Abstract
New influenza A viruses with pandemic potential periodically emerge due to viral genomic reassortment. In the face of pandemic threats, production of conventional egg-based vaccines is time consuming and of limited capacity. We have developed in this study a novel DNA vaccine in which viral hemagglutinin (HA) is bivalently targeted to MHC class II (MHC II) molecules on APCs. Following DNA vaccination, transfected cells secreted vaccine proteins that bound MHC II on APCs and initiated adaptive immune responses. A single DNA immunization induced within 8 d protective levels of strain-specific Abs and also cross-reactive T cells. During the Mexican flu pandemic, a targeted DNA vaccine (HA from A/California/07/2009) was generated within 3 wk after the HA sequences were published online. These results suggest that MHC II–targeted DNA vaccines could play a role in situations of pandemic threats. The vaccine principle should be extendable to other infectious diseases.
Introduction
Pandemic influenza emerges with random intervals due to viral genomic reassortment between different influenza viruses (1). New combinations of surface proteins could render the human population partially or completely naive against such influenza strains. Should a viral pandemic with the proportions of the 1918 Spanish flu (H1N1) reappear, existing egg-based strategies for development of new vaccines are likely to be insufficient both with respect to time needed for production and vaccine quantities (2). Even if assuming that enough eggs could be available for large scale production, highly pathogenic viruses have been demonstrated to be lethal for chicken embryos. Viral production in mammalian cell lines could solve this problem (3), but the manufacture is time consuming and of limited capacity. Recombinant protein subunit vaccination is also an unlikely alternative due to limitations in mass production. Hence, it is of utmost importance to develop novel influenza vaccines that can be rapidly produced upon pandemic threats.
DNA vaccines encoding viral subunits can be rapidly generated, and may present the timeline and production efficiency needed to prevent a pandemic. However, the induction of adequate immune responses against influenza require large doses of DNA even in mice, and often in combination with adjuvants or several booster injections (4–10). Also, DNA vaccines often have a tendency of skewing immune responses toward T cell immunity, whereas Abs are of major importance in influenza prevention. Thus, there is a need for novel DNA vaccines against influenza that rapidly can induce protective Abs after a single injection.
The immunogenicity of Ags can be increased by targeting of Ag to various surface molecules on APCs (11–23). Such targeting has typically been done to obtain increased MHC presentation of Ag and thus increased T cell responses (20, 21, 23). However, in some studies, APC targeting resulted in increased Ab responses (11–14). Notably, several reports have demonstrated that immunization with anti–MHC class II (MHC II) mAb chemically conjugated to avidin (12, 19), FITC (16), and hemagglutinin (HA) (17) enhanced Ab responses. However, Ab–Ag complexes are difficult to produce and suffer from high batch-to-batch variability (24). Use of Ab–Ag conjugates is thus not a plausible scenario for mass vaccination. Recombinant fusion proteins, with high batch-to-batch consistency, could be an alternative, but suffer from difficulties in mass production.
Perhaps solving the above problems, we (25–27) and others (20) have previously shown that DNA vaccination and APC targeting can be combined. Thus, DNA that encodes fusion proteins targeting Ags to MHC II molecules (25), CD40 (27), and chemokine receptors (20, 26) increased Ab and T cell responses in mice against idiotypic tumor-specific Ag and protection against a subsequent tumor challenge (20, 25–27). It was further demonstrated that DNA-injected and electroporated muscle cells secreted fusion proteins that were absorbed to MHC II molecules in the vaccinated mice (25). Importantly, bivalency of the DNA-encoded fusion protein increased immune responses over the monovalent form (26).
Given the previous encouraging results in mouse tumor models, we wanted to investigate whether the principle could be extended to the infectious setting of influenza. HA is the primary target for Abs that neutralize influenza virus (28) and is also the most abundantly expressed protein on the surface of influenza virions. To maintain conformational determinants necessary for generation of strong Ab responses, extracellular portions of HA were inserted into vaccine-encoding plasmids (25). We demonstrate in this study that a single injection of mice with the DNA vaccine induced a complete Ab-mediated protection against virus within 8 d. Furthermore, the vaccine induced T cells capable of mediating cross protection against other strains of influenza. The vaccine could be produced and administered within a few weeks, as demonstrated in this study for a vaccine encoding HA from the Mexican flu pandemic of 2009.
Materials and Methods
Molecular cloning
Vaccine molecules were constructed by inserting targeting units and antigenic units into the cloning sites of the previously described pLNOH2 CMV-based expression vector (25, 26). Targeting units were either a single-chain variable fragment (scFv) specific for MHC II molecules (I-Ed) or an scFv specific for the hapten 4-hydroxy-3-iodo-5-nitrophenylacetic acid (NIP) (negative control) (25). As the antigenic unit, a fragment encoding aa 18–541 of HA from influenza A/PR/8/34 was cloned using plasmid HAwt-pCMV (kind gift from Harald von Boehmer) as a template. The primers were designed with fixed restriction sites for SfiI on the 5′ and 3′ ends: HA185′, gag gcc tcg gtg gcc tgg aca caa tat gta tag gct acc and HA5413′, gga tcc ggc cct gca ggc ctc aca gtg aac tgg cga cag. A vaccine encoding only the antigenic unit (HA, aa 18–541) was also constructed. Internal HA BsmI sites were mutated (silent mutations) and a primer designed with a fixed restriction site for BsmI in the 5′ end (5′, ggt gtg cat tcg aca caa tat gta tag gct acc a). The 3′ end primer used was identical to that described above. An examination of secretion following transient transfections with HA inserted alone into the pLNOH vector showed, however, very low secretion as compared with the other vaccines. To obtain comparable levels of protein secretion, we inserted HA into a pLNOH2 vector encoding the HA [A/PR/8/34 (H1N1) (PR8)] leader sequence.
Western blot
Vaccine proteins, affinity purified from stably transfected NS0 cells, were run on a Novex 4–12% Tris-Glycine gel (Invitrogen) together with a SeeBlue Plus2 Prestained Standard (LC5925; Invitrogen), blotted (Immun-Blot PVDF membrane, 162-0177; Bio-Rad), and incubated with a biotinylated anti-HA Ab (H36-4-52; kind gift from Siegfried Weiss) (29) and streptavidin-HRP (RPN1231V; GE Healthcare). The membrane was developed with the ECL Western blotting analysis system (RPN2109; GE Healthcare) and analyzed on a Kodak Image station 200R with Molecular Imaging Software v 4.0.5 (Kodak).
Electron microscopy
Anti-NIP–HA proteins in supernatants from transfected NS0 cells were affinity purified (anti-HA mAb H36-4-52 coupled to CNBr-activated Sepharose 4 Fast Flow [GE Healthcare]) and stained for electron microscopy (EM) analysis as previously described (30, 31). Briefly, preparations of 1.0 μg/ml in borate-buffered saline (0.1 M H3BO3, 0.025 M Na2B4O7, and 0.075 M NaCl [pH 8.3]) were affixed to thin carbon membranes, stained with 1% uranyl formate, and mounted on 600-mesh copper grids for analysis. The sample grids were examined and photographed at a nominal magnification of ×100,000 at 100 kV on a JEOL JEM 1200EX electron microscope (JEOL).
ELISA for detection of HA vaccine proteins
One microgram DNA in a total of 50 μl OptiMEM (51985-026; Life Technologies) was mixed with 2.5 μl Lipofectamine 2000 (11668-019; Invitrogen) in 47.5 μl OptiMEM and incubated for 20 min at room temperature. After careful addition of the DNA/Lipofectamine solution to 293E cells (1 × 105/well), the transfectants were incubated for 48 h at 37°C and in a 5% CO2 humidified atmosphere. Supernatants were collected and centrifuged for 4 min at 13,000 rpm before being added in triplicates to ELISA plates (Costar 3590) that had been coated with either 2 μg/ml mouse anti-human IgG (CH3 domain) mAb MCA878G (AbD Serotec) or NIP-BSA. The plates were incubated for 1.5 h at room temperature, followed by incubation with biotinylated anti-HA (mAb H36-4-52) (1 μg/ml, 1 h) and streptavidin alkaline phosphatase (1:5000, 30 min; GE Healthcare). Plates were developed using phosphatase substrate (P4744-10G; Sigma Aldrich) dissolved in substrate buffer and read with a Tecan reader using the Magellan v5.03 program. For comparing secretion of HA alone to that of the other constructs, titrated amounts of supernatant were directly coated in duplicates onto 96-well plates, followed by detection with biotinylated mAb H36-4-52 as described above.
ELISA for detection of serum anti-HA Abs
Blood samples were collected from mice by puncture of the saphenous vein and sera isolated by two successive centrifugations for 5 min at 13,000 rpm. The 96-well plates were coated with inactivated PR8 virus (Charles River Laboratories) (1:1600 in PBS) or Pandemrix (Ag suspension with A/California/7/2009 [H1N1]v-like strain [X-179A]; GlaxoSmithKline) (1:100 in PBS), blocked with 0.1% BSA in PBS, and incubated overnight at 4°C with titrated amounts of sera from mice assayed individually (n = 6/group). Abs in sera were detected with biotinylated anti-IgG (A2429; Sigma-Aldrich), anti-IgG1a (553599; BD Pharmingen), anti-IgG2aa (553502; BD Pharmingen), anti-IgA (A4937; Sigma-Aldrich), anti-IgM (553515; BD Pharmingen), or anti-IgE (553419; BD Pharmingen), followed by streptavidin alkaline phosphatase and development as described above. Purified H36-4-52 mAb (IgG2a) was used as standard for quantification of IgG in the titration experiment (Fig. 2G). For other experiments, titers are given, defined as the last serum dilution giving an absorbance above background (mean absorbance for NaCl-vaccinated mice plus five times SEM).
