Abstract
The ten-eleven translocation family of proteins (Tet1/2/3, Tets) converts 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), which can be further oxidized and repaired by thymine DNA glycosylase (TDG), to influence gene transcription in embryonic and adult tissues. However the mechanisms of how Tets and TDG levels are regulated are unknown. We show that miR-29 can directly regulate Tet1-3 and TDG mRNA levels through binding to their 3’UTRs. miR-29 mimic decreases global 5hmC levels, a hallmark of Tet activity. Moreover, the mRNA levels for Tet3 and TDG are inversely correlated with the levels of miR-29 in aged mouse aorta implying that aging may affect methylation patterns via miRNA. In summary, our data show that Tets and TDG are direct targets of miR-29 and unravel a novel regulatory role for this miRNA in epigenetic DNA demethylation pathways.
Keywords: microRNA, epigenetics, Tet, TDG, 5hmC
Introduction
DNA methylation is a major epigenetic modification in the eukaryotic genome that regulates gene expression during many biological processes. Although the DNA methyltransferase (DNMT) family of enzymes establish and maintain DNA methylation, the Ten-eleven translocation (Tet) family of enzymes (Tet1, Tet2, and Tet3) are implicated in DNA demethylation and epigenetic control of gene expression (1–3). The Tet enzymes have dioxygenase activity and can convert 5-methyl cytosine (5mC) to 5hydroxymethylC (5hmC), 5-formyl cytosine (5fC), and 5-carboxylcytosine (5caC) (4, 5). These 5mC derivatives can be recognized and removed by the base excision repair machinery involving thymine DNA glycosylase (TDG) (4, 6) resulting in demethylation on once methylated cytosines. In addition to Tet/TDG role in active demethlyation, Tets also fine-tune epigenetic processes by binding to NANOG and synergistically enhance the efficiency of reprogramming (7) or binding to O-GlcNAc transferase to influence histone methyltransferase GlcNAcylation (8). TDG, on the other hand, has been shown to associate with DNMTs to maintain normal methylation patterns (9), or bind to transcription coactivators CBP/p300 for transcription modulation and base repair (10).
In the past decade, microRNAs (miRNAs) have emerged as important regulators of gene expression robustness. miRNAs predominantly target the 3’UTR of mRNAs, either destabilize the mRNA transcript or interfering with its translation into protein. miRNA can influence the epigenetic code by regulating histone acetylase and methytransferase (11–13) and by targeting transcription factors which may control Dnmt transcription (14). Recent evidence suggests that the miR-29 family of miRNAs can influence gene methylation patterns since miR-29 can bind to 3’UTR of the methyltransferases, DNMT3a and 3b, to maintain normal DNA methylation patterns (15). However, whether miRNAs regulate the pathway leading to DNA demethylation has not been studied. Here, we show that miR-29 can also control demethylation reactions by targeting the 3’UTRs of Tet-1,-2 and -3 and TDG and repressing their mRNA levels. miR-29 mimics decrease Tet and TDG mRNA levels and global 5hmC levels, while miR-29 inhibition increase their mRNA and 5hmC levels. These data suggest that miR-29 regulates two key emerging players in DNA demethylation and epigenetic control supporting a novel role of miRNAs in epigenetic regulation of gene expression.
Materials and Methods
Cell culture and miRNA treatment
Human dermal fibroblasts (HDF) and vascular smooth muscle cells (VSM) were obtained and cultured as described (16). 293 HEK cells were grown in DMEM with 10% fetal bovine serum. Cells were treated with miRIDIAN miRNA mimic, hairpin inhibitors or corresponding controls (Dharmacon, Chicago, IL) at 60 nM for 15 h unless specified otherwise, then were grown in complete media for 48h before harvest for RNA or protein studies. siRNA against human Tet3 was purchased from QIAGEN and transfected cells at 50 nM with RNAiMax (Invitrogen).
qPCR
Total RNA, including miRNA, was isolated by miRNAeasy isolation kit (Qiagen). After reverse transcription, qPCR was performed using Bio-Rad real-time thermal cycler and power SYBR green master mix (Applied Biosystem). mRNA levels were normalized by those of GAPDH and control levels.
