Abstract
As the main microtubule-organizing center in animal cells, the centrosome has a fundamental role in cell function. Surrounding the centrioles, the Pericentriolar material (PCM) provides a dynamic platform for nucleating microtubules. While the PCM’s importance is established, its amorphous electron-dense nature has made it refractory to structural investigation. By using SIM and STORM sub-diffraction resolution microscopies to visualize proteins critical for centrosome maturation, we demonstrate that the PCM is organized into two major structural domains: a layer juxtaposed to the centriole wall, and proteins extending further away from the centriole organized in a matrix. Analysis of Pericentrin-like protein (Plp) reveals that its C-terminus is positioned at the centriole wall, it radiates outward into the matrix and is organized in clusters having quasi-nine-fold symmetry. By RNAi we show that Plp fibrils are required for interphase recruitment and proper mitotic assembly of the PCM matrix.
Introduction
Microtubules are polymers of αβ-tubulin that have a fundamental role in cell function. In animal cells, the main microtubule organization center is the centrosome, an organelle that is involved in integral tasks within cells of diverse tissues. The centrosome helps establish the axis of cell division, a critical step in stem cell duplication and embryonic development, and is involved in determining the plane of cytokinesis, thus ensuring inheritance of an equal number of chromosomes by daughter cells. In ciliated cells centrosomes dock at the cell membrane and differentiate into basal bodies, which are essential for the formation of the axoneme, a structure critical for integrating signals via primary cilia and for facilitating movement as flagella1–3. Not surprisingly, mutations associated with several hereditary diseases have been mapped to genes whose products encode centrosomal proteins4.
In the current view5–7 the centrosome is composed of two structural elements: the centriole, a barrel-shaped cylinder encircled by microtubule blades and the PCM, described as an amorphous, electron-dense structure surrounding the centrioles. The primary role of the PCM is to anchor microtubules directly or through microtubule nucleating centers (γ-tubulin ring complexes; γTuRC) 8–11. During mitosis, in a process known as centrosome maturation12,13, the PCM increases in size and γTuRCs are recruited from the cytosol, thereby promoting microtubule nucleation. While proteomic analyses have revealed hundreds of centrosomal components1, 14–18, electron microscopy (EM) studies have only provided insights into the ultrastructural organization of centrioles5–7, 19–21 and basal bodies22–24 or γTuRCs10,25,26. By contrast, even with the use of electron tomography methods19,20,27,28, no organizational pattern of PCM components has been discerned.
Studies on salt-stripped centrosomes have revealed a fibrous scaffold running throughout the PCM made of 12–15 nm fibers29,30. Unfortunately, the molecular identity of these fibers has remained elusive and their role in organizing the PCM is unknown. Previous reports have also hypothesized the existence of a layer of proteins that attaches the PCM to the centriole, PCM tube9,31,32. But if this layer exists, its components and function are unclear.
Here, we re-evaluate the notion that the PCM is an amorphous structure. Using a combination of three-dimensional (3D) Structured Illumination Microscopy (SIM), Stochastic Optical Reconstruction Microscopy (STORM)33–35 sub-diffraction resolution imaging methods and 3D image processing, we have begun to quantitatively map the architecture of the PCM. By examining the distribution and orientation of centrosomal proteins critical for centrosome maturation1,36, we show that the PCM is composed of two major domains with distinct molecular composition and architecture. The PCM layer most proximal to the centriole wall is made of Plp fibrils that radiate outward from the centriole wall to the outer PCM matrix. RNAi experiments shows that Plp’s elongated structures provide a scaffold critical for the 3D organization of the outer PCM matrix.
Results
SIM and volume averaging of proteins critical for centrosome maturation identifies two distinct structural domains within the PCM
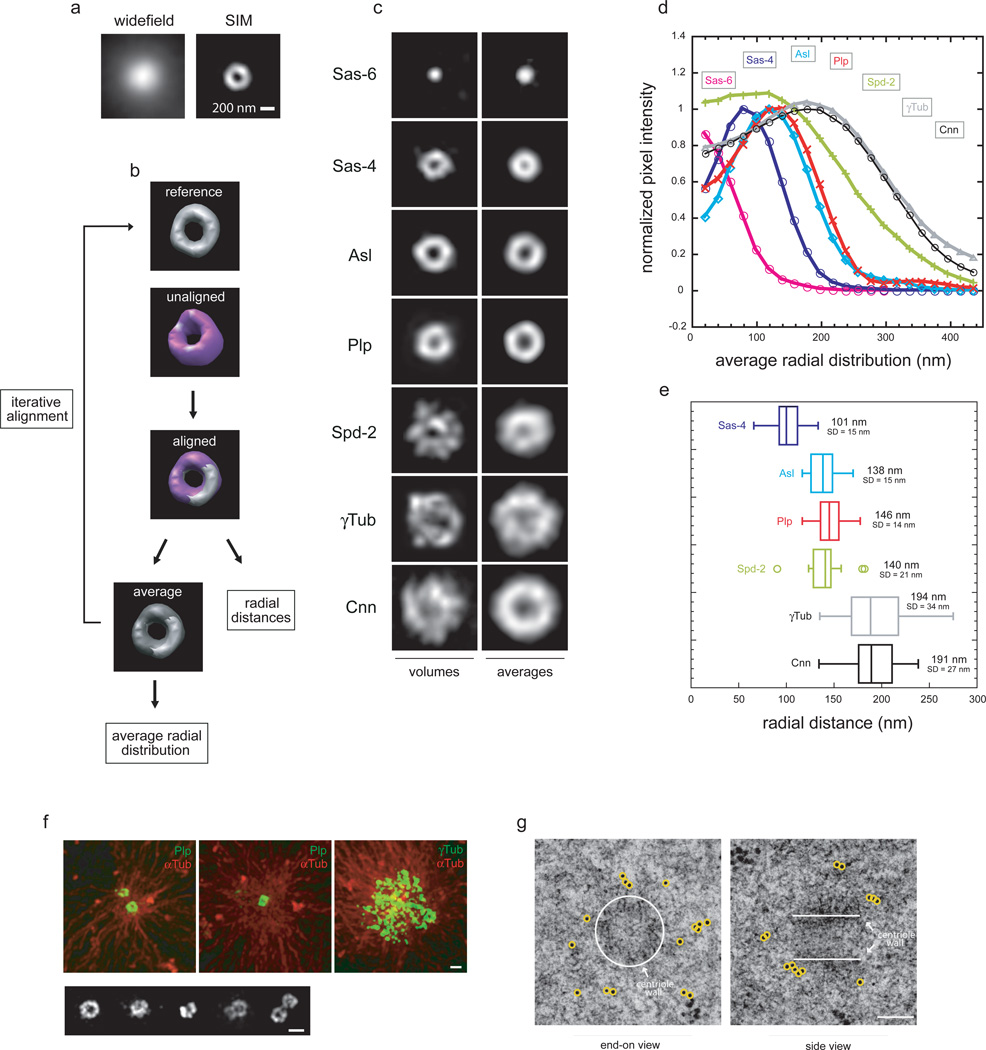
While the centrosome size is too small for conventional fluorescence microscopy to reveal significant structural details, SIM has sufficient resolution to image distinct centrosomal substructures37 (Fig.1a). For our analysis, we chose the complete set of Drosophila centrosomal proteins shown by genetic mutants and RNAi studies in several species to be critical for centrosome maturation, excluding regulatory components such as kinases1,36. As reference markers, we used proteins whose localizations have been mapped by EM in Drosophila and mostly conserved in other species: Sas-6 to label the inner centriolar region38,39, Sas-4 for the centriole wall40,41 and γTub as a PCM marker10.
Figure 1. 3D SIM of proteins critical for centrosome maturation identifies two distinct structural domains within the PCM.
a) Maximum intensity projections along the Z-axis of centrioles from S2 cells labeled with antibodies against centriolar proteins obtained either with widefield or 3D SIM microscopy.
b) Summary schematic of 3D sub-volume iterative alignment strategy based on cross-correlation in real space used for alignment and analysis of experimental 3D volumes. See methods for a detailed description.
c) 2D projections of the average aligned volumes. Sas-6 n=10 (antibodies anti-eGFP), Sas-4 n=88 (antibodies against a.a. 2–150), Asl n=21 (a.a. 958–972 of Asl), Plp n=82 (a.a. 1–381 of Plp-PB), Spd-2 n=19 (a.a. 375–695 of Asl), Cnn n=22 (a.a. 1–571 of Cnn), γTub n=33.
d) Fluorescence intensities profiles from the center of the centriole image outward measured from radially averaged 2D projections of average volumes of centrosomal proteins.
e) Radially averaged fluorescence intensities values obtained from individual centrosomal protein projected volumes were fit to an offset Gaussian function to calculate the center position and deviation of the distribution.
f) Centrosomes isolated from Drosophila syncytial blastoderm embryos were allowed to regrow microtubules with rhodamine-Tubulin and stained with rabbit anti-Plp (a.a. 1–381 of Plp-PB) or mouse anti-γTub antibodies recognized with anti-mouse 488. Bottom, image gallery of centrosomes from Drosophila embryos stained with rabbit anti-Plp NTD and anti-rabbit Alexa 488. Scale bar 500 nm.
f) High pressure frozen Drosophila syncytial blastoderm embryos embedded in resin were sectioned (75 nm) and stained with rabbit antibody anti-Plp NTD (a.a. 1–381) and anti-rabbit immunogold labeled secondary antibodies (10 nm beads). Scale 100 nm.
