Abstract
We describe the use of a targeted proteomics approach, Selected Reaction Monitoring (SRM) mass spectrometry, to detect and assess RNAi-mediated depletion or ‘knockdown’ of specific proteins from human cells and from Drosophila flies. This label-free approach does not require any specific reagents to confirm the depletion of RNAi target protein(s) in unfractionated cell or whole organism extracts. The protocol described here is general, can be developed rapidly and can be multiplexed to detect and measure multiple proteins at once. Furthermore, the methodology can be extended to any tandem mass spectrometer - making it widely accessible. This methodology will be applicable to a wide range of basic science and clinical questions where RNAi-mediated protein depletion needs to be verified, or where differences in relative abundance of target proteins need to be rapidly assessed between samples.
Keywords: Mass spectrometry, selected reaction monitoring, RNAi
INTRODUCTION
RNA interference (RNAi) has established itself as a particularly powerful methodology for large scale loss-of-function screens to identify genetic determinants of development, aging or disease processes such as cancer. The genes, proteins and pathways identified by RNAi screens can serve as an immediate focus for additional experiments to elucidate mechanisms of action or, in the case of disease, identify genes or proteins with the potential to serve as diagnostic or therapeutic biomarkers or targets. Libraries of RNAi reagents such as short hairpin (sh) and short interfering (si) RNAs are now available for many organisms to perform genome-scale screens or more targeted analyses of specific families of proteins, such as kinases 1-5.
Several practical approaches have been developed to validate RNAi screens and identify off-target and other confounding effects. These approaches include the use of multiple RNAi reagents predicted to target the same gene, and organism-specific ‘scrambled sequence’ or nonspecific RNAi controls; RT-PCR or Western blot assays to demonstrate reduced expression of the targeted RNA or protein; and biochemical or biological ‘rescue’ by expressing an RNAi-resistant target gene or cDNA 3,6. Direct verification of target protein depletion is desirable, as protein–rather than mRNA–depletion is the goal of many RNAi experiments. Direct analysis of protein abundance also allows researchers to account for the widely differing biological half-lives of proteins that may affect the interpretation of RNAi screen results.
Despite the need for protein-based verification in RNAi experiments, high quality antibodies are not available for most proteins even in well studied organisms. Moreover, proteins of high biological interest in any organism may be refractory to antibody development, and antibody-based analytical methods such as Western blotting are difficult to multiplex to enable the detection and quantitation of multiple proteins in single analyses. Mass spectrometry (MS) is a versatile alternative to detect and measure the difference in abundance of individual peptides from a digest of a protein mixture between conditions or states. Selected reaction monitoring (SRM) mass spectrometry, which has historically been used for small molecule analyses and is also well established for peptides absolute quantitation 7, represents a particularly promising approach to validate the depletion or ‘knockdown’ of a protein by RNAi. Due to its sensitivity and selectivity, SRM can detect specific target proteins, including low abundance proteins that may be missed or proteins intrinsically difficult to detect by ‘shotgun’ proteomic profiling methods 8-11. Moreover SRM can be applied to protein targets regardless of immunogenicity; can be used to detect and quantify specific proteins or modifications over a wide range of protein abundances with high accuracy 10,12; and can be multiplexed to detect and quantify multiple protein targets in single experiments. Additionally, multiplexing may be accomplished by implementing scheduling, where a larger number of transitions could be monitored by breaking the chromatographic space into windows.
Here, we describe a general SRM-based methodology that can be used to detect low abundance peptides that can be used as a proxy of a target protein and demonstrate the use of this method to measure RNAi-mediated depletion of specific proteins from human cells and from the fruit fly Drosophila melanogaster. We deliberately chose target proteins for these analyses that represent analytical challenges. The multi-functional human tumor suppressor protein TP53 is variably expressed and is modified across a wide range of residues by post-translational modifications that include phosphorylation, acetylation, ubiquitination, sumoylation, neddylation, methylation and glycosylation 13,14. Our two Drosophila target proteins, in contrast, are short accessory gland seminal fluid proteins that are among >130 seminal fluid proteins transferred from males to females during mating 15,16. Acp70A is a processed protein that in mature form is a 36 residue lysine- and arginine-rich peptide 16,17. Acp53C14a is a processed protein of 98 residues with no homology to known proteins, and for which there is no good antibody. WRN protein is a low-abundance, DNA binding protein that belongs to the RecQ family of helicases. Its loss causes Werner Syndrome, which is characterized by premature aging and genomic instability 18. Previous analysis by quantitative immune blots has shown that lymphoblastoid cells lines and fibroblasts contain ~ 6 × 104 WRN molecules per cell 19, and a more recent report using SRM estimates WRN abundance at < 5 × 102 copies per cell in U-2 OS cells 20.
In this work we demonstrate the development of a common SRM protocol to detect and identify differences in abundance for these four target proteins in the context of RNAi-mediated depletion experiments in human cells or in whole flies. Our results show excellent agreement between SRM and Western blot based detection and quantitation methods where a direct comparison was possible. Using different proteins as examples, we validate our peptide selection by utilizing publically available NIST spectral library for verification of our chosen transitions, using commercially available recombinant protein, and incorporating stable isotope labeled (SIL) peptides in the analysis.
EXPERIMENTAL PROCEDURES
Cell culture and fly rearing
The SV40-transformed human fibroblast cell lines GM639 and HEK-293T cell line were obtained from the Coriell Institute Cell Repositories (Camden, NJ). Cells were grown in Dulbecco-modified Eagle's medium (MediaTech CellGro, Manassas, VA) containing 4,500 mg/L glucose and supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), penicillin and streptomycin sulfate (100 U/ml and 100 mg/ml, respectively; Invitrogen, Carlsbad, CA) in a humidified 37°C, 7% incubator. Drosophila melanogaster RNAi lines were obtained from the Vienna Drosophila RNAi Center (VDRC; www.vdrc.at) 21. Flies were reared at 25°C on yeast-cornmeal-molasses medium.
siRNA-mediated depletion of TP53
TP53 depletions were performed using two methods, and differentially treated cells were used as indicated. Cells were initially transfected with a TP53-specific siRNA (Silencer Select s606) or Silencer Negative Control #1 siRNA (Applied Biosystems, Foster City, CA). In brief, cells were grown in 6-well plates (Nunc, Denmark) and transfected when 50% confluent with either 50 pmol or 100 pmol of siRNA using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Transfection efficiency was consistently >90% as estimated from the transfection of a Cy3-labeled control oligonucleotide of sequence 5'-Cy3-CCTAAGGTTAAGTCGCCCTCGCTCTAGCGAGGGCGACTTAACCTTAGG-3' (Operon, Valencia, CA) followed by fluorescence microscopy. Cells were harvested by addition of trypsin 40 hrs after transfection, and used for Western blot analysis and mass spectrometry as described in following sections.
shRNA-mediated depletion of TP53
Depletion of TP53 was additionally achieved by viral transduction of shp53 pLKO.1 puro plasmid (plasmid 19119; Addgene) or pLKO.1 expressing a scrambled shRNA with no known target sequence in the human genome (plasmid 1864, “scramble shRNA”; Addgene). The cells were grown under puromycin selection (1 μg/mL) for 48 hours prior to collection. The reduced level of protein was verified by Western blot.