Western blotting
Nuclear extracts were harvested using NE-PER nuclear extraction kit (Thermo Scientific) and equal lysates (10 ug) were loaded onto 7% SDS-PAGE. Western membranes were probed with a polyclonal antibody recognizing human Tet3 (Abicode). Hsp90 levels were used to normalize protein loading.
Luciferase 3’UTR luciferase reporter assay
3’UTRs of human Tet1-3 at indicated length were cloned behind Renilla luciferase gene in psiCHECK2TM dual luciferase reporter vector (Promega). Cells were transfected with 60 nM miRNA together with 0.1 ug luciferase vector for 24 h. Luciferase signals were normalized by signals from a constitutively expressed firefly luciferase gene and signals from control treated groups. Mutations in predicted miR-29 target sites were generated using quikchange lightning site-directed mutagenesis kit (Agilent Technologies).
5hmC dot-blot
1µg genomic DNA were denatured with 0.N NaOH and spotted on GE/Amesham Hybond-N+ membrane using 96-vacum apparatus. After UV cross-linking, membranes were probed with 5hmC antibody (Active Motif). To ensure equal spotting of total DNA on the membranes, the same blot was stained with 0.02% methylene blue in 0.3M sodium acetate (pH 5.5).
3’RACE
3’RACE was performed using FirstChoice® RLM-RACE kit (Invitrogen) according to manufacturer instructions. Briefly, total RNA were isolated and converted to cDNA using 3’RACE adaptor primer. PCR were performed using gene specific and adaptor primers, and products analyzed by agarose electrophoresis.
Microarray
Total RNA from HDF cells after 48 h treatment with 90 nM miR-29a inhibitor or control inhibitor were isolated and hybridized to GeneChip Human Gene 1.0 ST Array that covers 28,869 genes (Affymetrix) at Yale center for Genome analysis. Microarray analyses were performed as described (17).
Statistical Analysis
Comparisons between two groups were by unpaired student t test. Statistical analyses were performed using Prism 4 software (GraphPad). P values were two tailed and values < 0.05 were considered to be statistical significant.
RESULTS AND DISCUSSION
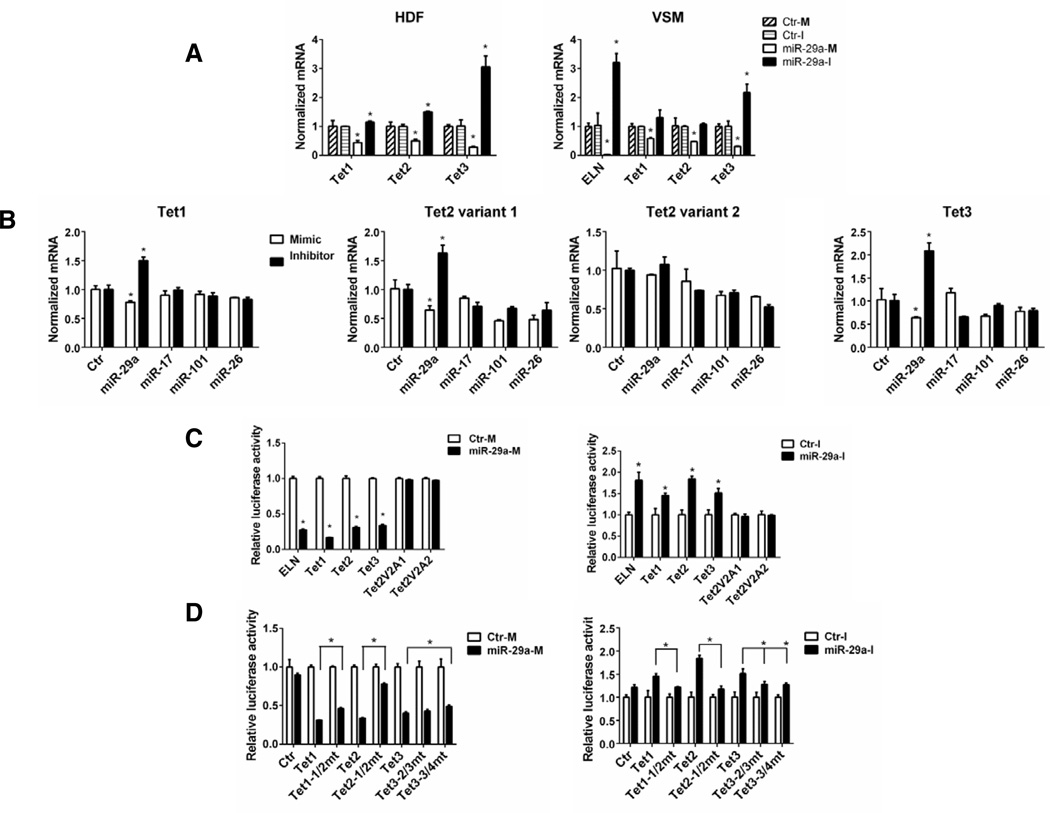