To assess PCM architecture in a functional but relatively stable organizational state12,13, we focused on metaphase centrosomes of S2 cells. It was immediately apparent that the organization of these proteins differs greatly, suggesting the existence of distinct molecular domains within the PCM (Fig.1c). To go beyond an anecdotal description, it was necessary to quantitatively compare the distributions of PCM components averaged over many centrosomes. Because centrosomes are randomly oriented in cells, averaging requires that they be brought into a standard orientation. This was accomplished through an iterative 3D sub-volume alignment and averaging strategy (Fig.1b), which uses a real space cross-correlation filtered in Fourier space to correct for non-isotropic resolution (XY plane vs Z axis). As initial reference for the 3D alignment we used representative centriole end-on views determined by the relative distribution of centriole markers (Sas-6, Sas-4 and γTub) in pairwise immunofluorescence experiments. After an initial round of alignment of the experimental volumes to the reference, the aligned volumes were averaged and used as a new reference for the next round. The final averaged 3D volumes were algebraically projected in the Z direction and the average radial distribution of fluorescence intensity from the center of the projected image outward was determined (Fig.1c,d). The peak position and the width of the distribution (σ) were determined by fitting to an offset Gaussian, while the standard deviations (SD) were derived from fits of radial averages calculated from the individual aligned volumes (Fig.1e, Supplementary Fig.1a).
Sas-6, a protein that localizes to the inner region provides a reference for the center of the centriole38,39,42,43. As expected, Sas-6 occupies a small region of the centriole (σ=72 nm). Sas-4, known to localize to the centriole wall40,41 has a peak position at r=101 nm (σ=60 nm), a measurement in agreement with the radius (80–100 nm) of Drosophila centrioles27. Surprisingly, when we analyzed the distributions of other components, they appeared to fall into two separate, but overlapping domains within the PCM (Fig.1d). One PCM domain, proximal to the centriole wall, is occupied by two components, Plp and Asterless (Asl), while the remaining proteins are concentrated further from the centriole and have a broad and matrix-like distribution. (Cnn r=191 nm, σ=133 nm; γTub r=194 nm, σ=126 nm. Fig.1c,d,e). The molecular distribution of the centriole-proximal proteins resembles a toroid and has a narrower distribution than the outer PCM layer proteins (Plp r=146 nm, σ=72 nm; Asl r=138 nm, σ=66 nm). The exception is Spd-2, which spans both PCM domains, having both a centriole-wall associated and a matrix component (r=140 nm, σ=121 nm. Fig.1c,d,e).
To confirm the existence of the distinct PCM layer proximal to the centriole wall, we imaged centrosomes isolated from Drosophila embryos. We focused on Plp, the Drosophila member of a protein family that shares a centrosomal-targeting sequence, the PACT domain44, since studies in flies and in vitro indicated that Plp plays a specific role in PCM-mediated microtubule nucleation45,46. While the PCM distal layer47 is expanded in embryos, the Plp toroids appear indistinguishable from the ones in cells (Fig.1f).
To visualize centrosomes directly we employed High Pressure Freezing–Freeze Substituted (HPF-FS) EM of Drosophila embryos gold labeled using anti-Plp antibodies (Fig.1g and Supplementary Fig.1b). This confirmed the distribution of Plp outside the centriole wall, but, because of the sparse gold-immunolabeling and the rather uniform appearance of the PCM, we were unable to identify a specific domain of the PCM enriched for Plp (Fig.1g).
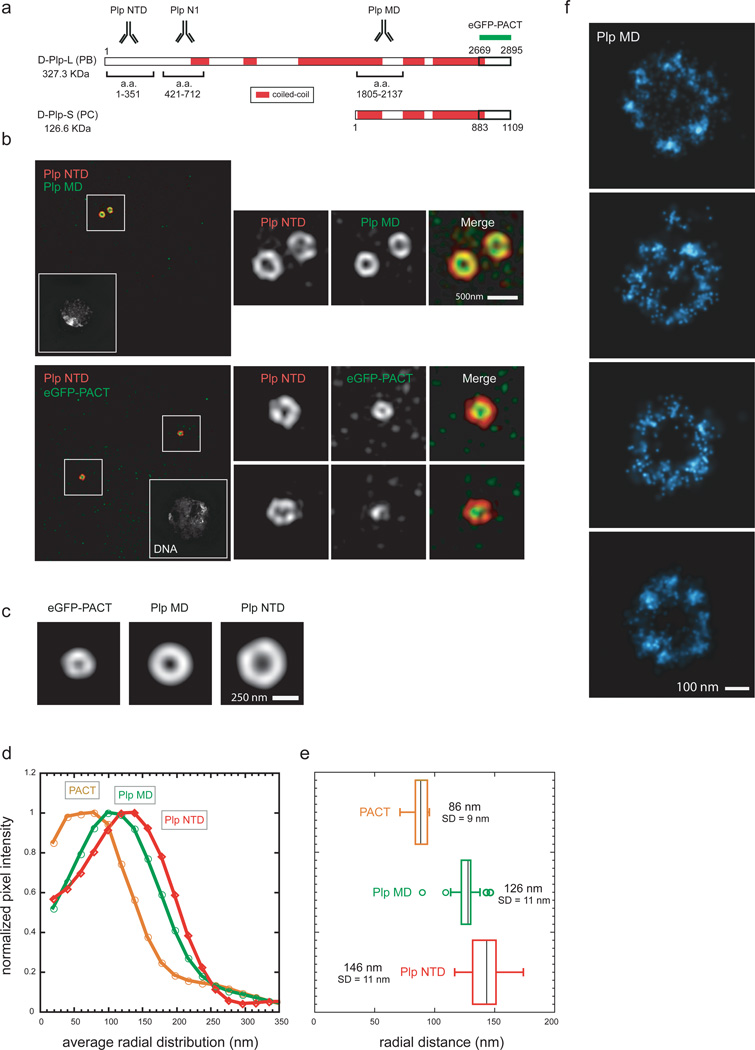
The molecular architecture of Plp
Since Plp is predicted to have several coiled-coil stretches and is thus likely to be extended (Fig.2a), we questioned whether its orientation was radial or longitudinal to the centriole wall. In addition to the antibody directed against the N-terminal region of Plp (NTD, Fig.1c), we labeled the middle region (MD) with another antibody46 and used an eGFP-PACT fusion44 to mark the C-terminal domain (CTD, Fig.2a). When cells were co-labeled with anti-Plp NTD and MD antibodies, or eGFP antibody to label the PACT domain, we observed that the CTD is distributed in a ring internal to the NTD, with the PACT signal overlapping with that of the centriole wall (Sas-4) (Fig.2b,c,d). The peak locations suggest that Plp CTD and NTD are ~60 nm apart (eGFP-PACT r=86 nm, σ=54 nm; Fig.2e), a distance far greater than the diameter of a globular protein with the same molecular weight as Plp-L (~9 nm), but smaller than the length of the predicted Plp coiled-coil regions (~107 nm)48, suggesting local foldbacks (Fig.2d,e). Analysis of anti-Plp-N1 antibody (Fig.2a) confirmed the Plp radial orientation (r=138 nm, σ=51 nm)45. Our data suggest that the extensive coiled-coil regions within Plp lead to a highly, but not completely extended conformation, radiating outward from their CTD at the centriole wall to their NTD embedded within the PCM matrix.
Figure 2. The molecular architecture of Pericentrin-like protein.
a) Amino-acid map of Drosophila Plp predicted from the Flybase database. Sequence prediction of coiled-coil conformation was performed with the software Coils using a window of 28 residues. Amino-acid stretches were considered coiled-coil if predicted with a probability of ≥70%.
b) Top: Wild type S2 cells co-stained with rabbit anti-Plp MD primary antibodies (a.a 1805–2137, anti-rabbit Alexa 488) and with guinea pig anti-Plp NTD primary antibodies (a.a. 1–381, anti-guinea pig Alexa 555).
