Abstract
The challenges posed by the analysis of mono-nucleotide mixtures by direct infusion electrospray ionization (ESI) were examined in the context of recent advances of MS technologies. In particular, we evaluated the merits of high-resolution mass analysis, multistep gas-phase dissociation, and ion mobility determinations for the characterization of species with very similar or identical elemental composition. The high resolving power afforded by a linear trap quadrupole (LTQ)-orbitrap allowed the complete differentiation of overlapping isotopic distributions produced by nucleotides that differed by a single mass unit. Resolving 12C signals from nearly overlapped 13C contributions provided the exact masses necessary to calculate matching elemental compositions for unambiguous formulae assignment. However, it was the ability to perform sequential steps of gas-phase dissociation (i.e., MSn-type analysis) that proved more valuable for discriminating between truly isobaric nucleotides, such as the AMP/dGMP and UMP/ψMP couples, which were differentiated in the mixture from their unique fragmentation patterns. The identification of diagnostic fragments enabled the deconvolution of dissociation spectra containing the products of coexisting isobars that could not be individually isolated in the mass-selection step. Approaches based on ion mobility spectrometry (IMS)-MS provided another dimension upon which isobaric nucleotides could be differentiated according to their distinctive mobility behaviors. Subtle structural variations, such as the different positions of an oxygen atom in AMP/dGMP or the glycosidic bond in UMP/ψMP, produced detectable differences in the respective ion mobility profiles, which enabled the differentiation of the isobaric couples in the mixture. Parallel activation of all ions emerging from the ion mobility element provided an additional dimension for differentiating these analytes on the basis of both mobility and fragmentation properties.
Nucleotides are the fundamental building blocks of nucleic acids and, as such, constitute the alphabet of the genetic code [1-3]. In addition, nucleotides function also as co-factors, energy carriers, metabolic regulators, second messengers, and vitamin components [1-3]. In the case of deoxyribonucleotides, numerous covalent modifications of the four fundamental units have been discovered, which are typically produced by damage processes [4-6], exposure to mutagens [7-9], and epigenetic modulation of gene expression/replication [10-12]. For this reason, modified DNA constituents represent valuable biomarkers in the elucidation of genotoxic mechanisms, the evaluation of risk factors, and the development of diagnostic tools [13-15]. In the case of ribonucleotides, up to 110 natural variants have been thus far documented in the RNA Modifications Database [16,17]. The majority of these post-transcriptional modifications are thought to determine function by modulating the stability of RNA structure and its ability to interact with cognate factors. For the most part, however, their biological significance and mechanism of action are still far from being understood. These considerations accentuate the critical need for proper analytical methods to support the characterization of natural/modified components and the elucidation of their structure-function relationships.
Since mass spectrometry (MS) was first employed for the characterization of nucleosides 50 years ago [18], its application to the analysis of nucleic acid components has followed in lockstep the evolution of this analytical platform over the years [19-22]. McCloskey and Crain pioneered many of the approaches employed in the field and established high standards for their application. Regardless the type of available instrumentation, a constant feature has been the utilization of enzymatic hydrolysis to reduce initial nucleic acid samples into complex mixtures of mono-nucleotides (e.g., by treatment with specific nucleases to cleave the phosphodiester backbone) or nucleosides (e.g., by further phosphatase reaction to remove the phosphate group) [23]. Liquid chromatography (LC) and capillary electrophoresis (CE) have been typically employed on- and off-line to achieve product separation and to minimize the adverse effects of salts on ionization (reviewed in [24,25]). Structural elucidation has been completed by utilizing different forms of tandem mass spectrometry (MS/MS) to obtain readily recognizable fragmentation patterns (reviewed in [21,22]). Together with 32P-postlabeling, immunochemical and fluorescence-based methods, these MS techniques constitute the foundations for the detection and quantification of nucleic acid adducts [26-28].
In this report, we examined the challenges of tackling nucleotide mixtures by direct infusion electrospray ionization (ESI) without the assistance of front-end separation techniques. With an eye on direct biomarker analysis and possible diagnostic applications, we evaluated the benefits of employing Fourier transform mass spectrometry [29-31] and ion mobility spectrometry-mass spectrometry (IMS-MS) [32-34] to unambiguously discriminate between analytes with very similar or identical elemental compositions. The high resolving power and sub-ppm accuracy afforded by linear trap quadrupole (LTQ)-orbitrap instruments [31,35] can typically provide the means to resolve complex analyte mixtures and to determine the elemental composition of small molecules and metabolites [36,37]. Further, this platform is capable of sequential stages of tandem mass spectrometry (MSn) to support detailed structural characterization of target analytes [38]. In contrast, IMS-MS experiments probe ions’ three-dimensional structure by determining their drift time as they travel across a low-pressure region of the instrument, which may be subjected to a continuous [39,40] or oscillating [41,42] electric field. Owing to the relationship between drift time and probability to interact with background gas, which is a function of structure and conformation, these determinations can lead to the correct differentiation of isomeric and chiral species [43-45]. A multi-pronged approach combining high resolving power, sequential fragmentation capabilities, and possible conformation discrimination affords an enormous potential in the analysis of complex analyte mixtures, which we have explored here for the identification of isobaric/isomeric nucleotides.
Experimental
Individual nucleosides 5′-monophosphate (nucleotides) employed in the study were purchased from Sigma-Aldrich (St Louis, MO) and used with no further purification. For this reason, stock solutions contained characteristic unidentified contaminants detected in all ESI spectra recorded in the study. Pseudouridine monophosphate (ψMP) was instead produced as a custom synthesis by Dharmacon Research Inc. (Lafayette, CO). Appropriate mixtures of these analytes can aptly mimic typical samples obtained by enzymatic digestion of DNA and RNA by the conventional nucleases employed in nucleic acid analysis [22,46]. Sample concentrations were determined by UV absorbance at 260 nm by using extinction coefficients reported previously [47]. Immediately before analysis, stock solutions were typically diluted to a final concentration of 9.0 μM of total NMPs in 100 mM ammonium acetate and 10% isopropanol, unless otherwise stated.
Samples were analyzed by direct infusion ESI on either a Thermo Fisher Scientific (West Palm Beach, CA) LTQ-Orbitrap Velos mass spectrometer, or a Waters (Milford, MA) Synapt G2 HDMS IMS mass spectrometer. All analyses were performed in nanoflow ESI mode by using quartz emitters produced in house by a Sutter Instruments Co. (Novato, CA) P2000 laser pipette puller. Up to 5μL samples were typically loaded onto each emitter by using a gel-loader pipette tip. A stainless steel wire was inserted in the back-end of the emitter to supply an ionizing voltage that ranged between 0.6 and 1.4 kV. Source temperature and desolvation conditions were adjusted by closely monitoring the incidence of ammonium adducts and water clusters [48].
For high-resolution determinations, the LTQ-Orbitrap instrument was calibrated by using an anion mixture that contained sodium dodecyl-sulfate, sodium taurocholate, and Ultramark. These standards enabled the calibration of the instrument over a range of m/z 50-2000 with a ~200 ppb mass accuracy. Tandem mass spectrometry (MS/MS) was accomplished by isolating the precursor ion of interest in the linear trap quadrupole (LTQ) element, which was then collided with N2 to activate fragmentation. The ensuing products were mass analyzed either in the LTQ or the orbitrap region of the instrument. Data were processed by using Xcalibur 2.1. Mass calculations and predictions of elemental composition were performed by using the Molecular Weight Calculator software [49] made available by the Pacific Northwest National Laboratory (PNNL).
Apparent drift time (tD) was determined by allowing ions to move through the travelling wave (Tri-WAVE) element of the IMS mass spectrometer [41], which were then transferred for mass analysis into the time-of-flight (TOF) stage operated in single reflectron mode. The instrument was calibrated by using a 2 mg/mL solution of cesium iodide in 50:50 water/methanol, which afforded a ~9 ppm mass accuracy. For comprehensive mixture analysis, the Tri-WAVE region was held at a pressure of approximately 4.40 mbar (uncalibrated gauge reading) by a 90 mL/min flow of N2 and 180 mL/min of He. It was operated with an approximately 650 m/s IMS wave velocity, a 40 V wave height, a 109 m/s transfer wave velocity, and a 2.0 V transfer wave height. Analysis of mass-selected isobars was performed by raising the cell pressure to ~4.60 mbar (uncalibrated gauge reading) with a flow of 140 mL/min N2 and 180 mL/min He. At the same time, IMS wave velocity was raised to ~700 m/s, transfer wave velocity to −600 m/s, and transfer wave height to 4.0 V. The observed mobility profiles were compared with theoretical Gaussian distributions obtained from the curve-fitting algorithm present in the PeakFit 4.1 package. In our hands, PeakFit deconvolution allowed for the discrimination of contiguous profiles with a resolving power of ~40, which exceeded the value of ~25 quoted for these types of experiments by the instrument’s manufacturer.
