Abstract
House flies disseminate numerous species of bacteria acquired during feeding and breeding activities in microbe-rich habitats. Previous house fly surveys have detected the pathogen Staphylococcus aureus Rosenbach 1884, which causes cutaneous and septic infections in mammals, and enterotoxic food poisoning. We assessed the fate of GFP-expressing S. aureus (GFP-S. aureus) in the house fly alimentary canal with microscopy and by culture of whole flies and excreta. Furthermore, the concurrent expression of the antimicrobial peptide gene defensin was measured in the crop, proventriculus, midgut, and fat body. As soon as 4 h postingestion (PI), GFP-S. aureus were visualized as cocci or diplococci in the hindgut and rectum of flies fed ≈105 colony forming units. Bacteria persisted up to 6 h PI but significantly decreased. Excretion of viable GFP-S. aureus peaked at 2 h PI and, although significantly less, continued up to 4 h PI. defensin was highly upregulated locally in the alimentary canal and systemically in fat body at 2, 4, and 6 h PI making this study the first to report, to our knowledge, an epithelial and systemic response to a bacterium with lysine-type peptidoglycan in flies exposed via feeding. While flies harbored S. aureus for up to 6 h PI, the highest probability of vectoring biologically relevant amounts of bacteria occurred 0–2 h PI. The combined effects of excretion, digestion and antimicrobial effectors likely contribute to loss of ingested bacteria. Nonetheless, house flies are relevant vectors for S. aureus up to 2 h PI and environmental reservoirs up to 6 h PI.
Keywords: bacteria, culture recovery, excreta, midgut, fat body
House flies (Musca domestica L.; Diptera: Muscidae) have been shown to harbor, transport, and vector numerous species of human and livestock pathogenic bacteria (Graczyk et al. 2001). Because larvae have a nutritional requirement for bacteria (Zurek et al. 2000), adult flies (particularly females) are attracted to septic substrates for oviposition (West 1951). Subsequently when adults search for sugar- or protein-rich foods in human or animal habitats, flies deposit live microbes via physical contact or excreta. In agricultural settings, bacteria transmitted by flies frequently are enteropathogens such as Escherichia coli O157:H7, Salmonella spp., and Campylobacter spp., which are picked up by flies during associations with animal waste or contaminated environments (Rosef and Kapperud 1983, Moriya et al. 1999, Szalanski et al. 2004, Kinde et al. 2005, Holt et al. 2007). However, in human habitats, house flies have been shown to harbor a wide range of pathogenic and nonpathogenic microbes from various taxa (Cohen et al. 1991; Fotedar et al. 1992a,b; Graczyk et al. 2001; Gupta et al. 2012). In this study, we chose to look at the fate of Staphylococcus aureus Rosenbach 1884 in flies, because this pathogen has been isolated in several of the surveys above and causes cutaneous, systemic, and enterotoxigenic diseases in humans and livestock (Ben Zakour et al. 2008, Fitzgerald 2012).
The fly’s vector potential for particular pathogens is affected by location of bacteria either on or in the fly, fly antimicrobial defenses, and inherent species-specific properties of bacteria that interact with either of these factors. For example, bacteria harbored externally may dry during flight and lose viability (Yap et al. 2008), but bacteria harbored internally face digestion and defensive responses from gut epithelia. Ingested bacteria usually are initially stored in the fly crop, where food can be predigested via salivary carbohydrases and/or regurgitated to liquefy the next meal (Lehane and Billingsley 1996). Once food is partitioned past the proventriculus to the midgut, it cannot be regurgitated and passes posteriorly via peristalsis. Along the entire midgut, food and bacteria are physically separated from the delicate epithelium by a type-II peritrophic matrix (PM); however, digestive enzymes and defensive effectors secreted by the epithelium can traverse the PM and act on ingested substrates (Richards and Richards 1977).
Epithelial immunity in the house fly gut has been understudied, but insight can be gleaned from studies on the model dipteran Drosophila melanogaster (Meigen), because fruit flies also commonly associate with bacteria during feeding and breeding activities. Numerous studies on the local epithelial response in the gut of naturally infected D. melanogaster show recognition of gram-negative bacteria via binding of diaminopimelic acid-type peptidoglycan (DAP-PGN) fragments to transmembrane peptidoglycan recognition receptors (PGRPs) (Stenbak et al. 2004, Vodovar et al. 2005, Liehl et al. 2006, Ryu et al. 2006, Buchon et al. 2009, Charroux and Royet 2010). These small subunits of DAP-PGN are able to cross the peritrophic matrix and stimulate immune cascades, which eventually produce antimicrobial peptides (AMPs), secreted bactericidal effector molecules (Lemaitre and Hoffmann 2007). In this study, the expression of defensin (def) was examined because this broad-spectrum AMP is particularly effective against gram-positive cocci with lysine-type PGN (LYS-PGN) (Lambert et al. 1989, Hoffmann and Hetru 1992, Yamada and Natori 1993). Defensin from the blow fly Lucilia sericata Meigen (= “lucifensin”) has shown bactericidal activity against pathogens such as methicillin-resistant S. aureus (MRSA) and, thus, may be the key effector for successful maggot therapy treatment of refractory wounds (Andersen et al. 2010, Cerovsky et al. 2010). Additionally, the spectrum activity of purified house fly Defensin was recently analyzed and revealed activity against bacteria with DAP- and LYS-PGN as well as fungi (Dang et al. 2010). However, both of these studies involved larvae and pupae, respectively, and did not determine local expression in the alimentary canal, proximal to bacteria. Thus, in our study we examined the spatial and temporal expression of def both locally in the gut and systemically in the fat body of adult house flies.