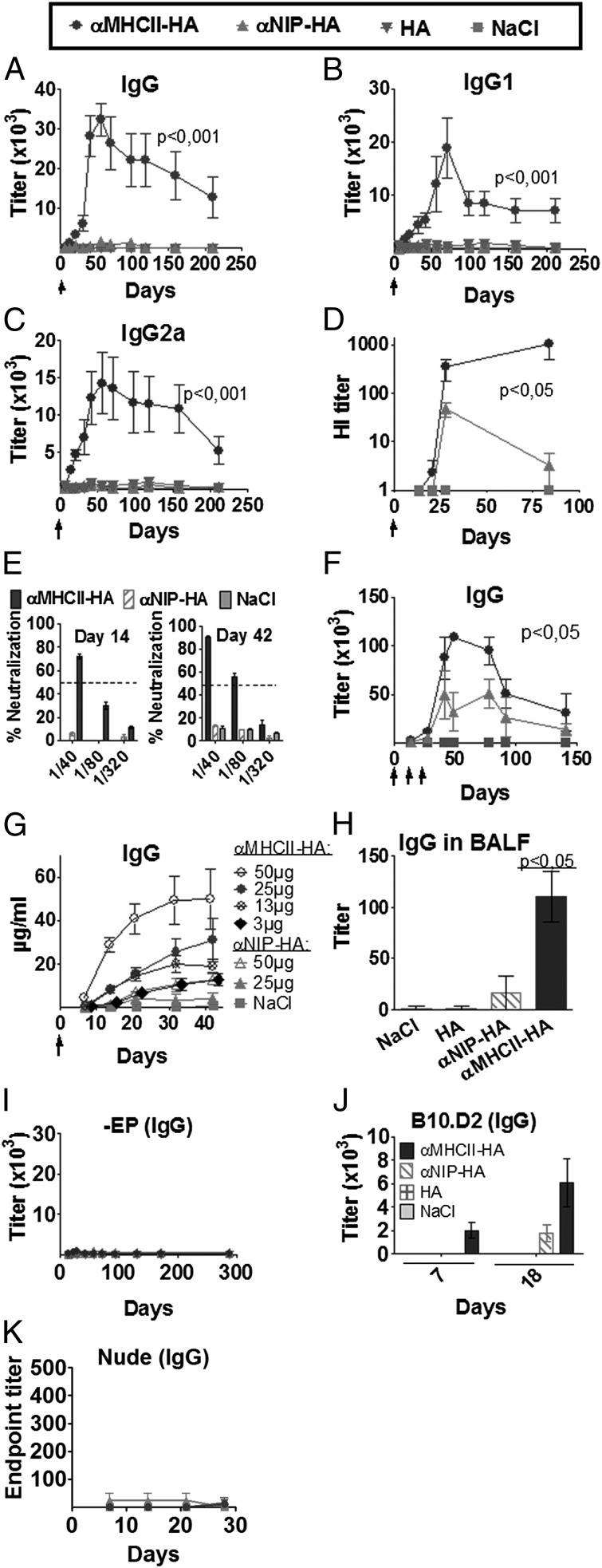
FIGURE 2.
DNA vaccine format that targets HA protein to MHC II increases Ab responses. (A–F) BALB/c mice were vaccinated intradermally with 25 μg of the indicated plasmids (key at top) (n = 6/group), followed by EP, as indicated by arrows (↑). Values given are mean ± SEM. Serum levels of total IgG (A), IgG1 (B), and IgG2a (C) anti-PR8 Abs, as measured in ELISA. (D) Serum HI titers. (E) Serum samples from days 14 and 42 after a single immunization were assayed in a microneutralization assay (PR8 virus) (n = 6/group). Dotted lines indicate threshold for positive neutralization. (F) BALB/c mice were immunized three times (↑) and tested for anti-HA Abs in ELISA (n = 6/group). (G) IgG anti-PR8 following a single immunization with various doses of DNA (n = 6/group). (H) IgG anti-PR8 in BALF from mice at day 125 after a single immunization (n = 5/group). (I) BALB/c mice were vaccinated intradermally with 25 μg DNA in the absence of EP (n = 6/group). Blood samples were collected at various time points and IgG anti-PR8 assayed in ELISA. (J) B10.D2 H-2d mice were immunized once with 25 μg DNA/EP (HA from PR8) or NaCl. Sera were assayed for IgG anti-HA Abs. (K) Athymic BALB/c nude mice were DNA/EP-immunized once with 25 μg of the plasmids indicated above (n = 4/group). Serum samples were examined for IgG anti-HA in ELISA using PR8 as coat. No significant differences between groups were observed.
Detection of anti-HA Abs in bronchoalveolar lavage fluid
Bronchoalveolar lavage fluids (BALF) were harvested from anesthetized (Hypnorm/Dormicum; 0.05 ml working solution/10 g s.c.) and exanguinated mice (n = 5/group) and added in triplicates to 96-well plates coated with PR8 as described above. Abs against PR8 were detected with alkaline phosphatase–conjugated anti-IgG Abs (A2429; Sigma-Aldrich). Plates were developed and read as described above.
Flow cytometry
MHC II I-Ed–transfected L-cell fibroblasts [CA36.2.1 (EβdEαk), CA36.1.3 (EβkEαk), and CA25.8.2 (Dd)] (32) were FcγR-blocked by incubation with 30% heat-aggregated rat serum and 0.1 mg/ml 2.4G2 mAb and then stained with affinity-purified vaccine proteins (10 μg/ml), biotinylated anti-HA Ab (H36-4-52; 1 μg/ml), and streptavidin-PE (1 μg/ml) (554061; BD Pharmingen). Cells were fixed with 2% paraformaldehyde and analyzed on an FACSCalibur flow cytometer (BD Biosciences).
Splenocytes from BALB/c and C57BL/6 mice were blocked as above and stained sequentially with vaccine proteins (10 μg/ml), biotinylated anti-human CH3 Abs (B3773; Sigma-Aldrich), and streptavidin-allophycocyanin (554067; BD Pharmingen). The staining solution also included PerCP-Cy5.5–conjugated anti-CD11b Abs (550993; BD Pharmingen), PE-conjugated CD11c Abs (553802; BD Pharmingen), FITC-conjugated anti-CD19 Abs (553785; BD Pharmingen), and Pacific Blue–conjugated CD4- (57-0042-82; eBioscience) and CD8-specific Abs (558106; BD Pharmingen) (CD4+/CD8+ constituted dump gate). Splenocytes were run on an LSRII flow cytometer (BD Biosciences) and data analyzed with the FlowJo software (version 7.6; Tree Star).
The peritoneal cavity of BALB/c mice was flushed with PBS, and single-cell suspensions prepared as above. Peritoneal cells and splenocytes (1 × 106 cells/well) were incubated at 37°C and 5% CO2 for 48 h with vaccine proteins (10 μg/ml) or medium and stained with allophycocyanin-conjugated mAbs against CD11b (553312; BD Pharmingen) or CD11c (550261; BD Pharmingen), FITC-conjugated mAbs against CD19 (553785; BD Pharmingen), and PE-conjugated mAbs against CD80 (1730-09L; Southern Biotechnology Associates) or CD86 (1915-09; Southern Biotechnology Associates). Pacific Blue–conjugated mAbs against CD8 (553033; BD Pharmingen) and CD4 (57-0042-82; eBioscience) were included, and positive cells were removed in a dump gate. Samples were fixed as above and run on an LSRII flow cytometer (BD Biosciences). Data were analyzed with the FlowJo software (version 10.0.5; Tree Star).
Hemagglutination-inhibition assay
Equal volumes of sera from individual mice (n = 6/group) were pooled and treated with receptor-destroying enzyme (II) (Denka Seiken) at 37°C for 20 h. The enzyme was deactivated by incubation at 56°C for 40 min. Sera were titrated and added in triplicates to 96-well plates. Diluted allantoic fluid containing 4 hemagglutinating units PR8 virus was added and plates incubated for 40 min at room temperature. Turkey RBCs (1%) were added to wells, and the plates were read for hemagglutination-inhibition (HI) 45 min later. HI was scored as the highest dilution of antiserum giving a complete inhibition of hemagglutination. Validity of results was confirmed by the positive control serum (influenza PR-8 antiserum; Charles River Laboratories) reaching its predicted titer and the negative control serum giving a titer of <23.
Microneutralization assay
Viral TCID50 was determined by the Reed-Muench method (33). Inclusion of TPCK trypsin did not significantly improve influenza infectivity and was therefore omitted in these assays. Equal serum volumes from individual mice (n = 6/group) were pooled and treated with receptor-destroying enzyme (II) as described above. Two-fold duplicate dilutions in virus diluent (DMEM supplemented with 1% bovine albumin fraction V, antibiotics, and 0.02 M HEPES) were set up in triplicates in 96-well plates. Four control wells of virus in diluent and diluent alone (cells only) were included on each plate. Fifty microliters 100 × TCID50 virus was added to each well, except the cells only wells, and plates were incubated for 2 h at 37°C in a 5% CO2 humidified atmosphere. Madin-Darby canine kidney cells (2 × 105) were added to each well, followed by incubation for 20 h at 37°C and 5% CO2. The monolayers were washed with PBS, fixed in cold 80% acetone for 10 min, and viral proteins detected by an ELISA using biotinylated mAb against the influenza nucleoprotein (HB65; American Type Culture Collection) and streptavidin-alkaline phosphatase. Plates were read as described above, with a Tecan reader using the Magellan v5.03 program.
Viruses
PR8 and A/California/07/09 (H1N1) (Cal07) were kindly provided by Dr. Anna Germundsson (The National Veterinary Institute, Oslo, Norway). The viruses were propagated by inoculating virus into the allantoic cavity of 10-d-old embryonated chicken eggs. Identity of the injected viruses was confirmed by sequencing of HA. Allantoic fluid was harvested and confirmed negative for bacterial contaminations. TCID50 was determined.
Mice
Six- to 8-wk-old female BALB/c mice (Taconic Farms), 6–10-wk-old B10.D2-H-2d (Harlan UK), and 6–8-wk-old female BALB/c Nude (Taconic Farms) were used. Animals were housed under minimal disease conditions. All animal experiments were approved by the National Committee for Animal Experiments (Oslo, Norway).
Vaccination and viral challenge
Mice were anesthetized by s.c. injection of Hypnorm/Dormicum (0.05 ml working solution/10 g) and shaved in the lower back region. Twenty-five microliters plasmids (purified from the Endofree Qiagen kit [Qiagen]), dissolved in NaCl, was injected intradermally on each flank of the mouse, immediately followed by skin electroporation (EP) with DermaVax (Cellectis) (34, 35). A total of 25 μg DNA, divided into two equal doses, was injected per mouse, except in the dose titration experiments (n = 6/group).