Time aligned parallel (TAP) dissociation of all the ions that had traveled with different tD through the Tri-WAVE element was performed in the transfer region present between the Tri-WAVE and the TOF analyzer [50,51]. Collision voltages ranging from 10 to 25 V were explored. Data were collected in IMS mode with the TOF analyzer operated in single reflectron mode. Heat-map representations of data obtained from full-range MS and TAP experiments were produced in the form of mass-over-charge ratio (m/z) vs. tD vs. relative intensity plots by the DriftScope v2.1 software provided with the Waters’ MassLynx 4.1 package. Characteristic TAP fragments were identified by examining longitudinal sections of heat-maps obtained from an entire sample, which corresponded to the tD of nucleotides of interest in the mixture.
Results and Discussion
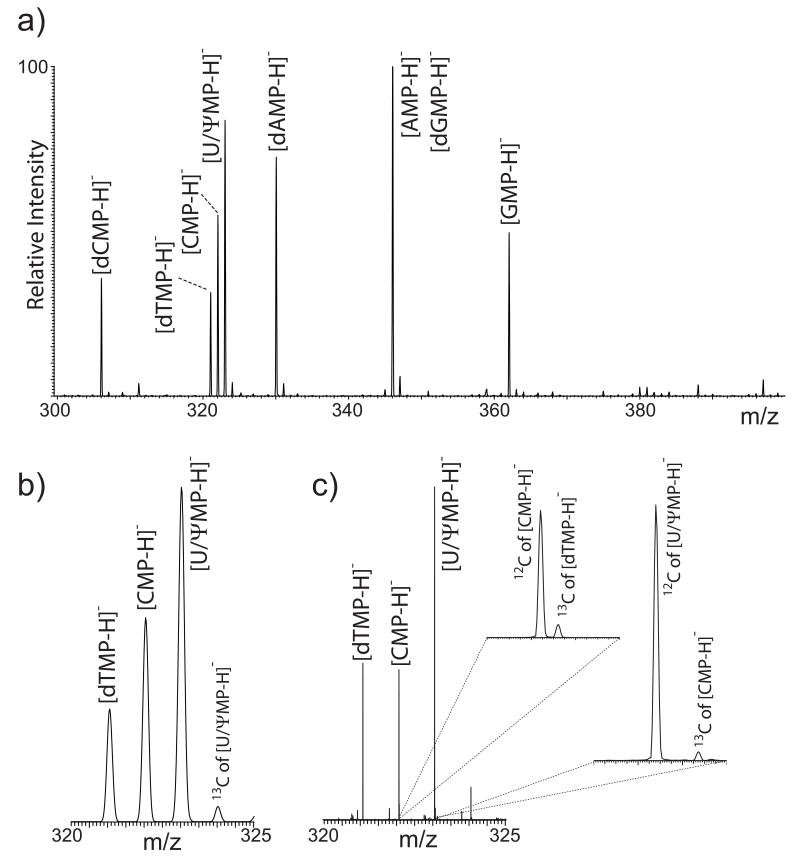
Resolving overlapping isotopic distributions in nucleotide mixtures. Nucleic acid components exhibit intrinsic properties that pose several challenges to direct mixture analysis, which cannot be readily overcome by the implementation of proper sample preparation or hyphenated separations. Table 1 provides the elemental composition and monoisotopic mass of the most abundant nucleotides present in natural DNA and RNA. The list also includes pseudouridine –long considered the “fifth ribonucleotide” for its broad biological distribution [52]– in which the uracil ring is linked to ribose through C5 rather than N1. A close examination reveals isobaric species with identical or very similar elemental compositions, which could lead to ambiguous data interpretation when analyzed as a mixture without front-end separation. The analytical challenges are exemplified by the direct infusion ESI spectrum of an equimolar solution of these nucleotides, which was recorded in the ion trap analyzer of the LTQ-orbitrap instrument (Fig. 1a). In negative ion mode, the analytes were detected as singly-charged ions produced by deprotonation of the respective phosphate groups. The majority of the observed signals were correctly assigned to individual nucleotides and respective ammonium, sodium, or potassium adducts. As a testament to the gentle character of ESI, no signals were detected for products corresponding to nucleobase losses, which are typically observed when more energetic ionization techniques are employed [18,21]. As expected from their identical elemental composition (Table 1), deprotonated AMP and dGMP were detected together as a single signal with a mass-to-charge ratio (m/z) of 346.08. Similarly, the isomeric species UMP and ψMP were observed together at m/z 323.08 and could not be differentiated. The results showed also that the spacing of one mass unit between adjacent nucleotide signals prevented the observation of full-fledged isotopic distributions, which remained blurred by extensive overlap. In the m/z 320-325 region, for example, only the abundant 12C signals of dTMP, CMP, and U/ψMP were readily recognizable, whereas corresponding 13C contributions were not (Fig. 1b).
Table 1.
Elemental compositions and monoisotopic masses of neutral 5′-phospho-nucleosides examined in the study, calculated by using the PNNL’s Molecular Weight Calculator [49].
| Name |
Elemental composition |
Monoisotopic mass (Da) |
|---|---|---|
| Guanosine monophosphate (GMP) | C10H14N5O8P | 363.05800 |
| Adenosine monophosphate (AMP) | C10H14N5O7P | 347.06308 |
| Cytidine monophosphate (CMP) | C9H14N3O8P | 323.05185 |
| Uridine monophosphate (UMP) | C9H13N2O9P | 324.03587 |
| Pseudouridinemonophosphate (ψMP) | C9H13N2O9P | 324.03587 |
| Deoxyguanosine monophosphate (dGMP) | C10H14N5O7P | 347.06308 |
| Deoxyadenosine monophosphate (dAMP) | C10H14N5O6P | 331.06817 |
| Deoxycytidine monophosphate (dCMP) | C9H14N3O7P | 307.05693 |
| Deoxythymidine monophosphate (dTMP) | C10H15N2O8P | 322.05660 |
Figure 1.
a) ESI-MS spectrum of the nucleotide mixture recorded in the LTQ analyzer; b) detail of the same spectrum showing isotopic distribution overlap between species that differ by a single mass unit; c) detail of the same region of an analogous ESI-MS spectrum recorded in the orbitrap analyzer. The enlarged insets demonstrate the ability of orbitrap experiments to resolve the 13C and 12C signals of contiguous nucleotides.
A different picture emerged when the nucleotide mixtures was examined at higher resolution in the orbitrap analyzer. In the m/z 320-325 region (Fig. 1c), the 12C of [CMP-H]− was fully resolved from the 13C of [dTMP-H]− (m/z 322.04406 and 322.05198, respectively) and the 12C of [U/ψMP-H]− from the 13C of [CMP-H]− (m/z 323.02795 and 323.04709) (Fig. 1 insets). The ~45k FWHM resolving power necessary to differentiate the 12C of [CMP-H]− from the 13C of [dTMP-H]− was easily exceed by the actual ~150k FWHM achieved in this determination (as compared to ~2.3k FWHM displayed by the ion trap data in Fig. 1b). Resolving overlapped isotopic signals ensured that each mass assignments and corresponding intensities were not affected in any way by the contributions of adjacent species. However, it is not difficult to see how the significant 13C contribution of an overlapping neighbor could potentially overshadow the presence of a weaker 12C signal or skew its mass assignment from low resolution determinations. For analytes of this size, the ~200 ppb accuracy afforded by external calibration enabled the unambiguous assignment of elemental compositions, or limited the choice of possible compositions to a handful of alternatives. For example, when the experimental value of m/z 322.04406 was input in the Formula Finder function of the PNNL Molecular Weight Calculator [49], only two possible empirical formulae containing C, H, N, O, and P were obtained with 200 ppb tolerance, i.e., C8H7N10O3P and C9H13N3O8P. While the former was clearly made implausible by the number of nitrogen atoms, the latter matched correctly the composition of deprotonated CMP. It is important to note that, although these types of figures of merit have come to be expected from typical high-resolution platforms [29-31], their intrinsic inability to differentiate UMP/ψMP and AMP/dGMP served as a stark reminder of the challenge represented by true isobaric mixtures.