The objective of this study was to determine the fate of GFP-expressing S. aureus in house flies by microscopical examination of the location of bacteria in the gut over time, and by culture of viable bacteria from whole fly homogenate as well as excreta. Further, the concurrent expression of the AMP gene def was quantified to assess the house fly response to ingested bacteria. Examining the interaction of house flies and bacteria from the perspective of both organisms provided an opportunity to uniquely observe the dynamics of bacteria fate and ultimately vector potential for this pathogen.
Materials and Methods
House Fly Rearing
Adult house flies from a colony established in 2004 at Georgia Southern University were maintained on a diet of 2:2:1 powdered sugar: milk:egg at 30°C with a photoperiod of 12:12 (L:D) h. Flies were provided moistened wheat bran media for oviposition and larva development. Pupae collected from this colony were allowed to emerge in sterile petri dishes and separated in individual, sterile jars for bacterial feeding. House flies were determined to be free of S. aureus but did contain low numbers (<1,000 CFU [colony forming unit]) of bacteria carried over through pupariation including Bacillus spp.
S. aureus Culture
FP-expressing S. aureus RN6390 strain ALC 1743 with plasmid psk236 (GFP-S. aureus) was provided by A. Cheung (Dartmouth College) and maintained at 37°C in tryptic soy broth plus chloramphenicol (10 μg/ml) for all experiments (Fisher, Atlanta, GA). Liquid culture was inoculated with a single colony of GFP-S. aureus and shaken (120 rpm) at 37°C for 2–6 h, until the OD600 reached ≈0.60, which corresponded to ≈108 CFU/ml.
Fly Feeding
Within 24 h posteclosion, adult flies were segregated into individual jars and fed 5 μl 10% sterile sucrose to ensure proper nutrition to produce AMPs and reduce the amount of indigenous microbiota in the house fly digestive tract. Flies then were fasted overnight (8–12 h) and each fed a single 2 μl droplet of S. aureus, which were later quantified by serial dilution and plating in duplicate on chloramphenicol (10 μg/ml) tryptic soy agar (TSA). Mean counts (i.e., CFU in the 2 μl droplet) of bacteria fed to each fly in the following experiments were as follows: for epifluorescence microscopy, 2.0 ± 105 CFU; for culture-recovery, 6.75 ± 1.44 ×105 CFU; for excreta recovery, 3.2 ± 1.4 × 105 CFU; for qRTPCR, 4 × 105 CFU.
Epifluorescence Microscopy
Adult house flies (n = 15) were fed 2 μl-droplets of GFP-S. aureus, and at 2, 4, and 6 h postingestion (PI) five flies were randomly chosen for dissection and aseptic removal of the entire alimentary canal including proventriculus, crop, midgut, hindgut, and rectum. Guts were placed on microscope slides, and the location of GFP-S. aureus was examined using epifluorescent microscopy as previously described (McGaughey and Nayduch 2009).
Culture Recovery
Adult flies (n = 15 per n = 3 biological replicates) were fed a single 2 μl droplet of GFP-S. aureus, and at 2, 4, and 6 h PI, five flies were immobilized by chilling and surface-sanitized by submersion in 10% bleach followed by 70% alcohol for at least 10 min each. Flies were rinsed in sterile water, air-dried, and homogenized in 500 ml of sterilized 1× phosphate buffered saline (Fisher). Homogenate was serially diluted and cultured on chloramphenicol (10 μg/ml) TSA agar, which was incubated at 37°C overnight for enumeration of CFU/fly. Cultured dilutions not within 30–300 CFU were not considered to be in the acceptable countable range and were excluded. Mean CFU recoveries from three combined replicates had normal distributions and were analyzed by one-way analysis of variance (ANOVA) with Tukey’s honestly significant difference (HSD) posttest for log transformed CFU counts (GraphPad Prism five for Mac OSX, GraphPad Software, LA Jolla, CA, www.graphpad.com).
Excreta Recovery
Following the same feeding protocol, house flies (n = 5 per replicate, for three replicates) were put in sterile jars and fed 2 μl droplets of S. aureus. After consumption of bacteria, flies were transferred to sterile 35 mm petri dishes (Fisher). At 2, 3, and 4 h PI, flies were chilled at 4°C for temporary removal, and the dishes were washed with 500 μl sterile 1× PBS to collect fecal and vomit specks. The same flies were transferred to new petri dishes for the next 1 h interval, after which time the washes were repeated. Each wash was serially diluted, plated on TSA with chloramphenicol (10 μg/ml), and incubated overnight at 37°C for CFU enumeration. At each time point, only flies with recoverable CFU were included in the analysis. CFU recoveries from excreta did not fit Gaussian distribution, so nonparametric Kruskal–Wal-lis test was performed with Dunn’s multiple comparison posttest being used to compare mean excreta CFU recoveries between time intervals.