Groups of anesthetized mice (n = 4/group) were infected intranasally with titrated doses of influenza virus in allantoic fluid (1, 101, 102, 103, 104, or 105 TCID50) (n = 4/group) and LD50 determined in accordance with the Reed and Muench method (33). For assessing the effect of vaccination, anesthetized mice were infected with 5× LD50 PR8 (2.0 × 104 TCID) or Cal07 (1.0 × 106 TCID) in 10 μl/nostril. Mice were monitored for weight loss (n = 6/group), with an end point of 20% weight reduction, as required by the National Committee for Animal Experiments. Mice reaching the >20% weight loss were euthanized by cervical dislocation. In the experiment in which mice were challenged 10 mo after vaccination (n = 6/group, except for NaCl, in which n = 5), a group of previous PR8 survivors was included as a positive control (n = 3).
Quantitative PCR
Noses of mice (n = 4) were flushed with 1 ml PBS/BSA (2%) and RNA extracted using NucliSens easyMag (Biomèrieux). Quantitative RT-PCR was performed with samples in triplicates and with the following program (Qiagen OneStep RT-PCR kit): 50°C (30 min), 95°C (2 min), followed by 45 cycles of 95°C (15 s) and 55°C (30 s) using a Stratagene RealTime machine (Stratagene) with primers 5′-GAC CRA TCC TGT CAC CTC TGA C-3′ and 5′-AGG GCA TTY TGG ACA AAK CGT CTA-3′ and probe 5′-TGC AGT CCT CGC TCA CTG GGC ACG-3′ (MedProbe), all in accordance with the Centers for Disease Control and Prevention protocol (http://www.who.int/csr/resources/publications/swineflu/CDCrealtimeRTPCRprotocol_20090).
H&E staining
Lungs were collected at day 7 because this was the time point at which most mice vaccinated with NaCl or anti-NIP–HA had to be euthanized for humane reasons. The excised lungs were fixed in formalin, embedded in paraffin, and stained with H&E. Tissue sections were examined with a Leica DMRB microscope (Leica Microsystems; original magnification ×10/0.75). A human pathologist experienced in evaluations of lung tissues from patients deceased from severe influenza was consulted for interpretation of images. Mice were scored as displaying lung pathology if the following signs were observed: histiocytic alveolitis, interstitial edema, and thickening of alveolar septum.
ELISPOT assay
Multiscreen HTS plates (Millipore) were coated with 12 μg/ml anti-mouse IFN-γ (AN18) (36) and blocked with complete tissue culture medium (RPMI 1640 containing 10% FCS and supplements). Single-cell suspensions (n = 6/group) were prepared by mashing individual spleens through a cell strainer. The suspended cells were incubated 7 min on ice with ACT lysis buffer and washed three times with DMEM. The viable cells were added to multiscreen plates in 106, 5 × 105, and 2.5 × 105 cells/well and stimulated with HNTNGVTAACSHEG, IYSTVASSL, or an irrelevant control peptide (GYKDGNEYI) (0.8 μg/ml; ProImmune) before incubation with biotinylated anti-mouse IFN-γ (1 μg/ml; XMG1.2; BD Pharmingen) and streptavidin–alkaline phosphatase (1:3000; GE Healthcare). IFN-γ–producing cells were detected by the BCIP/NBT kit (Zymed Laboratories), and an automated analysis of spots was performed using the Zeiss KS ELISPOT system v 4.3.56 (Carl Zeiss).
Serum transfer
Mice were vaccinated once with 25 μg DNA/EP (n = 6/group). Two (Fig. 5C) or 4 wk (Fig. 7A) later, mice were anesthetized by an s.c. injection of Hypnorm/Dormicum (0.05 ml working solution/10 g) and blood harvested by cardiac puncture. Sera were collected by two successive 5-min centrifugations at 13,000 rpm. Pooled sera (n = 6) were transferred into naive BALB/c by i.v. injection (200 μl volume). Twenty-four hours later, mice were challenged with 5× LD50 PR8 and monitored for weight loss (n = 8/group).
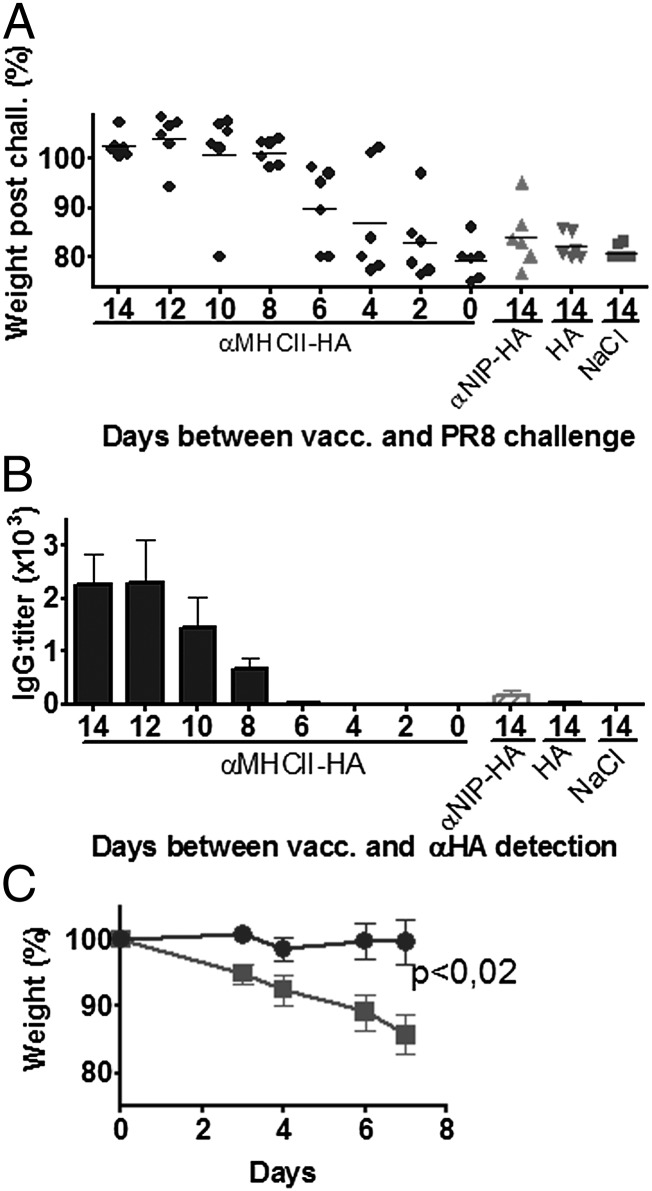
FIGURE 5.
A single MHC II–targeted HA DNA vaccine rapidly induces protective levels of Abs. (A and B) BALB/c mice were vaccinated with 25 μg anti–MHC II–HA (PR8) DNA/EP (black circles) either 14, 12, 10, 8, 6, 4, 2, or 0 d prior to a challenge with PR8 virus. Control groups [anti-NIP–HA (PR8)/EP (gray upward-facing triangles), HA (PR8)/EP (gray downward-facing triangles), and NaCl/EP (gray squares)] were only immunized at day 14 (n = 6/group). (A) Weight following viral challenge, data for day 7 are shown. (B) Immediately prior to challenge, serum samples were obtained, and anti-HA IgG was measured in a PR8 ELISA. (C) Protection by serum transfer. Total of 200 μl serum from BALB/c mice vaccinated with anti–MHC II–HA (PR8)/EP (gray circles) or NaCl/EP (gray squares) 14 d earlier (n = 6/group) were injected i.p. into naive BALB/c mice (n = 8/group). Twenty-four hours later, mice were challenged with PR8 virus and the weight monitored.
FIGURE 7.
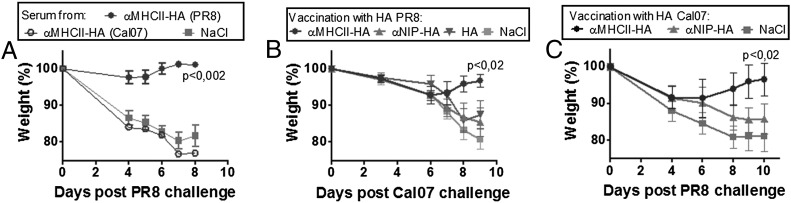
Vaccine-induced T cells, but not Abs, cross-protect between PR8 and Cal07 H1 viruses. (A) Pooled sera from mice vaccinated as indicated 4 wk earlier (n = 6/group) were injected i.p. into naive BALB/c mice (n = 8/group). Twenty-four hours later, recipients were challenged with PR8 and weight monitored. (B) Mice were vaccinated once with 25 μg of the indicated plasmids encoding HA from PR8 (n = 6/group), challenged with Cal07, and monitored for weight loss. (C) Mice were vaccinated once with 25 μg of the indicated plasmids encoding HA from Cal07 (n = 6/group), challenged with PR8, and monitored for weight loss.
T cell depletion
Mice were vaccinated once with 25 μg DNA/EP (n = 6/group). From day 12, groups of mice vaccinated with anti–MHC II–HA were injected every other day i.p. with 400 μg either purified anti-CD4 (GK1.5; American Type Culture Collection) (37) or anti-CD8 (TIB105; American Type Culture Collection) (38), both, or control mAbs (SRF8-B6 and Y13-238). On day 14, mice were challenged with PR8 and monitored for weight loss. At the day of termination postchallenge, mice were euthanized by cervical dislocation and spleens harvested for assessment of in vivo T cell depletion. Single-cell suspensions were prepared and blocked as above, followed by staining with Abs against CD3 (PE-conjugated, 1530-09; Southern Biotechnology Associates), CD8 (allophycocyanin-conjugated, 1550-11; Southern Biotechnology Associates), CD4 (FITC-conjugated, 1540-02; Southern Biotechnology Associates), or isotype-matched controls (PE-conjugated hamster IgG1 [553972; BD Biosciences], allophycocyanin-conjugated rat IgG2a [553932; BD Biosciences], FITC-conjugated rat IgG2a [11214C; BD Pharmingen]). The degree of depletion was calculated to be ≈85% for CD4+ T cells and ≈93% for CD8+ T cells, respectively. Representative results of stained splenocytes from mice depleted with Abs against CD4 or CD8 are shown in Supplemental Fig. 2A and 2B.
