Abstract
Background
Asthma exacerbation and other respiratory symptoms are associated with exposure to air pollution. Since environment affects gene methylation, it is hypothesized that asthmatic responses to pollution are mediated through methylation.
Materials & methods
We study the possibility that airborne particulate matter affects gene methylation in the asthma pathway. We measured methylation array data in clinic visits of 141 subjects from the Normative Aging Study. Black carbon and sulfate measures from a central monitoring site were recorded and 30-day averages were calculated for each clinic visit. Genespecific methylation scores were calculated for the genes in the asthma pathway, and the association between the methylation in the asthma pathway and the pollution measures was analyzed using sparse Canonical Correlation Analysis.
Results
The analysis found that exposures to black carbon and sulfate were significantly associated with the methylation pattern in the asthma pathway (p-values 0.05 and 0.02, accordingly). Specific genes that contributed to this association were identified.
Conclusion
These results suggest that the effect of air pollution on asthmatic and respiratory responses may be mediated through gene methylation.
Keywords: sulfate, black carbon, epigenetics, gene-specific methylation scores, pathway analysis, sulfate
Introduction
Air pollution has been associated with asthma exacerbations for decades, including emergency hospital visits[1–3], transitory reductions in lung function[4–6], and respiratory symptoms[7–9]. Allergic responses have also been associated with pollutant exposure[10]. While a number of pollutants have been associated with these outcomes, particulate air pollution has been prominent among them. In particular Diesel exhaust has been shown to be an adjuvant enhancing response to allergen challenges[11–15].
Other studies have looked at longer-term exposures to air pollutants and respiratory outcomes, including lung function.[16,17] The Swiss Cohort Study on Air Pollution and Lung Diseases in Adults (SAPALDIA) used geocoded exposure to particles at residences and reported that higher particle exposure was associated with more rapid decline in lung function.[18] Similarly changes in particle exposure have been associated with changes in symptom prevalence, including wheeze.[19]
Other studies have reported associations between exposure to pollution, particularly from traffic and prevalence or incidence of asthma. [20–23]
Recent interest has also focused on examining different components of particles to assess their relative toxicity. For example, black carbon (BC), a marker of traffic particles, has been shown to be more strongly associated with blood pressure in older adults,[24] but both sulfate, a marker of secondary particles, and BC showed similar effects on flow mediated dilatation of the brachial artery.[25] Similarly traffic pollutants are often more commonly cited as associated with asthma,[22] but sometimes PM2.5 (particulate matter of diameter smaller than 2.5 micrometer) has been stronger than black carbon,[21,22] and sulfates also appear to play a role.[26] While there are many components of particles, BC and sulfate are reasonable surrogates for two of the more important components.
The mechanisms by which air pollution may be producing these effects are still being explored. DNA methylation, the best understood of the epigenetic mechanisms, is the covalent addition of methyl groups to cytosine to form 5-methyl-cytosine (5mC). Methylation of promoter regions and other regulatory sequences tends to reversibly repress gene transcription. As such, DNA methylation is now recognized as an important regulator of transcription. Since DNA methylation is responsive to environment cues, but changes less rapidly than mRNA or protein levels, it represents an attractive, potentially more stable marker of response to environmental factors. Recent investigations have indicated that exposure to particular matter or other airborne pollutants modifies DNA methylation in peripheral blood DNA of exposed individuals.[27,28] We hypothesized that exposure to BC and sulfate particles were associated with changes in methylation of genes along the asthma pathway. The asthma pathway is composed of a set of genes, coding for proteins involved in asthmatic response. We investigated this using methylation microarray data from a subset of 132 older men who were participants in the Normative Aging Study, and were not asthmatic at the time of the examination.