Tissue-Specific Analyses of def Temporal Expression
Flies (n = 30 per replicate, with two parallel biological replicates) were individually housed in glass jars and fed 2 μl GFP-S. aureus. Tissues (i.e., proventriculus, midgut, crop, and fat body) were aseptically dissected from 10 flies at 2, 4, and 6 h PI and pooled by tissue for RNA extraction (Ribopure kit; Ambion, Austin, TX) and cDNA synthesis (Quantitect kit; Qiagen, Valencia, CA) following manufacturers’ protocols. Quantitative polymerase chain reaction (PCR) was performed with three technical replicates for each primer set on cDNA (quantitative reverse-transcriptase PCR, qRTPCR) using primers for two reference genes, tubulin and rps18, and def (Table 1), with the 5-Prime SYBR-ROX kit (Fisher) following manufacturer’s instructions with 250 nM of each primer and 1:40 diluted cDNA template (1 μl). PCR cycling was performed on an Eppendorf Realplex thermal cycler with the following parameters: 2 min at 95°C, 40 cycles of 59°C for 20 s, 68°C for 15 s, 95°C for 15 s, followed by 2 min at 68°C. Threshold cycles (CT) for each reaction were collected and analyzed using the Relative Expression Software Tool (REST; Pfaffl et al. 2002), which allows for group wise comparative statistical analyses of relative expression while accounting for primer-efficiency differences.
Table 1.
Primer sequences for qRTPCR
| Gene | Accession no. | Primer sequence |
|---|---|---|
| rps18 | ES608249.1 | Fwd: 5′-GTTGGTATTGCCATGACCGCCATT-3′ Rev: 5′-ATGGGTTGGAGATGATGGTGACGA-3′ |
| tubulin | ES608434.1 | Fwd: 5′-GCTTGCTGCATGTTGTATCGTGGT-3′ Rev: 5′-CGAATTGAATGGTGCGCTTGGTCT-3′ |
| defensin | DQ384634.1 | Fwd: 5′-CAATTTCGTCCATGGAGCTGATGC-3′ Rev: 5′-ACCGCTCAACAAATCGCAAGTAGC-3′ |
Results
Temporal and Spatial Fate of GFP-S. aureus in the Gut
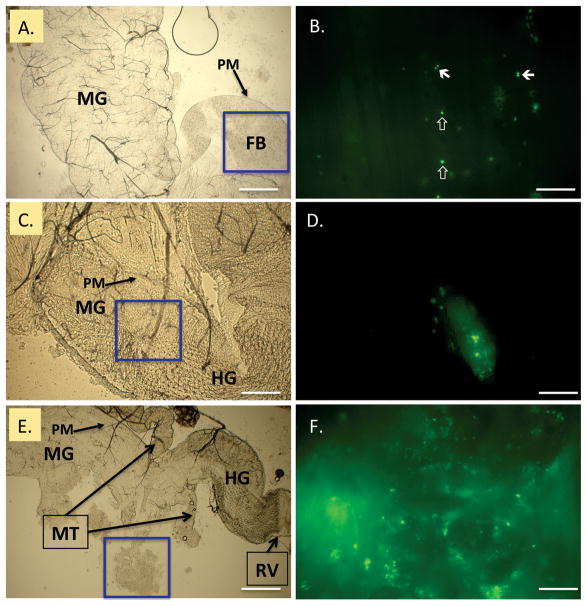
Epifluorescence microscopy was used to detect viable GFP-S. aureus in distinct regions of the digestive tract including the crop, midgut, hindgut, and rectum at 2 h intervals for 6 h PI. At 2 h PI, viable bacteria were visualized in the crop and midgut, and in the midgut, bacteria were restricted to the inner PM (Fig. 1A, B). At 2 h PI and throughout the observation period, cells appeared not as typical staphylococcal clusters but as single cocci or diplococci (Fig. 1B). At 4 h PI, noticeably fewer viable bacteria were observed expressing GFP, and clumps of remaining bacteria moved distally, presumably by peristalsis, toward the rectum. Clumps of bacteria cells were observed in the distal midgut (Fig. 1C, D) and hindgut/rectum (data not shown). Although bacteria were observed at 6 h PI, few viable cells were found in the crop. At 6 h PI, clumps of GFP-S. aureus mostly were seen in the hindgut (Fig. 1E, F) and rectum (not shown). Similar patterns of progression of bacteria in the gut of five flies examined at each time point were noted, and the only variability observed was a slight difference in the number of viable bacteria observed in the crop of flies at 2 or 4 h PI. All flies examined had viable GFP-S. aureus in the hindgut and rectum area as soon as 4 h PI.
Fig. 1.
GFP-expressing Staphylococcus aureus in the house fly alimentary canal. Flies were fed 2.0 × 105 CFU, and bacteria were examined in dissected alimentary canals at 2, 4, and 6 h postingestion (PI). Epifluorescent microscopy (right, B, D, F) of blue boxed regions in bright field panels (left, A, C, E) revealed bacteria throughout the alimentary canal. Bacteria mainly appeared as single coccic and diplococcic forms as soon as 2 h (B., closed and open arrows, respectively), and were contained within food boluses (FB) in the peritrophic matrix (PM) of the midgut. Viable bacteria were visualized in the distal midgut (MG) and hindgut (HG) at 4 h PI. At 6 h PI, clumps of lysed and viable bacteria were released from the terminal open end of the PM, near the midgut/hindgut junction (E, boxed, freed during preparation and magnified for detail in F). MT = Malpighian tubules; RV = rectal valve. Scale bars: A, C, E, 200 μm; B, D, F, 20 μm. (Online figure in color.)
