Abstract
Background
Targeted liposome-polycation-DNA complex (LPD), mainly conjugated with antibodies using functionalized PEG derivatives, is an effective nanovector for systemic delivery of small interference RNA (siRNA). However, there are few studies reporting the effect of different conjugation linkers on LPD for gene silencing. To clarify the influence of antibody conjugation linkers on LPD, we prepared two different immunoliposomes to deliver siRNA in which DSPE-PEG-COOH and DSPE-PEG-MAL, the commonly used PEG derivative linkers, were used to conjugate anti-EGFR Fab’ with the liposome.
Methods
First, 600 μg of anti-EGFR Fab’ was conjugated with 28.35 μL of a micelle solution containing DSPE-PEG-MAL or DSPE-PEG-COOH, and then post inserted into the prepared LPD. Various liposome parameters, including particle size, zeta potential, stability, and encapsulation efficiency were evaluated, and the targeting ability and gene silencing activity of TLPD-FPC (DSPE-PEG-COOH conjugated with Fab’) was compared with that of TLPD-FPM (DSPE-PEG-MAL conjugated with Fab’) in SMMC-7721 hepatocellular carcinoma cells.
Results
There was no significant difference in particle size between the two TLPDs, but the zeta potential was significantly different. Further, although there was no significant difference in siRNA encapsulation efficiency, cell viability, or serum stability between TLPD-FPM and TLPD-FPC, cellular uptake of TLPD-FPM was significantly greater than that of TLPD-FPC in EGFR-overexpressing SMMC-7721 cells. The luciferase gene silencing efficiency of TLPD-FPM was approximately three-fold high than that of TLPD-FPC.
Conclusion
Different conjugation linkers whereby antibodies are conjugated with LPD can affect the physicochemical properties of LPD and antibody conjugation efficiency, thus directly affecting the gene silencing effect of TLPD. Immunoliposomes prepared by DSPE-PEG-MAL conjugation with anti-EGFR Fab’ are more effective than TLPD containing DSPE-PEG-COOH in targeting hepatocellular carcinoma cells for siRNA delivery.
Keywords: liposome-polycation-DNA, anti-EGFR Fab’, immunoliposomes, small interfering RNA delivery, conjugation technology, hepatocellular carcinoma
Introduction
In the late 1990s, the discovery of RNA interference by Fire et al1 opened up an entirely new feld of “gene” therapy. Since then, RNA interference has been demonstrated by clinical trials as a potential therapy for different kinds of human diseases.3–5 These small interfering RNA (siRNA), 21–23 nucleotides in length, result in silencing of the corresponding gene by downregulation of the complementary messenger RNA for the selected specific sequence.6–8 However, it is a challenging task to systemically deliver siRNAs into target cells, because siRNAs themselves are unstable in the bloodstream and cannot penetrate cellular membranes.9 To overcome these obstacles, a number of delivery systems have been developed, including lipid-based systems, polymer-based systems, peptide conjugates, and single-chain fragment variable antibody fusion protein systems.10–15
Among the various systems studied, the lipid-based delivery system has received more attention as a promising carrier for siRNA delivery owing to its simplicity of production, relatively low immunogenicity, and absence of oncogenicity.16–19 The liposome-polycation-DNA complex (LPD) system, prepared by condensing the siRNA and DNA with protamine into a compact complex followed by coating with cationic liposomes, is an effective lipidic nanovector for systemic siRNA delivery, and was developed in 1996 by Lee and Huang.2 To overcome the kinetic and physical barriers to systemic siRNA delivery, it is necessary to develop PEGylated immunoliposomes conjugated with targeting ligands.20,21 Huang et al22,23 developed an anisamide-targeted LPD system and used it successfully to deliver siRNA to tumor cells overexpressing the sigma receptor. To extend further the application of this LPD system, we adopted the anti-epidermal growth factor receptor(EGFR) Fab’ as the targeting ligand for LPD, knowing that it has higher specificity and affinity than the small molecule ligand, and is thus able to provide flexible conjugation sites for crosslinking of nanoparticles. LPD modified with anti-EGFR Fab’ has been demonstrated to possess potent gene silencing activity in EGFR overexpressing breast cancer and hepatocellular carcinoma.24,25 In our previous study, we investigated the effect of antibody type and amount on the essential physicochemical properties and targeting efficiency of targeted LPD conjugated with anti-EGFR antibody (TLPD), and obtained an optimized siRNA delivery system. However, it remains unclear whether the antibody conjugation linker via which the anti-EGFR Fab’ is conjugated to LPD would also affect the physicochemical properties and targeting ability of LPD and further influence the gene delivery effect of LPD. Few studies have demonstrated whether different conjugation linkers would affect the essential physicochemical properties and targeting efficiency of immunoliposomes in siRNA delivery.