Bottom: S2 cells expressing eGFP-PACT co-stained with mouse anti-eGFP monoclonal antibody (anti-mouse Alexa 488) and with rabbit anti-Plp NTD primary antibodies (a.a. 1–381, anti-rabbit Alexa 555). DNA is labeled with DAPI stain. Scale bar 1µM.
c) 2D projections of the average aligned volumes. Plp NTD=82; Plp MD n=29; eGFP-PACT n=9.
d) Fluorescence intensity profiles from the center of the centriole image outward measured from radially averaged 2D projections of average volumes of centrosomal proteins.
e) Radially averaged fluorescence intensities values obtained from individual centrosomal protein projected volumes were fit to an offset Gaussian to calculate the center position and deviation of the distribution. Scale bar 200 nm.
f) STORM image of centrioles from S2 cells stained with rabbit anti-Plp MD antibody (a.a 1805–2137) and anti-rabbit secondary antibodies conjugated with Alexa 647/Alexa 405 dye pairs.
To reveal more detailed structural features we employed STORM microscopy, a single molecule localization technique with a resolution up to 10 nm in the X-Y plane and 20 nm in Z-axis34,49. To study the architecture of all Plp isoforms we analyzed centrioles stained with MD antibodies46 and discovered unexpected characteristics of Plp organization. First, Plp molecules are distributed in distinct molecular clusters around the centriole (Fig.2f). By counting the number of single-molecule activation events in a cluster (average=135±71, n=9) and comparing it to the expected number from a single labeled antibody (average=27±7, n=30), it is likely that each cluster contains multiple Plp molecules. This suggests that Plp forms molecular fibrils extending into the PCM matrix, comprising a scaffold similar to spokes of a wheel. While the number and size of Plp clusters was somewhat variable likely due to the heterogeneous labeling density of secondary antibodies50, Plp clusters appear roughly symmetrical, suggesting an organization dominated by the nine-fold symmetry of the centriole. Staining with Plp NTD antibody highlighted Plp structures similar to the MD antibody, but sometimes with an elongated distribution, probably because of flexibility of Plp fibrils and/or deformation induced by fixation (Supplementary Fig.3b).
Although Asl is 1/3 the size of Plp, it also had an extended conformation (Supplementary Fig.2a,b) with its CTD near the centriole wall (r=97 nm, σ=58 nm) and its NTD embedded in the PCM matrix (r=138nm, σ=66 nm; Supplementary Fig.2c,d). This finding is consistent with recent in vitro studies suggesting an interaction between Sas-4 NT region and Asl CTD51. While Cnn is also predicted to have long stretches of coiled-coil, it is organized in a branched matrix, since antibodies against non-overlapping regions show a similar distribution (Supplementary Fig.2f). Interestingly, anti-Spd-2 antibodies raised against both the MD and the CTD (Supplementary Fig.2g,h) also show a similar pattern thus confirming that the distribution of Spd-2 is distinct from that of Plp and Asl.
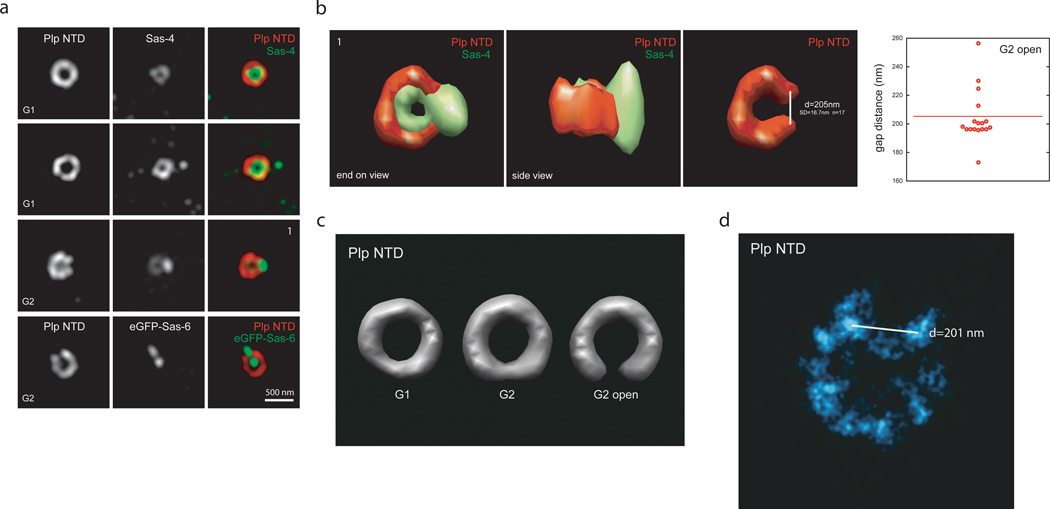
Plp fibrils associated with mother centrioles form a gap where daughter centriole assembles
Since Plp fibrils appeared to be symmetrically oriented around the centriole we questioned how its architecture changes during centriole duplication when there is a substantial break in symmetry. To distinguish between stages of centriole duplication, we used Sas-6 and Sas-4, which are recruited in the early phases (Fig.3a)39,41. We note that the central tube protein Sas-6 can be used as a marker for all centrioles because, unlike in mammalian cells, Drosophila centrioles always contain Sas-6. While analyzing the Plp images, we observed that Plp structures in G2 cells frequently have a gap, which co-localized with the site of emergence of the new centriole (Fig.3a). The largest Plp gap correlated with the accumulation of a high density of Sas-4 on the daughter centrioles (Fig.3a,b). To reveal structural differences between different populations of Plp stained centrioles in G2, subclass averaging was performed. This allowed us to clearly distinguish a population of Plp structures with a ~ 200 nm gap, a space large enough to accommodate a nascent centriole. In contrast, during earlier phases of the cell cycle Plp forms a complete ring without a noticeable gap or a ring with a smaller gap (Fig.3b,c). When STORM was used to further probe this change in Plp architecture, we observed that centrioles labeled with Plp NTD antibodies had a roughly symmetrical cluster distribution, but appeared to lack one cluster, suggesting the existence of a molecular gate that is opened and/or disassembled during construction of the daughter centriole (Fig.3d)
Figure 3. Plp fibrils associated with mother centrioles form a gap where daughter centriole assembles.
a) Centrosome from wild type S2 cells co-stained with rabbit anti-Plp NTD antibodies (a.a. 1–381, anti-rabbit 555) together with mouse anti-Sas-4 antibodies (anti-mouse 488). Bottom panel. Centrosomes from S2 cells expressing eGFP-Sas-6 co-stained with mouse anti-GFP antibodies (anti-mouse 488).
b) Volume rendering of G2 centriole from panel a) stained with mouse anti-Sas-4 and rabbit anti-Plp NTD shown from end-on and side views.
c) Gap distance measurements obtained from 3D SIM volumes of a subpopulation of G2 centrosomes stained with rabbit Plp NTD antibody.
d) Rendering of volumes averages of G1 and G2 centrosomes stained with anti-Plp NTD antibody. G2 centrosomes were divided into two separate populations for sub-class averaging of centrioles with an open gap or partially open gap.
e) STORM image of centrioles from S2 cells stained with rabbit anti-Plp NTD antibody (a.a. 1–381, left) and anti-rabbit secondary antibodies conjugated with Alexa 647/Alexa 405 dye pairs. Given the cluster variability, we used a blinded study to quantify how often a missing cluster could be recognized (58%) in STORM images of mother centrioles stained with anti-Plp antibody from cells blocked in G2.