Characterization of isobaric mixtures by tandem mass spectrometry
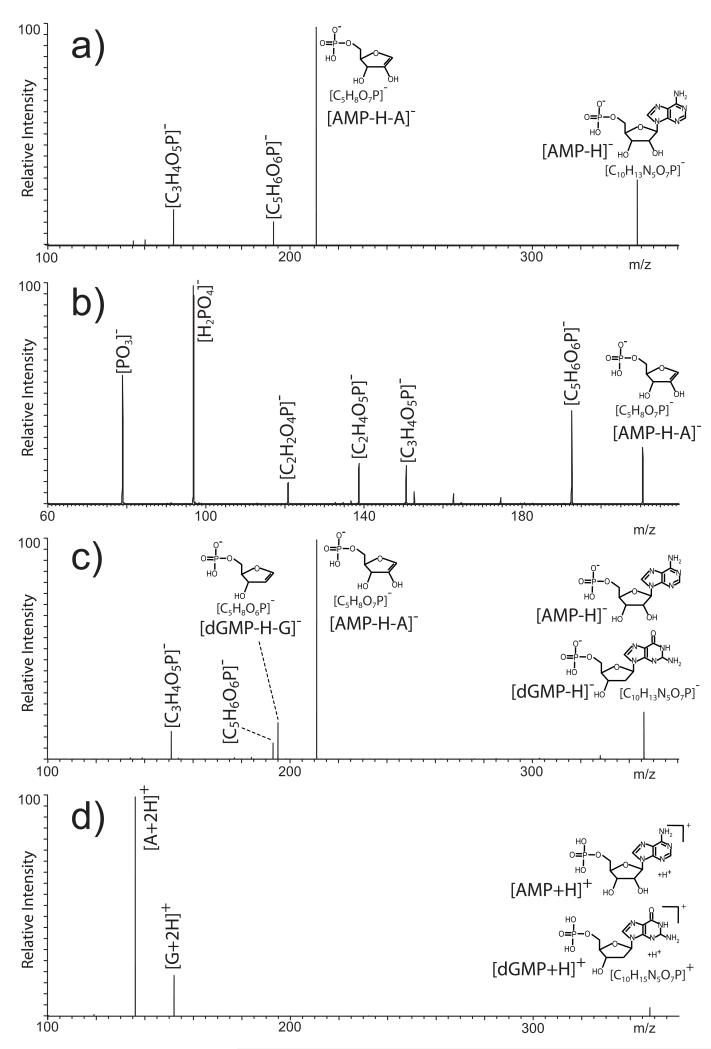
We next investigated the possibility of achieving unambiguous differentiation of isobaric populations by direct gas-phase dissociation of mixed precursors with no mutual isolation. When analyzed separately, isobaric mono-nucleotides can be unambiguously identified from fragments produced during ionization or subsequent gas-phase activation (reviewed in [21,22]). The rather labile nature of the N-glycosidic bond makes nucleobase elimination the most favorable fragmentation event observed for deprotonated nucleotides [22,53]. Accordingly, the fragments produced by this process represented the predominant signals observed in the MS/MS spectra of nearly all nucleotides in the study. In the case of [AMP-H]−, the base peak consisted of the 5′-phosphoribose moiety generated by the loss of adenine, whereas weaker fragments were produced by further loss of water and cleavage of the ribose structure (Fig. 2a). When the 5′-phospho-ribose product was in turn isolated and activated in an MS3 experiment, second-generation fragments were produced by ring cleavages and losses of water and phosphate (Fig. 2b), which confirmed the initial assignment of this precursor ion as the base-loss product. In similar fashion, the dNMPs in the study generated the equivalent 5′-phospho-2′-deoxyribose product that was readily confirmed by MS3 analysis (not shown). In apparent conflict with the known stabilizing properties of the 2′-hydroxyl group in multiply-charged oligo-ribonucleotides [22,53], no significant differences were noted in the propensity to lose base from 2′-deoxy- vs. ribonucleotides under the selected experimental conditions. This process has been the object of intensive investigation for its possible link with backbone dissociation and associated effects on the overall stability of oligonucleotides during ionization [54-56] and gas-phase activation [57-59]. In the polymeric form, the leading mechanisms involve the putative participation of the 3′-phosphate and other functional groups of contiguous units [53], which are absent in the individual 5′-phospho-nucleosides examined here. The lack of the necessary structural context is likely responsible for the surprisingly similar behaviors manifested by the two classes of nucleotides.
Figure 2.
a) Negative ion mode MS/MS spectrum of deprotonated AMP obtained from a separate sample; b) MS3 spectrum obtained by isolation and activation the 5′-phosphoribose fragment produced in the previous dissociation step (i.e., m/z 346.08 → 211.02 →); c) negative ion mode MS/MS spectrum of coexisting AMP/dGMP in the nucleotide mixture; d) positive ion mode MS/MS spectrum of protonated AMP/dGMP from the nucleotide mixture.
It is important to note that no signals were detected in negative ion mode for the nucleobase moieties themselves (i.e., B−), which might be expected to represent the complementary fragments produced by cleavage of the N-glycosidic bond. In contrast, fragments corresponding to protonated bases (i.e., BH +2) were readily observed in positive ion mode for all nucleotides in the study, save for [dTMP+H]+ and [U/ψMP+H]+ (not shown). As a first approximation, the relatively basic character of the nucleobases, which is substantiated by their gas-phase proton affinities, could help explain the ability to observe their corresponding protonated forms upon dissociation. Even the absence of protonated T and U in the respective spectra could be ascribed to their low rank in the relative scale of proton affinities (i.e. G > C > A >> T ≈ U) [60-62]. However, acid-base properties alone cannot explain the results obtained from the deprotonated analytes. Indeed, it has been noted that loss of neutral base (i.e., BH) is the predominant dissociation channel for 5′-phospho-2′-deoxynucleosides [63,64], whereas 3′-phoshonucleosides, dinucleotides, and larger oligonucleotides display a greater propensity to eliminate the deprotonated form B− [53,65,66]. These observations further emphasize the importance of the structural context in determining the mechanism of base loss and, by extension, the exact form in which the lost fragment may be detected.
The information gleaned from individual nucleotides provided the basis for discriminating the isobaric couples in the mixture. The resolving power available in the isolation step of MS/MS experiments is typically insufficient to enable the separate activation of precursor ions with very similar molecular masses, let alone that of isomers with identical elemental composition. Therefore, spectra generated by heterogeneous precursor populations are bound to contain combinations of fragments reflecting the characteristic structural features of all coexisting components. This fact was clearly highlighted, for example, by the product ion spectrum of the precursors at m/z 346.08 in the nucleotide mixture (Fig. 1a). Consistent with the presence of coexisting AMP and dGMP populations, products that could be readily attributed to either precursor were readily detected, such as those corresponding to loss of water from the pentose moiety (i.e., [C5H6O6P]−) and opening of the purine’s five-member ring (i.e., [C3H4O5P]−). At the same time, however, the unique fragments generated by base elimination were also observed, such as the 5′-phosphoribose moiety released from deprotonated AMP and the equivalent 5′-phospho-2′-deoxyribose from deprotonated dGMP (Fig. 2c), which were confirmed in separate MS3 experiments. Therefore, the ability to obtain unique diagnostic fragments, which could be readily traced to the respective precursor, provided the means for confirming unambiguously the presence of both AMP in dGMP components in the heterogeneous precursor.