Protein depletion in Drosophila
The VDRC fly line #52425 21 contains a chromosome III P-element insertion from which an inverted repeat (IR)-Acp70A is expressed under the control of the UAS promoter. To generate RNAi flies, line #52425 males (genotype: w1118; +/+; UAS-IR-Acp70A/UAS-IR-Acp70A) were crossed to virgin females harboring the GAL4 transcription factor gene under control of a Drosophila tubulin promoter (genotype: w1118/w1118; +/+; tubulin-GAL4/TM3, Sb). The tubulin-GAL4 driver causes UAS-IR expression, resulting in RNAi-mediated knockdown of the Acp70A target gene in all adult tissues 22. Knockdown males were identified by the lack of the Sb marker; control males inherited and expressed the Sb marker. RNAi-mediated depletion of a second transferred and readily identifiable accessory gland protein, Acp53C14a 15,23, was also performed using the corresponding VDRC line for this gene, #44789, in crosses and sample preparation in parallel with Acp70A flies.
shRNA-mediated depletion of WRN
Depletion of WRN protein from human GM639-cc1 cells was previously described by Mao et al 24. In brief, cells that stably express the lentiviral shRNA plasmid were grown under puromycin selection (1 μg/mL) for 48 hours prior to collection. Decreased protein expression was verified by Western blot. Controls included cells transduced with pLKO.1 vector DNA alone or with pLKO.1 expressing a scrambled shRNA with no known target sequence in the human genome (plasmid 1864, “scramble shRNA”; Addgene).
Generation of expression constructs and tetracycline-inducible cell lines
The tet-inducible expression vector for N-terminally SH-tagged human WRN, pFTSH-WRN (originally named pcDNA5/FRT/TO/SH/GW/WRN), was a gift from Dr. Alessandro Vindigni (St. Louis University, MO, USA). It was constructed by Gateway cloning using human WRN ORF in pDONR223 vector for LR recombination with Flp-In tet-inducible expression vector pcDNA5/FRT/TO/SH/GW 25.
The Flp-In T-REx-293 cells (Invitrogen, NY, USA) containing genomic FRT sites and stably expressing the tet repressor were maintained in DMEM (4.5 g/L glucose, 10% FBS, 2mM L-glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin) containing 100 μg/ml zeocin and 15 μg/ml blasticidin. For generation of tet-inducible WRN expressing cell line, 1.5 × 106 Flp-In T-REx-293 cells were seeded in 6-cm Petri dish in culture medium without zeocin for 16-24 hours, co-transfected with 1 μg of pFTSH-WRN and 2 μg of the Flp-recombinase expression vector pOG44 (Invitrogen, NY, USA) using calcium phosphate transfection. Two days after transfection, cells were reseeded in a 10-cm Petri dish and selected in DMEM containing 100 μg/ml hygromycin and 15 μg/ml blasticidin for 2 weeks, with medium change every 3-4 days. Pooled hygromycin-resistant cells were treated with doxycycline, a tetracycline analog with a longer half-life, as inducing agent at 1 μg/ml for at least 24 hours and inducible expression of SH-tagged WRN was assessed by Western blotting and IHC using anti-HA antibodies (Covance, NJ, USA).
Stable isotope labeled peptides
Mass labeled peptides from TP53 sequence were purchased at AQUA QuantPro quality level were purchased from Thermo Fisher Scientific (Thermo Fisher Scientific Production team, Ulm, Germany): ALPNNTSSSPQP[Lys(13C6; 15N2)], 100.00% measured purity and ELNEALEL[Lys(13C6; 15N2)], 99.01% measured purity. Peptides were provided in solution of 5 pmol/ul, and serially diluted to generate the limit of quantitation and detection (LOQ/LOD) curves.
SRM methods development
SRM methods development was done using the freely available Skyline software package (http://proteome.gs.washington.edu/software/skyline) 26. In brief, protein sequences were trypsin-digested in silico, and all predicted y-ion fragments from y3 to yn-1 for peptides of 7-25 amino acids were used to experimentally validate ion ratios and predicted retention times27. Methods were exported directly from Skyline to a ThermoFisher TSQ Quantum Ultra triple quadrupole (QqQ) mass spectrometer to guide data acquisition. Step-by-step tutorials for method refinement can be found on Skyline website.
The reference sequence of human tumor protein 53 (TP53, p53; UniProt accession number P04637; Swiss Institute of Bioinformatics) was used to predict transitions as described above. Two experimentally verified peptides, ALPNNTSSSPQPK (residues 307-319) at m/z 670.8 and ELNEALELK (residues 342-351) at m/z 529.8, were monitored together with two internal control peptides derived from GAPDH (accession P04406): IISNASCTTNCLAPLAK (residues 146-162, 5 fragments) at m/z 917.5 and VPTANVSVVDLTCR (residues 235-248, 6 fragments) at m/z 765.9. Recombinant TP53 (BD Pharmingen, San Jose, CA) protein was used during methods development to select peptides, identify the most useful transitions and confirm predicted chromatographic retention times. The five most abundant transitions for each peptide were selected for final data acquisition, and were monitored using an unscheduled protocol in three analytical replicates. Additionally, stable isotope labeled form of each peptide was purchased for validation and quantitation, as noted previously.
The mature Drosophila accessory gland seminar fluid protein Acp70A protein product is a 36 residue lysine- and arginine-rich peptide from which only the tryptic peptide LNLGPAWGGR at m/z 520.8 could be readily detected 23. As a positive control we used the readily detected semi-tryptic peptide KEDMLLGVSNFK, at m/z 690.8, derived from accessory gland protein Acp62F 23. All y-ions, from y3 through yn-1 were monitored for selected peptides.
Two semi-tryptic peptides were used to monitor Acp53C14a in SRM analysis of strain #44798: AISSELDHYLR at m/z 652.3 and SSELDHYLR at m/z 560.3. Both peptides were consistently identified in previous experiments. Although the entire sequence of the m/z 560.3 peptide is contained within the m/z 652.3 peptide, both peptides provided good confirmation of depletions in which peptide KEDMLLGVSNFK, from control protein Acp62F, was used as an internal control.
The reference sequence of human Werner protein (WRN; RecQ3; RecQL2; UniProt accession number Q14191; Swiss Institute of Bioinformatics) was used to predict transitions as described above. Three fully tryptic, experimentally verified peptides, CTETWSLNSLVK (residues 170-181), m/z 719.3558, LLSAVDILGEK (residues 965-975), m/z 579.3424, and AAMLAPLLEVIK (residues 1208-1219), m/z 634.8860 were monitored concurrently with the GAPDH control peptides referenced above.