Our previous results have shown that anti-miR 29a treatment of human dermal fibroblast (HDF) and vascular smooth cells (VSM) markedly upregulates elastin mRNA levels and this anti-miR reduces levels of miR-29a, -b and –c (16) To examine the broad effects of miR-29 inhibition on global gene expression patterns, HDF were treated for 48hr with anti-miR 29a and the levels of gene expression analyzed by Affymetrix microarrays. Treatment with anti-miR 29a significantly increased the expression levels of only 33 transcripts implying that endogenous miR-29 in HDF represses a relatively small set of transcripts (supplementary Fig. 1 and GEO access), suggesting they are likely to be physiological targets of miR-29. Amongst the genes elevated with anti-miR 29 were Tet1 and Tet3, whose levels were increased by more than 50% after miR-29 inhibitor treatment. Indeed, in the 3’UTR there are two potential miR-29 target sequences in Tet-1 and Tet-2 and five potential sites in Tet-3 (Figure 1). To validate the microarray data, three different donors of HDF and VSM were analyzed by qPCR. Indeed, miR-29a mimics decreased the levels of three Tets and miR-29a inhibition increased all three Tets in HDF (Fig. 1A, left) and markedly upregulated Tet3 in VSM (Fig. 2A, right). qPCR assays using primers that do not distinguish between Tet2 variants showed that the miR-29 inhibitor had no effect on Tet2 mRNA levels in VSM (Fig. 1A, right).
Figure 1. Predicted binding sites for miR-29 in the 3'UTRs of Tet genes and TDG.
The length of the 3'UTR is listed with target sites for miR-29a in bold. Mutants generated are listed below the WT sequence and mutations indicated by nucleotides underlined.
Figure 2. Tet1-3 are direct targets of miR-29a.
A: HDF or VSM were treated with miR-29a mimic (M) or inhibitor (I) or controls (Ctr-M or Ctr-I) for 48 h and mRNA were analyzed for Tet expression. B: HDF were treated with indicated miRNA mimic/inhibitor for 48 h, and Tet mRNA levels were analyzed. C: HEK 293 cells (left panel) or HDF (right panel) were co-transfected with luciferase reporter constructs for the 3'UTRs of ELN, Tet1-3 and Tet2 variants and control mimics (Ctr-M), miR-29-M or miR-29a-I. The Tet2V2A1 and Tet2V2A2 constructs lack consensus sites for miR-29 as shown in Fig 1. D: HEK 293 cells (left panel) or HDF (right panel) were co-transfected with WT or mutant luciferase reporter constructs and Ctr-M, miR-29-M, Ctr-I or miR-29-I. Tet mutants (Tet1-1/2, Tet2-1/2, Tet3-2/3 and Tet3-3/4) are depicted in Fig 1. Data are representative of 3 independent experiments. In all cases, data are mean ± SEM, *p<0.05 vs. control or otherwise indicated.