Materials and Methods
The Normative Aging Study (NAS) is a longitudinal study of aging in Eastern Massachusetts established in 1963 by the Veterans Administration (VA). Community-dwelling men from the greater Boston metropolitan area were screened at entry and accepted into the study if they had no history of heart disease, hypertension, diabetes mellitus, cancer, peptic ulcer, gout, recurrent asthma, bronchitis, or sinusitis. Between 1963 and 1968, a total of 2,280 men were enrolled, ranging in age from 21 to 80 years (mean = 42 years) at entry. Since their enrollment, the participants have undergone comprehensive clinical examinations at 3-5 year intervals. As part of these examinations, DNA has been extracted from leukocytes and stored in all visits since 1999. We conducted a genome wide scan of the promoter regions of 19,000 genes on 141 subjects from the NAS. The subjects were selected based on having sufficient DNA for the assay, while leaving DNA for subsequent studies. Since we were specifically interested in the effect of particulate matter exposure on methylation in the asthma pathway, we excluded 9 subjects who were diagnosed as asthmatic at the time of the examination. Informed consent has been obtained from the participants involved and the study was approved by the institutional review board (IRB) at the participating institutions.
Chromatin immunoprecipitation (Chip) and DNA Methylation Microarray- Chip on Chip
Samples were hybridized to the RefSeq 385K Promoter tiling array (Roche NimbleGen, Madison, WI) representing the promoter regions of all well-characterized genes in the RefSeq database (RefSeq genes with NM Prefix), as well as all the UCSC-annotated CpG islands. The array coverage is based on 50-75mer probes with approximately 100bp spacing, dependent on the sequence composition of the region. Sample immunoprecipitation, labeling, hybridization and data extraction were performed according to standard procedures optimized by Roche-NimbleGen, as previously reported.[29]
Five μg high-quality genomic DNA was isolated from blood buffy coat using QiAmp DNA blood kits (QIAGEN, Hilden, Germany) and digested with 24U Mse I (5′-T▼TAA) to produce small fragments (200 - 1,000bp). Fragmented DNA was heat denatured to produce single-stranded DNA and immunoprecipitatedusing anti-5-methyl cytidine (Abcam-ab10805) monoclonal mouse antibodies. Methylated DNA immunoprecipitated (MeDIP) fragments were heat-denatured for 10min at 95°C, and immediately cooled on ice. Immune complexes were captured with Protein-A agarose beads (Invitrogen-15918-014) and washed to remove nonspecifically-bound material. Following elution of bound complexes, MeDIP samples were purified with phenol-chloroform:isoamyl alcohol and ethanol precipitation in a −80°C freezer for 30min. After centrifugation, the supernatant was carefully removed, the pellet washed with cold 70% ethanol and centrifuged again to remove remaining supernatant. MeDIP samples were completely dried and resuspended in 30μl 10mM TrisHCl (pH 8.5). Fragments were amplified by whole-genome amplification (GenomePlex® Complete Whole Genome Amplification (WGA2) Kit, Sigma-Aldrich). Experimental and total DNA samples were denatured and labeled by Klenow fragment-catalyzed primer extension using Cy dyes (Cy3 and Cy5) coupled 9mer primers. The labeled experimental IP and total DNAs were co-hybridized to the array for 16-20 hours, washed, and scanned by the Roche NimbleGen Service Laboratory (Reykjavík, Iceland). The intensity ratio of IP to total DNA was used to identify DNA methylation.
PM2.5 and Components (BC and Sulfate)
Continuous black carbon (BC) and particulate matter of sizes smaller than 2.5 micrometer (PM2.5) concentrations were measured at a Harvard School of Public Health monitoring site located at Countway Library (10 Shattuck Street, Boston, MA), 1 km from the clinical examination site, and average values for the month before the blood draw were computed. BC, a marker for traffic particles weighted toward Diesel particles, was measured using an aethalometer (Magee Scientific, Berkeley, CA), and PM2.5 was measured using a Tapered Element Oscillating Microbalance (model 1400A; Rupprecht&Pataschnick Co., East Greenbush, NY), operated at 50 degrees with two 4 liter per minute PM2.5 impactors before the inlet. From September 25, 1999 to February 2, 2004, particulate sulfate was measured using the Harvard/EPA Denuder System (HEADS), which samples inorganic gaseous and particulate species in air. From January 1, 2003 through 2007, daily particulate filter samples were analyzed by X-ray fluorescence (XRF) spectroscopy for elemental components. From these samples, we multiplied the mass of sulfur by three to obtain the mass of sulfate. For days when both HEADS impactors and XRF were in operation, we used linear regression and determined that the measurements had a slope of 1 and R2 greater than 0.9, indicating a high correlation between the two monitoring methods. Hence, XRF measurements were used during this period of overlap. Sulfate particles are secondary, long range particles primarily from coal burning powerplants.