Assigned peptides were cross-correlated to MS spectral libraries from the National Institute of Standards and Technology (NIST) 28, downloaded from http://peptide.nist.gov.
MS Sample preparation
Whole cell extracts from RNAi-depleted or control GM639 cells containing 500 μg total protein were diluted to 1 ml with ammonium bicarbonate buffer (50 mM, pH 8). Samples were reduced by the addition of 10 μL of 500 mM dithiothreitol followed by incubation at 60°C for 30 min, then alkylated by the addition of 15 μL 500 mM iodoacetamide at room temperature for 30 min in the dark. The resulting protein samples were digested with trypsin (50 μg/sample; Promega, Madison, WI) at 37°C for 1 h. Trypsin digests were stopped by adding 10 μL of formic acid (90%, J.T.Baker, Phillipsburg NJ), cleared of insoluble debris by centrifugation at 14,000 rpm for 10 min at 4°C in a tabletop microcentrifuge (Eppendorf 5417R) and then run on Oasis MCX mixed phase columns (Waters Corporation, Milford, MA) according to manufacturer's instructions in order to remove detergent. Samples were resuspended in 0.1% formic acid in water and stored at −80°C until analysis.
Drosophila extracts were prepared by homogenizing 5 whole RNAi depleted or control male flies in 70 μL of 50 mM ammonium bicarbonate, then centrifuging for 5 min at 18,000 g to clear insoluble debris. The protein content of supernatants containing soluble accessory gland proteins 23 was quantified using a BCA assay (Pierce), and 40 μg aliquots were denatured, reduced, alkylated and digested with trypsin as previously described 29.
Liquid Chromatography
Fused silica capillary tubing (75 μm i.d., Polymicro Technologies) was pulled to a 5 μm tip, then packed with 25 cm of Jupiter Proteo reversed-phase chromatography material (Phenomonex, Torrance, CA). Nanoflow liquid chromatography was performed using an Eksigent nanoLC-1D system (Dublin, CA). Samples were injected into a 5 μL loop using an autosampler, and washed directly onto the column. Solvents A and B for gradient elutions were water/acetonitrile (95%/5% v/v), and water/acetonitrile (20%/80% v/v), respectively. Both contained 0.1% formic acid. In order to better separate GM639 lysates, solvent B was increased from 3% to 15% acetonitrile over 8 min, followed by an increase to 35% over the subsequent 52 min; a step to 90% for 10 min; then a return to Solvent B at 3% for 20 min. The total gradient time was 90 min at a flow rate of 350 nL/min. Drosophila peptides were eluted using a 60 min linear gradient of 5% to 25% solvent B.
Mass spectrometry
Eluting peptides were ionized via electrospray with the emitter held at 2.4 kV using a homebuilt ESI source, and directed into a Thermo Scientific TSQ Quantum Ultra triple quadrupole mass spectrometer. Transitions developed in Skyline were used to monitor precursor-fragment ion pairs defined previously. Resolution in Q1 and Q3 was 0.7 in all experiments,. Skyline was used to calculate peak areas by integrating total ion chromatograms. In order to normalize signal intensity, total peak areas for individual peptides from each target protein were divided by total peak areas for each internal control peptide. Differences in protein abundance were calculated by dividing the normalized peak areas of experimental samples by the analogous peak areas of control samples in the same experiment.
Standard curve generation
Heavy peptides were added to a buffer solution of 50 mM ammonium bicarbonate (pH 7.5) at the final concentration of 1 amol/μL, 10 amol/μL, 100 amol/μL, 1 fmol/μL, 10 fmol/μL, 100 fmol/μL, and 1 pmol/μL. 5 μL of each sample was injected onto column for the analysis, and three analytical replicates were taken.
Heavy peptides of these concentrations were then added to the digest of whole cell lysate from GM639-cc1 cells, and analyzed as described above. Peak area values were extracted using Skyline.
Western blot analyses
Whole cell extracts of GM639 and HEK-293T cells were prepared by resuspending cells in lysis buffer (15 mM Tris pH 8, 15 mM NaCl, 60 mM KCl, 1 mM EDTA, 0.5 mM EGTA, 0.5 mM spermidine, 0.5% NP-40 supplemented with Sigma protease inhibitor cocktail 1) followed by incubation on ice for 30 min. Nucleic acids were digested by adding Benzonase nuclease (EMD Biosciences, Darmstadt, Germany; 0.375 U/μL) and incubating for 15 min on ice, followed by the addition of DNase I (Sigma-Aldrich, St. Louis, MO; 180 U/mL) and incubation at 37°C for 15 min with periodic mixing. Protein concentrations of extracts were determined by Bradford assay (Bio-Rad, Hercules, CA), and 50 μg aliquots were resolved by SDS-PAGE electrophoresis (Invitrogen, Nu-PAGE) prior to transfer onto PVDF membrane (Bio-Rad, Richmond, CA, USA). TP53 protein was detected with using a mouse monoclonal anti-p53 primary antibody (AHO0112, Invitrogen). Endogenous WRN protein was detected using a mouse monoclonal anti WRN primary antibody (W0393, Sigma), inducible tagged WRN was detected using a mouse monoclonal anti-HA antibody (MMS-101P, Covance). GAPDH, an internal control protein, was detected using a mouse monoclonal anti-GAPDH primary antibody (MAB374, Millipore). Bound primary antibodies were detected using Alexa Fluor® 647 donkey anti-mouse polyclonal antibody (A-31571, Invitrogen), and quantified on an Alpha Innotech FluorChemQ Imager (Alpha Innotech Corporation, San Leandro, CA).
Western blot analyses of Drosophila samples were performed using extracts from RNAi-expressing and control males of line #52425. Triplicate samples from depleted and control flies containing 40 μg of reduced protein were separated on 15% polyacryl-amide gels. One set of replicates was stained with Coomassie blue to verify equal protein abundance in samples from RNAi and control flies (data not shown). The second and third sets of replicates were Western blotted to identify target proteins: one replicate was probed with a 1:2,000 dilution of an anti-Acp70A antibody 30, the other with a 1:4,000 dilution of an anti-Acp62F antibody. Both primary antibodies were generously provided by M. F. Wolfner (Cornell University). Bound primary antibodies were detected and quantified by incubating membranes with a 1:10,000 dilution of a goat anti-rabbit secondary antibody (Jackson ImmunoResearch) followed by enhanced chemiluminescence detection (GE Healthcare).
RESULTS
We developed a general SRM protocol (Figure 1) to detect and quantify RNAi-mediated depletion of four target proteins: the human TP53 tumor suppressor protein, the human Werner (WRN) protein, and the Drosophila accessory gland seminal fluid proteins Apc70A and Apc53C14a. To improve measurement precision we selected organism-specific internal controls to account for sample preparation and sample loading differences. We used the human glycolytic enzyme glucose-6-phosphate dehydrogenase (GAPDH), and the Drosophila accessory gland protein Apc62F.