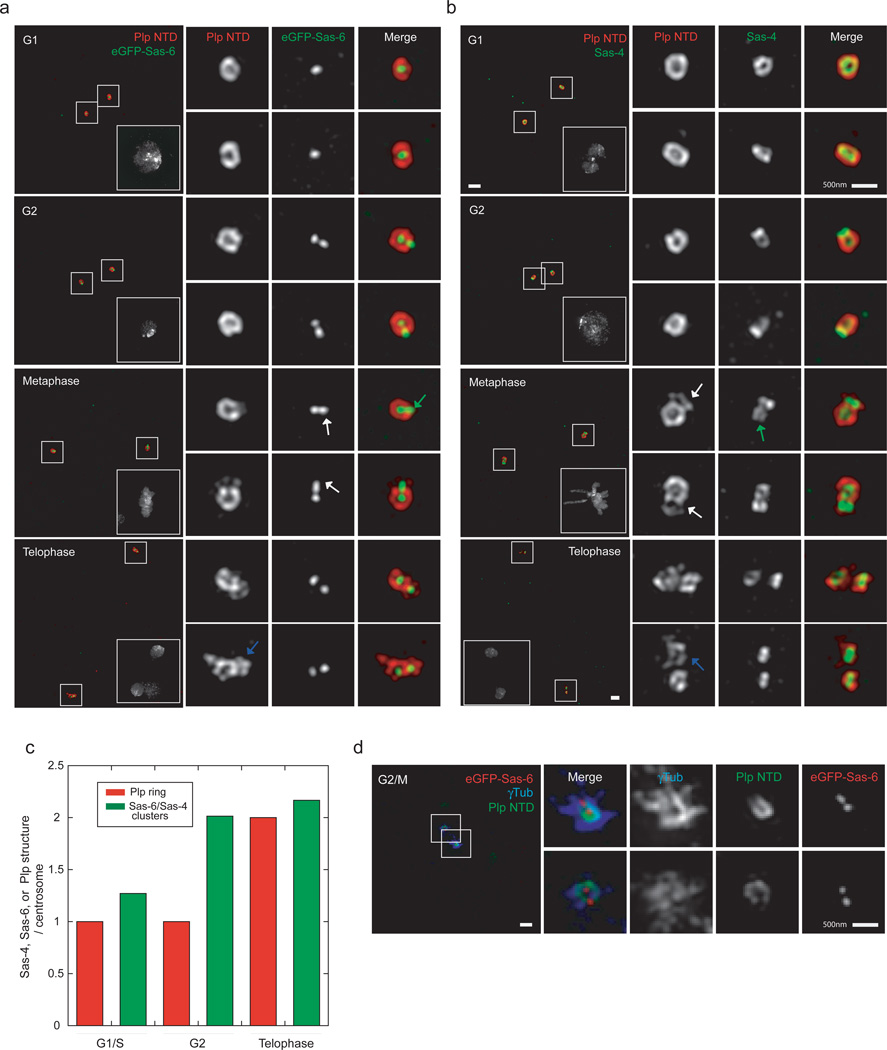
Plp is associated exclusively with mother centrioles until metaphase
To better understand the role that Plp fibrils have in PCM maturation, we expanded our SIM analysis to visualize Plp throughout the cell cycle. Our images show that while Plp encircles the mother centrioles before and after duplication, by metaphase most daughters have either no Plp or a partially assembled Plp toroid (Fig.4,4c). Consistently, the outer PCM marked by γTub is organized mostly around the mother centrioles, as in mammalian cells (Fig.4d)5,53,54. Only by telophase does Plp recruitment to daughters appear complete (Fig.4a,b). This behavior differs from that of Sas-6 and Sas-4 which are recruited to daughter centrioles early in S/G2 (Figs.4a,b). A similar, asymmetric distribution of Plp was also observed in isolated embryonic centrosomes (Supplementary Fig.3a), suggesting that Plp’s time of recruitment is consistent from embryos to somatic cells.
Figure 4. Plp is associated exclusively with mother centrioles until metaphase.
a) S2 cells expressing eGFP-Sas-6 stained with mouse anti-GFP antibodies (anti-mouse 488) and with rabbit anti-Plp NTD (a.a. 1–381, anti-rabbit 555). DNA is labeled with DAPI stain. Scale bar 1µM. White arrows point at the lack of or partial recruitment of Plp on daughter centrioles at metaphase. Grey arrows on the telophase panels show complete recruitment of Plp on daughter centrioles.
b) Wild type S2 cells stained with mouse anti-Sas-4 antibodies (anti-mouse 488) and rabbit anti-Plp NTD antibodies (a.a. 1–381, anti-rabbit 555). DNA is labeled with DAPI stain. Scale bar 1µM. White arrows point at the lack of or partial recruitment of Plp on daughter centrioles at metaphase. Notice the green arrow on the metaphase panel showing the separation of Sas-4 stained centrioles compared to G2 cells. Grey arrows on the telophase panels show complete recruitment of Plp on daughter centrioles.
c) Quantification of the number of Sas-4, Sas-6 clusters or Plp rings per centrosomes per S2 cells. (G1 n=26, G2=29, Telophase =10).
d) S2 cells expressing eGFP-Sas-6 co-stained with guinea pig anti-Plp NTD antibodies (a.a. 1–381, anti-guinea pig 405) and rabbit anti-GFP antibodies (anti-rabbit 488) and mouse anti γTubulin (anti-mouse 555). Scale bar 1µM.
Plp is required for the initial recruitment and proper 3D assembly of the PCM matrix layer
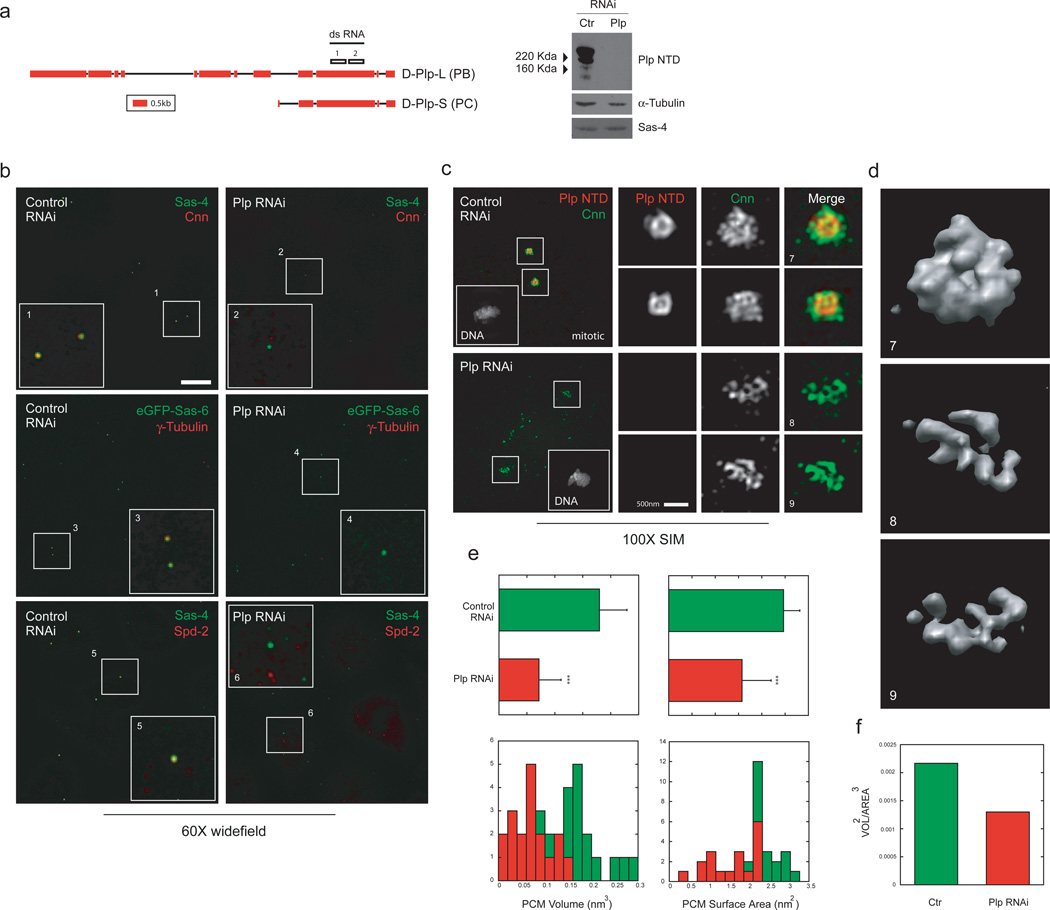
Our observation that Plp fibrils form a scaffold around the mother centriole strongly implicates Plp in regulating PCM organization. Studies in Drosophila have previously shown that Plp is involved in recruiting PCM proteins to the centrioles in mitosis45,46,55. Since Plp organization does not change substantially during centrosome maturation, its role remains unclear. Interestingly, while studying PCM architecture in cells by SIM, we observed a layer of outer PCM markers (Cnn, γTub) that in interphase centrosomes was closely associated with Plp density. Previous studies have suggested that this close in shell of PCM distal proteins (Fig.5b, Supplementary Figs.3d,4a,b), is not a major platform for MT-nucleating material56. Since Cnn likely recruits γ-tubulin complexes57, we suspected that Plp fibers might influence centrosome maturation by organizing the layer of Cnn molecules on interphase mother centrioles. Remarkably, when we deplete Plp with RNAi in S2 cells, the interphase layer of matrix components disappears completely from Sas-4 positive centrioles (Fig.5b). By contrast, in metaphase cells depletion of Plp leads to a significant reduction, disorganization and fragmentation of the outer PCM matrix stained by Cnn, but rarely a complete loss (Fig.5c,e,f). These experiments suggest that Plp is required to recruit the first layer of outer PCM proteins, and to organize its proper 3D assembly, but is not essential for PCM expansion. Interestingly, RNAi directed against the NTD of Plp had a minor effect on γTub recruitment and organization, suggesting that shorter Plp isoforms are sufficient to provide a scaffold to stabilize the PCM matrix (Supplementary Fig.3c,d).