Statistical Methods
Normalization and Pre-processing of DNA Methylation Data
We normalize the raw methylation intensities (log2 green vs red channel ratio) for each probe by subtracting the overall median and then dividing by the probe's GC-content specific standard deviation which is the standard deviation of all the probes whose sequence have the same number of G and C nucleotides as the target probe. We then smooth the normalized scores using a local linear kernel smoother (Fan and Gijbels, 1996) over the probe locations. For a given gene, the gene methylation score is then calculated by taking the area under the smoothed curve, truncated at zero, over a 500bp window around the transcription start site of the gene, and then dividing by percentage of CpGdinucleotides in the DNA sequence within the window and the number of probes having positive scores within the window.
Pathway analysis
We identified the genes associated with the asthma pathway using KEGG [30,31]. A list of the genes in the pathway is given in table 4.
Table 4.
The genes in the asthma pathway, listed in the KEGG website.
| Gene symbols |
|---|
| LOC100133583 |
| HLA-DMA |
| HLA-DMB |
| HLA-DOA |
| HLA-DOB |
| HLA-DPA1 |
| HLA-DPB1 |
| HLA-DQA1 |
| HLA-DQA2 |
| HLA-DQB1 |
| HLA-DRA |
| HLA-DRB1 |
| HLA-DRB3 |
| HLA-DRB4 |
| HLA-DRB5 |
| IL4 |
| CD40LG |
| CD40 |
| FCER1A |
| MS4A2 |
| FCER1G |
| IL9 |
| IL10 |
| IL13 |
| IL5 |
| TNF |
| IL3 |
| PRG2 |
| RNASE3 |
| EPX |
| CCL11 |
Our goal was to examine whether either particle component, black carbon (BC) or power plan particles (SO4) or PM2.5 in general, was associated with differences in methylation patterns along the entire pathway, rather than for specific genes.
Since we recognize that all of the genes in a given pathway may not contribute to the particle association, we adopted a sparse stepwise Canonical Correlation Analysis (step-CCA) strategy, as described in ref. [32], allowing us to analyze the association between a single exposure variable (BC or sulfate) and a set of outcomes (gene methylations in the asthma pathway) while identifying specific genes associated with the exposure.
The sparse CCA approach requires the selection of a tuning parameter that controls the size of the selected set of genes. We used the empirical-CIC [33], a score that compares a computed measure of model fit, to its null distribution, and selects the set of genes that yields the largest improvement in model fit when compared to the expected measure under the null.
Because methylation varies with age, and smoking potentially affects methylation in the asthma pathway, we adjusted for age and smoking by taking the methylation scores of each of the genes in the pathway and regressed them against age and smoking status. We did the same for the exposure variables. Subsequent analyses looked at the association of the residuals of methylation and the residuals of exposure.
Results
Table 1 shows a summary of the characteristics of the participants in the study (n = 132). These characteristics were almost identical when calculated for the subset of the 85 participants who had sulfate measures for the day of their appropriate clinic visit.
Table 1.
Summary of relevant characteristics of participant in the methylation study (n = 132). The sulfate exposure is calculated over n = 85 participant.
| Characteristic | Measure |
|---|---|
| Age (years) | Range 56 – 88, median 73 |
| Smoking status | |
| Never smoked | 50 people |
| Current smoker | 7 people |
| Former smoker | 75 people |
| Take any medication | 77 people |
| BC, 30 days averaged exposure | Mean 0.85 μg/m3 |
| Sulfate, 30 days averaged exposure | Mean 3.05 μg/m3) |
The methylation pattern in the asthma pathway was significantly associated both with exposure to black carbon (BC) (p=0.048) and sulfate (p=0.017).
The genes that contributed to the association with BC exposure included genes coding for components in the major histocompatibility complex, class II (MHCII) such as HLA-DOB; for IgE receptors such as FCER1A(Fc Fragment IgE Receptor, high affinity 1, alpha subunit) and FCER1G(Fc Fragment IgE Receptor, high affinity 1, gamma subunit); for interleukins (IL-9); and for the eosinophil granule major basic protein (MBP, also known as PRG2 [PRoteoGlycan 2]). The genes that contributed to the association with sulfate exposure included genes coding MHCII, such as HLA-DPA1; for IgE receptors such as FCER1G(Fc Fragment IgE Receptor, high affinity 1, gamma subunit); for cytokines such as IL-10, Eosinophil Cationic Protein (ECP, also known as Ribonuclease A Family 3 [RNASE3]), and eotaxin (also known as chemokine CC-motif Ligand 11 [CCL11]). Interestingly, only one gene (FCER1G) was included in both groups of genes associated with black carbon and sulfates.