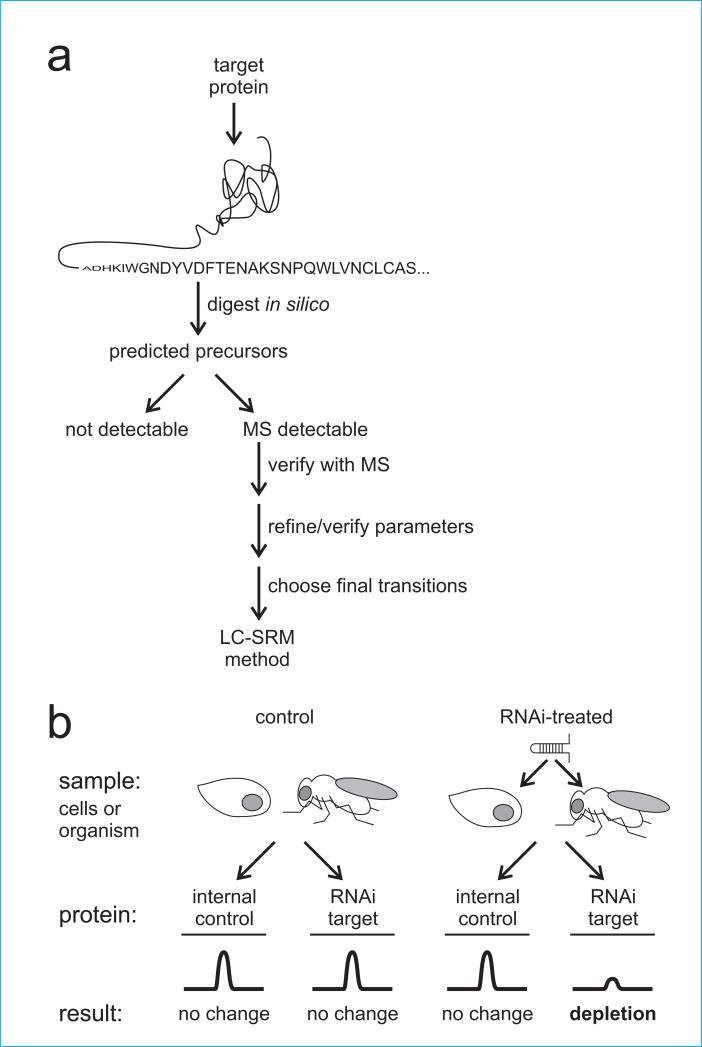
Figure 1.
Outline of selected reaction monitoring mass spectrometry (SRM) workflow. (a) SRM protocol used to detect and quantify a specific target protein versus a control. (b) Experiment design of an SRM experiment to detect and quantify depletion of a target protein in an RNAi experiment using cells or whole organisms.
SRM analysis of unfractionated cell extracts from a human fibroblast cell line GM639 depleted of TP53 by transfecting a TP53-specific siRNA revealed TP53 depletions ranging from 70-93% in independent experiments across several peptides. There was excellent agreement between the extent of depletion derived by measuring peak area ratios from individual peptides in SRM data (Figure 2), and the extent of depletion estimated by Western blot analysis (Figure 3).
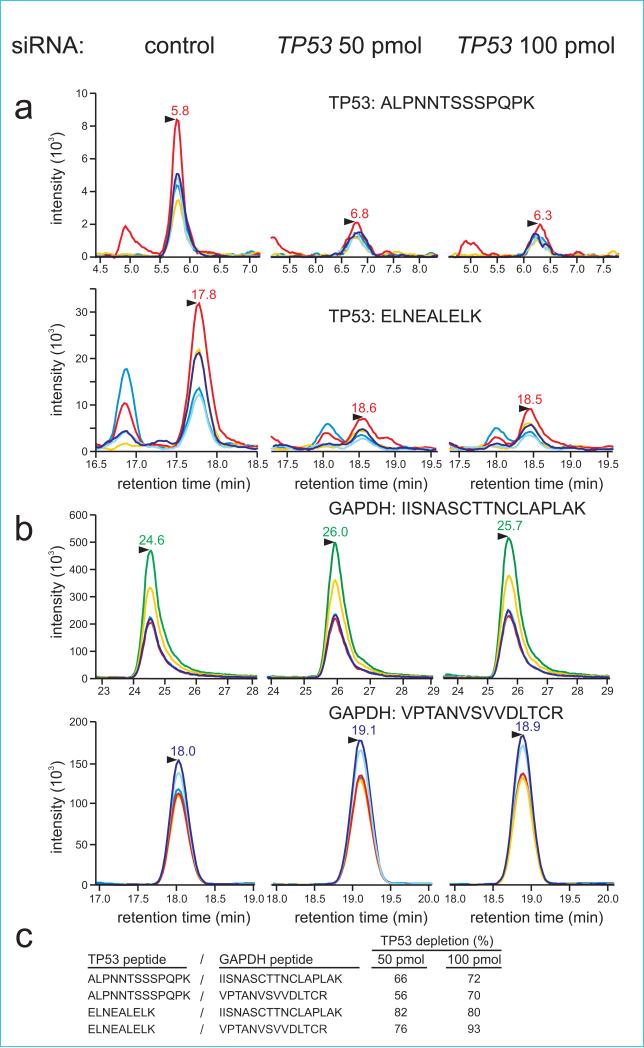
Figure 2.
RNAi-mediated TP53 depletion quantified by Selected Reaction Monitoring (SRM) mass spectrometry. The chromatograms show sets of transitions for specific peptides derived from target protein TP53 (a) or loading control GAPDH (b) in human cell RNAi depletion experiments. Samples were treated with a control RNAi or with 50 or 100 pmol of a TP53-specific siRNA prior to SRM analysis. Specific peptides detected and quantified from TP53 were ALPNNTSSSPQPK and ELNEALELK, and from GAPDH VPTANVSVVDLTCR and IISNASCTTNCLAPLAK. (c) TP53 depletion was quantified from individual peak area ratios determined for all measured peptides and normalized against an internal standard GAPDH as described in Methods. Normalized peptide intensities in depleted and control samples were used to estimate target protein depletion in the RNAi experiment depicted in panels (a) and (b).