Culture Recovery of GFP-S. aureus From Whole House Flies and Excreta
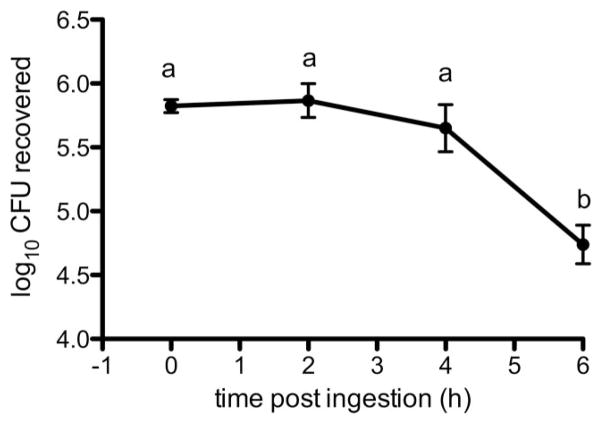
GFP-S. aureus was cultured from whole fly homogenate at all time points (Fig. 2), and a significant effect of time on bacterial recovery was observed (F(3,29) = 9.262; P = 0.0002). Between 0 and 2 h PI, bacteria recovery from whole flies did not change significantly, but interestingly, we recovered a mean of 7,908 CFU from fly excreta during the same time interval (12/15 flies; Fig. 3). Throughout the 6 h collection period, recoverable numbers of bacteria from homogenized flies decreased, but significant differences occurred between both 2 and 4 h PI and 6 h PI (mean CFU at 6 h = 8.0 × 104; Tukey’s HSD, P < 0.05; Fig. 2). Although 10/15 and 12/15 flies continued excreting bacteria at three and 4 h PI, respectively, CFU counts were nearly two orders of magnitude less than those at 2 h PI. Kruskal–Wallis analysis revealed a significant effect of time on mean CFU in excreta (df = 2; H = 10.02; P = 0.0067), and recoverable CFU of GFP-S. aureus in excreta at 4 h PI differed significantly from 2 h PI (Dunn’s test; P < 0.05). Of note, while all 15 flies excreted recoverable bacteria during at least one of the time intervals, the amount of bacteria excreted varied greatly. The fewest CFUs recovered were 10 cells at 2 h PI, which also was the only time this fly excreted bacteria. The largest total number of CFU recovered was 30,470 CFU from one fly over the entire collection period; this fly also had the largest single excreta recovery, which was 30,000 CFU at 2 h PI. Overall, the average CFU per fly excreta (±SD) was 6,646 ± 9,302, which further demonstrates the great fly-to-fly variability in recovery of bacteria from excreta.
Fig. 2.
Recovery of Staphylococcus aureus from whole fly homogenate. Flies were fed 6.75 ± 1.44 × 105 CFU bacteria, and mean CFUs recovered for three replicate experiments are shown (n = 15 flies per replicate). Mean recoveries of bacteria from whole fly homogenate are shown ± SE. Different letters represent statistical significance (Tukey’s; P < 0.05).
Fig. 3.
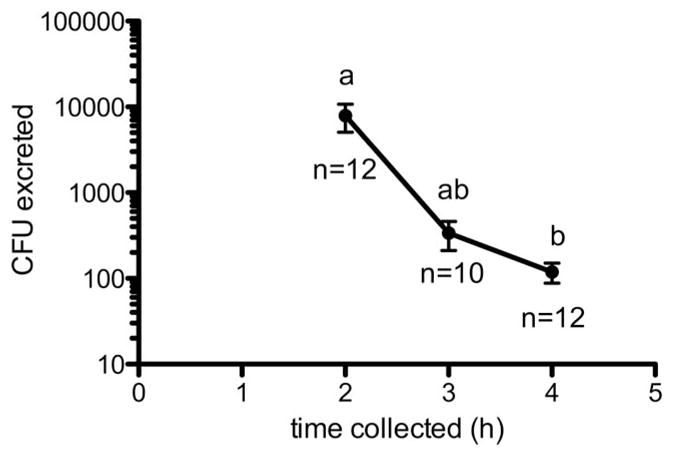
Recovery of Staphylococcus aureus from house fly excreta. Flies were fed a mean 3.2 ± 1.4 × 105 CFU bacteria in three separate experiments. Replicates were combined (n = 15 total flies). Mean recoveries of bacteria from excreta droplets (vomit and feces) are shown ± SE. Different letters above means represent statistical significance (Dunn’s test; P < 0.05). Numbers of flies that excreted at that time point are indicated below means.