Table 2 and 3 presents the results of the pathway analysis with sulfate and BC as exposures, respectively. In each of the tables weights are given for each of the genes, where larger weight in absolute values is interpreted as larger association between the exposures to the methylation of the gene, more specifically positive values are interpreted as increased (hyper-) methylation, and negative values as decreased (hypo-) methylation. In addition to the weights, the Canonical Correlation between the selected set of genes and the exposure is provided.
Table 2.
Results of the step-CCA algorithm applied on the Asthma pathway with BC as exposure. The selected genes and their weights are provided. The correlation between the gene methylation scores and exposure to BC is 0.47 and the P-value is 0.048.
| Gene | Weight |
|---|---|
| HLA-DOB | 0.49 |
| CD40LG | 0.37 |
| FCER1A | 0.58 |
| FCER1G | −0.33 |
| IL9 | −0.28 |
| IL13 | 0.37 |
| MBP (PRG2) | 0.49 |
Table 3.
Results of the step-CCA algorithm applied on the Asthma pathway with Sulfate as exposure. The selected genes and their weights are provided. The correlation between the gene methylation scores and exposure to sulfate is 0.56 and the P-value is 0.017.
| Gene | Weight |
|---|---|
| HLA-DPA1 | −0.28 |
| FCER1G | −0.38 |
| IL10 | 0.27 |
| ECP (RNASE3) | −0.37 |
| EOTAXIN (CCL11) | 0.74 |
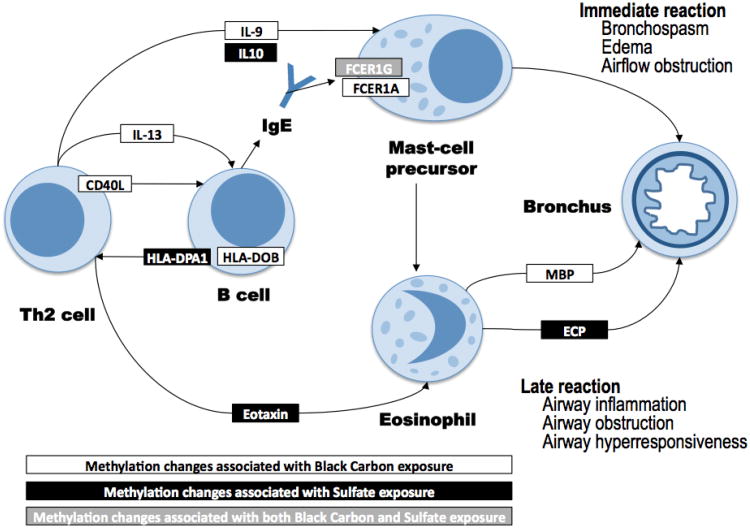
Figure 1 shows the roles within the KEGG asthma pathway (has-05310) of the proteins encoded by the genes found to be differentially methylated in relation to BC and sulfate levels. The pathway that we present is adapted from the asthma KEGG pathway to show only cells that circulate in blood as well as only the genes identified in our analysis. As described there, the asthma pathway consists of genes coding for proteins involved in asthmatic response. Asthmatic response begins with allergens encountering antigen presenting cells (APC) that line the airway, which are then activated. Then various proteins (that form the asthma pathway) are involved in a chain reaction by which first native T-cells are differentiated into TH2 cells, that in turn stimulate the formation of IgE by B-cells, that are then bind to mast cells, leading to the release of histamines and leukotrienes and forming the “immediate reaction”. Mast cells, again through mediation of proteins from the asthma pathway, also contribute to the recruitment of eosinophils. The activation of esodinophils leads to “late reaction” of chronic airway inflammation.
Figure 1.
The proteins coded by the genes identified in the analysis as associated with sulfate and black carbon exposures, and their roles within the KEGG asthma pathway.