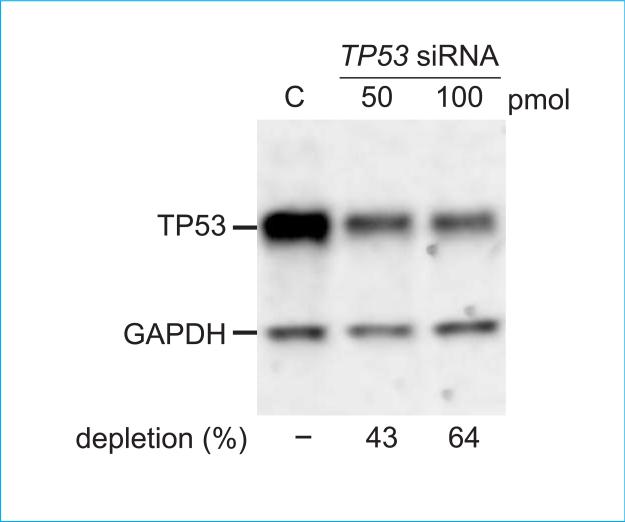
Figure 3.
Western blot verification of TP53 depletion from human cells. Cells were transfected with TP53-specific or control (C) siRNAs prior to preparing whole cell extracts. TP53 and GAPDH were detected by Western blot analysis. Band intensities for TP53 versus GAPDH were normalized against the blot background, then used to estimate percent TP53 depletion as a function of siRNA dose.
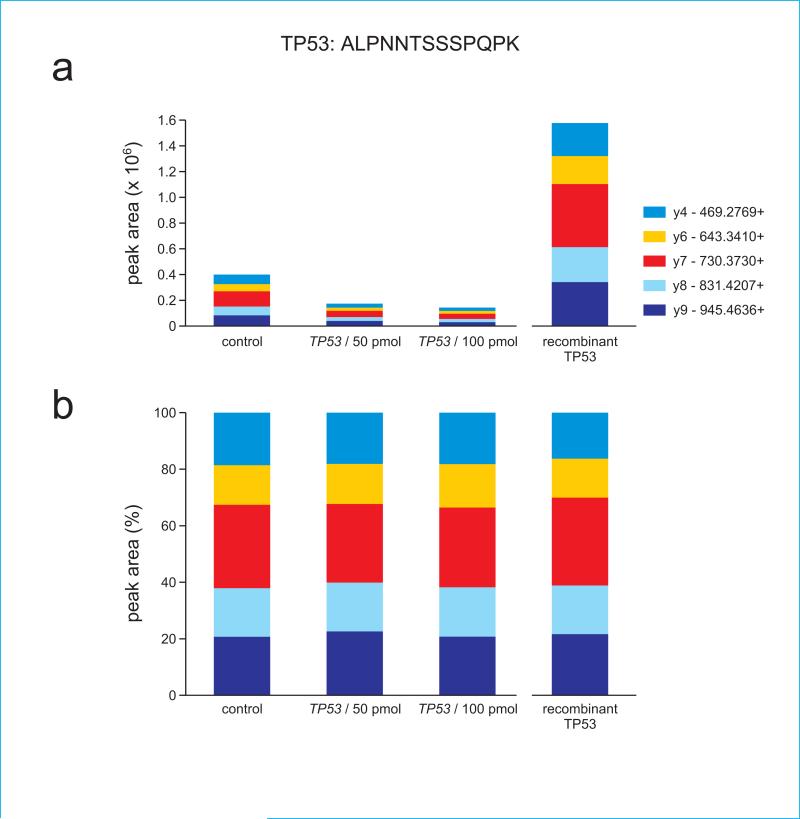
In addition to Western blot analysis, we purchased the recombinant protein and stable isotope labeled peptide standards to corroborate our findings in TP53. We also determined the limit of quantitation for each of the labeled peptides by isotope dilution curves, and we provide ion fragment ratio data for every measured peptide. These are presented in Figure 4, and show how individual ion fragments contribute to the total signal received from a given precursor. This confirms that we specifically identify the peptides of interest using our method by comparing the fragmentation patterns of our analyte in whole cell lysate to peptides from a recombinant protein. Recombinant human TP53 protein was used to verify peptide elution times and confirm the relative ratios of contributing fragments. The consistency in absolute and normalized ion ratios for the TP53 and GAPDH tryptic peptides in independent siRNA depletion experiments are illustrated in Figure 4. These results demonstrate the proportionally consistent contribution of different fragments to the total peak area regardless of absolute peptide abundance in the given sample, and confirm that all measured transitions were derived from TP53 target peptides as opposed to contaminants.
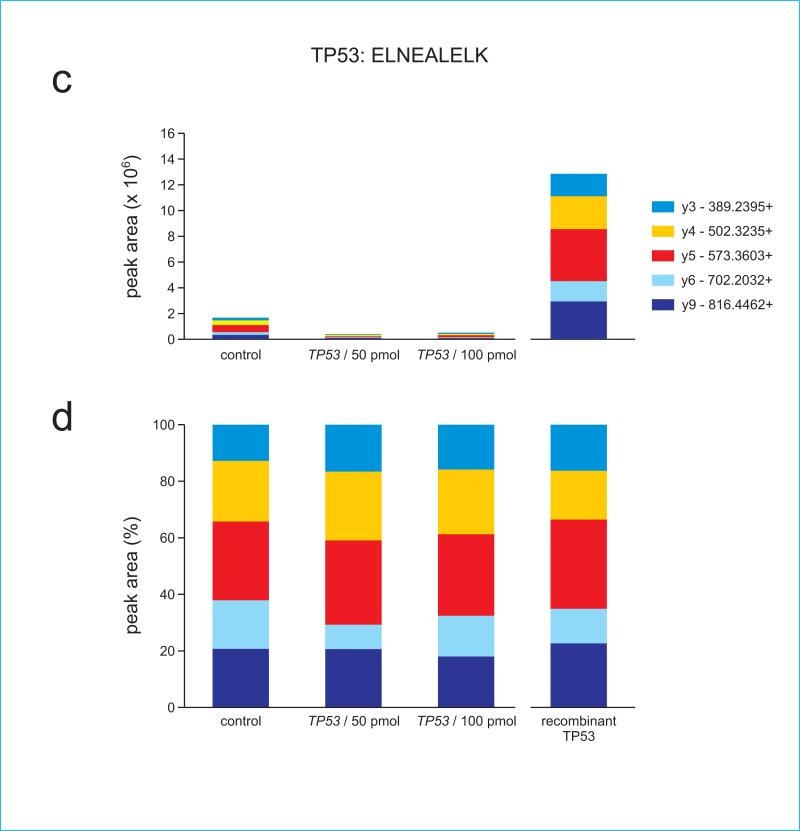
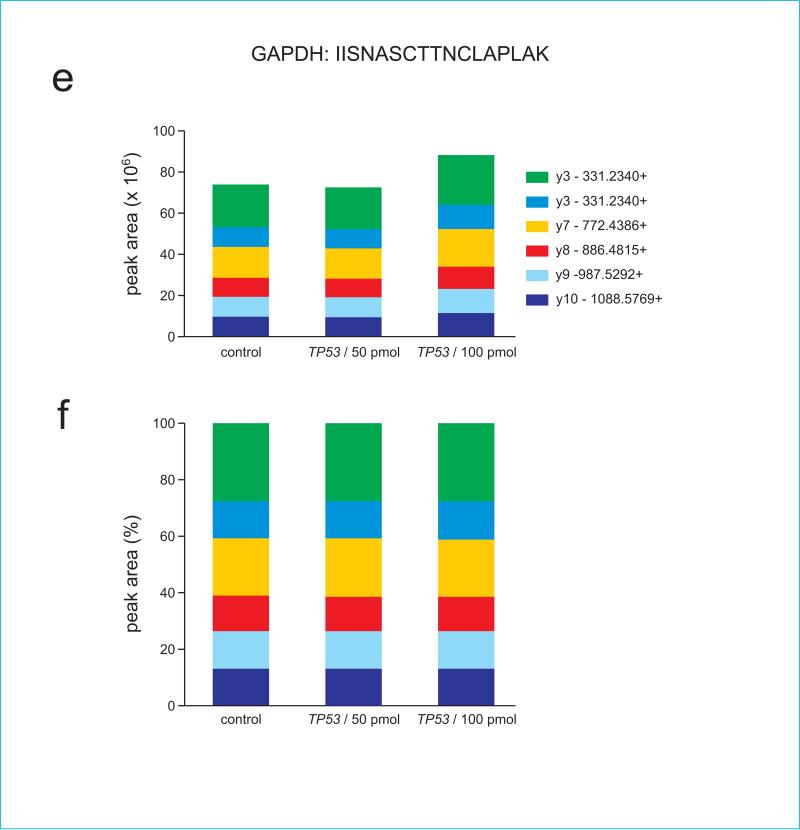
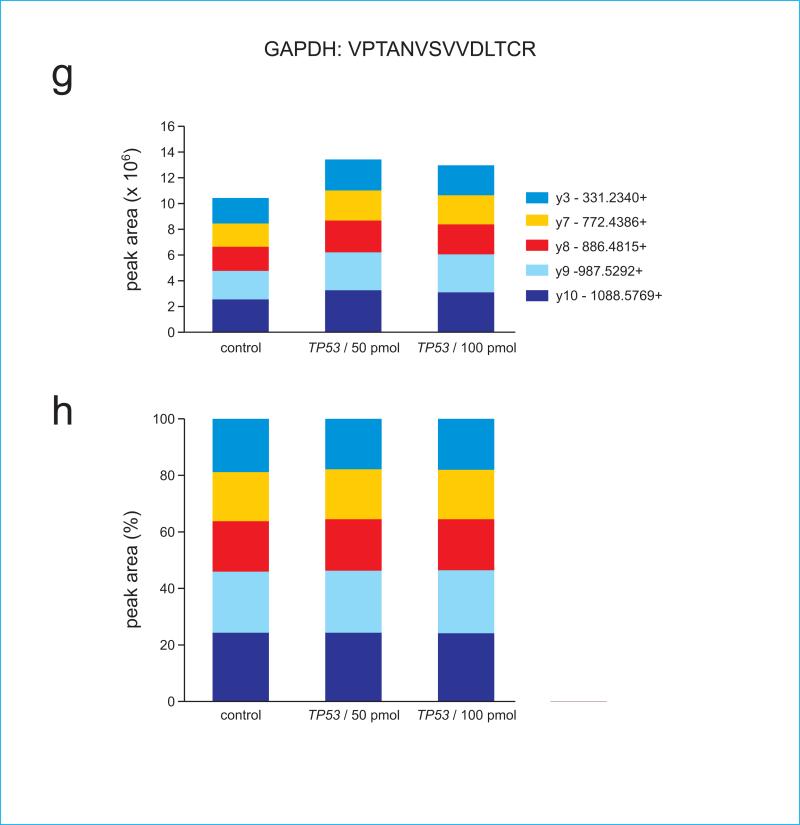
Figure 4.
Target protein-specific ion ratios are maintained in samples having different protein abundances. Absolute (a) and normalized (b) ion ratios from TP53 peptide ALPNNSSSPQPK are shown for samples having substantially different amounts of TP53 protein. The first three samples in each panel are unfractionated cell extracts, whereas the final sample (right) in each panel consisted of purified recombinant human TP53 protein. siRNA key indicates transfection of cell samples with a control siRNA or with two different concentrations of TP53-specific siRNA.
Panels (c) and (d) show ion ratios for TP53 peptide ELNEALELK, (e) and (f) show ion ratios for GAPDH peptide IISNASCTTNCLAPLAK, and (g) and (h) show ion ratios for GAPDH peptide VPTANVSVVDLTCR. Note that because GAPDH peptides are not present in TP53, their signal is absent in the recombinant TP53 digest.