Temporal and Spatial Expression of def in Flies Fed GFP-S. aureus
Tissue-specific qRTPCR analyses of flies fed GFP-S. aureus were analyzed separately, as replicates could not be combined because of inter-replicate variability. def was upregulated in all tissues examined at 2, 4, and 6 h postingestion, in comparison to teneral flies that served as calibrators (Fig. 4). REST analysis revealed that expression differed significantly from that of teneral flies in proventriculus, crop, and fat body at all time points in both replicates (P < 0.05). However, upregulation in the midgut differed from baseline only at 2 h PI in the first biological replicate while upregulation was observed at all time points in the second biological replicate (Fig. 4; P < 0.05). The range of def upregulation in tissues of flies fed GFP-S. aureus was between ≈180 to >16,000 times the teneral level (Fig. 4).
Fig. 4.
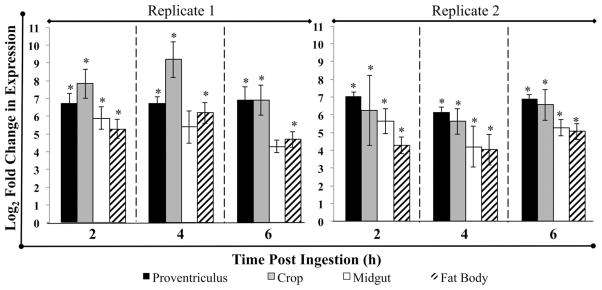
qRT-PCR analysis of tissue-specific expression of defensin in house flies fed GFP-Staphylococcus aureus. Fold changes in mRNA expression of defensin (def) were calculated using REST-MCS software and calibrated to expression levels in teneral adults and the reference genes rps18 and tubulin. Log2 fold change in expression levels is shown for each tissue, time and biological replicate, and error bars represent SE of technical replicates. Asterisks denote significant upregulation of def from baseline expression levels (P < 0.05).
Discussion
One goal of this study was to determine the fate of the pathogen S. aureus in the house fly using microscopy and culture recovery (from whole flies and excreta). These methods, along with the use of a GFP-expressing bacterial strain, allowed us to closely monitor the location and viability of bacteria over time. In addition, we concurrently assessed the expression of the antimicrobial peptide gene def both locally in the gut, proximal to bacteria, and systemically in the fat body. The combination of these experiments gave us a distinct spatial and temporal view of house fly-microbe interactions from the perspective of both organisms.
Spatial observation of bacteria in the house fly alimentary canal helped in determining critical time points to be included in subsequent culture-recovery experiments and provided insight into the mode by which flies could putatively excrete bacteria. Up to 2 h PI, viable cells of GFP-S. aureus were observed in the crop and midgut, primarily as diplococci and cocci. Thus, we suspect at early time points post ingestion, vomitus is the likely means of excretion of the large amounts of viable bacteria, similar to what has been demonstrated with other bacteria such as Aeromonas hydrophila (Chester) Stainer (McGaughey and Nayduch 2009) and Enterococcus faecalis (Andrewes and Horder) Schleifer (Doud and Zurek 2012). At later time points, bacteria more likely were dispersed in feces, because as soon as 4 h PI we observed bacteria in the hindgut and rectum. However, we observed lysis, clumping, and free GFP in several regions of the gut, which indicates that fly feces should contain fewer numbers of bacteria compared with vomitus at 2 h PI. While excreta were not collected at 6 h PI, our microscopical examinations indicated that the number of bacteria shed at this time point would continue to temporally decrease, because clumps of bacteria and GFP were observed in the hindgut/rectum (Fig. 1F).
Culture recovery of bacteria from whole flies and excreta over time provided information not only on the vectoring and dissemination of S. aureus by flies, but also the extent to which flies may serve as environmental reservoirs for this pathogen. At 2 h PI, bacteria recovery from whole flies was not significantly different than the number of CFU fed (Fig. 2), yet a mean of ≈8,000 CFU (with up to 30,000 CFU) bacteria were cultured from fly excreta during that same time interval (Fig. 3), which equates to 1–10% of the amount of bacteria fed. It is important to note that recovery of an apparently large number of CFU from excreta may be artifactual interpretation, because of the discrepancy in what comprises a “CFU” between fed cultures and recovered in vivo S. aureus. In liquid culture, S. aureus were in clusters of 2 to >12 cells (data not shown), but in vivo we observed only cocci and diplococci (Fig. 1). Thus, because this morphological inconsistency exists, all recoveries from whole fly homogenate and excreta could reflect an apparent, but not actual, increase in CFU. We can further infer that the differences between fed and recovered amounts (whole fly or excreta) is actually greater than what was measured. Future experiments could be designed to sonicate or vortex liquid cultures before feeding to flies, with the alternate caveat being that this type of pretreatment would not represent the morphology of S. aureus that flies encounter in natural conditions.