Pathway information adapted from the KEGG Asthma pathway, Hsa-05310
The figure shows that BC and sulfate exposure affect via altered DNA methylation the control of genes expressed on several blood cell types, including Th2 cells, B cells, mast-cells and their precursors, and eosinophils. Also, most of the differentially-methylated genes were either part of the Th2/B-cell signaling or contributed to controlling eosinophils and late reactions such as airway inflammation, obstruction, and hyper-responsivenness (Figure 1).
Discussion
Both BC, primarily from Diesel exhaust in Boston, and Sulfate particles, primarily from coal burning powerplants, were associated with changes in the methylation of genes in the asthma pathway. This finding provides a potential mechanism for the reported association of traffic pollution, particularly Diesel exhaust, and especially the particles, and the exacerbation of asthma. Diesel particles have been found to be an adjuvant for atopic responses,[11,13,34] and heavily traffic roads, particularly with high bus or truck traffic, have been linked to respiratory symptoms and asthma.[20,22,35] The genes modified by BC exposure include IgE signaling and eosinophil control, which fits in well with those observations. Hence altered epigenetic control of the immunoregulatory cells may be in the causal pathway between black carbon and asthmic responses. The specific genes identified suggest target proteins for toxicologic studies of Diesel particles.
Equally interesting, we found that sulfate particles, which are primarily from coal burning power plants, are also associated with altered methylation in the asthma pathway. While most attention has focused on traffic exposure and asthma in recent years, it is important to remember that a number panel and ER studies [36–38] reported sulfate associated with asthma exacerbation, and there are reported drops in pulmonary function and increased symptoms in asthmatic children following exposure to episodes of high ozone and sulfate particles.[39] While the genes whose methylation is associated with sulfate exposure were mostly different from those influenced by BC, they also included IgE receptors, cytokines, and eosinophil control. Hence reduced asthma severity may be an underappreciated benefit of control of sulfate precursors.
Another study linked exposure to air pollution and methylation in a gene related to asthma. [40] and found that increased exposure to AAP is associated with hypermethylation of the Foxp3 locus, a regulatory T cell that with impaired function in asthmatics. Note, however, that this gene is not a member of the asthma pathway that we used.
A major limitation of this study is the cells that were used, which were peripheral leukocytes. Target tissue cells are difficult to obtain in human studies, but differential methylation by cell type can occur, raising questions about their relevance. In this case, white cells are key players in asthma, and particularly acute exacerbations, and the methylation targets identified, eosinophils, mast cells, cytokines, IgE, are all central to asthma. While BAL cells would perhaps have greater relevance, peripheral cells circulate through the lung, and are exposed there to many of the same air pollution derived signals that resident cells are. Support for this comes also from the association of air pollution with increased markers of inflammation in peripheral blood, such as C reactive protein, fibrinogen, sICAM and sVCAM, IL6, etc. Hence this limitation seems less of a concern in the case of asthma and air pollution.
Another limitation is the population being studied—elderly men. The differential diagnosis of asthma in the elderly is difficult, and many immunologic responses decline with age. Hence the same pattern of methylation changes may not be seen in younger subjects. Further, it is possible that the observed association does not hold in the same manner in females, as our investigation was conducted in a male-only cohort. Nevertheless, the finding that DNA methylation in the asthmatic pathway is modified by exposure to black carbon and sulfates is unlikely to be a response only seen in the male elderly, although the magnitude of response, and specific genes affected, may differ with age and gender. In addition, many of the participants take medications, mainly to treat high blood pressure, and it is possible that the treated condition is affected by air pollution, making drugs possible confounders in the association between pollution and methylation. However, since asthmatic subjects are excluded, it is unlikely that the drugs used are associated with methylation in the asthma pathway. Finally, a limitation is the small sample size (n = 132 for BC, and n = 85 for sulfate association analysis). These results should be validated in a separate study.
Executive summary.
Introduction
Air pollution has been association with asthma exacerbations.
In particular, Diesel exhaust had been association with allergic responses.
Two prominent components of particulate matter are black carbon (BC), a marker of traffic, particularly Diesel, particles, and sulfate, a marker of secondary particles, especially from power plants.
The mechanisms by which air pollution produces allergic and asthmatic effects are still being explored.
The asthma pathway is composed of a set of genes, coding for proteins involved in asthmatic responses.