Figure 5. Plp is required for the initial recruitment and proper 3D assembly of the PCM distal layer.
a) Left. Plp genomic region obtained from the Flybase database (5’ and 3’UTR regions not displayed). The position of the sequences on Plp exons used for dsRNA production is annotated on the sequence. Right, western blot on extracts obtained from S2 cells treated for control or Plp dsRNA probed with rabbit anti-Plp NTD antibody.
b) S2 cells treated with control dsRNA or dsRNA specific for Plp exon 10 were stained with either anti-Cnn antibodies (anti-mouse 488) or mouse anti-γTubulin antibodies or rabbit anti-Spd2 antibodies (anti-rabbit or mouse 555). Widefield images were collected with a 60X objective and deconvolved with a non-linear positivity-constrained iterative algorithm. DNA was labeled with DAPI stain. Scale bar 10µM
c) Mitotic S2 cells treated with dsRNA specific for Plp exon 10 were stained with guinea pig anti-Plp NTD (anti-guinea pig 555 and rabbit anti-Cnn 1–571 (anti-rabbit 488). DNA was labeled with DAPI stain. SIM Images were acquired in the camera linear range of response and are displayed with identical intensity settings. Scale bar 1µM.
d) Volume rendering of control and Plp RNAi S2 cells shown in panel b. Scale bar 500 nm.
e) Volumes obtained from stained S2 cells as in panel b were quantified for the volume occupied and their surface area with Chimera software (control n=16, Plp Rnai n=18).
f) Volumes were approximated to a spherical object to measure Vol^2/Area^3.
The molecular architecture of Kendrin/Pericentrin is similar to that of Plp
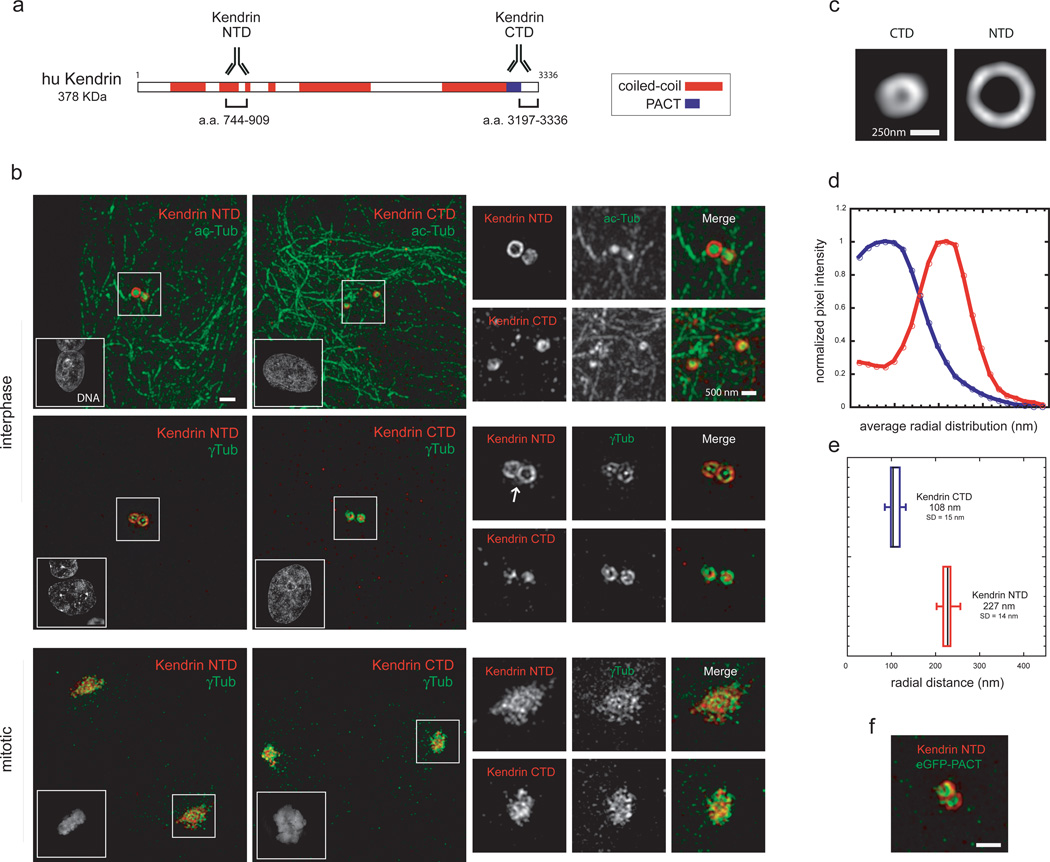
In mammalian species, the PACT family includes two large, mostly coiled-coil proteins, Kendrin/Pericentrin and CG-NAP. Localization and functional studies suggest that Kendrin is the likely functional homologue of Plp58,59,60,61. To assess whether the molecular architecture of Plp is conserved, we thus studied Kendrin organization by SIM. HRPE cells were stained with antibodies directed against the NTD and CTD of Kendrin, together with centriolar and PCM markers (acetylated tubulin and γTubulin; Figs.6a,b). Kendrin NTD revealed a similar, albeit more complex distribution compared to Plp (Fig.6b). Two different populations of Kendrin appear to coexist: one tightly associated to centrioles with a toroid shape, reminiscent of Plp organization; another, distributed diffusely and matrix-like (Fig.6b). Analysis of Kendrin distribution (Fig.6c) indicates that is an elongated and radially oriented molecule similarly to Plp (CTD r=108 nm, σ=77 nm; NTD r=227 nm, σ=56 nm; Fig.6 d,e,f). In mitosis, however, the lattice-reticular organization is prominent62. Since Kendrin antibodies directed against both ends of the molecule have similar distributions in mitosis, we conclude that the CTDs of most Kendrin molecules are not attached to centrioles. Our data show that Plp architecture is mostly conserved in humans, but that other forms of organization have been acquired by its homologue.
Figure 6. The molecular architecture of Kendrin/Pericentrin is similar to that of Plp.
a) Linear map of the amino-acid sequence of human Kendrin predicted from the UCSC database. The antibodies on the cartoon show the location of Plp protein sequence used for immunization. Sequence prediction of coiled-coil conformation was performed with the software Coils using a window of 28 residues. Amino-acid stretches were considered coiled-coil if predicted with a probability of ≥70%.
b) Human RPE cells were labeled with antibodies against Kendrin NTD (a.a. 744–909, anti-rabbit 555) and Kendrin CTD (a.a. 3197–3336, anti-rabbit 555) with mouse acetylated-Tubulin or mouse anti γTubulin antibodies (anti-mouse 488). Nuclei were labeled with DAPI stain. The white arrow points at the diffused population of Kendrin in interphase. Scale bar 1 µm.
c) 2D projections of the average aligned volumes (Kendrin NTD n=16, Kendrin CTD n=15).
d) Fluorescence intensities profiles from the center of the centriole image outward measured from radially averaged 2D projections of average volumes of centrosomal proteins.
e) Radially averaged fluorescence intensities values obtained from individual centrosomal protein projected volumes were fitted to an offset mirrored Gaussian function to calculate the center position of the distribution.
e) HeLa cells expressing eGFP-PACT were co-stained with antibodies against Kendrin NTD (a.a. 744–909, anti-rabbit 555) and anti-GFP (anti-mouse 488). Scale bar 500 nm.
Discussion
The two-layer structural and functional organization of the PCM revealed by sub-diffraction resolution fluorescence microscopy
To tackle the complex and long-standing problem of the organization of the PCM, we combined the strengths of two sub-diffraction fluorescence microscopy approaches. Where SIM provides a robust method for simultaneous co-localization of multiple proteins in 3D, STORM provides substantially higher resolution, but has stringent requirements for labeling density34,50. Thus once the general organizational framework was established by SIM, STORM microscopy was used to obtain a considerably higher resolution structure of a key component revealing unexpected details. Together, the two methods provided complementary and mutually supportive information that was more insightful than the information provided by immuno-EM. Importantly, alignment and averaging of 3D volumes provided robust quantitative metrics of the spatial distribution of relevant centrosomal components.