Def was upregulated both locally, proximal to bacteria (i.e., crop, proventriculus, and midgut), and systemically (e.g., fat body) in flies. Systemic def expression may be stimulated by diffusing dimers of lysine-type peptidoglycan (LYS-PGN, comprising cell wall of gram-positive cocci) or by primary recognition via receptors on gut epithelia followed by secondary communication with the fat body, mediated by signaling molecules (Hao et al. 2003, Gendrin et al. 2009). In D. melanogaster gut AMP responses are entirely mediated through DAP-PGN recognition by trans-membrane PGRPs and the Imd pathway (Lhocine et al. 2008, Leulier and Royet 2009). How the house fly epithelial cells recognize LYS-PGN remains unknown. Because Defensin is classically associated with the Toll pathway systemically, where LYS-PGN would act upstream of Toll and not directly bind transmembrane PGRPs on the fat body (Wang et al. 2006), it would be interesting to determine if gut epithelial PGRPs have different affinity for PGN, or perhaps if cross activation of Toll and Imd occurs in the house fly gut as seen systemically in D. melanogaster (Tanji and Ip 2005, Tanji et al. 2007). Nonetheless, this is the first demonstration, to our knowledge, of epithelial def expression in the alimentary canal of cyclorrhaphous Diptera in response to feeding LYS-PGN bacteria. Def transcripts have been detected locally in the alimentary canal of other flies. For example, blood feeding alone stimulates def upregulation in the stable fly Stomoxys calcitrans L. (Munks et al. 2001), and DAP-PGN gram-negative bacteria like E. coli or even protozoan parasites induce def expression in the gut, proventriculus, and fat body of Glossina morsitans Westwood (Hao et al. 2001, 2003). Ongoing studies in our laboratory have determined that Defensin is upregulated in the gut of the house fly on both mRNA and protein levels in response to ingestion of both DAP-PGN and LYS-PGN expressing bacteria and protozoan parasites, but not in teneral flies that contain low levels of bacteria (<1,000 CFU) carried over from pupariation (D.N. et al., unpublished data). We have not yet determined whether all paralogs of def or, alternatively, transcript variants, were detected in our qRTPCR assays, but future work is aimed at a house fly transcriptome that should help elucidate the regulation of def transcription.
House fly Defensin may be one of the primary effectors contributing to observed GFP-S. aureus lysis and loss in the house fly gut because MRSA lysis by Defensin from other filth flies has been demonstrated (Andersen et al. 2010). Bacteria lysis in vivo likely results from the joint action of several effectors (e.g., other AMPs), digestive enzymes, and pH changes in concert with mechanical removal via peristalsis (Lehane and Billingsley 1996). Nonetheless, survival of bacteria in the crop or rectum, for oral or fecal excretion, substantiates house fly vector potential for this organism. The mean CFU excreted from flies in our study was >6,000, which is within the clinically significant range. For example, 103 CFU of S. aureus can establish infection in 100% of guinea pig animal models (Vaudaux et al. 2002) and 3 × 102 CFU was found to be the minimal infectious dose for mice to establish long-term infection (John et al. 2011). Further, this is a sufficient inoculum for human or animal foods that could lead to enterotoxic food borne illness. The Food and Drug Administration (FDA 2012) has determined that only 105 CFU S. aureus per gram of food produces enough toxin for gastrointestinal illness (Adams and Moss 2008). Considering the generation time can be as short as ≈30 min in optimal conditions (Okuma et al. 2002) and the average inoculum deposited by flies in our study, generation of sufficient endotoxin could occur within 2 h after fly excreta contamination of food items. In summary, although flies only harbor S. aureus for a short period of time, and lysis of bacteria may occur via AMPs like Defensin and other processes in the gut, vector and dissemination potential for this pathogen is possible, albeit transiently.
Acknowledgments
We thank C. Evett for assistance with qPCR optimization and tissue collection, and M. Lee for reviewing this manuscript. This work was supported by R15 Academic Research Enhancement Award (AREA) 1R15AI084029-01 from the National Institutes of Health awarded to D.N. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
References Cited
- Adams MR, Moss MO. Food microbiology. 3. RSC Publishing; Cambridge, United Kingdom: 2008. [Google Scholar]
- Andersen AS, Sandvang D, Schnorr KM, Kruse T, Neve S, Joergensen B, Karlsmark T, Krogfelt KA. A novel approach to the antimicrobial activity of maggot debridement therapy. J Antimicrob Chemother. 2010;65:1646–1654. doi: 10.1093/jac/dkq165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Zakour NL, Guinane CM, Fitzgerald JR. Pathogenomics of the staphylococci: insights into niche adaptation and the emergence of new virulent strains. FEMS Microbiol Lett. 2008;289:1–12. doi: 10.1111/j.1574-6968.2008.01384.x. [DOI] [PubMed] [Google Scholar]
- Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe. 2009;5:200–211. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Cerovsky V, Zdarek J, Fucik V, Monincova L, Voburka Z, Bem R. Lucifensin, the long-sought antimicrobial factor of medicinal maggots of the blowfly Lucilia sericata. Cell Mol Life Sci. 2010;67:455–466. doi: 10.1007/s00018-009-0194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B, Royet J. Drosophila immune response: from systemic antimicrobial peptide production in fat body cells to local defense in the intestinal tract. Fly (Austin) 2010;4:40–47. doi: 10.4161/fly.4.1.10810. [DOI] [PubMed] [Google Scholar]
- Cohen D, Green M, Block C, Slepon R, Ambar R, Wasserman SS, Levine MM. Reduction of transmission of shigellosis by control of houseflies (Musca domestica) Lancet. 1991;337:993–997. doi: 10.1016/0140-6736(91)92657-n. [DOI] [PubMed] [Google Scholar]
- Dang XL, Wang YS, Huang YD, Yu XQ, Zhang WQ. Purification and characterization of an antimicrobial peptide, insect defensin, from immunized house fly (Diptera: Muscidae) J Med Entomol. 2010;47:1141–1145. doi: 10.1603/ME10016. [DOI] [PubMed] [Google Scholar]
- Doud CW, Zurek L. Enterococcus faecalis OG1RF:pMV158 survives and proliferates in the house fly digestive tract. J Med Entomol. 2012;49:150–155. doi: 10.1603/me11167. [DOI] [PubMed] [Google Scholar]
- (FDA) Food and Drug Administration. Bad bug book: introduction to foodborne pathogenic microorganisms and natural toxins. 2. Center for Food Safety and Applied Nutrition, FDA; Silver Spring, MD: 2012. [Google Scholar]
- Fitzgerald JR. Livestock-associated Staphylococcus aureus: origin, evolution and public health threat. Trends Microbiol. 2012;20:192–198. doi: 10.1016/j.tim.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Fotedar R, Banerjee U, Samantray JC, Shriniwas Vector potential of hospital houseflies with special reference to Klebsiella species. Epidemiol Infect. 1992a;109:143–147. [PMC free article] [PubMed] [Google Scholar]
- Fotedar R, Banerjee U, Singh Shriniwas S, Verma AK. The housefly (Musca domestica) as a carrier of pathogenic microorganisms in a hospital environment. J Hosp Infect. 1992b;20:209–215. doi: 10.1016/0195-6701(92)90089-5. [DOI] [PubMed] [Google Scholar]
- Gendrin M, Welchman DP, Poidevin M, Herve M, Lemaitre B. Long-range activation of systemic immunity through peptidoglycan diffusion in Drosophila. PLoS Pathog. 2009;5:e1000694. doi: 10.1371/journal.ppat.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graczyk TK, Knight R, Gilman RH, Cranfield MR. The role of non-biting flies in the epidemiology of human infectious diseases. Microbes Infect. 2001;3:231–235. doi: 10.1016/s1286-4579(01)01371-5. [DOI] [PubMed] [Google Scholar]
- Gupta AK, Nayduch D, Verma P, Shah B, Ghate HV, Patole MS, Shouche YS. Phylogenetic characterization of bacteria in the gut of house flies (Musca domestica L.) FEMS Microbiol Ecol. 2012;79:581–593. doi: 10.1111/j.1574-6941.2011.01248.x. [DOI] [PubMed] [Google Scholar]
- Hao Z, Kasumba I, Aksoy S. Proventriculus (cardia) plays a crucial role in immunity in tsetse fly (Diptera: Glossinidiae) Insect Biochem Mol Biol. 2003;33:1155–1164. doi: 10.1016/j.ibmb.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Hao Z, Kasumba I, Lehane MJ, Gibson WC, Kwon J, Aksoy S. Tsetse immune responses and trypanosome transmission: implications for the development of tsetse-based strategies to reduce trypanosomiasis. Proc Natl Acad Sci USA. 2001;98:12648–12653. doi: 10.1073/pnas.221363798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JA, Hetru C. Insect defensins: inducible antibacterial peptides. Immunol Today. 1992;13:411–415. doi: 10.1016/0167-5699(92)90092-L. [DOI] [PubMed] [Google Scholar]
- Holt PS, Geden CJ, Moore RW, Gast RK. Isolation of Salmonella enterica serovar Enteritidis from houseflies (Musca domestica) found in rooms containing Salmonella serovar Enteritidis-challenged hens. Appl Environ Microbiol. 2007;73:6030–6035. doi: 10.1128/AEM.00803-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John AK, Schmaler M, Khanna N, Landmann R. Reversible daptomycin tolerance of adherent staphylococci in an implant infection model. Antimicrob Agents Chemother. 2011;55:3510–3516. doi: 10.1128/AAC.00172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinde H, Castellan DM, Kerr D, Campbell J, Breitmeyer R, Ardans A. Longitudinal monitoring of two commercial layer flocks and their environments for Salmonella enterica serovar enteritidis and other Salmonellae. Avian Dis. 2005;49:189–194. doi: 10.1637/7228-062704R. [DOI] [PubMed] [Google Scholar]
- Lambert J, Keppi E, Dimarcq JL, Wicker C, Reichhart JM, Dunbar B, Lepage P, Van Dorsselaer A, Hoffmann J, Fothergill J, et al. Insect immunity: isolation from immune blood of the dipteran Phormia terranovae of two insect antibacterial peptides with sequence homology to rabbit lung macrophage bactericidal peptides. Proc Natl Acad Sci USA. 1989;86:262–266. doi: 10.1073/pnas.86.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehane MJ, Billingsley PF. Biology of the insect midgut. Chapman & Hall; London, United Kingdom: 1996. [Google Scholar]