DNA methylation, the covalent addition of methyl group to the cytosine nucleotide, is an important regulator of gene transcription, and it is responsive to environmental cues.
We hypothesized that exposure to BC and sulfate particles were associated with changes in methylation of genes in the asthma pathway.
Results
Exposure to BC was associated with methylation of 7 genes from the asthma pathway (p-value = 0.05), and exposure to sulfate was associated with methylation of 5 genes from the pathway (p-value = 0.02).
A single gene was affected by both BC and sulfate exposure.
The genes associated with air pollution exposure are related to both short and long term allergic responses related to asthma.
Discussion
Our findings suggest that altered epigenetic control of the immunoregulatory cells may be in the causal pathway between black carbon and asthmic responses.
A limitation of this study is the use of peripheral leukocytes to measure methylation, rather than bronchoalveolar lavage (BAL) cell. However, specifically in the case of asthma, it is reasonable, since peripheral cells circulate through the lungs.
Possible limitations of this study are relatively small sample size, and the elderly population.
Methods
132 non-asthmatic male participants from the Normative Aging Study (NAS), residing in the Boston area, were assessed for methylation status in peripheral leukocytes using a NimbleGen promoter array.
30 days averaged BC (for n=132 participants) and sulfate (for n = 85 participants) exposure measures from a central location in Boston were calculated for each participant.
A sparse Canonical Correlation Analysis were conducted to study the association between exposure to BC and sulfate, and the methylation genes from the asthma pathway, and to identify specific genes that are directly associated with air pollution.
References
- 1.Schwartz J, Slater D, Larson TV, Pierson WE, Koenig JQ. Particulate Air Pollution and Hospital Emergency Room Visits for Asthma in Seattle. American Journal of Respiratory and Critical Care Medicine. 1993;147(4):826–831. doi: 10.1164/ajrccm/147.4.826. [DOI] [PubMed] [Google Scholar]
- 2.Fusco D, Forastiere F, Michelozzi P, et al. Air pollution and hospital admissions for respiratory conditions in Rome, Italy. The European respiratory journal: official journal of the European Society for Clinical Respiratory Physiology. 2001;17(6):1143–50. doi: 10.1183/09031936.01.00005501. [DOI] [PubMed] [Google Scholar]
- 3.Morgan G, Corbett S, Wlodarczyk J. Air pollution and hospital admissions in Sydney, Australia, 1990 to 1994. American Journal of Public Health. 1998;88(12):1761–1766. doi: 10.2105/ajph.88.12.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peters A, Goldstein IF, Beyer U, et al. Acute Health Effects of Exposure to High Levels of Air Pollution in Eastern Europe. Am J Epidemiol. 1996;144(6):570–581. doi: 10.1093/oxfordjournals.aje.a008967. [DOI] [PubMed] [Google Scholar]
- 5.Delfino RJ, Staimer N, Tjoa T, et al. Personal and ambient air pollution exposures and lung function decrements in children with asthma. Environmental health perspectives. 2008;116(4):550–8. doi: 10.1289/ehp.10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kinney PL, Ware JH, Spengler JD, Dockery DW, Speizer FE, Ferris BG. Short-term Pulmonary Function Change in Association with Ozone Levels. American Journal of Respiratory and Critical Care Medicine. 1989;139(1):56–61. doi: 10.1164/ajrccm/139.1.56. [DOI] [PubMed] [Google Scholar]
- 7.Dockery DW, Speizer FE, Stram DO, Ware JH, Spengler JD, Ferris BG. Effects of Inhalable Particles on Respiratory Health of Children. American Journal of Respiratory and Critical Care Medicine. 1989;139(3):587–594. doi: 10.1164/ajrccm/139.3.587. [DOI] [PubMed] [Google Scholar]
- 8.Pope CA, Dockery DW, Spengler JD, Raizenne ME. Respiratory Health and PM10 Pollution: A Daily Time Series Analysis. American Journal of Respiratory and Critical Care Medicine. 1991;144(3 Pt 1):668–674. doi: 10.1164/ajrccm/144.3_Pt_1.668. [DOI] [PubMed] [Google Scholar]