T cell transfer
Mice were vaccinated once with 25 μg DNA/EP. Ten days later, mice were euthanized by cervical neck dislocation, and spleens and draining lymph nodes (LNs) were collected. Single-cell suspensions from spleen were prepared as described above, whereas LNs were directly mashed through a cell strainer and washed three times in DMEM. T cells were negatively selected by using the Dynabeads Mouse Pan B (B220) kit (114.41D; Invitrogen) and stained for FACS analyzes as described above with Abs against CD8a (Pacific Blue–conjugated, 558106; BD Biosciences), CD4 (allophycocyanin-conjugated, 1540-11; Southern Biotechnology Associates), CD45R/B220 (FITC-conjugated, 1665-02; Southern Biotechnology Associates), and CD3 (PE-conjugated, 1530-09C; Southern Biotechnology Associates). The purity of the T cell solutions used for transfer was found to be 95–98% (see Supplemental Fig. 3C–E for representative FACS stainings). The purified T cells (5 × 106/mouse) were injected i.v. into naive BALB/c mice (n = 6/group). Twenty-four hours later, mice were challenged with 5× LD50 of PR8 and monitored for weight loss.
Statistical analyzes
Statistical analyzes of Ab responses in sera were performed using one-way ANOVA and Bonferroni multiple comparison test. All other analyses were performed using the nonparametric Mann–Whitney U test (GraphPad Software). For experiments with viral challenge, the statistical testing was performed on data from day 7. The α level was set to 0.05 for all analyzes.
Results
Construction and characterization of the MHC II–targeted influenza vaccine
The DNA-encoded vaccine proteins are homodimers, each chain consisting of: 1) a targeting unit; 2) a dimerization unit derived from the hinge and CH3 exons of human IgG3; and 3) an antigenic unit (Fig. 1A). The antigenic unit, HA of PR8, was truncated at aa 18 and 541 to remove transmembrane sequences that could otherwise have caused problems with secretion. The shortened HA (aa 18–541) gene was fused via the hinge and CH3 exons of human IgG3 domain (25) to the scFv of an mAb specific for a mouse MHC II molecule (I-Ed) expressed on B cells, macrophages, and dendritic cells. The dimeric vaccine is thus denoted anti–MHC II–HA. The construct was inserted into the pLNOH2 vector (39) and expressed under the control of a CMV promoter and an Ig-derived signal sequence. To measure the effect of MHC II targeting, we prepared a control vaccine identical to anti–MHC II–HA, but in which the MHC II–specific scFv was exchanged with an scFv specific for the synthetic hapten NIP (anti-NIP–HA). A standard vaccine containing the HA subunit (aa 18–541) alone served as an additional control (HA).
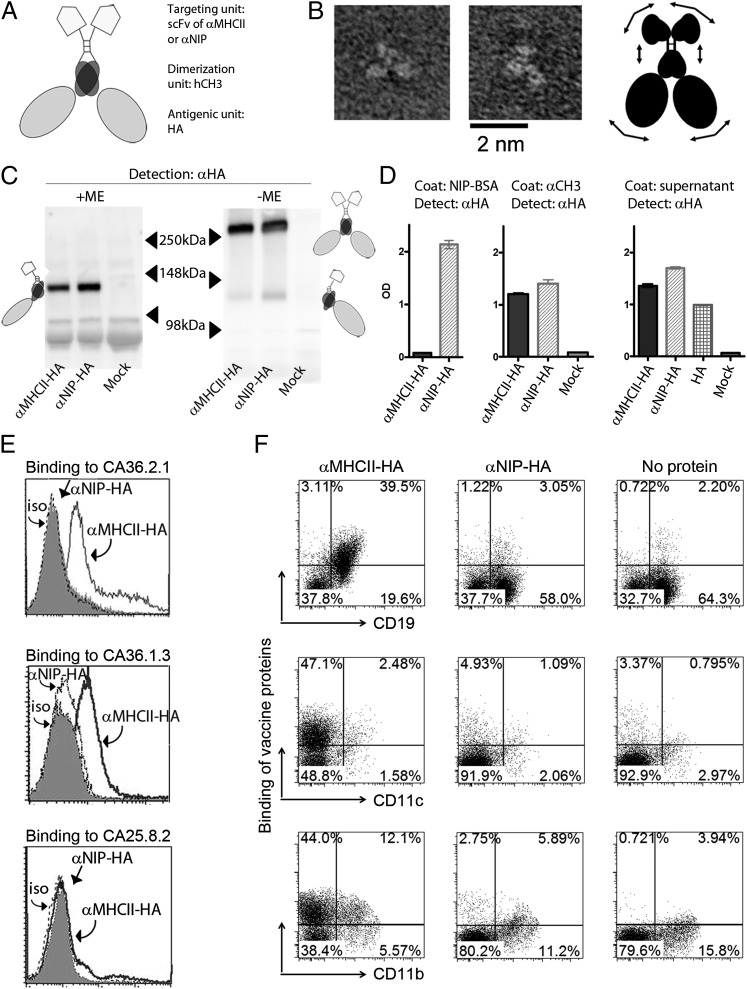
FIGURE 1.
Characterization of HA-containing vaccine molecules. (A) Schematic structure of dimeric vaccine protein. Each chain has an scFv (either anti–MHC II or anti-NIP) as targeting unit and a truncated HA in the antigenic unit, the two being connected by a shortened Ig hinge and hγ3 CH3 domain. (B) Purified NIP-specific HA vaccine proteins as imaged by transmission EM. Representative examples. A cartoon indicating the structure of the vaccine molecule, as suggested by the images, is shown in the right panel. Arrows suggest flexibility. (C) Supernatants of transfected 293E cells were examined by Western blotting with anti-HA mAb under reducing (+ME) or nonreducing (−ME) conditions. Dimers, monomers, and molecular sizes (molecular mass) are indicated. Vaccine proteins (HA from PR8) are indicated below lanes. (D) Binding of vaccine proteins to NIP-BSA or anti-CH3 mAb in sandwich ELISA. An ELISA in which supernatants from 293E transfections were used as coat is shown in the right panel. (E) Binding of vaccine proteins to MHC-transfected L-cell fibroblasts [CA36.2.1 (EβdEαk), CA36.1.3 (EβkEαk) and CA25.8.2 (Dd)]. In (C)–(E), vaccine proteins were detected by anti-HA mAb. (F) Binding of vaccine proteins to CD19+, CD11c+, and CD11b+ splenocytes from BALB/c mice. Vaccine proteins (y-axis) were detected by anti-CH3 mAb.
By EM, the monomeric HA subunits of the dimeric vaccine protein appeared to be independently arrayed without evidence of mutual interaction, in contrast to the trimeric association of membrane HA on influenza virions. The vaccine proteins displayed an inherent flexibility, allowing for some variation of subunit orientation (Fig. 1B).
Cells transfected in vitro with either the anti–MHC II–HA or anti-NIP–HA constructs secreted dimeric vaccine proteins with expected sizes and properties (Fig. 1C, 1D). However, transfection of 293E cells with the plasmid encoding truncated HA alone yielded only low amounts of protein. In an attempt to increase the amount of secreted HA proteins, the Ig-derived signal peptide for this construct was replaced with the naturally occurring HA signal peptide. This modification resulted in increased secretion by transfected cells, comparable to that observed for the other constructs (Fig. 1D, right panel). The anti–MHC II targeting unit was demonstrated to specifically bind MHC II–transfected fibroblasts (Fig. 1E) and CD19+ B cells, CD11c+ dendritic cells, and CD11b+ macrophages from BALB/c spleens (Fig. 1F). As expected, anti-MHC–HA did not bind splenocytes from C57BL/6 mice that do not express I-E MHC II molecules (data not shown).
Previous reports have demonstrated that ligation of MHC II molecules results in signaling. However, the downstream effects appear to differ with cell type and developmental stage, as well as with specificity of the MHC II ligand (40–43). We therefore tested if anti–MHC II–HA added to ex vivo peritoneal cells or splenocytes could upregulate expression of costimulatory CD80 and CD86 on CD19+, CD11b+, and CD11c+ cells. No effect was observed on splenocytes (data not shown) or B cells from the peritoneal cavity (Supplemental Fig. 1). However, anti–MHC II–HA enhanced expression of CD80 and CD86 on CD11b+ and CD11c+ peritoneal cells, but the magnitude of upregulation differed for the two cell types (Supplemental Fig. 1). Similar results were obtained from two mice (Supplemental Fig. 1). Surprisingly, the nonspecific vaccine molecules (anti-NIP–HA) had a partial activating effect compared with medium alone, perhaps due to the HA or CH3 moieties of the molecule. However, anti–MHC II–HA was overall superior at inducing CD80 or CD86 expression compared with anti-NIP–HA. These results are generally consistent with previous reports demonstrating that ligation of MHC II molecules can signal APCs (40–43).
Targeting of HA to MHC II molecules enhances Ab responses
BALB/c mice were injected once intradermally with the different DNA vaccines, immediately followed by EP of the injected site. EP increases the transfection efficiency of DNA (35, 44), resulting in increased vaccine production and local inflammation (45, 46). Vaccination with anti–MHC II–HA induced significantly higher amounts of Abs than did either anti-NIP–HA or HA, as detected in ELISAs against influenza virus (PR8). Increased Ab levels were observed by day 14, peaked around day 50, and persisted at a plateau level for at least 210 d. Increases were observed for total IgG, IgG1, and IgG2a (Fig. 2A–C), whereas IgA, IgE, or IgM were not detected above background levels. The effect of MHC II targeting was even more pronounced when assaying sera by HI assays (Fig. 2D) and was also reflected by increased amounts of neutralizing Abs (Fig. 2E). Thus, targeting of HA to MHC II molecules seemed to not only increase the levels of induced Abs, but also to qualitatively increase their capacity for neutralization.