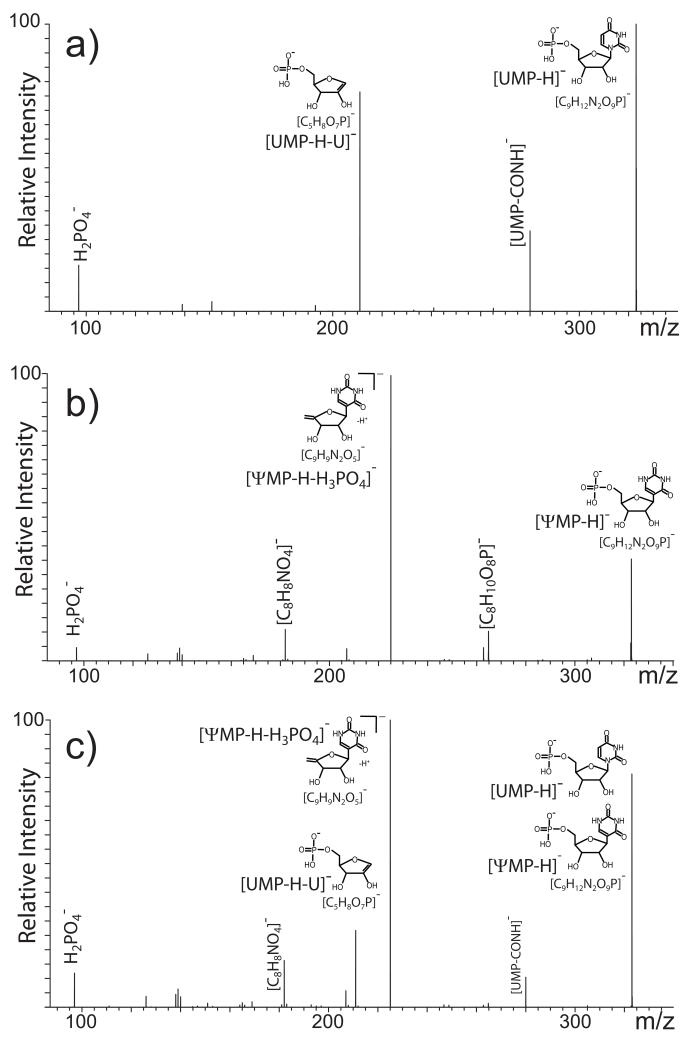
In the case of AMP/dGMP, the different placement of the oxygen on the pentose or purine system made it possible for the same dissociation process –nucleobase elimination- to produce unique diagnostic fragments for isobar discrimination. Base loss, however, may not be necessarily sufficient to differentiate more subtle structural variations exhibited by true isomeric species, which must translate into distinctive fragmentation pathways to lead to viable diagnostic products. A case in point was provided by the UMP and ψMP components that underwent remarkably different dissociations processes. Analyzed individually, the former displayed the typical 5′-phosphoribose fragment from cleavage of the N-glycosidic bond, in addition to a species produced by formal elimination of CONH from the pyrimidine ring (Fig. 3a) [53,67]. In contrast, no fragments associated with base loss were observed for deprotonated ψMP (Fig. 3b). Instead, an intense signal was detected for the nucleoside moiety left by cleavage of the phosphate group (i.e., [C9H9N2O5]−, Fig. 3b), which was not observed for any other species under the same experimental conditions. The absence of base loss is consistent with the fact that, unique among the nucleotides in the study, the ring of ψMP is connected to the pentose by a C-glycosidic bond that enjoys greater stability than that of typical N-glycosidic bonds [68]. When the mixed precursor at m/z 324.04 was activated, the resulting spectrum contained all products observed for the individual isomers (Fig. 3c). However, the detection of fragments generated by mutually exclusive pathways, such as the 5′-phosphoribose and phosphate-loss product, readily confirmed the presence of both UMP and ψMP in the selected m/z 324.04 precursor. Therefore, also in this case, the availability of unique diagnostic products provided the key for differentiating isomeric populations coexisting in the same precursor ion.
Figure 3.
Negative ion mode MS/MS spectra obtained from a) an individual UMP sample; b) individual ψMP; c) coexisting UMP/ψMP in the nucleotide mixture.
Discrimination of isobaric mixtures by ion mobility determinations
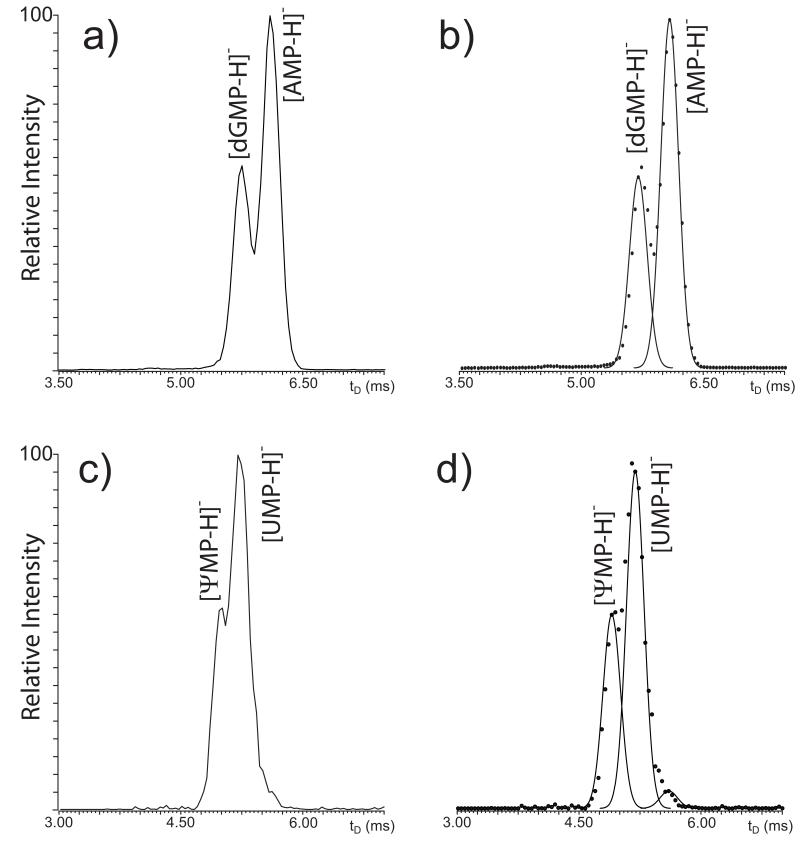
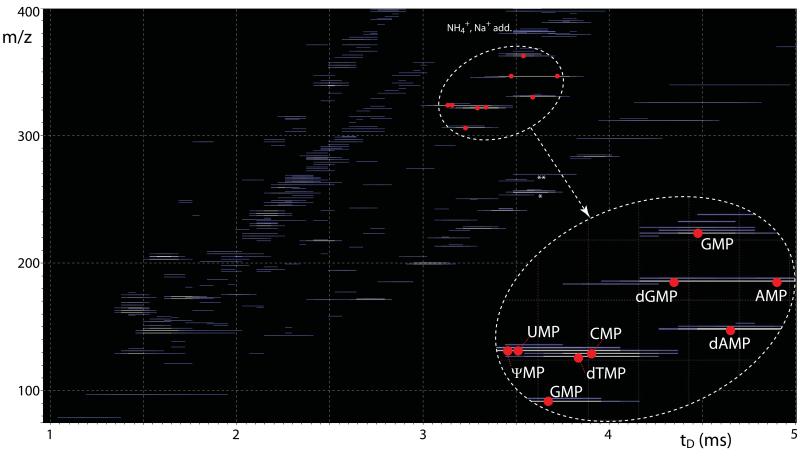
Gas-phase activation can access structural information in the form of the identity of fundamental units, functional groups, and bonds, which can be derived directly from the masses of observed fragments. In contrast, IMS-MS experiments can access structural information in the form of the placement of structures and functional groups in the three dimensions, which define the collisional cross-section obtained from apparent drift time (tD) determinations. In the case of isolated mono-deoxyribonucleotides, excellent correlation was previously reported between experimental collisional cross-sections and theoretical values calculated from their 3D structures [69]. Although the study did not cover mono-ribonucleotides or discuss mixture analysis, this observation suggested that IMS-MS may be capable of tackling mixtures of species with identical elemental composition, but different 3D structures. For this reason, the mixed AMP/dGMP precursor at m/z 346.08 was isolated in the mass-selective quadrupole and then injected in the travelling wave (Tri-WAVE) element of the instrument. The corresponding mobility profile, plotted as relative intensity vs. apparent tD, showed two maxima separated by a minimum, consistent with the presence of at least two main components with different propensity to undergo low-energy interactions with background gas (Fig. 4a). When curve-fitting was employed to differentiate the individual contributions (see Experimental), the presence of two distinct populations was more clearly delineated (Fig. 4b). The tD values afforded by such populations matched those determined individually for deprotonated AMP and dGMP (i.e., 6.08 and 5.74 ms, respectively), thus confirming the identity assignments. In similar fashion, the mobility profile recorded for the UMP/ψMP mixed precursor displayed a major signal with a smaller side peak (Fig. 4c). The curve-fitting algorithm helped distinguish two components with different tD values (Fig. 4d) that matched those determined from individual samples (i.e., 5.24 and 5.04 ms for deprotonated UMP and ψMP, respectively). Therefore, these experiments demonstrated the ability of IMS-MS to resolve the effects of significant structural differences, such as the distinctive placement of a discrete functional group in the AMP/dGMP isobaric couple, but also those of more subtle variations, such as the different position of the bond between nucleobase and pentose in UMP/ψMP. The mobility of isobaric couples was determined not only in series, which implied mass-selection and analysis of each mixed precursor in separate experiments, but also in parallel by recording the arrival time of all ions in the same experiment without mass selection. Dispensing with individual isolation steps, all ions were transmitted at once through the quadrupole stage, allowed to disperse across the Tri-WAVE, and finally analyzed in the time-of-flight analyzer. The recorded m/z was plotted against the corresponding tD in two-dimensional maps in which the relative intensity was expressed by color-coding. The resulting heat-maps, which provided a very comprehensive representation of the complexity of the sample mixture, allowed for each individual ion to be uniquely identified by its respective mass and mobility characteristics. The recorded signals corresponded not only to the analytes of interest, but also to their salt adducts and unidentified background contaminants (Fig. 5). As highlighted in the enlarged region, discrete signals for all individual nucleotides, including the isobaric components, could be unambiguously recognized from their characteristic coordinates. Indeed, the map showed that an identical elemental composition placed AMP and dGMP on the same m/z coordinate, but their structural differences translated into a broad dispersion on the tD dimension. Owing to the more modest structural variations between UMP and ψMP, their dispersion on the tD dimension was not as pronounced as the one between AMP and dGMP, but still sufficient to differentiate them unambiguously. It is important to note that, if necessary, ion mobility profiles in the format shown in Fig. 4a and c could be readily extracted from the original data to enable curve-fitting analysis and to facilitate the recognition of partially resolved species.