- 9.Delfino RJ, Coate BD, Zeiger RS, Seltzer JM, Street DH, Koutrakis P. Daily asthma severity in relation to personal ozone exposure and outdoor fungal spores. American journal of respiratory and critical care medicine. 1996;154(3 Pt 1):633–41. doi: 10.1164/ajrccm.154.3.8810598. [DOI] [PubMed] [Google Scholar]
- 10.Boutin-Forzano S, Hammou Y, Gouitaa M, Charpin D. Air pollution and atopy. European annals of allergy and clinical immunology. 37(1):11–16. [PubMed] [Google Scholar]
- 11.Bastain TM, Gilliland FD, Li YF, Saxon A, Diaz-Sanchez D. Intraindividual reproducibility of nasal allergic responses to diesel exhaust particles indicates a susceptible phenotype. Clinical immunology (Orlando, Fla) 2003;109(2):130–6. doi: 10.1016/s1521-6616(03)00168-2. [DOI] [PubMed] [Google Scholar]
- 12.Diaz-Sanchez D, Garcia MP, Wang M, Jyrala M, Saxon A. Nasal challenge with diesel exhaust particles can induce sensitization to a neoallergen in the human mucosa ☆☆☆★. Journal of Allergy and Clinical Immunology. 1999;104(6):1183–1188. doi: 10.1016/s0091-6749(99)70011-4. [DOI] [PubMed] [Google Scholar]
- 13*.Diaz-Sanchez D, Penichet-Garcia M, Saxon A. Diesel exhaust particles directly induce activated mast cells to degranulate and increase histamine levels and symptom severity. The Journal of allergy and clinical immunology. 2000;106(6):1140–6. doi: 10.1067/mai.2000.111144. Showed that Diesel exhaust particles activated mast cells degranulation. [DOI] [PubMed] [Google Scholar]
- 14.Fahy O, Hammad H, Sénéchal S, et al. Synergistic effect of diesel organic extracts and allergen Der p 1 on the release of chemokines by peripheral blood mononuclear cells from allergic subjects: involvement of the map kinase pathway. American journal of respiratory cell and molecular biology. 2000;23(2):247–54. doi: 10.1165/ajrcmb.23.2.4116. [DOI] [PubMed] [Google Scholar]
- 15.Fujimaki H, Yamamoto S, Kurokawa Y. Effect of diesel exhaust on immune responses in C57BL/6 mice intranasally immunized with pollen antigen. Journal of UOEH. 2005;27(1):11–24. doi: 10.7888/juoeh.27.11. [DOI] [PubMed] [Google Scholar]
- 16.Lubinski W, Toczyska I, Chcialowski A, Płusa T. Influence of air pollution on pulmonary function in healthy young men from different regions of Poland. Annals of agricultural and environmental medicine: AAEM. 2005;12(1):1–4. [PubMed] [Google Scholar]
- 17.Rojas-Martinez R, Perez-Padilla R, Olaiz-Fernandez G, et al. Lung function growth in children with long-term exposure to air pollutants in Mexico City. American journal of respiratory and critical care medicine. 2007;176(4):377–84. doi: 10.1164/rccm.200510-1678OC. [DOI] [PubMed] [Google Scholar]
- 18.Downs SH, Schindler C, Liu LJS, et al. Reduced Exposure to PM10 and Attenuated Age-Related Decline in Lung Function. The New England journal of medicine. 357(23):2338–2347. doi: 10.1056/NEJMoa073625. [DOI] [PubMed] [Google Scholar]
- 19.Bayer-Oglesby L, Grize L, Gassner M, et al. Decline of Ambient Air Pollution Levels and Improved Respiratory Health in Swiss Children. Environmental Health Perspectives. 2005;113(11):1632–1637. doi: 10.1289/ehp.8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin S, Munsie JP, Hwang SA, Fitzgerald E, Cayo MR. Childhood asthma hospitalization and residential exposure to state route traffic. Environmental research. 2002;88(2):73–81. doi: 10.1006/enrs.2001.4303. [DOI] [PubMed] [Google Scholar]
- 21.Carlsten C, Dybuncio A, Becker A, Chan-Yeung M, Brauer M. Traffic-related air pollution and incident asthma in a high-risk birth cohort. Occupational and environmental medicine. 2011;68(4):291–5. doi: 10.1136/oem.2010.055152. [DOI] [PubMed] [Google Scholar]