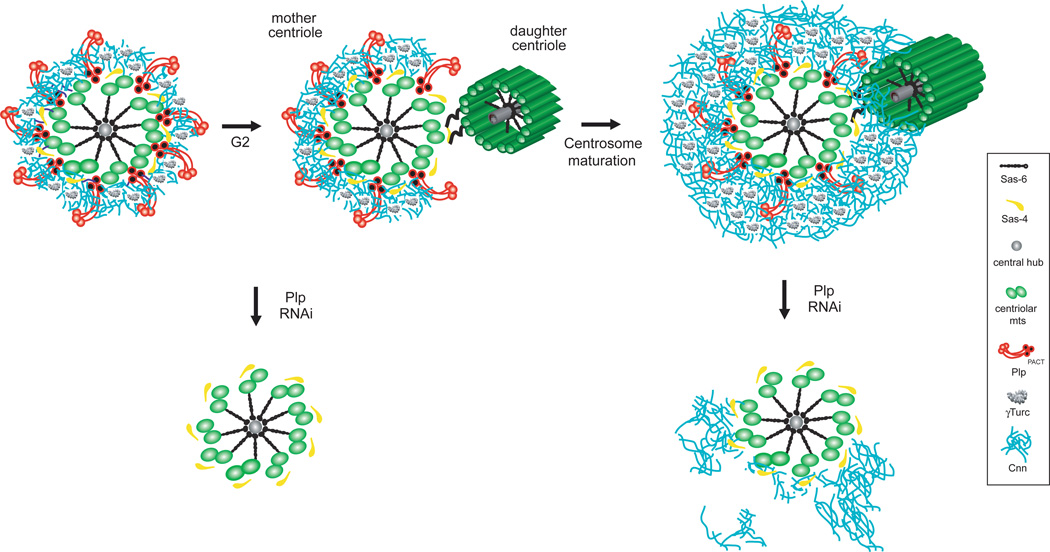
Previous studies had begun to dispel the notion of a uniform amorphous PCM organization around the centriole9,31,32,63. Here, we devised a strategy to study PCM architecture and demonstrated that the PCM is composed of two distinct structural layers that provide separate functionality and further show that the inner layer protein Plp plays an important role in PCM organization by forming a molecular scaffold for the PCM matrix. As shown by overlapping radial distributions between Sas-4 and Plp, Plp likely binds to the centriole wall through its C-terminal PACT domain and shows a symmetrical organization derived from the nine fold symmetry of the centriole. Plp is organized in quite extended fibrils that function in recruiting and organizing the outer PCM matrix. This occurs through the initial recruitment of a shell of Cnn molecules on interphase centrioles. A second PCM matrix layer extending further from the centriole wall has the core functionality of microtubule organization, tethering γTuRC to a Cnn matrix that can expand when needed (Fig.7). Since overexpression of eGFP-Cnn in interphase cells does not increase the size of the PCM outer layer (Supplementary Fig.4c), a level of regulation must be in place to promote Cnn matrix organization during centrosome maturation. The observation that some Cnn and γTub are being recruited to mitotic centrosomes without Plp might indicate that the mitotic drive to expand the PCM is so strong that either undetectable amounts of residual Plp or interactions with another component are sufficient to initiate formation of a reticular structure64. While our data show a density overlap between Plp and Spd-2 in metaphase –and previous experiments suggested a biochemical interaction47 – Spd-2 can be recruited on daughter centrioles before Plp (Supplementary Fig.2h). Altogether these observations suggest that Plp and Spd-2 are indirectly linked in the PCM and might have distinct roles in building the PCM.
Figure 7. Pericentrin-like protein forms elongated fibrils that extend radially from the centriole wall to support the 3D organization of the PCM.
During centrosome maturation, the PCM is organized in two distinct structural domains: a layer juxtaposed to the centriole wall, and proteins extending further away from the centriole organized in a matrix. In this proximal PCM domain, we found elongated Plp fibrils that are anchored with the PACT domain to the centriole wall and with their N-terminus extending outwards. Plp is exclusively associated with mother centrioles until metaphase and form a gap where daughter centriole assembles. During centrosome maturation, Plp facilitates the proper 3D assembly of the PCM distal layer by organizing a shell of Cnn molecules that is in place around the wall of mother centrioles from the interphase cell cycle stage.
Interestingly, we show that Asl, while providing a separate function as a platform for centriole duplication51, is located in the same PCM domain and shares a similar orientation to Plp. Our RNAi data show that neither Plp nor Asl –similarly to previous studies51– are required for the recruitment of the other component (not shown).
The molecular architecture of Plp fibrils
Plp is expressed in several isoforms and splicing variants45,46,65. How are these differently sized molecules organized? Our domain mapping experiments suggest that they share a similar orientation. Indeed, when cells were depleted of the longer splicing variants of Plp, the distribution of the shorter isoforms did not visibly change (Supplementary Fig.3d). Moreover, with STORM imaging we observed the Plp cluster distribution with the MD antibody, which recognizes all isoforms. This observation suggests that the different Plp molecules are in close proximity, possibly self-associating or organized in a multi-molecular fiber with other components.
Our data show that Plp undergoes a remarkable change in architecture by forming a molecular gate on mother centrioles whose opening is correlated with daughter centriole formation. It is unlikely that the antibody lacks accessibility to the antigen, because when we examine metaphase cells in which Sas-4 positive daughter centrioles are completely formed and more distant from their mothers than during G2, the Plp ring appears to be still open (Fig.4b). Since we observe a lack of density in SIM images and we observe that one cluster is missing in the area of the gap in STORM images, we favor a mechanism by which disassembly of a Plp fibril —either by an active mechanism or by occlusion of its binding site by a novel protein— and not the rearrangement of an individual fibril, determines the location of the gap. While we observe a reduction in centrioles in cells depleted of Plp (not shown), Plp structural organization appears inconsistent with a gatekeeping role during duplication.
In conclusion, this study expands and changes our understanding of the PCM architecture by providing a fundamental map of its organization, revealing a functional framework to dissect how PCM organization is achieved. Furthermore, it demonstrates that multi-modal sub-diffraction resolution imaging combined with quantitative analyses provides a powerful and general strategy for studying sub-organellar organization.
Our data, along with independent studies using SIM published in this issue of NCB66 and elsewhere67, demonstrate the highly conserved nature of PCM organization.
Material and Methods
RNA interference and cell culture
RNA interference experiments were performed as previously described68,69. For in vitro transcription of the dsRNA oligonucleotides, DNA templates were amplified from S2 cell cDNA using the following set of gene-specific primers: Plp exon 10 construct 1 (forward), 5′- ACTTTAGTTCATGGATCTCGGC −3′; (reverse), 5′- GAGCTACTGGGAGAGAAACAGC-3′. Plp exon 10 construct 2 (forward), 5′- CGCTGGTCTACCAGAAGAGG −3′; (reverse), 5′- CTCACCGCCAATGGTTAACT −3′;Plp exon 1 (forward), 5′- CTTGGTCTCCAGTTGGGTGT −3′; (reverse), 5′- TGAGGTGACGTACGAAGCTG −3.
Primer sequences for the genes Asl were described elsewhere70. Cells were depleted of centrosomal proteins by RNAi using a dsRNA treatment for 7 to 14 days (40 µg of dsRNA applied on days 0, 4, 7, 11, 14). For experiments in mammalian cells, HeLa Tet-on cells (Clontech) were transfected with a plasmid encoding eGFP-PACT (Mm amino-acids 3095–3336) with Amaxa nucleofector II (Lonza). After 24–48h of induction with doxocyclin (300 ng/ml), cells were fixed and stained as described below.
Immunofluorescence microscopy and antibodies
S2 cells were plated for 2.5h on 35 mm glass bottom Delta T dishes (Bioptechs) coated with concanavalin-A (Sigma) to promote cell spreading. Cells were washed in PBS, fixed for 30 minutes in methanol pre-chilled to −20°C, rehydrated in PBS and blocked in a solution of PBS/0.02% Tween (PBST) with 3% BSA. Immunostaining of Drosophila embryonic centrosomes was performed as previously described [27]. For a complete list of antibodies and relative references used in this study see Supplementary Fig.1a. Dilutions used are specified in parenthesis: rabbit and guinea-pig anti-Plp a.a. 1–381 (1:3000, 1:1000); rabbit anti-Plp a.a. 1805–2137 (1:500); mouse anti α-tubulin (1:1,000 DM1α, Sigma); rabbit anti-Spd-2 a.a. 375–695 (1:500); rabbit anti-Spd-2 a.a. 915–1036 (1:500); mouse anti-Sas-4 a.a 2–150 (1:50); rabbit anti-Asl NT a.a. 958–972 and CT a.a 21–40 (1:100); rabbit anti-Cnn 1–574 (1:1000); rabbit anti-Cnn 271–1034 (1:500); rabbit anti-Cnn 744–909 (1:500); anti-GFP (1:100, Invitrogen); mouse anti-γTubulin (1:100, Sigma clone GTU88); rabbit anti-Kendrin a.a. 744–909 (1:1000); rabbit anti-Kendrin a.a. 3197–3336 (Santacruz); anti-acetylated Tubulin (6–11B-1, Sigma). Fluorescent secondary antibodies Alexa 488,555 (Invitrogen), dylight-405 (Jackson) were used at a final concentration of 1:500. Cells were counterstained with DAPI (300 nM, Invitrogen) in PBST for 10 min. Samples were mounted in a PBS 50% glycerol 2w/v propyl gallate solution.