Computational analysis predicts several miRNAs other than miR-29 may have binding sites in the 3’UTR of Tet transcripts, including miRNAs -17, -101 and -26. However, among the miRNAs tested, only the miR-29a mimic consistently decreased Tet1 and Tet3 transcripts while the miR-29a inhibitor increased Tet1 and Tet3 levels consistent with a pattern of direct targeting (Fig. 2B, left and right panels). The effects of miR-17, -101 and -26 were variable in multiple experiments. Deeper analysis of Tet2 revealed two forms of the Tet2 transcript, a Tet2 variant 1 (Tet2) with consensus miR-29a site or a shorter form Tet2 variant 2 (Tet2V2) with alternative polyadenylation sites (named Tet2V2A1 and Tet2V2A2) lacking putative binding sites for miR-29a. Indeed, qPCR experiments using primers to distinguish these two variants showed only Tet2, but not Tet2V2, has the potential to be directly targeted by miR-29a (Fig. 2B, middle panels). 3’RACE studies as well as careful examinations of 3’UTR sequences of Tets suggested that HDF cells tend to use alternative poly-adenylation signals and produce Tet transcripts with shorter 3’UTRs which still contain regions for miR-29 binding (Fig, 1 and Supplementary Fig. 1). Transcripts can be processed at one or multiple poly (A) signals to generate 3’UTR of varied length, depending on the cell type and circumstances (18, 19). The number of binding sites seems to influence the sensitivity of Tet transcripts by miR-29 as Tet3 has the most predicted sites and is consistently to be the most significantly regulated Tet by miR-29 in HDF and VSM (Fig. 2A)
In order to examine whether Tets are direct targets of miR-29, the 3’UTRs of selected length for Tet1, 2 and 3 (Fig. 1) were cloned into dual luciferase reporter constructs. Transfection with the control luciferase reporter without a cloned in 3’UTR (empty vector) did not affect luciferase activity (Fig. 2D). miR-29a mimic treatment decreased luciferase activities by more than 50% using the 3’UTR containing a well described miR-29 target, ELN (16, 20) and also reduced Tet1, Tet2 as well as Tet3, but not in the two Tet2V2 constructs (Fig. 2C, left). Likewise, miR-29a inhibitor increased luciferase activities in constructs containing ELN, Tet1, Tet2 and Tet3, but not in Tet2V2 (Fig. 1C, right). When two nucleotides in miR-29 binding sites in Tet1 and Tet2 3’UTR were mutated from “GU” to “CA” (see Fig 1 for construct), the effects of miR-29a mimic or inhibitor on these 3’UTRs were significantly attenuated (Fig. 2D). However, there are five putative miR-29 binding sites in the first 1963bp of Tet3 3’UTR and mutating sites 2 and 3 (Tet3-2/3mt) or sites 3 and 4 (Tet3-3/4mt) can only marginally rescue the effects of miR-29a mimic or inhibitor, suggesting the involvement of other sites or cooperativity between the sites (Fig. 2D).