For assessing the effect of repeated immunizations, mice were vaccinated three times with 14-d intervals. The levels of anti-PR8 Abs increased by several orders of magnitude (Fig. 2F). The increased potency of anti–MHC II–HA vaccination was further reflected in a dose-response experiment. Thus, a single immunization with 3 μg DNA of anti–MHC II–HA induced Ab levels equivalent to that of 50 μg DNA of anti-NIP–HA (Fig. 2G). The increased Ab responses following vaccination with anti–MHC II–HA were not limited to sera, as a single vaccination with anti–MHC II–HA also induced increased IgG titers in BALF compared with controls (Fig. 2H). It should be emphasized that EP of the DNA-injected site was necessary for the induction of enhanced Ab levels (Fig. 2I). The enhanced induction of Ab responses following vaccination with anti–MHC II–HA was not a peculiarity of the Th2-prone BALB/c mice, because similar findings were observed in the Th1-prone B10.D2 strain (Fig. 2J). Furthermore, the induction of Ab responses was dependent on the presence of T cells because BALB/c nude mice, which lack a thymus, failed to respond to anti–MHC II–HA vaccination (Fig. 2K). In summary, the hierarchical anti–MHC II–HA > anti-NIP–HA > > HA vaccine efficacy for Ab induction suggests an immune-potentiating effect of both MHC II targeting and bivalency of Ag.
Targeting of HA to MHC II molecules induces complete protection against influenza
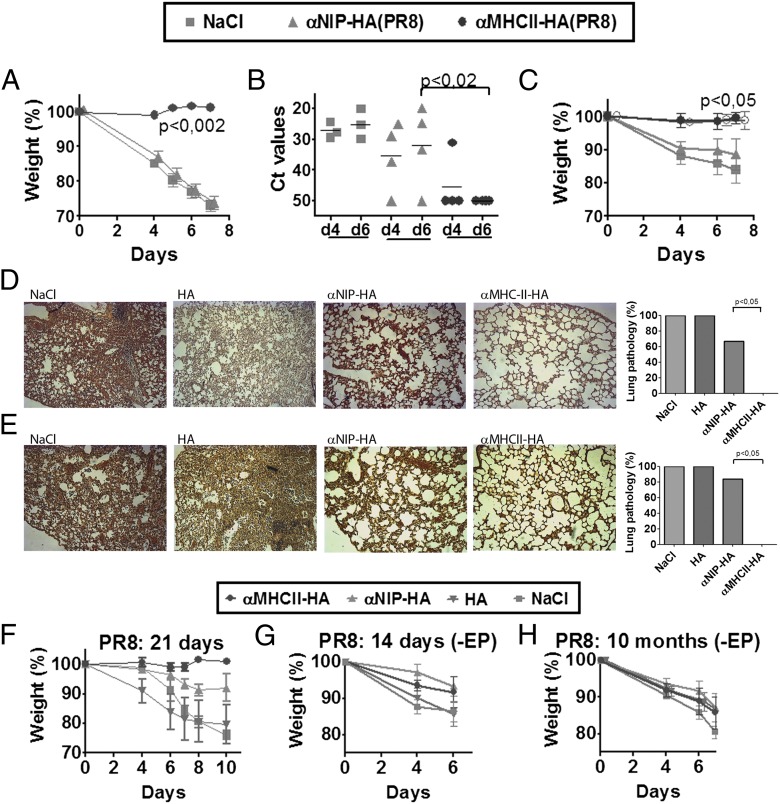
Given the strong Ab responses elicited by DNA immunization with anti–MHC II–HA, we next tested whether such vaccination conferred protection against a viral challenge. Mice were vaccinated once and challenged either 14 d (Fig. 3A, 3B) or 10 mo (Fig. 3C) later with a lethal dose of PR8 virus. Mice that received saline or anti-NIP–HA rapidly lost weight and had to be euthanized by day 7 (Fig. 3A, 3C). In contrast, mice vaccinated with anti–MHC II–HA maintained their weight and showed no signs of disease. PCR analysis of viral loads in nasal washes showed that mice immunized with anti–MHC II–HA for the most part had cleared the virus infection by day 4 and completely so by day 6 (Fig. 3B). Lungs harvested from saline- or HA-vaccinated mice displayed histiocytic alveolitis and interstitial pneumonia. In contrast, mice vaccinated with anti–MHC II–HA had healthy lungs. Anti-NIP–HA–vaccinated mice also displayed lung pathology, but less severe than that of the NaCl and HA mice, consistent with partial clearance of virus in some mice (Fig. 3D, 3E). Anti–MHC II–HA DNA vaccination was also protective in B10.D2 mice, demonstrating that the observed immune responses were not strain specific (Fig. 3F). Vaccination in the absence of EP failed to induce Abs and protection (Fig. 3G, 3H).
FIGURE 3.
MHC II–targeted HA vaccine delivered as DNA provide short- and long-term protection against a lethal challenge with influenza virus. (A and B) BALB/c mice were immunized once with 25 μg plasmids/EP as indicated (key at top) (n = 6/group), challenged 14 d later with a lethal dose of influenza virus (PR8), and monitored for weight loss (A) and virus (B) in nasal washes at days 4 and 6 after challenge (quantitative RT-PCR) (n = 4/group). (C) Mice were immunized once 10 mo earlier with the indicated plasmids (25 μg/EP) (n = 6/group), challenged with PR8, and monitored for weight loss. Mice surviving a previous challenge with PR8 influenza virus was included as positive control (n = 3) (○). (D and E) Representative HE-stained sections of lungs (original magnification ×10) collected 7 d postchallenge. Mice (n = 6/group) were vaccinated once as indicated (25 μg plasmids/EP) and challenged 14 d (D) or 10 mo (E) later with PR8. Frequency of lung pathology (histiocytic alveolitis, interstitial edema, and thickening of alveolar septum) is graphically summarized in the right panels. p < 0.05 for anti–MHC II versus anti-NIP–HA, HA, and NaCl. (F) B10.D2 H-2d mice were immunized once with 25 μg DNA/EP (HA from PR8) or NaCl. Three weeks after vaccination, mice were challenged with PR8 virus and monitored for weight loss (n = 5/group). BALB/c mice vaccinated in the absence of EP were challenged 14 d (G) or 10 mo (H) later with a lethal dose of influenza PR8 and monitored for weight loss. Data in (G) and (H) are not significant. Ct, Threshold cycle.
Targeted DNA HA vaccines can be produced and employed within weeks
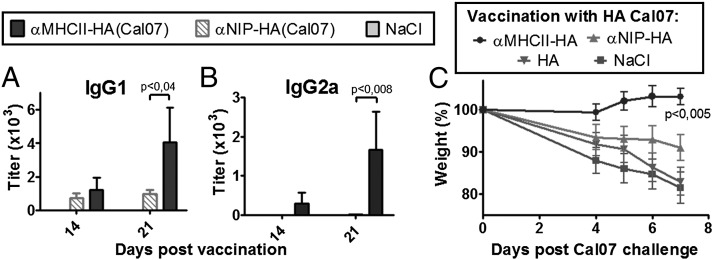
During the spring of 2009, the reassortant H1N1 Mexican swine flu emerged as a pandemic threat. We used this occasion to test how rapid an MHC II–targeted vaccine for this virus could be established. As soon as the HA sequence was available online, we inserted this sequence, Cal07, into the vaccine-encoding plasmids, replacing the PR8 HA. Within 3 wk, mice were vaccinated with the tailored Cal07 vaccine and monitored for Ab responses (Fig. 4A, 4B). MHC II targeting improved IgG1 and IgG2a Ab responses against Cal07. In a separate experiment, mice were challenged 14 d after a single vaccination with the virulent Cal07 virus (Fig. 4C). Again, MHC II targeting was essential for complete protection against disease. Thus, the targeted DNA vaccine strategy works for at least two different H1 strains of influenza virus (PR8 and Cal07), and tailored vaccines can be generated and tested in model systems within weeks of an influenza outbreak.
FIGURE 4.
Extension to Cal07. (A–C) BALB/c mice were DNA/EP vaccinated with the indicated plasmids encoding HA from Cal07 (n = 6/group). Serum levels of IgG1 (A) and IgG2a (B) anti-HA Abs measured in a Cal07 ELISA. Values given are mean ± SEM. (C) BALB/c mice were DNA/EP vaccinated once with 25 μg of the indicated plasmids encoding HA from Cal07 (n = 6/group), challenged 14 d later by a lethal dose of Cal07, and monitored for weight loss.
Mice are protected against influenza within 8 d after a single vaccination, and protection can be transferred with serum
To determine how fast an MHC II–targeted immunization with HA could confer protection, mice were vaccinated with anti–MHC II–HA 14, 12, 10, 8, 6, 4, 2, or 0 d prior to a lethal challenge with PR8. Eight days after vaccination, mice had acquired protective immunity against viral challenge (Fig. 5A). Protection occurred at the same time as Abs rapidly increased in sera (Fig. 5B), indicating a correlation between protection and Ab responses.
To directly test if Abs conferred protection, sera from six mice immunized 14 d earlier with anti–MHC II–HA were transferred into naive mice that were then challenged with PR8 virus and monitored for development of disease. Transfer of 0.2 ml serum completely protected against a viral challenge (Fig. 5C).
Targeting of HA to MHC II molecules enhances T cell responses that contribute to protection against viral challenge
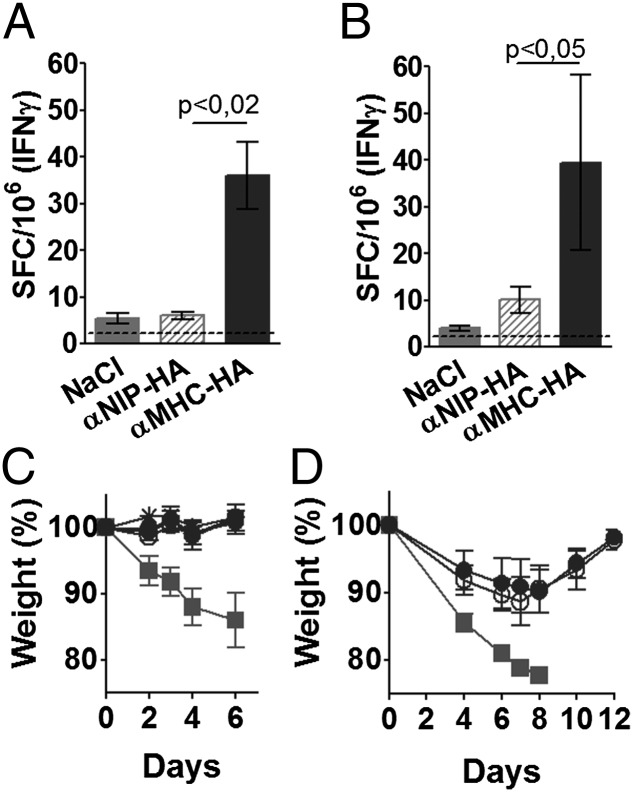
For assessment of T cell responses, splenocytes from BALB/c mice, immunized once with either the APC-targeted anti–MHC II–HA or nontargeted controls, were stimulated with a class II–restricted peptide [HNTNGVTAACSHEG (47)], a class I–restricted peptide [IYSTVASSL (48)], or a control peptide. Targeting to MHC II molecules on APCs enhanced the levels of HA-reactive IFN-γ–producing T cells (Fig. 6A, 6B).