We also demonstrate a linear quantitative range for the spiked-in peptides over six orders of magnitude (Supplementary Figure 1). Supplementary Figure 2 shows that when added to a complex background of whole cell extract (WCE), the peptides show a higher LOQ, but still maintain a similar pattern of ion fragments at lower spiked-in concentrations, confirming detection. Importantly, as demonstrated in panels (a) and (h), the endogenous level of the peptide is well within the quantitative range within two orders of magnitude. Therefore, signal reduction by 99% would still fall within the LOQ.
All five proteins were trypsin-digested in silico using Skyline, a software tool for SRM method development and data analysis 26. The resulting predicted tryptic peptides were sorted to identify peptides of 7-25 residues that had a high likelihood of being detected by MS. Detection of these peptides was then verified to identify the subset of peptides that gave strong MS signal intensities. Specific transitions (i.e., sets of specific precursors and fragmentation products) were then selected for each target peptide and verified to demonstrate that target and control proteins could be reliably measured by MS in unfractionated cell extracts.
The reproducibility of SRM and Western blot quantitation were compared by performing technical replicates for each sample and method. For SRM we used 6 technical replicates in which 4 different peptides were analyzed and the relative signal intensities were compared. Western blot analyses of TP53 protein using the same samples allowed us to assess the reproducibility of TP53 depletion relative to a within-lane loading control (GAPDH; Supplementary Figure 3a). Both methods detected comparable levels of protein depletion, with SRM-based measures having the lower coefficient of variation (CV)(Supplementary Figure 3b).
An independent set of TP53 depletions was performed to demonstrate that the depletion levels could be verified using synthetic SIL peptides. The depletion was achieved by the lentiviral shRNA plasmid transduction, and cells were grown under puromycin selection (1 μg/mL) prior to analysis. The results indicate that knockdown was much higher, and estimated to be approximately 90% by quantitation of fluorescent signal in Western blots (Supplementary Figure 4) and by using SRM (Supplementary Figure 5). Ion fragment analysis for all peptides is presented in Supplementary Figure 6.
In this set of experiments, in addition to normalization to internal standard GAPDH, SIL peptides were used as a verification of the level of depletion. Our results indicate that both normalization measurements show high level of agreement within 10% for all measured peptides.
