27:927–940. doi: 10.1016/j.immuni.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyanishi M, Tada K, Koike M, Uchiyama Y, Kitamura T, Nagata S. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450:435–439. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- 15.Ichimura T, Asseldonk EJ, Humphreys BD, Gunaratnam L, Duffield JS, Bonventre JV. Kidney injury molecule-1 is a phosphatidylserine receptor that confers a phagocytic phenotype on epithelial cells. J Clin Invest. 2008;118:1657–1668. doi: 10.1172/JCI34487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hazel TG, Nathans D, Lau LF. A gene inducible by serum growth factors encodes a member of the steroid and thyroid hormone receptor superfamily. Proc Natl Acad Sci U S A. 1988;85:8444–8448. doi: 10.1073/pnas.85.22.8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calnan BJ, Szychowski S, Chan FK, Cado D, Winoto A. A role for the orphan steroid receptor Nur77 in apoptosis accompanying antigen-induced negative selection. Immunity. 1995;3:273–282. doi: 10.1016/1074-7613(95)90113-2. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki S, Suzuki N, Mirtsos C, Horacek T, Lye E, Noh SK, Ho A, Bouchard D, Mak TW, Yeh WC. Nur77 as a survival factor in tumor necrosis factor signaling. Proc Natl Acad Sci U S A. 2003;100:8276–8280. doi: 10.1073/pnas.0932598100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolluri SK, Bruey-Sedano N, Cao X, Lin B, Lin F, Han YH, Dawson MI, Zhang XK. Mitogenic effect of orphan receptor TR3 and its regulation by MEKK1 in lung cancer cells. Mol Cell Biol. 2003;23:8651–8667. doi: 10.1128/MCB.23.23.8651-8667.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Lin B, Agadir A, Liu R, Dawson MI, Reed JC, Fontana JA, Bost F, Hobbs PD, Zheng Y, Chen GQ, Shroot B, Mercola D, Zhang XK. Molecular determinants of AHPN (CD437)-induced growth arrest and apoptosis in human lung cancer cell lines. Mol Cell Biol. 1998;18:4719–4731. doi: 10.1128/mcb.18.8.4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chintharlapalli S, Burghardt R, Papineni S, Ramaiah S, Yoon K, Safe S. Activation of Nur77 by selected 1,1-Bis(3'-indolyl)-1-(p-substituted phenyl)methanes induces apoptosis through nuclear pathways. J Biol Chem. 2005;280:24903–24914. doi: 10.1074/jbc.M500107200. [DOI] [PubMed] [Google Scholar]
- 22.Lin B, Kolluri SK, Lin F, Liu W, Han YH, Cao X, Dawson MI, Reed JC, Zhang XK. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell. 2004;116:527–540. doi: 10.1016/s0092-8674(04)00162-x. [DOI] [PubMed] [Google Scholar]
- 23.Thompson J, Winoto A. During negative selection, Nur77 family proteins translocate to mitochondria where they associate with Bcl-2 and expose its proapoptotic BH3 domain. J Exp Med. 2008;205:1029–1036. doi: 10.1084/jem.20080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balasubramanian S, Jansen M, Valerius MT, Humphreys BD, Strom TB. Orphan nuclear receptor nur77 promotes acute kidney injury and renal epithelial apoptosis. J Am Soc Nephrol. 2012;23:674–686. doi: 10.1681/ASN.2011070646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuang AA, Cado D, Winoto A. Nur77 transcription activity correlates with its apoptotic function in vivo. Eur J Immunol. 1999;29:3722–3728. doi: 10.1002/(SICI)1521-4141(199911)29:11<3722::AID-IMMU3722>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 26.Davis IJ, Hazel TG, Lau LF. Transcriptional activation by Nur77, a growth factor-inducible member of the steroid hormone receptor superfamily. Mol Endocrinol. 1991;5:854–859. doi: 10.1210/mend-5-6-854. [DOI] [PubMed] [Google Scholar]
- 27.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 28.Yeo MG, Yoo YG, Choi HS, Pak YK, Lee MO. Negative cross-talk between Nur77 and small heterodimer partner and its role in apoptotic cell death of hepatoma cells. Mol Endocrinol. 2005;19:950–963. doi: 10.1210/me.2004-0209. [DOI] [PubMed] [Google Scholar]