FIGURE 6.
Protective T cell immunity induced by a single MHC II–targeted HA (PR8) DNA vaccine. (A and B) Mice were immunized once with 25 μg DNA/EP as indicated (n = 6/group). Fourteen days later, spleens cells were stimulated with either an MHC II–restricted (HNTNGVTAACSHEG) (A) or a class I–restricted HA peptide (IYSTVASSL) (B) and analyzed by ELISPOT. Dotted lines indicate maximum levels observed after stimulation with an irrelevant control peptide (GYKDGNEYI). (C) Mice were immunized with 25 μg anti–MHC II–HA (PR8)/EP (n = 6/group). From day 12 and until the termination of the experiment, mice were injected every other day i.p. with 400 μg of either anti-CD4 (open circles) or anti-CD8 (open triangles) mAbs, both (+), or control mAb (closed circles). A group of control mice were immunized with NaCl/EP (gray squares). On day 14, mice were challenged with PR8 and monitored for weight development. See Supplemental Fig. 2A and 2B for efficiency of depletion. (D) Mice were immunized with 25 μg anti-MHC II–HA (PR8)/EP (n = 6/group). Ten days later, T cells were purified by negative selection from spleens (open circles) and draining LNs (closed circles) and injected i.v. into naive BALB/c mice (n = 6/group). A group of control mice received T cells from mice vaccinated with NaCl/EP (gray squares). Twenty-four hours later, recipients were challenged with PR8 and monitored for weight development. See Supplemental Fig. 3A–E for survival curves and efficacy of negative selection. SFC, spot-forming cell.
For an assessment of T cell contributions to protection, mice vaccinated once with anti–MHC II–HA (PR8) were treated from day 12 with injections of depleting Abs against CD8, CD4, or both. The efficiencies of depletion were determined to reach 85 and 93% for CD4+ and CD8+ T cells, respectively (see Supplemental Fig. 2A, 2B for representative FACS stainings). The absence of weight loss in T cell–depleted mice challenged with virus on day 14 (Fig. 6C) was not surprising because Abs alone protected against influenza, as demonstrated by serum transfer (Fig. 5C). Thus, even though HA-specific T cells were required for the induction of anti-HA Abs (Fig. 2K), they were not needed for the elimination of virus. However, this experiment did not exclude a protective role of T cells because the protective anti-HA Abs present in this experimental setting could have disguised their importance. We therefore transferred B cell–depleted LN and spleen cells from immunized mice into naive recipients that were then challenged with virus. T cell recipients gradually lost some weight for the first 7 d, but then recovered, indicating that the anti–MHC II–HA-induced HA-specific T cells independently can confer protection against PR8 influenza virus (Fig. 6D, Supplemental Fig. 3A–E). Delayed protection is typical of T cell–mediated protection against influenza (49) because the initial entry of virus into host cells will not be prevented by T cells. In conclusion, a single anti–MHC II–HA DNA vaccination induces protective levels of both Abs and T cells (Figs. 5C, 6D).
Targeted DNA HA vaccines induces T cells that cross-protect between H1 strains
Anti-HA Abs generally do not cross-react between different strains of influenza virus. Consistent with this, sera from mice vaccinated with anti–MHC II–HA (PR8) failed to recognize Cal07 virus in a microneutralization assay (Supplemental Fig. 4), and transfer of sera from mice vaccinated with anti–MHC II–HA (Cal07) did not protect recipients against a challenge with PR8 (Fig. 7A). However, despite the lack of serological cross-reactivity, there is considerable sequence homology between the HAs of these two H1 strains, and they share two out of three HA T cell epitopes described in BALB/c mice, a CD8 epitope (aa 533–541/Kd) and a CD4 epitope (aa 111–119/I-Ed) (PR8 numbering) (47, 48, 50). Therefore, because anti–MHC II–HA (PR8) vaccination induced T cell–mediated protection against homologous PR8, we asked whether MHC II–targeted vaccination could induce T cell–mediated cross-protection between the two different H1 viruses. Mice were vaccinated with anti–MHC II–HA (PR8) and then challenged with the Cal07 virus. Mice initially showed moderate weight loss, followed by a regain of weight (Fig. 7B). Conversely, mice vaccinated with anti–MHC II–HA (Cal07) and challenged with PR8 lost some weight but then recovered (Fig. 7C). In both cases, the protection was dependent on MHC II targeting. Because Abs did not cross-protect, it is highly likely that T cells confer vaccine-elicited cross-protection between the two H1 strains. Immunization with HA alone was not included in all of the experiments with viral heterochallenge because such immunization did not confer protection even against homologous challenge (Figs. 3D–F, 4C).
Discussion
In response to pandemic influenza, it is important to quickly produce large quantities of vaccines that rapidly can induce protective Abs. We describe in this study a DNA vaccine against influenza, in which transfected cells serve as production factories for secreted dimeric molecules that target HA to MHC II molecules on mouse APC. A single immunization induced within 8 d large amounts of strain-specific Abs that protected mice against challenges with the homologous H1 virus. Ab production and protection was long-lasting. In addition, the vaccine enhanced the induction of T cells that cross-protected between different H1 strains of influenza.
Several factors could contribute to the accelerated and heightened immunogenicity of the MHC II–targeted HA DNA vaccine, given intradermally in combination with EP. Previous studies with a similar DNA vaccine (25), together with the present data, suggest the following mechanism: 1) EP of the injection site increases the number of locally transfected cells (31, 32), resulting in increased secretion of vaccine proteins (25). 2) The secreted vaccine proteins bind MHC II molecules on APC. 3) Such binding can result in increased expression of costimulatory CD80/CD86 molecules on APC, consistent with previous reports demonstrating that cross-linking of MHC II molecules induce APC maturation (42, 43). 4) APCs primed with anti–MHC II–HA vaccine proteins are found in draining LNs of immunized mice (25). 5) Targeting of vaccine proteins to APC results in a more efficient stimulation of CD4+ Th cells in vivo and in vitro, probably due to increased peptide loading of MHC II molecules (25). 6) The dimeric vaccine proteins expressing two antigenic HA units may more extensively cross link BCRs, resulting in more efficient B cell stimulation.
The specificity of anti-HA Abs obtained by the MHC II–targeted DNA vaccine resembled that observed after conventional egg-based vaccines in that Abs were strain specific. Interestingly, the enhancing effect of MHC II targeting was especially pronounced in HI and microneutralization assays, indicating that the induced Abs preferentially were directed against the head of the HA molecule. As a mechanism for such a skewing of Ab specificity for HA, anti–MHC II–HA proteins could bridge a synaptic cleft (51) between MHC II+ APC and HA-specific B cells, resulting in the head of HA being oriented toward the HA-specific BCR. Alternatively, the anti–MHC II–HA proteins could cross link BCR and MHC II molecules on single HA-specific B cells, resulting in enhanced B cell stimulation. Regardless of mechanism, HA-specific B cells needed help from T cells for production of anti-HA Abs.
The current vaccine molecules were targeted to MHC II molecules on APCs. This choice was based upon the previous findings by others that anti–MHC II Abs conjugated to Ags induce strong Ab and T cells responses against the Ag (12, 17, 19, 52). There may indeed exist even better targets on APC than MHC II for induction of strong Ab responses, and further studies in this respect are clearly warranted.
An important aspect of the present vaccine design is the inclusion of large protein Ags, such as HA, with an intact conformation. This is likely to be important for two reasons: 1) elicitation of B cell (Ab) responses against conformationally dependent antigenic determinants require large Ags; and 2) long polypeptides have an increased chance of generating peptides that fit any of the polymorphic MHC binding sites found within a species, facilitating the generation of T cell responses in any individual. In this respect, the described vaccine molecules can accommodate a range of large viral and bacterial protein Ags, the hitherto largest being the truncated HA (541 aa) used in this study.
Trimerization of HA has been described a prerequisite for transport from ER to the Golgi complex (53, 54). Despite this, we demonstrate efficient secretion of monomeric HA inserted into the fusion vaccine. A monomeric structure of HA was supported by EM that revealed single, nonaggregated, dimerized vaccine proteins in which HA appeared monomeric and flexible in their orientation within the dimer. Secretion could be facilitated by the Ig-like structure of the vaccine molecule, with monomeric HA trailing the aminoterminal scFv-CH3. Despite the monomeric structure of HA in a dimeric vaccine protein, the targeted fusion vaccine rapidly induced large amounts of Abs that upon serum transfer protected naive mice against influenza. These results demonstrate that the monomeric HA in a dimeric context displays relevant antigenic determinants corresponding to those expressed by virus. However, the HA vaccine proteins failed to agglutinate erythrocytes, indicating that they lacked the degree of multimerization required for detectable binding sialic acid in the hemagglutination assay (55). Alternatively, monomeric HA expressed in the dimeric fusion protein could have lost their binding site for sialic acid.
Vaccination with the nontargeted anti-NIP–HA control resulted in increased anti-HA immune responses compared with that elicited by HA alone. The enhanced immunogenicity of anti-NIP–HA, although inferior to that of anti–MHC II–HA, may be attributed to bivalency of HA and thus an ability to cross link anti-HA BCR. In addition, efficient secretion and the xenogeneic dimerization unit could both have contributed. Further studies are needed to assess the immune-enhancing properties inherent to the dimeric vaccine backbone, in the absence of targeting.