Figure 4.
Ion mobility profiles obtained from the precursor ions at m/z 346.17 a) and 324.04 c). Panel b) and d) are the respective curve-fitted plots generated by the deconvolution function of PeakFit 4.1 (see Experimental). Analyte identity was assigned by comparing the apparent drift time (tD) of the deconvoluted signals with those obtained in separate experiments from isolated nucleotides.
Figure 5.
Comprehensive representation of IMS-MS data obtained from the entire nucleotide mixture. In the heat-map, the m/z of each ion is plotted against its apparent tD, whereas its relative intensity is expressed by progressively lighter colors for the more abundant species. Red dots mark the maxima corresponding to the various nucleotides. The enlarged inset highlights the separation between mixture components, which was achieved in the tD and m/z dimension of the plot. No signal corresponding to typical nucleotide fragments was recognized in experiments completed without ion activation. A variety of salt adducts were observed with typical incremental masses recognizable on the m/z dimension. Asterisks mark unidentified background contaminants present in the initial stock solutions, which were detected in all ESI-MS analyses (see Experimental).
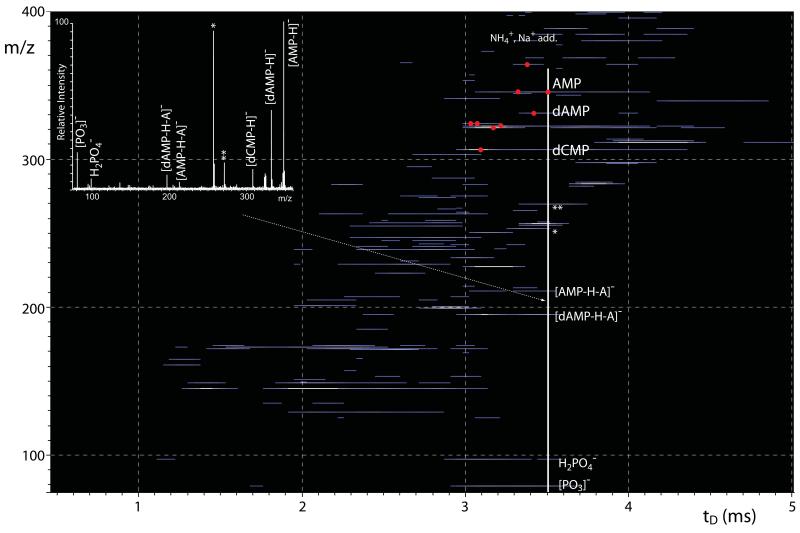
We also evaluated the possible benefits of combining the knowledge of diagnostic fragments from gas-phase dissociation experiments with the unique information gleaned from ion mobility determinations to achieve a more stringent differentiation of isobaric species. Both types of information were obtained in the same experiment by performing the parallel activation of all ions after dispersion in the time dimension in the Tri-WAVE and before mass analysis in the TOF. The fragments generated at this stage maintained the tD value of the respective precursor ion, but were now spread on the m/z dimension of the heat-map (Fig. 6). The data afforded by time aligned parallel (TAP) [50,51] dissociation were analyzed by extracting longitudinal sections of the map, which amounted to actual product ion spectra displaying all precursor ions with the same tD and their respective fragments, in the conventional format of relative intensity vs. m/z. Given that ions tend to exhibit relatively broad tD profiles (see for example Fig. 4a and c), the extracted TAP spectra are likely to contain the products of multiple precursors. For this reason, the longitudinal section taken at 3.6 ms (i.e., solid vertical line in Fig. 6) included the maximum of deprotonated AMP, but impinged also onto the broad profiles of dAMP and dCMP, as well as those of unidentified background species recorded also in the absence of activation (Fig. 5). The corresponding spectrum displayed deprotonated AMP, dAMP, and dCMP, in addition to familiar fragments observed in their separate MS/MS spectra, such as the 5′-phosphoribose and 5′-phospho-2′-deoxyribose moieties and phosphate fragments (Fig. 6 inset). As taught us by the analysis of the MS/MS spectra of mixed precursors, which also faced the presence of multiple precursors and corresponding product ions (e.g., Fig. 2c, d and 3c), knowledge of the fragmentation behavior of individual components was employed here to complete the interpretation of extracted TAP data.
Figure 6.
Heat-map representation of time aligned parallel (TAP) dissociation products provided by the entire nucleotide mixture. The data were collected by using a transfer voltage of 25 V (see Experimental). The inset represents the mass spectrum extracted at 3.6 ms of the plot (white solid line). The spectrum contains quasi-molecular ions that shared the same tD across the Tri-WAVE, as well as their respective fragments produced in the transfer region between the Tri-WAVE and the TOF analyzer. Such fragments were not observed in analogous experiments performed. Asterisks mark ubiquitous unidentified contaminants (see Experimental).
Effects of isotopic overlap on the quantification of nucleotide mixtures
The implementation of typical quantification strategies may be severely hampered by the additive effects of unresolved signals on the recorded ion currents. Resolving the isotopic distributions of adjacent components can effectively stave off possible errors associated with 13C/12C overlaps by discriminating unwanted contributions. As discussed earlier, however, mass resolution is intrinsically powerless for discriminating isobaric components detected at exactly the same m/z. A possible avenue was provided by the ability of IMS-MS to disperse isobars in the tD dimension, which enabled the differentiation of the isobaric couples in the study (Fig. 4). The curve-fitting algorithm employed to deconvolute partially resolved species was used also to calculate the areas under the respective peaks for quantification purposes. Following this approach, recognizable dGMP curves were still attainable at concentrations as low as ~5 nM with total sample consumption of ~500 × 10−18 mol (estimated from a typical nanospray flow rate of ~20nL/min[70] and ~5 min acquisition).