- 22.McConnell R, Islam T, Shankardass K, et al. Childhood incident asthma and traffic-related air pollution at home and school. Environmental health perspectives. 2010;118(7):1021–6. doi: 10.1289/ehp.0901232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Jerrett M, Shankardass K, Berhane K, et al. Traffic-related air pollution and asthma onset in children: a prospective cohort study with individual exposure measurement. Environmental health perspectives. 2008;116(10):1433–8. doi: 10.1289/ehp.10968. Showed that traffic-related air pollution contributes to new-onset asthma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mordukhovich I, Wilker E, Suh H, et al. Black carbon exposure, oxidative stress genes, and blood pressure in a repeated-measures study. Environmental health perspectives. 2009;117(11):1767–72. doi: 10.1289/ehp.0900591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Neill MS, Veves A, Zanobetti A, et al. Diabetes enhances vulnerability to particulate air pollution-associated impairment in vascular reactivity and endothelial function. Circulation. 2005;111(22):2913–20. doi: 10.1161/CIRCULATIONAHA.104.517110. [DOI] [PubMed] [Google Scholar]
- 26.Peters A, Dockery DW, Heinrich J, Wichmann HE. Medication use modifies the health effects of particulate sulfate air pollution in children with asthma. Environmental health perspectives. 1997;105(4):430–5. doi: 10.1289/ehp.97105430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bollati V, Baccarelli A, Hou L, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer research. 2007;67(3):876–80. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 28*.Baccarelli A, Wright RO, Bollati V, et al. Rapid DNA methylation changes after exposure to traffic particles. American journal of respiratory and critical care medicine. 2009;179(7):572–8. doi: 10.1164/rccm.200807-1097OC. Showed that DNA methylation was affected by exposure to benzene particles. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selzer RR, Richmond TA, Pofahl NJ, et al. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes, chromosomes & cancer. 2005;44(3):305–19. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]
- 30.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic acids research. 2012;40(Database issue):D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sofer T, Maity A, Coull B, Baccarelli A, Schwartz J, Lin X. Multivariate Gene Selection and Testing in Studying the Exposure Effects on a Gene Set. Statistics in Biosciences. 2012 doi: 10.1007/s12561-012-9072-7. In print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sofer T. Covariance Transfored Sparse Canonical Correlation Analysis. Submitted [Google Scholar]
- 34.Bateson TF, Schwartz J. Children's response to air pollutants. Journal of toxicology and environmental health Part A. 2008;71(3):238–43. doi: 10.1080/15287390701598234. [DOI] [PubMed] [Google Scholar]
- 35.Patel MM, Quinn JW, Jung KH, et al. Traffic density and stationary sources of air pollution associated with wheeze, asthma, and immunoglobulin E from birth to age 5 years among New York City children. Environmental research. 2011;111(8):1222–9. doi: 10.1016/j.envres.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ostro BD, Lipsett MJ, Wiener MB, Selner JC. Asthmatic responses to airborne acid aerosols. American journal of public health. 1991;81(6):694–702. doi: 10.2105/ajph.81.6.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bates DV, Sizto R. Air pollution and hospital admissions in Southern Ontario: the acid summer haze effect. Environmental research. 1987;43(2):317–31. doi: 10.1016/s0013-9351(87)80032-4. [DOI] [PubMed] [Google Scholar]
- 38.Bates DV, Baker-Anderson M, Sizto R. Asthma attack periodicity: a study of hospital emergency visits in Vancouver. Environmental research. 1990;51(1):51–70. doi: 10.1016/s0013-9351(05)80182-3. [DOI] [PubMed] [Google Scholar]
- 39.Thurston GD, Lippmann M, Scott MB, Fine JM. Summertime haze air pollution and children with asthma. American journal of respiratory and critical care medicine. 1997;155(2):654–60. doi: 10.1164/ajrccm.155.2.9032209. [DOI] [PubMed] [Google Scholar]
- 40.Nadeau K, McDonald-Hyman C, Noth EM, et al. Ambient air pollution impairs regulatory T-cell function in asthma. The Journal of allergy and clinical immunology. 2010;126(4):845–852.e10. doi: 10.1016/j.jaci.2010.08.008. [DOI] [PubMed] [Google Scholar]