3D SIM microscope image acquisition
For 3D SIM, images were recorded as previously described [37]. Solid-state or diode laser illumination (405 nm, 488 nm, 560 nm and 647 nm) were coherence-scrambled by a holographic diffuser before being coupled into a multi-mode fiber optic cable and passed through a diffraction grating. The objective used was plan-apochromatic 100x, 1.4 NA, oil-immersion (Olympus) with immersion oil of refractive index 1.515. For multi-color fluorescence experiments, the color channels were recorded sequentially onto separate CCD cameras. Emitted light from the sample passes through a set of four dichroic mirrors, which direct light of different wavelengths into four independently controlled cameras. Images are acquired with iXon 87 1MgHz back-illuminated EMCCD cameras, with 512 × 512 pixels operating either in conventional mode or EM gain mode fixed at 170 electrons per count. Depending on the antibody-fluorophore combination we used exposure times between 10–500 ms yielding 1,000 to 10,000 counts in a raw image of 16-bit dynamic range. Dichroics used in the emission path were: “GFP” EM Filter band: 525/50; “Cy5” EM: 670/30; “Rhodamin” EM: 585/40; “Dapi” EM: 447/60 (Semrock). Custom Python scripts that apply translation, rotation, and both isotropic and anisotropic scaling were used to align separate cameras. The alignment parameters were obtained from measurements with 100 nm multi-wavelength fluorescent beads (Invitrogen) taken with the same camera setup as the biological samples. Raw images were processed to reconstruct high-resolution information as previously described [37]
3D volume iterative alignment, distance measurement and volume rendering
3D SIM volumes of antibody-stained centrioles were aligned to reference volumes using 3D cross-correlation in real space. To handle the non-uniform 3D resolution of SIM images, for every orientation, the data are filtered in Fourier space such that only the common regions of Fourier space were compared, prior to calculation of the real space correlation (UCSF Priism software). Initial references were selected from large experimental datasets (>100 stacks per centrosomal protein) chosen for being optimally perpendicular to the centriole axis. For alignment, volumes roughly perpendicular to the imaging axis were selected to reduce measurement errors due to anisotropic resolution, although the volume alignment works so well that this could likely be relaxed. Once aligned to the reference, the volumes were averaged forming a new reference. This iterative process typically converged in 2–3 cycles, likely due to the high signal to noise of the SIM data. After volume alignment, we observed an average ~1.3x reduction in SD measurements of peak positions for all centrosomal components compared to projections of not aligned data. Radial averages about the center of mass were calculated from 2D algebraic projections of the N-most central sections of the average (used for determining the peak center, overall distribution, and width, σ) or individual 3D volumes (used for determining the variance in peak position). Fluorescence distributions were fit to an offset Gaussian function y0+A/(σ *sqrt(π/2))*(exp(−2*((r-rc)/σ)2)) with A, y0, xc and w as parameters to fit.
While there is an expected wavelength dependence of both magnification and resolution, the magnitude of these effects is quite small. Distances between beads visualized at both 525 nm and 585 nm reveal differences of only ~ 0.8%. To control for possible effects of wavelength dependent resolution, we stained S2 cells with antibodies against two different antigens (Sas-4 and Plp) using in the first experiment secondary anti-mouse 488 and anti-rabbit 555, and in a second experiment anti-mouse 555 and anti-rabbit 488. As expected, we did not observe a significant difference in ring size for either antigens in the two different experimental schemes (Plp NTD-488: 151nm, SD 13, n=32; Plp NTD-555: 142nm, SD=12nm, n=22; Sas-4–488 :100nm SD=16nm, n=46; Sas-4–555: 104 nm, SD=12nm, n=22). Distance measurements of Plp ring opening were performed with the distance measurement function in Priism software (UCSF). 3D MRC files were volume rendered with Chimera software (UCSF).
High Pressure freezing-freeze substitution of Drosophila Embryos
Drosophila embryos were aged for 2–3h in a humidified chamber, dechorionated for 3min in a solution of 50% bleach and high-pressure frozen as previously described71. Fly embryos were embedded in either epon resin to assess successful freezing and fixation procedure or LR white resin to perform EM immunochemistry (Supplementary Fig.1b). Tissue sections of 75 nm were stained with anti-Plp NTD antibody used at a concentration of 1:50 for 1h a RT. Secondary anti-rabbit gold particle conjugated (10 nm beads, Ted Pella) were used to detect the anti-Plp antibody. Grids were post-stained for 1 minute in 1% uranyl acetate dissolved in dH2O. After rinse, the grids were post-stained with lead citrate for 3min. Data were acquired on a Tecnai 12 TEM operating at 120 kV using a Gatan Ultrascan 1000 digital camera.
STORM microscope and image acquisition
STORM experiments were performed on a custom built microscope as previously described72. The microscope is based on a Nikon Ti-E inverted microscope with the Perfect-Focusing System (PFS). Four activation/imaging lasers (Stradus 405-100, Stradus 488-50, Stradus 642-110, Vortran Laser Technology; Sapphire 561-200-CW, Coherent) are combined using dichroic mirrors, aligned, expanded, and focused to the back focal plane of the objective (Olympus 100x UPlanSApo NA 1.4). The Stradus lasers are controlled directly by the computer whereas the Sapphire 561 nm laser is shuttered using an Acoustic Optical Modulator (Crystal Technology). A quadband dichroic mirror (zt405/488/561/640rpc, Chroma) and a band pass filter (ET705/70m, Chroma) separate the fluorescence emission from the excitation light. The images were recorded at a frame rate of 57 Hz on an EMCCD camera (Ixon+ DU897E–CS0-BV, Andor). The typical power for the lasers at the back port of the microscope was 30 mW for the 642 nm imaging laser and 0.5–5 µW for the other lasers. During image acquisition, the axial drift of the microscope stage was stabilized by PFS. To correct for the lateral stage drift, we installed an LED light as the transmitted light source for the microscope. In every 10 frames of STORM image acquisition, we turned on the LED for one frame with all the lasers off so that a bright-field image of the sample was recorded. The lateral stage drift was then calculated from correlation analysis of these bright-field images and subtracted from the molecule positions during analysis. Both data acquisition and analysis were performed using custom written software. Photoswitchable dye pairs A405-A647, was used for STORM imaging with a ratio of 0.34 A647 molecules per antibody. During the imaging process, the 405 nm activation laser was used to activate a small fraction of the A647 reporters at a time, and individual activated fluorophores were imaged and localized with a 657 nm laser. To determine how many localization points one antibody can generate, we identified scattered clusters of points in regions of cells surrounding by, but away from the centrosome. Because these points come from localizations without any centrosome structure, these clusters should be the result of non-specifically bound antibodies. The majority of these clusters should reflect only one antibody molecule, since antibody non-specific binding level is minimal.
Analysis of PCM organization and centriole number
3D SIM volumes of centrosomes were acquired with identical exposure time, gain and laser intensity to ensure proper comparison in control and Plp RNAi depleted cells. After applying identical thresholds, pericentriolar volume and surface area were measured by Chimera software (UCSF) using the volume data measurement function.
Statistics
Mean values of different observations were compared by Student t-test assuming unequal variance. In figures, statistical significance is indicated with asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001), and error bars indicate SD.
Supplementary Material
Acknowledgements
We especially thank John Sedat, Lukman Winoto and Chris Weisiger for invaluable assistance with the OMX and Sam Li for advice and help with the 3D alignment strategy, Dan Buster and Katherine Model for comments and editing on the manuscript. We also would like to thank T. Avidor-reiss, J. Raff, M. Bettencourt-Dias, M. Gatti, T. Megraw, M. Takahashi, Y. Zheng, T. Davis and T. Kaufmann for generously sharing their antibodies. This work was funded by HHMI and NIH grant GM310627. B.H. receives the Searle Scholarship and the Packard Fellowship for Science and Engineering.
Footnotes
Author contribution
V.M. conceived the strategy, designed and performed experiments, analysed data and wrote the paper; B.K. and V.M., with input from others at UCSF, developed the 3D volume alignment and analysis procedure; K.M. performed the Immuno-EM experiments; B.C. helped with STORM data acquisition; F.K. helped with Immuno-staining experiments; G.C. provided reagents and constructs; B.H. advised on data analysis and STORM experiments and discussed results; D.A. advised on data analysis, discussed results and wrote the paper.