- 29.Yoo YG, Yeo MG, Kim DK, Park H, Lee MO. Novel function of orphan nuclear receptor Nur77 in stabilizing hypoxia-inducible factor-1alpha. J Biol Chem. 2004;279:53365–53373. doi: 10.1074/jbc.M408554200. [DOI] [PubMed] [Google Scholar]
- 30.Lee HT, Emala CW. Preconditioning and adenosine protect human proximal tubule cells in an in vitro model of ischemic injury. J Am Soc Nephrol. 2002;13:2753–2761. doi: 10.1097/01.asn.0000032421.79225.6e. [DOI] [PubMed] [Google Scholar]
- 31.Yeung T, Gilbert GE, Shi J, Silvius J, Kapus A, Grinstein S. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 32.Santiago C, Ballesteros A, Martinez-Munoz L, Mellado M, Kaplan GG, Freeman GJ, Casasnovas JM. Structures of T cell immunoglobulin mucin protein 4 show a metal-Ion-dependent ligand binding site where phosphatidylserine binds. Immunity. 2007;27:941–951. doi: 10.1016/j.immuni.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benmerah A, Bayrou M, Cerf-Bensussan N, Dautry-Varsat A. Inhibition of clathrin-coated pit assembly by an Eps15 mutant. J Cell Sci. 1999;112(Pt 9):1303–1311. doi: 10.1242/jcs.112.9.1303. [DOI] [PubMed] [Google Scholar]
- 34.Damke H, Baba T, Warnock DE, Schmid SL. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hunyady L, Baukal AJ, Gaborik Z, Olivares-Reyes JA, Bor M, Szaszak M, Lodge R, Catt KJ, Balla T. Differential PI 3-kinase dependence of early and late phases of recycling of the internalized AT1 angiotensin receptor. J Cell Biol. 2002;157:1211–1222. doi: 10.1083/jcb.200111013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y, Keen JH. Gyrating clathrin: highly dynamic clathrin structures involved in rapid receptor recycling. Traffic. 2008;9:2253–2264. doi: 10.1111/j.1600-0854.2008.00819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Souza AJ, Oak JS, Jordanhazy R, DeKruyff RH, Fruman DA, Kane LP. T cell Ig and mucin domain-1-mediated T cell activation requires recruitment and activation of phosphoinositide 3-kinase. J Immunol. 2008;180:6518–6526. doi: 10.4049/jimmunol.180.10.6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Binne LL, Scott ML, Rennert PD. Human TIM-1 associates with the TCR complex and up-regulates T cell activation signals. J Immunol. 2007;178:4342–4350. doi: 10.4049/jimmunol.178.7.4342. [DOI] [PubMed] [Google Scholar]
- 39.Altschuler Y, Barbas SM, Terlecky LJ, Tang K, Hardy S, Mostov KE, Schmid SL. Redundant and distinct functions for dynamin-1 and dynamin-2 isoforms. J Cell Biol. 1998;143:1871–1881. doi: 10.1083/jcb.143.7.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumanogoh A, Marukawa S, Suzuki K, Takegahara N, Watanabe C, Ch'ng E, Ishida I, Fujimura H, Sakoda S, Yoshida K, Kikutani H. Class IV semaphorin Sema4A enhances T-cell activation and interacts with Tim-2. Nature. 2002;419:629–633. doi: 10.1038/nature01037. [DOI] [PubMed] [Google Scholar]
- 41.Uchida Y, Hasegawa J, Chinnapen D, Inoue T, Okazaki S, Kato R, Wakatsuki S, Misaki R, Koike M, Uchiyama Y, Iemura S, Natsume T, Kuwahara R, Nakagawa T, Nishikawa K, Mukai K, Miyoshi E, Taniguchi N, Sheff D, Lencer WI, Taguchi T, Arai H. Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes. Proc Natl Acad Sci U S A. 2011;108:15846–15851. doi: 10.1073/pnas.1109101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 43.Pekarsky Y, Hallas C, Palamarchuk A, Koval A, Bullrich F, Hirata Y, Bichi R, Letofsky J, Croce CM. Akt phosphorylates and regulates the orphan nuclear receptor Nur77. Proc Natl Acad Sci U S A. 2001;98:3690–3694. doi: 10.1073/pnas.051003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riccardi C, Nicoletti I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat Protoc. 2006;1:1458–1461. doi: 10.1038/nprot.2006.238. [DOI] [PubMed] [Google Scholar]
- 45.Valcu M, Valcu CM. Data transformation practices in biomedical sciences. Nat Methods. 2011;8:104–105. doi: 10.1038/nmeth0211-104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.