Prior to addition of SIL peptides to the lysates, we determined the limit of detection (LOD) and limit of quantitation (LOQ) for each peptide both in loading buffer and in WCE. The data are presented in Supplementary Figures 1 and 2. Each measurement was recorded on absolute and logarithmic scale, and relative ion fragment contributions are also presented. The acquired SIL peptide data guided our decision to load protein amounts that fell well within our LOQ.
Human WRN protein has been previously shown to be a very low abundance protein in a number of cell lines 19,20, and we successfully used it in this study. We employed a series of cell preparations containing different relative amounts of WRN: a tetracycline-inducible Flp-In T-REx-293 cell line with and without induction, and a GM639 fibroblast cell line that stably expresses a non-targeting control shRNA and an anti-WRN shRNA (Supplementary Figure 7). We reliably show that we can identify peptides that are suitable for SRM, we can verify identity of these peptides by comparing ion fragmentation to publically available NIST peptide tandem mass spectral library, and we can quantify extent of knockdown even at low signal intensity. Supplementary Figure 8 shows chromatographic traces extracted directly from Skyline, demonstrating relative intensities of WRN peptides and control peptides from GAPDH. Supplementary Figure 9 shows that not only do our selected peptides match the library spectra within the dot product cut off of 0.9, but that with decreasing signal intensity the overall pattern of ion fragment contribution is largely maintained.
In vivo RNAi-mediated protein depletions were performed in Drosophila by crossing flies that contained target gene-specific UAS-IR RNAi cassettes under control of a tubulin-GAL4 driver. RNAi expression caused almost complete (>99%) knockdown of both targeted seminal fluid accessory proteins as demonstrated by SRM analyses and, in the case of Acp70A where an antibody was available, by Western blot analysis (Supplementary Figures10-12). There was again excellent concordance of depletion estimates of Acp70A by SRM and Western blot analysis. The potential residual Acp53c14 protein shown in Supplementary Figure 12 was shown to be an unrelated, co-eluting peak that could be distinguished from target fragments on the basis of ion fragment ratios. This illustrates the ability of SRM to discriminate between co-eluting target and contaminating proteins. For all analyses, relative ion fragment ratios were determined. Absolute, log-transformed, and normalized ion ratios are reported for all measurements, and demonstrate that even at lower signal intensities the ratios are preserved.
DISCUSSION
We have developed a general, targeted mass spectrometry approach based on selected reaction monitoring (SRM) to detect and quantify specific target proteins in unfractionated extracts from cells or whole organisms. The applicability of this method was demonstrated by using SRM analysis to detect and assess the abundance of target protein depletion relative to internal, organism-specific controls in cell-based and whole organism RNAi-mediated depletion experiments. Where good antibodies existed for both target and control proteins, we demonstrated excellent agreement between SRM and Western blot based quantitation of target protein depletion.
The SRM approach described here has several advantages when compared with antibody-based protein detection or quantitation methods. SRM is a transparent detection protocol that uses defined and experimentally validated precursor-fragment ion pairs for detection and quantitation. Immunologic approaches, in contrast, may depend on antibodies whose epitope(s) are often not known in any detail. Thus, an advantage of SRM for targeted peptide measurement is that it provides unambiguous structural specificity of a target peptide. Additionally, precursor-fragment ion pair transitions can be chosen to detect specific protein regions, isoforms or post-translational modifications. For example, we show that SRM can be used to detect and measure even very small proteins and peptides such as the 36-residue Apc70A protein that may be challenging targets for antibody development. Once a robust set of transitions are identified for a specific protein, they can be applied immediately in subsequent experiments and combined with additional target protein-specific transition sets to allow the simultaneous detection and quantitation of multiple proteins within single experiments and samples. Dozens of different proteins, represented by as many as a hundred or more specific transitions, can in principle be detected in single experiments using a common protocol and reagents.
In contrast to immunodetection-based methods such as Western blotting, information about full-length proteins is lost in SRM analyses. This occurs because proteolytic digestion is performed prior to MS analysis. However, as noted above, careful peptide selection and verification can be used to compensate for this potential disadvantage and may be used to distinguish between protein isoforms or post translational modifications that are difficult or impossible to detect by conventional immunologic methods. The protein quantitation performed here by SRM is analogous to quantitation in antibody-based blotting in that it is relative to a selected standard or “loading control”. This approach allows even small differences in protein abundance to be detected and precisely measured. We demonstrate that there is no appreciable difference when normalizing endogenous peptide signal intensities to other endogenous protein chosen as a control, rather than SIL forms of each peptide. Thus, our analysis agrees with Hoofnagle et al. 31, who had demonstrated that a single protein internal standard applied to all proteins performed as well as multiple protein-specific peptide internal standards.
Absolute quantitation can also be performed by SRM, as is the case with antibody-based detection, by using known controls with appropriate calibration 32.
The general workflow outlined in Figure 1 should be applicable to a wide range of proteins from diverse taxa. The instrumentation and software to enable large-scale SRM analyses are widely available and becoming increasingly commonplace in protein mass spectrometry laboratories. Thus SRM methods based on genomic data should facilitate quantitative proteomic analyses in well-characterized organisms where many good immunologic reagents are available, as well as other organisms of high biological or medical interest where few or no protein or immunologic resources exist. The validation of RNAi-mediated protein depletion will minimize false negatives and assist in the curation and use of increasingly large RNAi reagent libraries.
Supplementary Material
ACKNOWLEDGMENT
We thank Joe Gasper for help with fly Western blots, Gennifer Merrihew for help with MS methods development, Brendan MacLean for help with Skyline and helpful discussion, and Alden F.M. Hackmann for data assembly and graphics support.
Funding Sources
This work was supported by NIH awards R01 HD057974 (G.F. and M.M.), P41 RR011823 (M.M.), R01 DK069386 (M.M.), an award from NIH Genome Training grant T32 HG00035 (to V.G.) and P01 CA77852 to R.J.M., Jr.
ABBREVIATIONS
- CV
coefficient of variation
- LOD
limit of detection
- LOQ
limit of quantitation
- SIL
stable isotope labeled
- SRM
selected reaction monitoring
- QqQ
triple quadrupole mass spectrometer
- WCE
whole cell extract
Footnotes
Supporting Information. All skyline files incorporating RAW data files are provided. This material is available free of charge via the Internet at: http://proteome.gs.washington.edu/software/panorama/spionin_RNAi_SRM.html
Present Addresses Geoffrey D. Findlay 421 Biotechnology Building Cornell University Ithaca, NY 14853
Author Contributions
M.M. and D.T. proposed the use of targeted proteomics to assess protein depletion efficiency. V.G. and G.F. performed the knockdown and Western blot analysis. V.G., G.F., and D.T. performed the LC-SRM-MS experiments. M.M. and R.M. provided guidance for the project. All authors contributed to writing the manuscript and constructing Figures.