An alternative strategy for teasing out the distinct contributions of coexisting isobars was prompted by the knowledge of characteristic fragmentation patterns from multistep dissociation experiments. Based on the univocal precursor-product relationships discussed above, we performed consecutive reaction monitoring (CRM)[71] on the different isobaric components. For example, we employed them/z 346.08 → 211.02 transition (i.e., formation of 5′-phospho-ribose) versus the m/z 346.08 → 195.01 one (i.e., formation of 5′-phospho-2′-deoxyribose) to effectively discriminate AMP and dGMP, respectively. Monitoring each transition in separate experiments enabled the detection of each isobar from a 250 pM solution of total nucleotides with an estimated sample consumption of 75 × 10−18 mol (~2 0nL/min flow rate for ~15min acquisition). As a testament to the excellent selectivity of CRM approaches, no interference or crosstalk were noted when the concentrations of two isobaric species were subjected to wide swings in opposite directions. For example, acceptable signal was still recorded for the m/z 346.08 → 211.02 transition of AMP when the solution contained also a 105-excess of dGMP (i.e., a five orders of magnitude difference). The greater sensitivity afforded by CRM versus regular MS analysis was consistent with the ability of the former to minimize the possible effects of chemical noise and to boost the observed signal-to-noise ratios. It should be noted also that all determinations were performed in negative ion mode to take advantage of the strongly acidic nature of the 5′-phospho-nucleosides. Although the analytes provided detectable protonated ions as an instance of “wrong-way-round” electrospray ionization [72,73], all analyses completed in positive ion mode exhibited an approximately 30% decrease of the overall response across the board.
In mixture analysis, it may be practical to generate calibration curves for only a few selected components, while the remaining ones may be estimated from the ratios of their respective signal intensities. This approach, however, must take in account possible differences of signal response afforded by the species involved. This effect was in full display, for example, in Fig 1a and 4a, c, in which nucleotides produced signals of markedly different intensities notwithstanding their equimolar amounts in the sample (Experimental). At first glance, the observed responses seemed to contradict the dTTP > dATP > dCTP > dGTP scale of relative intensities reported earlier for an equimolar mixture of 5′-triphospho-2′-deoxynucleosides (dNTPs), which matched the order of hydophobicities exhibited by the respective nucleobases [75]. In our hands, however, a mixture of 5′-monospho-2′-deoxynucleosides (dNMPs) displayed a dAMP > dGMP > dCMP > dTMP scale of relative intensities (not shown), which confirmed the data in Fig. 1a without the additive effects of the AMP/dGMP isobaric overlap. The same scale was obtained also when the hydrophobic character of the ESI solvent was increased by switching from a 10% volume of 2-propanol to a 60% acetonitrile/20% 2-propanol, matching the system employed in the earlier study [75]. These results suggest that the discrepancy may be ascribable to more subtle differences between 5′-triphospho- and 5′-monophospho-nucleosides, which will require further investigation.
Conclusions
The challenges posed by the direct analysis of nucleotide mixtures without front-end separation can be overcome by the implementation of alternative and often complementary approaches. The value of high-resolution determinations was immediately confirmed by the ability to distinguish the nearly overlapping isotopic distributions of species that differed by a single mass unit. In fact, full-fledged isotopic distributions were readily recognizable by resolving the small deviations between the 13C and 12C signals of contiguous species, such as dTMP and CMP, or CMP and U/ψMP. In this way, it was possible not only to validate the low resolution assignments, but also to obtain accurate experimental masses for empirical formula predictions. Regardless the excellent resolving power, however, these types of determinations were intrinsically incapable of discriminating between truly (i.e., not nominally) isobaric species and assigning unambiguously their identity. In contrast, this task was effectively tackled by first identifying exclusive correlations between subtle structural variations and unique fragmentation patterns. At this stage, MSn analyses were used to establish that the position of an oxygen atom in AMP/dGMP, or the different stability of N- and C-glycosidic bonds in UMP/ψMP, translated into unique fragments that could be attributed exclusively to the respective precursor ion. In this way, the presence of a particular isobar was immediately recognizable by detecting its corresponding diagnostic products among the collection of fragments generated by the coexisting precursors. Identifying these univocal precursor-product relationships enabled also the implementation of CRM determinations to effectively quantify coexisting isobars without requiring their actual separation.
The study confirmed that the additional dimension afforded by IMS-MS represents a very attractive alternative for differentiating analytes of identical elemental compositions. The different position of an oxygen atom or glycosidic bond had significant effects on the extent of the interactions with background gas and, thus, on the observed mobility behaviors. Such effects were readily detectable from isobaric couples that had been mass-selected, or in the context of ion mixtures analyzed in their entirety without mass selection. Each approach demonstrated the ability to correctly differentiate the AMP/dGMP and UMP/ψMP couples considered here, with the notable distinction that the former required prior knowledge of the specific targets to be mass-selected, which was not necessary for the latter. The TAP dissociation experiments represented an auspicious first step towards achieving “MS/MS of everything” to enable broad range analyses in which the presence of isobaric targets may not be known a priory. In our case, the availability of multistep fragmentation data benefited the interpretation of TAP dissociation spectra from multiple precursors, thus providing a measure of the possible synergies between the platforms employed in the study. Further improvement of the tD-domain resolution and the development of algorithms to unambiguously recognize fragment-precursor correlations will be necessary to promote the diffusion of time-aligned techniques.
The continued interest in modified nucleic acid components as possible markers of DNA damage and disease represents a powerful rationale for the development of strategies aimed at the analysis of complex sample mixtures with minimum sample preparation. Typical hybridization methods are severely hampered by the lack of the complementary nucleotides necessary to sustain the faithful amplification of modification-bearing strands. In the case of unknown samples, the practical application of standalone chromatographic and electrophoretic approaches is typically constrained by the limited availability of genuine standards required for validating the observed retention/migration times. The shortcomings of established methods in the observation of covalent modifications advocate for a more widespread utilization of MS-based technologies. The approaches evaluated here have demonstrated great potential for the characterization of nucleotide mixtures with very similar or identical elemental composition. This work represents the necessary stepping stone for applications aiming at the analysis of whole-cell nucleic acid extracts and total digests from specific cellular compartments, which will be the objects of our future efforts.
Acknowledgements
This work was supported by The RNA Institute of the University at Albany and by the National Institutes of Health (GM064328-12). Helpful discussions with Dr. J. B. Mangrum are gratefully acknowledged.