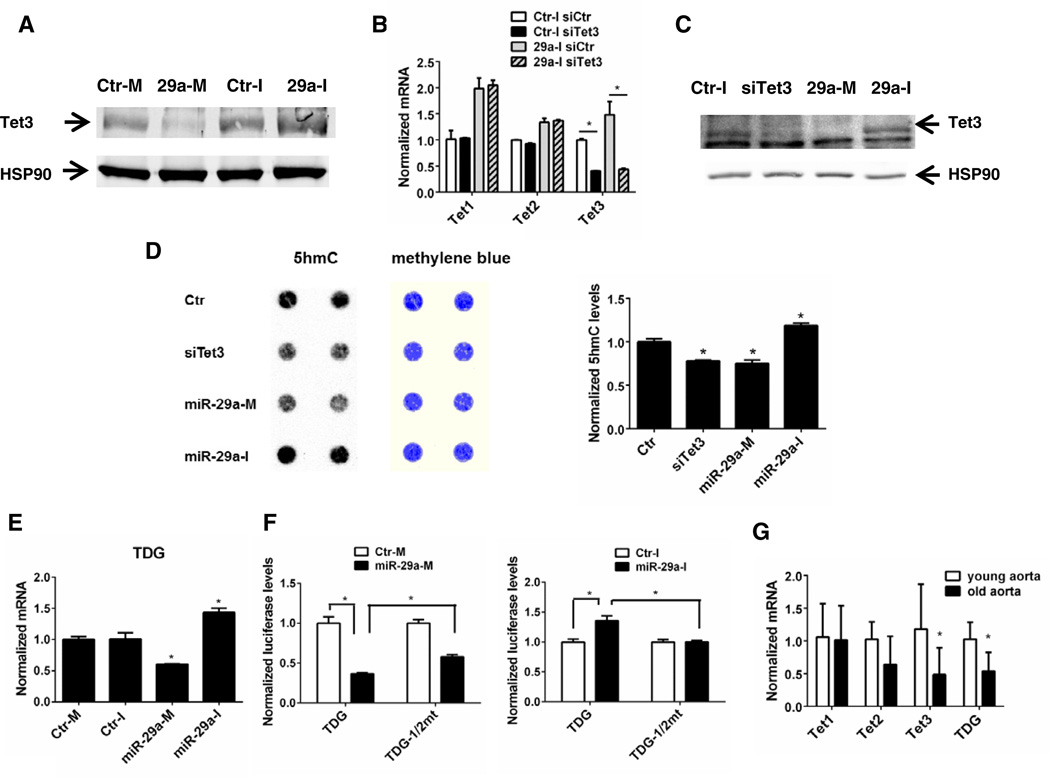
In order to examine Tet protein levels and potential functions of miR-29, nuclei were isolated from HDF treated with miR-29 mimic or inhibitor. We were unsuccessful in reproducibly detecting Tet1 or Tet2 protein levels with commercially available antibodies in isolated nuclear extracts. However, miR-29a mimic decreased while miR-29a inhibitor partially increased Tet3 protein levels (Fig. 3A). Treatment with a siRNA against Tet3 decreased Tet3 mRNA levels in both control and miR-29a inhibitor treated groups, but had no effect on Tet1 or Tet2 mRNAs (Fig. 3B). This Tet3 siRNA also decreased Tet3 protein levels, similar to that observed with miR-29a mimic (Fig. 3C). To examine if miR-29 can influence Tet activity, we assayed the Tet-dependent conversion of 5mC to 5hmC. To test this, genomic DNA was isolated after miR-29 mimic/inhibitor treatment for 48h and the levels of 5hmC determined by dot blotting. As seen in Fig 3D, knockdown of Tet3 as well as treatment with the miR-29a mimic modestly decreased global 5hmC levels, while the miR-29a inhibitor modestly increased global 5hmC levels (as quantified in the right panel). The inability to observe a complete loss or gain of 5hmc may be due to other Tet isoforms that lack target sites for miR-29, multiple Tet isoforms, and competing methylation reactions. In addition, there may be kinetic lag between the effects of miRNA mimics and inhibitors on mRNA, protein and integrated function. Tets are the only identified enzymes having abilities to oxidize 5mC to 5hmC (21), so the inverse correlation between miR-29 and 5hmC levels strongly support our hypothesis that miR-29 regulates Tet directly. Tet3 may be the major contributing Tet in HDF since knocking down Tet3 alone achieved a similar effect in reducing 5hmC as miR-29 mimic treatment. 5hmC can be further oxidized by Tets to 5fC and 5caC, which are excised by TDG and eventually repaired (4, 22). Loss of TDG is associated with aberrant methylation pattern and accumulation of 5fC and 5caC at gene regulatory elements (23, 24). Interestingly, TDG 3’UTR also contains two miR-29 binding sites and we identified that miR-29 mimic decreased TDG mRNA by 40%, while miR-29 inhibitor increased TDG mRNA by 43.7% in VSM (Fig 2E, similar data in HDF not shown). These effects are mediated by miR-29 binding to TDG 3’UTR because point-mutations of both predicted binding sites significantly reversed the repressive effect of miR-29 on luciferase activity (Fig. 3F). Therefore, our data demonstrated a novel regulatory role of miR-29 in active DNA demethylation, targeting both Tets and TDG.