REFERENCES
- 1.Hamilton B, Dong Y, Shindo M, Liu W, Odell I, Ruvkun G, Lee SS. A systematic RNAi screen for longevity genes in C. elegans. Genes Dev. 2005;19(13):1544–1555. doi: 10.1101/gad.1308205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen . PLoS. Genet. 2005;1(1):119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lord CJ, Martin SA, Ashworth A. RNA interference screening demystified. J. Clin. Pathol. 2009;62(3):195–200. doi: 10.1136/jcp.2008.058735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohr S, Bakal C, Perrimon N. Genomic screening with RNAi: results and challenges. Annu. Rev. Biochem. 2010;79:37–64. doi: 10.1146/annurev-biochem-060408-092949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mullenders J, Bernards R. Loss-of-function genetic screens as a tool to improve the diagnosis and treatment of cancer. Oncogene. 2009;28(50):4409–4420. doi: 10.1038/onc.2009.295. [DOI] [PubMed] [Google Scholar]
- 6.Echeverri CJ, Beachy PA, Baum B, Boutros M, Buchholz F, Chanda SK, Downward J, Ellenberg J, Fraser AG, Hacohen N, Hahn WC, Jackson AL, Kiger A, Linsley PS, Lum L, Ma Y, Mathey-Prevot B, Root DE, Sabatini DM, Taipale J, Perrimon N, Bernards R. Minimizing the risk of reporting false positives in large scale RNAi screens. Nat. Methods. 2006;3(10):777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- 7.Williams DK, Muddiman DC. Absolute quantification of C-reactive protein in human plasma derived from patients with epithelial ovarian cancer utilizing protein cleavage isotope dilution mass spectrometry. J. Proteome. Res. 2009;8(2):1085–1090. doi: 10.1021/pr800922p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, Krek W, Aebersold R, Domon B. High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol. Cell Proteomics. 2007;6(10):1809–1817. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Picotti P, Aebersold R, Domon B. The implications of proteolytic background for shotgun proteomics. Mol. Cell Proteomics. 2007;6(9):1589–1598. doi: 10.1074/mcp.M700029-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell. 2009;138(4):795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell Proteomics. 2006;5(4):573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Domon B, Aebersold R. Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 2010;28(7):710–721. doi: 10.1038/nbt.1661. [DOI] [PubMed] [Google Scholar]
- 13.Hollstein M, Hainaut P. Massively regulated genes: the example of TP53. J. Pathol. 2010;220(2):164–173. doi: 10.1002/path.2637. [DOI] [PubMed] [Google Scholar]
- 14.Collavin L, Lunardi A, Del SG. p53-family proteins and their regulators: hubs and spokes in tumor suppression. Cell Death. Differ. 2010;17(6):901–911. doi: 10.1038/cdd.2010.35. [DOI] [PubMed] [Google Scholar]
- 15.Wagstaff BJ, Begun DJ. Comparative genomics of accessory gland protein genes in Drosophila melanogaster and D. pseudoobscura. Mol. Biol. Evol. 2005;22(4):818–832. doi: 10.1093/molbev/msi067. [DOI] [PubMed] [Google Scholar]
- 16.Chen PS, Stumm-Zollinger E, Aigaki T, Balmer J, Bienz M, Bohlen P. A male accessory gland peptide that regulates reproductive behavior of female D. melanogaster. Cell. 1988;54(3):291–298. doi: 10.1016/0092-8674(88)90192-4. [DOI] [PubMed] [Google Scholar]
- 17.Peng J, Chen S, Busser S, Liu H, Honegger T, Kubli E. Gradual release of sperm bound sex peptide controls female postmating behavior in Drosophila. Curr. Biol. 2005;15(3):207–213. doi: 10.1016/j.cub.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 18.Monnat RJ, Jr., Saintigny Y. Werner syndrome protein--unwinding function to explain disease. Sci. Aging Knowledge. Environ. 2004;2004(13):re3. doi: 10.1126/sageke.2004.13.re3. [DOI] [PubMed] [Google Scholar]
- 19.Moser MJ, Kamath-Loeb AS, Jacob JE, Bennett SE, Oshima J, Monnat RJ., Jr. WRN helicase expression in Werner syndrome cell lines. Nucleic Acids Res. 2000;28(2):648–654. doi: 10.1093/nar/28.2.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. The quantitative proteome of a human cell line. Mol. Syst. Biol. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448(7150):151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 22.Ram KR, Wolfner MF. Sustained post-mating response in Drosophila melanogaster requires multiple seminal fluid proteins. PLoS. Genet. 2007;3(12):e238. doi: 10.1371/journal.pgen.0030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Findlay GD, Yi X, MacCoss MJ, Swanson WJ. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS. Biol. 2008;6(7):e178. doi: 10.1371/journal.pbio.0060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao FJ, Sidorova JM, Lauper JM, Emond MJ, Monnat RJ. The human WRN and BLM RecQ helicases differentially regulate cell proliferation and survival after chemotherapeutic DNA damage. Cancer Res. 2010;70(16):6548–6555. doi: 10.1158/0008-5472.CAN-10-0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wepf A, Glatter T, Schmidt A, Aebersold R, Gstaiger M. Quantitative interaction proteomics using mass spectrometry. Nat. Methods. 2009;6(3):203–205. doi: 10.1038/nmeth.1302. [DOI] [PubMed] [Google Scholar]
- 26.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bereman MS, MacLean B, Tomazela DM, Liebler DC, MacCoss MJ. The development of selected reaction monitoring methods for targeted proteomics via empirical refinement. Proteomics. 2012;12(8):1134–1141. doi: 10.1002/pmic.201200042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein SE, Rudnick PA. NIST Peptide Tandem Mass Spectral Libraries. Human Peptide Mass Spectral Reference Data, H.sapiens, ion trap, Official build date: Feb. 4, 2009. National Institute of Standards and Technology; Gaithersburg, MD: p. 20899. [Google Scholar]
- 29.Aagaard JE, Yi X, MacCoss MJ, Swanson WJ. Rapidly evolving zona pellucida domain proteins are a major component of the vitelline envelope of abalone eggs. Proc. Natl. Acad. Sci. U. S. A. 2006;103(46):17302–17307. doi: 10.1073/pnas.0603125103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ram KR, Wolfner MF. A network of interactions among seminal proteins underlies the long-term postmating response in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2009;106(36):15384–15389. doi: 10.1073/pnas.0902923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoofnagle AN, Becker JO, Oda MN, Cavigiolio G, Mayer P, Vaisar T. Multiple-reaction monitoring-mass spectrometric assays can accurately measure the relative protein abundance in complex mixtures. Clin. Chem. 2012;58(4):777–781. doi: 10.1373/clinchem.2011.173856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bronstrup M. Absolute quantification strategies in proteomics based on mass spectrometry. Expert. Rev. Proteomics. 2004;1(4):503–512. doi: 10.1586/14789450.1.4.503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.