References
- 1.Saenger N. Principles of Nucleic Acid Structure. Springer; 1984. [Google Scholar]
- 2.Bloomfied VA, Crothers DM, Tinoco I., Jr. Nucleic Acids - Structures, Properties, and Functions. University Science Books; 2000. [Google Scholar]
- 3.Neidle S. Nucleic Acid Structure and Recognition. Oxford University Press; USA: 2002. [Google Scholar]
- 4.Lijinsky W, Loo J, Ross AE. Mechanism of alkylation of nucleic acids by nitrosodimethylamine. Nature. 1968;218:1174–5. doi: 10.1038/2181174b0. [DOI] [PubMed] [Google Scholar]
- 5.Cadet J, Weinfeld M. Detecting DNA damage. Analytical chemistry. 1993;65:675A–682A. doi: 10.1021/ac00063a001. [DOI] [PubMed] [Google Scholar]
- 6.Stone MP, Huang H, Brown KL, Shanmugam G. Chemistry and structural biology of DNA damage and biological consequences. Chem. Biodivers. 2011;8:1571–1615. doi: 10.1002/cbdv.201100033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawley PD, Brookes P. Further studies on the alkylation of nucleic acids and their constituent nucleotides. Biochem. J. 1963;89:127–138. doi: 10.1042/bj0890127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maher VM, Lesko SA, Jr, Straat PA, Ts’o PO. Mutagenic action, loss of transforming activity, and inhibition of deoxyribonucleic acid template activity in vitro caused by chemical linkage of carcinogenic polycyclic hydrocarbons to deoxyribonucleic acid. J. Bacteriol. 1971;108:202–212. doi: 10.1128/jb.108.1.202-212.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swenberg JA, Lu K, Moeller BC, Gao L, Upton PB, Nakamura J, Starr TB. Endogenous versus Exogenous DNA Adducts: Their Role in Carcinogenesis, Epidemiology, and Risk Assessment. Toxicol Sci. 2011;120:S130–S145. doi: 10.1093/toxsci/kfq371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holliday R. DNA methylation and epigenetic mechanisms. Cell Biophys. 1989;15:15–20. doi: 10.1007/BF02991575. [DOI] [PubMed] [Google Scholar]
- 11.Lewin B. The mystique of epigenetics. Cell. 1998;93:301–303. doi: 10.1016/s0092-8674(00)81154-x. [DOI] [PubMed] [Google Scholar]
- 12.Robertson KD. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20:3139–3155. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- 13.Wogan GN. Markers of exposure to carcinogens. Environ. Health Perspect. 1989;81:9–17. doi: 10.1289/ehp.89819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farmer PB, Singh R. Use of DNA adducts to identify human health risk from exposure to hazardous environmental pollutants: The increasing role of mass spectrometry in assessing biologically effective doses of genotoxic carcinogens. Mutation Research/Reviews in Mutation Research. 2008;659:68–76. doi: 10.1016/j.mrrev.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Banoub JH, Limbach PA. Mass Spectrometry of Nucleosides and Nucleic Acids. CRC Press Inc.; 2010. [Google Scholar]
- 16.Limbach PA, Crain PF, McCloskey JA. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–96. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FAP, Fabris D, Agris PF. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biemann K, McCloskey JA. Application of mass spectrometry to structure problems. VI. Nucleosides. J. Am. Chem. Soc. 1962;84:2005–2007. [Google Scholar]
- 19.McCloskey JA. Characterization of nucleosides by mass spectrometry. Nucleic Acids Symp Ser. 1979:s109–13. doi: 10.1002/chin.197941058. [DOI] [PubMed] [Google Scholar]
- 20.McCloskey JA. In: Mass Spectrometry in the Health and Life Sciences. Burlingame AL, Castagnoli N, editors. Elsevier; 1985. pp. 521–546. [Google Scholar]
- 21.Crain PF. Mass spectrometric techniques in nucleic acid research. Mass Spectrom Rev. 1990;9:505–554. [Google Scholar]
- 22.Nordhoff E, Kirpekar F, Roepstorff P. Mass spectrometry of nucleic acids. Mass Spectrom. Rev. 1996;15:67–138. doi: 10.1002/(SICI)1098-2787(1996)15:2<67::AID-MAS1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 23.Crain PF. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol. 1990;193:782–90. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- 24.Apruzzese WA, Vouros P. Analysis of DNA adducts by capillary methods coupled to mass spectrometry: a perspective. Journal of chromatography. 1998;794:97–108. doi: 10.1016/s0021-9673(97)00820-0. [DOI] [PubMed] [Google Scholar]
- 25.Esmans E, Broes D, Hoes I, Lemière F, Vanhoutte K. Liquid chromatography-mass spectrometry in nucleoside, nucleotide and modified nucleotide characterization. Journal of Chromatography A. 1998;794:109–127. [Google Scholar]
- 26.Chiarelli MP, Lay JO. Mass spectrometry for the analysis of carcinogen-DNA adducts. Mass Spectrometry Reviews. 1992;11:447–493. [Google Scholar]
- 27.Farmer PB, Sweetman GM. Mass Spectrometric Detection of Carcinogen Adducts. J. Mass Spectrom. 1995;30:1369–1379. [Google Scholar]
- 28.Koivisto P, Peltonen K. Analytical methods in DNA and protein adduct analysis. Anal Bioanal Chem. 2010;398:2563–2572. doi: 10.1007/s00216-010-4217-3. [DOI] [PubMed] [Google Scholar]
- 29.Marshall AG, Hendrickson CL, Jackson GS. Fourier transform ion cyclotron resonance mass spectrometry: a primer. Mass Spectrom. Rev. 1998;17:1–35. doi: 10.1002/(SICI)1098-2787(1998)17:1<1::AID-MAS1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 30.Makarov A. Electrostatic Axially Harmonic Orbital Trapping: A High-Performance Technique of Mass Analysis. Anal. Chem. 2000;72:1156–1162. doi: 10.1021/ac991131p. [DOI] [PubMed] [Google Scholar]
- 31.Scigelova M, Hornshaw M, Giannakopulos A, Makarov A. Fourier Transform Mass Spectrometry. Mol Cell Proteomics. 2011:10. doi: 10.1074/mcp.M111.009431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Von Helden G, Wyttenbach T, Bowers MT. Conformation of macromolecules in the gas phase: use of matrix-assisted laser desorption methods in ion chromatography. Science (New York, N.Y) 1995;267:1483–5. doi: 10.1126/science.267.5203.1483. [DOI] [PubMed] [Google Scholar]
- 33.Clemmer DE, Jarrold MF. Ion mobility measurements and their applications to cluster biomolecules. J. Mass Spectrom. 1997;32:577–592. [Google Scholar]
- 34.Verbeck GF, Ruotolo BT, Sawyer HA, Gillig KJ, Russell DH. A fundamental introduction to ion mobility mass spectrometry applied to the analysis of biomolecules. J Biomol Tech. 2002;13:56–61. [PMC free article] [PubMed] [Google Scholar]
- 35.Makarov A, Denisov E, Kholomeev A, Balschun W, Lange O, Strupat K, Horning S. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal. Chem. 2006;78:2113–2120. doi: 10.1021/ac0518811. [DOI] [PubMed] [Google Scholar]
- 36.Xu Y, Heilier J-F, Madalinski G, Genin E, Ezan E, Tabet J-C, Junot C. Evaluation of accurate mass and relative isotopic abundance measurements in the LTQ-orbitrap mass spectrometer for further metabolomics database building. Anal. Chem. 2010;82:5490–5501. doi: 10.1021/ac100271j. [DOI] [PubMed] [Google Scholar]
- 37.Lommen A, Gerssen A, Oosterink JE, Kools HJ, Ruiz-Aracama A, Peters RJB, Mol HGJ. Ultra-fast searching assists in evaluating sub-ppm mass accuracy enhancement in U-HPLC/Orbitrap MS data. Metabolomics. 2011;7:15–24. doi: 10.1007/s11306-010-0230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collings BA, Campbell JM, Mao D, Douglas DJ. A combined linear ion trap time-of-flight system with improved performance and MSn capabilities. Rapid Communications in Mass Spectrometry. 2001;15:1777–1795. doi: 10.1002/rcm.440. [DOI] [PubMed] [Google Scholar]
- 39.Revercomb HE, Mason EA. Theory of plasma chromatography/gaseous electrophoresis. Review. Anal. Chem. 1975;47:970–983. [Google Scholar]
- 40.Von Helden G, Hsu M-T, Kemper PR, Bowers MT. Structures of carbon cluster ions from 3 to 60 atoms: Linears to rings to fullerenes. The Journal of Chemical Physics. 1991;95:3835–3837. [Google Scholar]
- 41.Giles K, Pringle SD, Worthington KR, Little D, Wildgoose JL, Bateman RH. Applications of a travelling wave-based radio-frequency-only stacked ring ion guide. Rapid Commun. Mass Spectrom. 2004;18:2401–2414. doi: 10.1002/rcm.1641. [DOI] [PubMed] [Google Scholar]