Figure 3. miR-29 regulates Tet3 protein, global 5hmC and TDG levels.
A-D: HDF were treated with indicated miRNA, siRNA against Tet3 (siTet3) or in certain cases both for 48 h. Nuclear extracts were isolated and western-blotted for Tet3 protein levels (A&C); mRNA were examined by qPCR for Tet3 knock-down efficiency and specificity (B); total genomic DNA were isolated and dot-blotted with antibodies for 5hmC, which are normalized by methylene blue staining for total DNA (D). E: total RNA from miRNA-treated VSM were isolated and analyzed for TDG mRNA. F: left, HEK 293 cells were transfected with TDG luciferase reporter constructs and Ctr-M or miR-29-M; right, HDF cells were transfected with TDG luciferase reporter constructs and Ctr-I or miR-29-I. G: RNA from old (22 months) and young (2 months) mouse (C57BL/6) aorta (n=6) were analyzed for Tet and TDG expression. Data are representative of 3 independent experiments. In all cases, data are mean ± SEM, *p<0.05 vs. control or otherwise indicated.
TDG can also bind to DNA methylation enzymes to maintain normal methylation patterns (9). miR-29 members were shown to target to Dnmt3a and 3b through complementary binding to their 3'-UTRs and enforced expression of miR-29 can restore normal activities of Dnmts and normal DNA methylation pattern in certain cancer cell lines (15). We have verified that miR-29 can regulate Dnmt3a and 3b in both HDF and VSM (data not shown), but the functional consequences in vascular cells remain to be determined. DNA methylation and demethylation are very dynamic and fine-tuned events and can coexist in the same promoter; i.e. - 5hmC and 5mC can be both enriched in certain promoters in embryonic stem cells (3, 25); Dmnts and Tets can both increase at intermediate reprogramming stage of induced pluripotent stem (iPS) cells generation, leading to increased production of both base modifications (26). However, the overall effects of miR-29 on DNA methylation/demethylation patterns remain largely unknown and may be tightly controlled by effectors regulating miR-29 expression. Previous work has shown that miR-29 levels are low during embryogenesis, but increase into adulthood and during aging (27–29). As a corollary to this both Tet3 and TDG levels were significantly lower in aged mouse aortae (22 months) when compared to young mouse aortae (2 months; Fig. 3G), suggesting that Tet and TDG may be reciprocally regulated by miR-29 in vivo.
In the past several years, intense research delineating the exciting range of functions of Tet/TDG in epigenetic demethylation reactions has shown their roles in crucial cellular events such as stemness, lineage determination, cell type specific functions and aging. Our data has defined an important regulatory role of miR-29 in modulating these key emerging players may yield insights into processes and diseases associated with alterations DNA methylation/demethylation.
Supplementary Material
Highlights.
Ten-eleven translocation (TET) enzymes convert 5-mC to 5-hmC.
5hmC can be repaired by thymine DNA glycosylase (TDG).
miR-29a targets to Tet1-3 and TDG mRNA and influences global 5hmC.
miR-29 can modulate epigenetics by targeting Tets and TDG.