In the present experiments, DNA/EP vaccination with HA alone induced immune responses that were only barely detectable above background. This may appear surprising because others have previously observed more pronounced Ab responses, and protection against viral challenge, following DNA vaccination with HA. The discrepancy might be explained by the current study employing only a single immunization, a low injection volume, and a low DNA dose. By contrast, previous studies by others describe protection first after several immunizations (6, 7, 10), larger volumes (7, 9, 10, 56, 57), and larger amounts of DNA (9, 56–58). Another factor that may explain the discrepancy is that we have used a truncated HA in which most of the transmembrane and intracellular parts have been deleted. Others (6, 9, 10, 56–58) have used complete HA or selected influenza epitopes (7) for immunizations. Thus, the discrepancy might be explained by secreted HA (presumably monomeric) being less immunogenic than HA in a membrane-bound (presumably trimeric) form. Finally, the in-house pLNOH2 vector used in the present studies has not been optimized for high Ag expression in transfected cells, and it might well be that we would have observed a response to (truncated) HA with an improved vector. With an optimized commercial vector, we have indeed observed increased immune responses (G. Grodeland, unpublished observations). Although not yet tested, we find it likely that the anti–MHC II–HA > anti-NIP–HA > HA hierarchy will hold true also with optimized vectors. If so, optimized vectors may further enhance the dose-sparing effect observed with MHC II–targeted DNA vaccines.
In addition to Abs, the MHC II–targeted HA vaccine enhanced T cell responses that cross-protected between different H1 strains of influenza, even in the absence of anti-HA Abs. However, the protection was not as efficient as with Abs because challenged mice suffered some weight loss prior to recovery. Because T cell epitopes are often conserved between different strains of influenza, vaccine-induced T cell immunity may have the advantage of conferring some cross-protection against different influenza viruses, even in the event of an antigenic shift.
An MHC II–targeted DNA vaccine for a novel influenza strain (Cal07) was generated and tested within 3 wk after the HA sequences were available online. Moreover, large amounts of DNA plasmids can be rapidly produced, compared with the slow and resource-demanding production of conventional egg-based influenza vaccines. Thus, in the face of pandemic threats, the technology described in this study should allow for rapid generation of large amounts of vaccine that induce protection in vaccinees within days. For human application, concerns may be raised with regard to the safety of DNA vaccination. Although DNA vaccines are not yet approved for prophylactic vaccination of humans, they have been licensed for rhabdo virus in rainbow trout (59) and for the West Nile virus in horses (60). Therapeutic DNA vaccines delivered with EP have been experimentally tested in cancer patients without serious side effects (61), and several clinical trials with therapeutic DNA vaccines for cancer are currently ongoing in humans (62, 63). It will be important to test if the triad of DNA injection, EP, and MHC II–targeted fusion protein vaccines can improve the efficacy of DNA vaccination in larger animals and humans. In that respect, the DNA dose-sparing effect observed with MHC II–targeted HA vaccines in mice is encouraging.
Acknowledgments
We thank Cellectis for providing the electroporator and Helge Scott for advices in microscopic evaluation of lung sections. We also thank Elisabeth Vikse for technical help and Even Fossum and Keith Thompson for manuscript comments. Mice were housed at the Department of Comparative Medicine, Oslo University Hospital, and at the animal facility at the Norwegian Institute of Public Health, Oslo, Norway.
This work was supported by funding from Global Health and Vaccination Research (The Research Council of Norway) and Helse Sør-Øst (Regional Health Authority), Norway.
The online version of this article contains supplemental material.
- BALF
- bronchoalveolar lavage fluids
- Cal07
- A/California/07/09 (H1N1)
- EM
- electron microscopy
- EP
- electroporation
- HA
- hemagglutinin
- HI
- hemagglutination-inhibition
- LN
- lymph node
- MHC II
- MHC class II
- NIP
- 4-hydroxy-3-iodo-5-nitrophenylacetic acid
- PR8
- A/PR/8/34 (H1N1)
- scFv
- single-chain variable fragment.
Disclosures
G.G., A.B.F., and B.B. are inventors on patent applications filed on the vaccine molecules according to institutional rules through the Technology Transfer Offices of the University of Oslo and Oslo University Hospital. A.B.F. is part-time Chief Scientific Officer of Vaccibody. B.B. is head of the Scientific Panel of Vaccibody. The other authors have no financial conflicts of interest.
References
- 1.Gatherer D. 2009. The 2009 H1N1 influenza outbreak in its historical context. J. Clin. Virol. 45: 174–178 [DOI] [PubMed] [Google Scholar]
- 2.Bavagnoli L., Maga G. 2011. The 2009 influenza pandemic: promising lessons for antiviral therapy for future outbreaks. Curr. Med. Chem. 18: 5466–5475 [DOI] [PubMed] [Google Scholar]
- 3.Perdue M. L., Arnold F., Li S., Donabedian A., Cioce V., Warf T., Huebner R. 2011. The future of cell culture-based influenza vaccine production. Expert Rev. Vaccines 10: 1183–1194 [DOI] [PubMed] [Google Scholar]
- 4.Keitel W. A., Atmar R. L. 2009. Vaccines for pandemic influenza: summary of recent clinical trials. Curr. Top. Microbiol. Immunol. 333: 431–451 [DOI] [PubMed] [Google Scholar]
- 5.Puig Barberà J., González Vidal D. 2007. MF59-adjuvanted subunit influenza vaccine: an improved interpandemic influenza vaccine for vulnerable populations. Expert Rev. Vaccines 6: 659–665 [DOI] [PubMed] [Google Scholar]
- 6.Song J. M., Kim Y. C., O E., Compans R. W., Prausnitz M. R., Kang S. M. 2012. DNA vaccination in the skin using microneedles improves protection against influenza. Mol. Ther. 20: 1472–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei H., Lenz S. D., Thompson D. H., Pogranichniy R. M. 2012. DNA-vaccine platform development against H1N1 subtype of swine influenza A viruses. Viral Immunol. 25: 297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie H., Liu T., Chen H., Huang X., Ye Z. 2007. Evaluating the vaccine potential of an influenza A viral hemagglutinin and matrix double insertion DNA plasmid. Vaccine 25: 7649–7655 [DOI] [PubMed] [Google Scholar]
- 9.Zhou F., Wang G., Buchy P., Cai Z., Chen H., Chen Z., Cheng G., Wan X. F., Deubel V., Zhou P. 2012. A triclade DNA vaccine designed on the basis of a comprehensive serologic study elicits neutralizing antibody responses against all clades and subclades of highly pathogenic avian influenza H5N1 viruses. J. Virol. 86: 6970–6978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin S. C., Lin Y. F., Chong P., Wu S. C. 2012. Broader neutralizing antibodies against H5N1 viruses using prime-boost immunization of hyperglycosylated hemagglutinin DNA and virus-like particles. PLoS ONE 7: e39075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawamura H., Berzofsky J. A. 1986. Enhancement of antigenic potency in vitro and immunogenicity in vivo by coupling the antigen to anti-immunoglobulin. J. Immunol. 136: 58–65 [PubMed] [Google Scholar]
- 12.Carayanniotis G., Barber B. H. 1987. Adjuvant-free IgG responses induced with antigen coupled to antibodies against class II MHC. Nature 327: 59–61 [DOI] [PubMed] [Google Scholar]
- 13.Snider D. P., Segal D. M. 1987. Targeted antigen presentation using crosslinked antibody heteroaggregates. J. Immunol. 139: 1609–1616 [PubMed] [Google Scholar]
- 14.Casten L. A., Pierce S. K. 1988. Receptor-mediated B cell antigen processing. Increased antigenicity of a globular protein covalently coupled to antibodies specific for B cell surface structures. J. Immunol. 140: 404–410 [PubMed] [Google Scholar]
- 15.Lees A., Morris S. C., Thyphronitis G., Holmes J. M., Inman J. K., Finkelman F. D. 1990. Rapid stimulation of large specific antibody responses with conjugates of antigen and anti-IgD antibody. J. Immunol. 145: 3594–3600 [PubMed] [Google Scholar]
- 16.Mjaaland S., Fossum S. 1990. Modulation of immune responses with monoclonal antibodies. I. Effects on regional lymph node morphology and on anti-hapten responses to haptenized monoclonal antibodies. Eur. J. Immunol. 20: 1457–1461 [DOI] [PubMed] [Google Scholar]
- 17.Skea D. L., Douglas A. R., Skehel J. J., Barber B. H. 1993. The immunotargeting approach to adjuvant-independent immunization with influenza haemagglutinin. Vaccine 11: 994–1002 [DOI] [PubMed] [Google Scholar]
- 18.Baier G., Baier-Bitterlich G., Looney D. J., Altman A. 1995. Immunogenic targeting of recombinant peptide vaccines to human antigen-presenting cells by chimeric anti-HLA-DR and anti-surface immunoglobulin D antibody Fab fragments in vitro. J. Virol. 69: 2357–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Estrada A., McDermott M. R., Underdown B. J., Snider D. P. 1995. Intestinal immunization of mice with antigen conjugated to anti-MHC class II antibodies. Vaccine 13: 901–907 [DOI] [PubMed] [Google Scholar]
- 20.Biragyn A., Tani K., Grimm M. C., Weeks S., Kwak L. W. 1999. Genetic fusion of chemokines to a self tumor antigen induces protective, T-cell dependent antitumor immunity. Nat. Biotechnol. 17: 253–258 [DOI] [PubMed] [Google Scholar]
- 21.Lunde E., Munthe L. A., Vabø A., Sandlie I., Bogen B. 1999. Antibodies engineered with IgD specificity efficiently deliver integrated T-cell epitopes for antigen presentation by B cells. Nat. Biotechnol. 17: 670–675 [DOI] [PubMed] [Google Scholar]