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Leulier F, Royet J. Maintaining immune homeostasis in fly gut. Nat Immunol. 2009;10:936–938. doi: 10.1038/ni0909-936. [DOI] [PubMed] [Google Scholar]
- Lhocine N, Ribeiro PS, Buchon N, Wepf A, Wilson R, Tenev T, Lemaitre B, Gstaiger M, Meier P, Leulier F. PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe. 2008;4:147–158. doi: 10.1016/j.chom.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Liehl P, Blight M, Vodovar N, Boccard F, Lemaitre B. Prevalence of local immune response against oral infection in a Drosophila/Pseudomonas infection model. PLoS Pathog. 2006;2:e56. doi: 10.1371/journal.ppat.0020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaughey J, Nayduch D. Temporal and spatial fate of GFP-expressing motile and nonmotile Aeromonas hydrophila in the house fly digestive tract. J Med Entomol. 2009;46:123–130. doi: 10.1603/033.046.0116. [DOI] [PubMed] [Google Scholar]
- Moriya K, Fujibayashi T, Yoshihara T, Matsuda A, Sumi N, Umezaki N, Kurahashi H, Agui N, Wada A, Watanabe H. Verotoxin-producing Escherichia coli O157:H7 carried by the housefly in Japan. Med Vet Entomol. 1999;13:214–216. doi: 10.1046/j.1365-2915.1999.00161.x. [DOI] [PubMed] [Google Scholar]
- Munks RJ, Hamilton JV, Lehane SM, Lehane MJ. Regulation of midgut defensin production in the blood-sucking insect Stomoxys calcitrans. Insect Mol Biol. 2001;10:561–571. doi: 10.1046/j.0962-1075.2001.00296.x. [DOI] [PubMed] [Google Scholar]
- Okuma K, Iwakawa K, Turnidge JD, Grubb WB, Bell JM, O’Brien FG, Coombs GW, Pearman JW, Tenover FC, Kapi M, et al. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J Clin Microbiol. 2002;40:4289–4294. doi: 10.1128/JCM.40.11.4289-4294.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AG, Richards PA. The peritrophic membranes of insects. Annu Rev Entomol. 1977;22:219–240. doi: 10.1146/annurev.en.22.010177.001251. [DOI] [PubMed] [Google Scholar]
- Rosef O, Kapperud G. House flies (Musca domestica) as possible vectors of Campylobacter fetus subsp. jejuni. Appl Environ Microbiol. 1983;45:381–383. doi: 10.1128/aem.45.2.381-383.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu JH, Ha EM, Oh CT, Seol JH, Brey PT, Jin I, Lee DG, Kim J, Lee D, Lee WJ. An essential complementary role of NF-κB pathway to microbicidal oxidants in Drosophila gut immunity. EMBO J. 2006;25:3693–3701. doi: 10.1038/sj.emboj.7601233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenbak CR, Ryu JH, Leulier F, Pili–Floury S, Parquet C, Herve M, Chaput C, Boneca IG, Lee WJ, Lemaitre B, Mengin–Lecreulx D. Peptidogly-can molecular requirements allowing detection by the Drosophila immune deficiency pathway. J Immunol. 2004;173:7339–7348. doi: 10.4049/jimmunol.173.12.7339. [DOI] [PubMed] [Google Scholar]
- Szalanski AL, Owens CB, McKay T, Steelman CD. Detection of Campylobacter and Escherichia coli O157:H7 from filth flies by polymerase chain reaction. Med Vet Entomol. 2004;18:241–246. doi: 10.1111/j.0269-283X.2004.00502.x. [DOI] [PubMed] [Google Scholar]
- Tanji T, Ip YT. Regulators of the Toll and Imd pathways in the Drosophila innate immune response. Trends Immunol. 2005;26:193–198. doi: 10.1016/j.it.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Tanji T, Hu X, Weber AN, Ip YT. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol Cell Biol. 2007;27:4578–4588. doi: 10.1128/MCB.01814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaudaux P, Francois P, Bisognano C, Schrenzel J, Lew DP. Comparison of levofloxacin, alatrofloxacin, and vancomycin for prophylaxis and treatment of experimental foreign-body-associated infection by methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:1503–1509. doi: 10.1128/AAC.46.5.1503-1509.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodovar N, Vinals M, Liehl P, Basset A, Degrouard J, Spellman P, Boccard F, Lemaitre B. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc Natl Acad Sci USA. 2005;102:11414–11419. doi: 10.1073/pnas.0502240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Weber AN, Atilano ML, Filipe SR, Gay NJ, Ligoxygakis P. Sensing of gram-positive bacteria in Drosophila: GNBP1 is needed to process and present peptidoglycan to PGRP-SA. EMBO J. 2006;25:5005–5014. doi: 10.1038/sj.emboj.7601363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West LS. The housefly: its natural history, medical importance, and control. Comstock Publ. Co; Ithaca, NY: 1951. [Google Scholar]
- Yamada K, Natori S. Purification, sequence and antibacterial activity of two novel sapecin homologues from Sarcophaga embryonic cells: similarity of sapecin B to charybdotoxin. Biochem J. 1993;291(Pt. 1):275–279. doi: 10.1042/bj2910275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Kalpana M, Lee HL. Wings of the common house fly (Musca domestica L.): importance in mechanical transmission of Vibrio cholerae. Trop Biomed. 2008;25:1–8. [PubMed] [Google Scholar]
- Zurek L, Schal C, Watson DW. Diversity and contribution of the intestinal bacterial community to the development of Musca domestica (Diptera: Muscidae) larvae. J Med Entomol. 2000;37:924–928. doi: 10.1603/0022-2585-37.6.924. [DOI] [PubMed] [Google Scholar]