Acknowledgements
We thank Drs. Jun Lu and JiJun Cheng (Department of Genetics, Yale University), for helpful discussions and help in trouble-shooting dot-blot experiments. This work was supported in part by the National Institutes of Health (HL64793, HL61371, HL081190, HL096670, PO1 1070205) and the Kiev Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
MicroarrayGEOaccess:
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=dvyjxyeyaygyctk&acc=GSE45564
References
- 1.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG, Deng Z, Szabo PE, Pfeifer GP, Li J, Xu GL. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 2.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS, Rambow F, Bassi MR, Bruno T, Fanciulli M, Renner C, Klein-Szanto AJ, Matsumoto Y, Kobi D, Davidson I, Alberti C, Larue L, Bellacosa A. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, Das S, Levasseur DN, Li Z, Xu M, Reik W, Silva JC, Wang J. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495:370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, Shih AH, Levine RL, Bernard O, Mercher T, Solary E, Urh M, Daniels DL, Fuks F. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. The EMBO journal. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li YQ, Zhou PZ, Zheng XD, Walsh CP, Xu GL. Association of Dnmt3a and thymine DNA glycosylase links DNA methylation with base-excision repair. Nucleic acids research. 2007;35:390–400. doi: 10.1093/nar/gkl1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Molecular cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 11.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature genetics. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca EA, Lawler S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer research. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- 13.Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. The Journal of biological chemistry. 2008;283:9836–9843. doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- 14.Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, Li E, Serrano M, Millar S, Hannon G, Blasco MA. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nature structural & molecular biology. 2008;15:268–279. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang P, Huang A, Ferruzzi J, Mecham RP, Starcher BC, Tellides G, Humphrey JD, Giordano FJ, Niklason LE, Sessa WC. Inhibition of microRNA-29 enhances elastin levels in cells haploinsufficient for elastin and in bioengineered vessels--brief report. Arteriosclerosis, thrombosis, and vascular biology. 2012;32:756–759. doi: 10.1161/ATVBAHA.111.238113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang P, Huang A, Morales-Ruiz M, Starcher BC, Huang Y, Sessa WC, Niklason LE, Giordano FJ. Engineered zinc-finger proteins can compensate genetic haploinsufficiency by transcriptional activation of the wild-type allele: application to Willams-Beuren syndrome and supravalvular aortic stenosis. Human gene therapy. 2012;23:1186–1199. doi: 10.1089/hum.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3' end processing regulation. Nucleic acids research. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen L, Zhang Y. 5-Hydroxymethylcytosine: generation, fate, and genomic distribution. Current opinion in cell biology. 2013 doi: 10.1016/j.ceb.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. The Journal of biological chemistry. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen L, Wu H, Diep D, Yamaguchi S, D'Alessio AC, Fung HL, Zhang K, Zhang Y. Genome-wide Analysis Reveals TET- and TDG-Dependent 5-Methylcytosine Oxidation Dynamics. Cell. 2013 doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, Steinacher R, Jiricny J, Bird A, Schar P. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 25.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 26.Gao Y, Chen J, Li K, Wu T, Huang B, Liu W, Kou X, Zhang Y, Huang H, Jiang Y, Yao C, Liu X, Lu Z, Xu Z, Kang L, Wang H, Cai T, Gao S. Replacement of Oct4 by Tet1 during iPSC Induction Reveals an Important Role of DNA Methylation and Hydroxymethylation in Reprogramming. Cell stem cell. 2013;12:453–469. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Boon RA, Seeger T, Heydt S, Fischer A, Hergenreider E, Horrevoets AJ, Vinciguerra M, Rosenthal N, Sciacca S, Pilato M, van Heijningen P, Essers J, Brandes RP, Zeiher AM, Dimmeler S. MicroRNA-29 in aortic dilation: implications for aneurysm formation. Circulation research. 2011;109:1115–1119. doi: 10.1161/CIRCRESAHA.111.255737. [DOI] [PubMed] [Google Scholar]
- 28.Carraro G, El-Hashash A, Guidolin D, Tiozzo C, Turcatel G, Young BM, De Langhe SP, Bellusci S, Shi W, Parnigotto PP, Warburton D. miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-Cadherin distribution. Developmental biology. 2009;333:238–250. doi: 10.1016/j.ydbio.2009.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez I, Cazalla D, Almstead LL, Steitz JA, DiMaio D. miR-29 and miR-30 regulate B-Myb expression during cellular senescence. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:522–527. doi: 10.1073/pnas.1017346108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.