References
- 1.Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8(6):451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 2.Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis duplication and inherent asymmetries. Nat Cell Biol. 2011;13(10):1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azimzadeh J, Marshall WF. Building the centriole. Curr Biol. 2010;20(18):R816–R825. doi: 10.1016/j.cub.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bettencourt-Dias M, et al. Centrosomes and cilia in human disease. Trends Genet. 2011;27(8):307–315. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vorobjev IA, Chentsov Yu S. Centrioles in the cell cycle. I. Epithelial cells. J Cell Biol. 1982;93(3):938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rieder CL. The centrosome cycle in PtK2 cells: asymmetric distribution and structural changes in the pericentriolar material. Biol. Cell. 1982;44:117–132. [Google Scholar]
- 7.Bornens M, et al. Structural and chemical characterization of isolated centrosomes. Cell Motil Cytoskeleton. 1987;8(3):238–249. doi: 10.1002/cm.970080305. [DOI] [PubMed] [Google Scholar]
- 8.Mogensen MM, et al. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci. 2000;113(( Pt 17)):3013–3023. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- 9.Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14(1):25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- 10.Moritz M, et al. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature. 1995;378(6557):638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- 11.Kollman JM, et al. Microtubule nucleation by gamma-tubulin complexes. Nat Rev Mol Cell Biol. 2011;12(11):709–721. doi: 10.1038/nrm3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palazzo RE, et al. Centrosome maturation. Curr Top Dev Biol. 2000;49:449–470. doi: 10.1016/s0070-2153(99)49021-0. [DOI] [PubMed] [Google Scholar]
- 13.Decker M, et al. Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr Biol. 2011;21(15):1259–1267. doi: 10.1016/j.cub.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Andersen JS, et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426(6966):570–6964. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 15.Muller H, et al. Proteomic and functional analysis of the mitotic Drosophila centrosome. EMBO J. 2010;29(19):3344–3357. doi: 10.1038/emboj.2010.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li JB, et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117(4):541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 17.Kilburn CL, et al. New Tetrahymena basal body protein components identify basal body domain structure. J Cell Biol. 2007;178(6):905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller LC, et al. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol. 2005;15(12):1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 19.Pelletier L, et al. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444(7119):619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- 20.Guichard P, et al. Procentriole assembly revealed by cryo-electron tomography. EMBO J. 2010;29(9):1565–1572. doi: 10.1038/emboj.2010.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paintrand M, et al. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J Struct Biol. 1992;108(2):107–128. doi: 10.1016/1047-8477(92)90011-x. [DOI] [PubMed] [Google Scholar]
- 22.Li S, et al. Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. EMBO J. 2012;31(3):552–562. doi: 10.1038/emboj.2011.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson RG. The three-dimensional structure of the basal body from the rhesus monkey oviduct. J Cell Biol. 1972;54(2):246–265. doi: 10.1083/jcb.54.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Toole ET, et al. Three-dimensional organization of basal bodies from wild-type and delta-tubulin deletion strains of Chlamydomonas reinhardtii. Mol Biol Cell. 2003;14(7):2999–3012. doi: 10.1091/mbc.E02-11-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiese C, Zheng Y. A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nat Cell Biol. 2000;2(6):358–364. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- 26.Keating TJ, Borisy GG. Immunostructural evidence for the template mechanism of microtubule nucleation. Nat Cell Biol. 2000;2(6):352–357. doi: 10.1038/35014045. [DOI] [PubMed] [Google Scholar]
- 27.Moritz M, et al. Three-dimensional structural characterization of centrosomes from early Drosophila embryos. J Cell Biol. 1995;130(5):1149–1159. doi: 10.1083/jcb.130.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ibrahim R, et al. Electron tomography study of isolated human centrioles. Microsc Res Tech. 2009;72(1):42–48. doi: 10.1002/jemt.20637. [DOI] [PubMed] [Google Scholar]
- 29.Moritz M, et al. Recruitment of the gamma-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J Cell Biol. 1998;142(3):775–786. doi: 10.1083/jcb.142.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnackenberg BJ, et al. Reconstitution of microtubule nucleation potential in centrosomes isolated from Spisula solidissima oocytes. J Cell Sci. 2000;113(Pt 6):943–953. doi: 10.1242/jcs.113.6.943. [DOI] [PubMed] [Google Scholar]
- 31.Ou YY, et al. Higher order structure of the PCM adjacent to the centriole. Cell Motil Cytoskeleton. 2003;55(2):125–133. doi: 10.1002/cm.10115. [DOI] [PubMed] [Google Scholar]
- 32.Ou Y, Rattner JB. A subset of centrosomal proteins are arranged in a tubular conformation that is reproduced during centrosome duplication. Cell Motil Cytoskeleton. 2000;47(1):13–24. doi: 10.1002/1097-0169(200009)47:1<13::AID-CM2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 33.Gustafsson MG, et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94(12):4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang B, et al. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319(5864):810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang B, Bates M, Zhuang X. Super-resolution fluorescence microscopy. Annu Rev Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gogendeau D, Basto R. Centrioles in flies: the exception to the rule? Semin Cell Dev Biol. 2010;21(2):163–173. doi: 10.1016/j.semcdb.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Schermelleh L, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320(5881):1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakazawa Y, et al. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr Biol. 2007;17(24):2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 39.Rodrigues-Martins A, et al. DSAS-6 organizes a tube-like centriole precursorand its absence suggests modularity in centriole assembly. Curr Biol. 2007;17(17):1465–1472. doi: 10.1016/j.cub.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 40.Gopalakrishnan J, et al. Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat Commun. 2011;2:359. doi: 10.1038/ncomms1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirkham M, et al. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 2003;112(4):575–587. doi: 10.1016/s0092-8674(03)00117-x. [DOI] [PubMed] [Google Scholar]
- 42.van Breugel M, et al. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331(6021):1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- 43.Kitagawa D, et al. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011;144(3):364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gillingham AK, Munro S. The PACT domain a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 2000;1(6):524–529. doi: 10.1093/embo-reports/kvd105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawaguchi S, Zheng Y. Characterization of a Drosophila centrosome protein CP309 that shares homology with Kendrin and CG-NAP. Mol Biol Cell. 2004;15(1):37–45. doi: 10.1091/mbc.E03-03-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez-Campos M, et al. The Drosophila pericentrin-like protein is essential for cilia/flagella function but appears to be dispensable for mitosis. J Cell Biol. 2004;165(5):673–683. doi: 10.1083/jcb.200402130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conduit PT, et al. Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr Biol. 2010;20(24):2178–2186. doi: 10.1016/j.cub.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 48.Erickson HP. Size shape of protein molecules at the nanometer level determined by sedimentation gel filtrationand electron microscopy. Biol Proced Online. 2009;11:32–51. doi: 10.1007/s12575-009-9008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu K, Babcock HP, Zhuang X. Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat Methods. 2012;9(2):185–188. doi: 10.1038/nmeth.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lau L, et al. STED Microscopy with Optimized Labeling Density Reveals 9-Fold Arrangement of a Centriole Protein. Biophys J. 2012;102(12):2926–2935. doi: 10.1016/j.bpj.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dzhindzhev NS, et al. Asterless is a scaffold for the onset of centriole assembly. Nature. 2010;467(7316):714–718. doi: 10.1038/nature09445. [DOI] [PubMed] [Google Scholar]
- 52.Rogers GC, et al. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J Cell Biol. 2009;184(2):225–239. doi: 10.1083/jcb.200808049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loncarek J, et al. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol. 2008;10(3):322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang WJ, et al. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J Cell Biol. 2011;193(4):727–739. doi: 10.1083/jcb.201101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobbelaere J, et al. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 2008;6((9)):e224. doi: 10.1371/journal.pbio.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rogers GC, et al. A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol Biol Cell. 2008;19(7):3163–3178. doi: 10.1091/mbc.E07-10-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi YK, et al. CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J Cell Biol. 2010;191(6):1089–1095. doi: 10.1083/jcb.201007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miyoshi K, et al. Pericentrin a centrosomal protein related to microcephalic primordial dwarfism is required for olfactory cilia assembly in mice. FASEB J. 2009;23(10):3289–3297. doi: 10.1096/fj.08-124420. [DOI] [PubMed] [Google Scholar]
- 59.Takahashi M, et al. Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol Biol Cell. 2002;13(9):3235–3245. doi: 10.1091/mbc.E02-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delaval B, Doxsey SJ. Pericentrin in cellular function and disease. J Cell Biol. 2010;188(2):181–190. doi: 10.1083/jcb.200908114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rauch A, et al. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science. 2008;319(5864):816–819. doi: 10.1126/science.1151174. [DOI] [PubMed] [Google Scholar]
- 62.Dictenberg JB, et al. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol. 1998;141(1):163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ou Y, Zhang M, Rattner JB. The centrosome: The centriole-PCM coalition. Cell Motil Cytoskeleton. 2004;57(1):1–7. doi: 10.1002/cm.10154. [DOI] [PubMed] [Google Scholar]
- 64.Mahen R, Venkitaraman AR. Pattern formation in centrosome assembly. Curr Opin Cell Biol. 2012 doi: 10.1016/j.ceb.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 65.Flory MR, Davis TN. The centrosomal proteins pericentrin and kendrin are encoded by alternatively spliced products of one gene. Genomics. 2003;82(3):401–405. doi: 10.1016/s0888-7543(03)00119-8. [DOI] [PubMed] [Google Scholar]
- 66.Lawo S, Hasegan M, Gupta GD, Pelletier L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nature Cell Biology XXX. 2012 doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- 67.Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D–structured illumination microscopy provides novel insight into architecture of human centrosomes. Biology Open 000. :1–12. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mennella V, et al. Motor domain phosphorylation regulation of the Drosophila kinesin 13. KLP10A J Cell Biol. 2009;186(4):481–490. doi: 10.1083/jcb.200902113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rogers SL, et al. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J Cell Biol. 2003;162(6):1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goshima G, et al. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316(5823):417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McDonald KL. Electron microscopy and EM immunocytochemistry. Methods Cell Biol. 1994;44:411–44. doi: 10.1016/s0091-679x(08)60926-7. [DOI] [PubMed] [Google Scholar]
- 72.Beaudoin GM, 3rd, et al. Afadin a ras/rap effector that controls cadherin function promotes spine and excitatory synapse density in the hippocampus. J Neurosci. 2012;32(1):99–110. doi: 10.1523/JNEUROSCI.4565-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.