- 42.Shvartsburg AA, Smith RD. Fundamentals of traveling wave ion mobility spectrometry. Anal. Chem. 2008;80:9689–9699. doi: 10.1021/ac8016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dwivedi P, Wu C, Matz LM, Clowers BH, Siems WF, Hill HH., Jr Gas-phase chiral separations by ion mobility spectrometry. Anal. Chem. 2006;78:8200–8206. doi: 10.1021/ac0608772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Enders JR, McLean JA. Chiral and structural analysis of biomolecules using mass spectrometry and ion mobility-mass spectrometry. Chirality 21 Suppl. 2009;1:E253–264. doi: 10.1002/chir.20806. [DOI] [PubMed] [Google Scholar]
- 45.Lapthorn C, Pullen F, Chowdhry BZ. Ion mobility spectrometry-mass spectrometry (IMS-MS) of small molecules: Separating and assigning structures to ions. Mass Spectrom Rev. 2013;32:43–71. doi: 10.1002/mas.21349. [DOI] [PubMed] [Google Scholar]
- 46.Limbach PA. Indirect mass spectrometric methods for characterizing and sequencing oligonucleotides. Mass Spectrom. Rev. 1996;15:297–336. doi: 10.1002/(SICI)1098-2787(1996)15:5<297::AID-MAS2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 47.Gray DM, Hung SH, Johnson KH. Absorption and circular dichroism spectroscopy of nucleic acid duplexes and triplexes. Meth. Enzymol. 1995;246:19–34. doi: 10.1016/0076-6879(95)46005-5. [DOI] [PubMed] [Google Scholar]
- 48.Fabris D, Turner KB, Hagan NA. In: Mass Spectrometry of Nucleosides and Nucleic Acids 303-327. Banoub JH, Limbach PA, editors. CRC Press; 2009. [Google Scholar]
- 49.Monroe M. Molecular Weight Calculator, v. 6.49. 2012. http://ncrr.pnl.gov/software/ [Google Scholar]
- 50.Damen CWN, Chen W, Chakraborty AB, Van Oosterhout M, Mazzeo JR, Gebler JC, Schellens JHM, Rosing H, Beijnen JH. Electrospray ionization quadrupole ion-mobility time-of-flight mass spectrometry as a tool to distinguish the lot-to-lot heterogeneity in N-glycosylation profile of the therapeutic monoclonal antibody trastuzumab. J. Am. Soc. Mass Spectrom. 2009;20:2021–2033. doi: 10.1016/j.jasms.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 51.Castro-Perez J, Roddy TP, Nibbering NMM, Shah V, McLaren DG, Previs S, Attygalle AB, Herath K, Chen Z, Wang S-P, Mitnaul L, Hubbard BK, Vreeken RJ, Johns DG, Hankemeier T. Localization of fatty acyl and double bond positions in phosphatidylcholines using a dual stage CID fragmentation coupled with ion mobility mass spectrometry. J. Am. Soc. Mass Spectrom. 2011;22:1552–1567. doi: 10.1007/s13361-011-0172-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 53.Wu J, ey SA. Gas-phase fragmentation of oligonucleotide ions. International Journal of Mass Spectrometry. 2004;237:197–241. [Google Scholar]
- 54.Gross J, Leisner A, Hillenkamp F, Hahner S, Karas M, Schäfer J, Lützenkirchen F, Nordhoff E. Investigations of the metastable decay of DNA under ultraviolet matrix-assisted laser desorption/ionization conditions with post-source decay analysis and hydrogen/deuterium exchange. J. Am. Soc. Mass Spectrom. 1998;9:866–878. doi: 10.1016/S1044-0305(98)00058-0. [DOI] [PubMed] [Google Scholar]
- 55.Gross J, Hillenkamp F, Wan KX, Gross ML. Metastable decay of negatively charged oligodeoxynucleotides analyzed with ultraviolet matrix-assisted laser desorption/ionization post-source decay and deuterium exchange. J. Am. Soc. Mass Spectrom. 2001;12:180–192. doi: 10.1016/S1044-0305(00)00209-9. [DOI] [PubMed] [Google Scholar]
- 56.Wan KX, Gross J, Hillenkamp F, Gross ML. Fragmentation mechanisms of oligodeoxynucleotides studied by H/D exchange and electrospray ionization tandem mass spectrometry. J Am Soc Mass Spectrom. 2001;12:193–205. doi: 10.1016/S1044-0305(00)00208-7. [DOI] [PubMed] [Google Scholar]
- 57.McLuckey SA, Habibi-Goudarzi S. Decompositions of multiply charged oligonucleotide anions. J. Am. Chem. Soc. 1993;115:12085–95. [Google Scholar]
- 58.Schurch S, Bernal-Mendez E, Leumann CJ. Electrospray tandem mass spectrometry of mixed-sequence RNA/DNA oligonucleotides. J. Am. Soc. Mass Spectrom. 2002;13:936–45. doi: 10.1016/S1044-0305(02)00413-0. [DOI] [PubMed] [Google Scholar]
- 59.Tromp JM, Schurch S. Gas-phase dissociation of oligoribonucleotides and their analogs studied by electrospray ionization tandem mass spectrometry. J Am Soc Mass Spectrom. 2005;16:1262–8. doi: 10.1016/j.jasms.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 60.Greco F, Liguori A, Sindona G, Uccella N. Gas-Phase Proton Affinity of Deoxyribonuclosides and Related Nucleobases by Fast Atom Bombardment Tandem Mass Spectrometry. J. Am. Chem. Soc. 1990;112:9092–96. [Google Scholar]
- 61.Liguori A, Napoli A, Sindona G, Cooks RG. Determination of substituent effects on the proton affinities of natural nucleosides by the kinetic method. Rapid Communications in Mass Spectrometry. 1994;8:89–93. doi: 10.1002/rcm.1290080117. [DOI] [PubMed] [Google Scholar]
- 62.Pan S, Verhoeven K, Lee JK. Investigation of the initial fragmentation of oligodeoxynucleotides in a quadrupole ion trap: charge level-related base loss. J. Am. Soc. Mass Spectrom. 2005;16:1853–1865. doi: 10.1016/j.jasms.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 63.Cerny RL, Tomer KB, Gross ML, Grotjahn L. Fast atom bombardment combined with tandem mass spectrometry for determining structures of small oligonucleotides. Anal Biochem. 1987;165:175–82. doi: 10.1016/0003-2697(87)90217-x. [DOI] [PubMed] [Google Scholar]
- 64.Habibi-Goudarzi S, McLuckey SA. Ion trap collisional activation of the deprotonated deoxymononucleoside and deoxydinucleoside phosphates. J. Am. Soc. Mass Spectrom. 1995;6:102–113. doi: 10.1016/S1044-0305(94)00108-C. [DOI] [PubMed] [Google Scholar]
- 65.Phillips DR, McCloskey JA. A comprehensive study of the low energy collision-induced dissociation of dinucleoside monophosphates. International Journal of Mass Spectrometry and Ion Processes. 1993;128:61–82. [Google Scholar]
- 66.Rodgers MT, Campbell S, Marzluff EM, Beauchamp JL. Site-specific protonation directs low-energy dissociation pathways of dinucleotides in the gas phase. International Journal of Mass Spectrometry and Ion Processes. 1995;148:1–23. [Google Scholar]
- 67.Neri N, Sindona G, Uccella N. Slow Degradations of Oligonucleotide Diesterophosphate Anions. The Mass Spectrometric Method of Fast Atom Bombardment-Mike-CID. Gazz. Chim. Ital. 1983;113:197–202. [Google Scholar]
- 68.Pomerantz SC, McCloskey JA. Detection of the common RNA nucleoside pseudouridine in mixtures of oligonucleotides by mass spectrometry. Anal. Chem. 2005;77:4687–4697. doi: 10.1021/ac058023p. [DOI] [PubMed] [Google Scholar]
- 69.Gidden J, Bowers MT. Gas-Phase Conformations of Deprotonated and Protonated Mononucleotides Determined by Ion Mobility and Theoretical Modeling. J. Phys. Chem. B. 2003;107:12829–12837. [Google Scholar]
- 70.Wilm M, Mann M. Analytical properties of the nanoelectrospray ion source. Anal. Chem. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 71.Tomer KB, Guenat CR, Deterding LJ. Consecutive reaction monitoring in a four-sector mass spectrometer: MS4 and one step beyond. Anal. Chem. 1988;60:2232–2236. doi: 10.1021/ac00171a014. [DOI] [PubMed] [Google Scholar]
- 72.Kelly MA, Vestling MM, Fenselau C. Electrospray of proteins: a comparison of positive-ion and negative-ion mass spectra at high and low pH. Org. Mass Spectrom. 1992;27:1143–1147. [Google Scholar]
- 73.Zhou S, Cook KD. Protonation in electrospray mass spectrometry:wrong-way-round or right-way-round? J. Am. Soc. Mass Spectrom. 2000;11:961–966. doi: 10.1016/S1044-0305(00)00174-4. [DOI] [PubMed] [Google Scholar]
- 74.Green-Church KB, Limbach PA. Mononucleotide gas-phase proton affinities as determined by the kinetic method. J Am Soc Mass Spectrom. 2000;11:24–32. doi: 10.1016/S1044-0305(99)00116-6. [DOI] [PubMed] [Google Scholar]
- 75.Null AP, Nepomuceno AI, Muddiman DC. Implications of hydrophobicity and free energy of solvation for characterization of nucleic acids by electrospray ionization mass spectrometry. Anal Chem. 2003;75:1331–9. doi: 10.1021/ac026217o. [DOI] [PubMed] [Google Scholar]