- 22.Caminschi I., Proietto A. I., Ahmet F., Kitsoulis S., Shin Teh J., Lo J. C., Rizzitelli A., Wu L., Vremec D., van Dommelen S. L., et al. 2008. The dendritic cell subtype-restricted C-type lectin Clec9A is a target for vaccine enhancement. Blood 112: 3264–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charalambous A., Oks M., Nchinda G., Yamazaki S., Steinman R. M. 2006. Dendritic cell targeting of survivin protein in a xenogeneic form elicits strong CD4+ T cell immunity to mouse survivin. J. Immunol. 177: 8410–8421 [DOI] [PubMed] [Google Scholar]
- 24.Flamar A. L., Zurawski S., Scholz F., Gayet I., Ni L., Li X. H., Klechevsky E., Quinn J., Oh S., Kaplan D. H., et al. 2012. Noncovalent assembly of anti-dendritic cell antibodies and antigens for evoking immune responses in vitro and in vivo. J. Immunol. 189: 2645–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fredriksen A. B., Sandlie I., Bogen B. 2006. DNA vaccines increase immunogenicity of idiotypic tumor antigen by targeting novel fusion proteins to antigen-presenting cells. Mol. Ther. 13: 776–785 [DOI] [PubMed] [Google Scholar]
- 26.Fredriksen A. B., Bogen B. 2007. Chemokine-idiotype fusion DNA vaccines are potentiated by bivalency and xenogeneic sequences. Blood 110: 1797–1805 [DOI] [PubMed] [Google Scholar]
- 27.Schjetne K. W., Fredriksen A. B., Bogen B. 2007. Delivery of antigen to CD40 induces protective immune responses against tumors. J. Immunol. 178: 4169–4176 [DOI] [PubMed] [Google Scholar]
- 28.Hobson D., Curry R. L., Beare A. S., Ward-Gardner A. 1972. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J. Hyg. (Lond.) 70: 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Staudt L. M., Gerhard W. 1983. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. I. Significant variation in repertoire expression between individual mice. J. Exp. Med. 157: 687–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roux K. H. 1989. Immunoelectron microscopy of idiotype-anti-idiotype complexes. Methods Enzymol. 178: 130–144 [DOI] [PubMed] [Google Scholar]
- 31.Roux K. H. 1996. Negative-Stain Immunoelectron-Microscopic Analysis of Small Macromolecules of Immunologic Significance. Methods 10: 247–256 [DOI] [PubMed] [Google Scholar]
- 32.Shastri N., Malissen B., Hood L. 1985. Ia-transfected L-cell fibroblasts present a lysozyme peptide but not the native protein to lysozyme-specific T cells. Proc. Natl. Acad. Sci. USA 82: 5885–5889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reed L. J., Muench H. 1938. Simple method of estimating fifty percent endpoints. Am. J. Hyg. 27: 493–497 [Google Scholar]
- 34.Roos A. K., Moreno S., Leder C., Pavlenko M., King A., Pisa P. 2006. Enhancement of cellular immune response to a prostate cancer DNA vaccine by intradermal electroporation. Mol. Ther. 13: 320–327 [DOI] [PubMed] [Google Scholar]
- 35.Roos A. K., Eriksson F., Walters D. C., Pisa P., King A. D. 2009. Optimization of skin electroporation in mice to increase tolerability of DNA vaccine delivery to patients. Mol. Ther. 17: 1637–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prat M., Gribaudo G., Comoglio P. M., Cavallo G., Landolfo S. 1984. Monoclonal antibodies against murine gamma interferon. Proc. Natl. Acad. Sci. USA 81: 4515–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Z., Day Y. J., Toufektsian M. C., Xu Y., Ramos S. I., Marshall M. A., French B. A., Linden J. 2006. Myocardial infarct-sparing effect of adenosine A2A receptor activation is due to its action on CD4+ T lymphocytes. Circulation 114: 2056–2064 [DOI] [PubMed] [Google Scholar]
- 38.Andreasson K., Eriksson M., Tegerstedt K., Ramqvist T., Dalianis T. 2010. CD4+ and CD8+ T cells can act separately in tumour rejection after immunization with murine pneumotropic virus chimeric Her2/neu virus-like particles. PLoS ONE 5: e11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Norderhaug L., Olafsen T., Michaelsen T. E., Sandlie I. 1997. Versatile vectors for transient and stable expression of recombinant antibody molecules in mammalian cells. J. Immunol. Methods 204: 77–87 [DOI] [PubMed] [Google Scholar]
- 40.Altomonte M., Pucillo C., Damante G., Maio M. 1993. Cross-linking of HLA class II antigens modulates the release of tumor necrosis factor-alpha by the EBV-B lymphoblastoid cell line JY. J. Immunol. 151: 5115–5122 [PubMed] [Google Scholar]
- 41.Andreae S., Buisson S., Triebel F. 2003. MHC class II signal transduction in human dendritic cells induced by a natural ligand, the LAG-3 protein (CD223). Blood 102: 2130–2137 [DOI] [PubMed] [Google Scholar]
- 42.Baleeiro R. B., Wiesmüller K. H., Dähne L., Lademann J., Barbuto J. A., Walden P. 2013. Direct activation of human dendritic cells by particle-bound but not soluble MHC class II ligand. PLoS ONE 8: e63039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lokshin A. E., Kalinski P., Sassi R. R., Mailliard R. B., Müller-Berghaus J., Storkus W. J., Peng X., Marrangoni A. M., Edwards R. P., Gorelik E. 2002. Differential regulation of maturation and apoptosis of human monocyte-derived dendritic cells mediated by MHC class II. Int. Immunol. 14: 1027–1037 [DOI] [PubMed] [Google Scholar]
- 44.Aihara H., Miyazaki J. 1998. Gene transfer into muscle by electroporation in vivo. Nat. Biotechnol. 16: 867–870 [DOI] [PubMed] [Google Scholar]
- 45.Peng B., Zhao Y., Xu L., Xu Y. 2007. Electric pulses applied prior to intramuscular DNA vaccination greatly improve the vaccine immunogenicity. Vaccine 25: 2064–2073 [DOI] [PubMed] [Google Scholar]
- 46.Roos A. K., Eriksson F., Timmons J. A., Gerhardt J., Nyman U., Gudmundsdotter L., Bråve A., Wahren B., Pisa P. 2009. Skin electroporation: effects on transgene expression, DNA persistence and local tissue environment. PLoS ONE 4: e7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott B., Liblau R., Degermann S., Marconi L. A., Ogata L., Caton A. J., McDevitt H. O., Lo D. 1994. A role for non-MHC genetic polymorphism in susceptibility to spontaneous autoimmunity. Immunity 1: 73–83 [DOI] [PubMed] [Google Scholar]
- 48.Tamura M., Kuwano K., Kurane I., Ennis F. A. 1998. Definition of amino acid residues on the epitope responsible for recognition by influenza A virus H1-specific, H2-specific, and H1- and H2-cross-reactive murine cytotoxic T-lymphocyte clones. J. Virol. 72: 9404–9406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo H., Santiago F., Lambert K., Takimoto T., Topham D. J. 2011. T cell-mediated protection against lethal 2009 pandemic H1N1 influenza virus infection in a mouse model. J. Virol. 85: 448–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hackett C. J., Dietzschold B., Gerhard W., Ghrist B., Knorr R., Gillessen D., Melchers F. 1983. Influenza virus site recognized by a murine helper T cell specific for H1 strains. Localization to a nine amino acid sequence in the hemagglutinin molecule. J. Exp. Med. 158: 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Batista F. D., Iber D., Neuberger M. S. 2001. B cells acquire antigen from target cells after synapse formation. Nature 411: 489–494 [DOI] [PubMed] [Google Scholar]
- 52.Berg S. F., Mjaaland S., Fossum S. 1994. Comparing macrophages and dendritic leukocytes as antigen-presenting cells for humoral responses in vivo by antigen targeting. Eur. J. Immunol. 24: 1262–1268 [DOI] [PubMed] [Google Scholar]
- 53.Boulay F., Doms R. W., Webster R. G., Helenius A. 1988. Posttranslational oligomerization and cooperative acid activation of mixed influenza hemagglutinin trimers. J. Cell Biol. 106: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Copeland C. S., Doms R. W., Bolzau E. M., Webster R. G., Helenius A. 1986. Assembly of influenza hemagglutinin trimers and its role in intracellular transport. J. Cell Biol. 103: 1179–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stevens J., Blixt O., Glaser L., Taubenberger J. K., Palese P., Paulson J. C., Wilson I. A. 2006. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J. Mol. Biol. 355: 1143–1155 [DOI] [PubMed] [Google Scholar]
- 56.Tan L., Lu H., Zhang D., Tian M., Hu B., Wang Z., Jin N. 2010. Protection against H1N1 influenza challenge by a DNA vaccine expressing H3/H1 subtype hemagglutinin combined with MHC class II-restricted epitopes. Virol. J. 7: 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan L., Lu H., Zhang D., Wang K., Tian M., Liu C., Liu Y., Hu B., Jin N. 2011. Efficacy of seasonal pandemic influenza hemagglutinin DNA vaccines delivered by electroporation against a seasonal H1N1 virus challenge in mice. Sci China Life Sci 54: 293–299 [DOI] [PubMed] [Google Scholar]
- 58.Xu K., Ling Z. Y., Sun L., Xu Y., Bian C., He Y., Lu W., Chen Z., Sun B. 2011. Broad humoral and cellular immunity elicited by a bivalent DNA vaccine encoding HA and NP genes from an H5N1 virus. Viral Immunol. 24: 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Einer-Jensen K., Delgado L., Lorenzen E., Bovo G., Evensen O., Lapatra S., Lorenzen N. 2009. Dual DNA vaccination of rainbow trout (Oncorhynchus mykiss) against two different rhabdoviruses, VHSV and IHNV, induces specific divalent protection. Vaccine 27: 1248–1253 [DOI] [PubMed] [Google Scholar]
- 60.Kutzler M. A., Weiner D. B. 2008. DNA vaccines: ready for prime time? Nat. Rev. Genet. 9: 776–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fioretti D., Iurescia S., Fazio V. M., Rinaldi M. 2010. DNA vaccines: developing new strategies against cancer. J. Biomed. Biotechnol. 2010: 174378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vasan S., Schlesinger S. J., Huang Y., Hurley A., Lombardo A., Chen Z., Than S., Adesanya P., Bunce C., Boaz M., et al. 2010. Phase 1 safety and immunogenicity evaluation of ADVAX, a multigenic, DNA-based clade C/B’ HIV-1 candidate vaccine. PLoS ONE 5: e8617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin J. E., Pierson T. C., Hubka S., Rucker S., Gordon I. J., Enama M. E., Andrews C. A., Xu Q., Davis B. S., Nason M., et al. 2007. A West Nile virus DNA vaccine induces neutralizing antibody in healthy adults during a phase 1 clinical trial. J. Infect. Dis. 196: 1732–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]