Since the procedures for producing highly specific monoclonal antibodies became well established, a number of methods have been reported for coupling antibodies to the surface of stealth liposomes. To obtain specific targeting liposomes with the ability to escape the reticuloendothelial system, functionalized poly(ethylene glycol) PEG derivatives have been successfully employed in the preparation of immunoliposomes. DSPE-PEG-MAL (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethyleneglycol)-2000] (ammonium salt)) and DSPE-PEG-COOH (distearoyl-N-(3-carboxypropionoyl poly(ethylene glycol) succinyl)phosphatidylethanolamine) have different characteristics and are commonly used as functionalized PEG derivatives to conjugate liposomes with antibodies. For DSPE-PEG-MAL, antibody molecules are thiolated first by 2-iminothiolane (Traut’s reagent) to generate a sulfhydryl group and then reacted with maleimide groups on PEG termini. However, antibody molecules thiolated using Traut’s reagent can lose substantial amounts of available thiol to recyclize in hours by attacking the amidine carbon. Therefore, once thiolated, the product should be used immediately to avoid significant loss of activity.26 For DSPE-PEG-COOH, antibody molecules are reacted with a carboxyl functional group on PEG termini, so that the antibody does not undergo prior ligand modification, thus reducing the risk of denaturation and loss of its specific activity. Maruyama et al27 used DSPE-PEG-COOH conjugation with a monoclonal immunoglobulin G antibody to prepare the immunoliposome, which had been demonstrated to bind effectively to the designated target site in vivo.
Because LPD is a complex colloidal delivery system, physicochemical characteristics, including particle size and surface charge, play an important role in determining the final destination of the complex in vivo. Different PEG linkers through which the antibody is conjugated to the liposome may lead to different antibody conjugation efficiency and colloidal properties, which finally can result in different gene silencing effects. Although there are different features and extensive applications in antibody conjugation of the two linkers, there are few studies reporting which immunoliposome prepared by the two functionalized PEG linkers is more efficient in silencing siRNA expression.
To determine which PEG linker is more efficient in delivering siRNA, we prepared two immunoliposomes modified with two functionalized PEG derivatives (DSPE-PEG-COOH or DSPE-PEG-MAL) conjugated with anti-EGFR Fab’ via a post-insertion approach. SMMC-7721 hepatocellular carcinoma cells in which EGFR is overexpressed were chosen as the model cell line to determine the specificity of the delivery system. To assess the effect of the two antibody conjugation technologies on siRNA delivery, the essential physicochemical properties and targeting efficiency of the two immunoliposomes were compared in vitro.
Materials and methods
Materials
Cholesterol and all lipids were obtained from Avanti Polar Lipids (Alabaster, AL, USA). 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), N-hydroxysuccinimide (NHS), 2-(N-morpholino)ethanesulfonic acid (MES), calf thymus DNA (for hybridization), and protamine sulfate salt (Grade X from salmon) were from Sigma-Aldrich (St Louis, MO, USA). Traut’s reagent was from Pierce (Oud Beijerland, The Netherlands). The anti-EGFR Fab’ was prepared as we have described previously28 and supplied by the National Engineering Research Center for Antibody Medicine (Shanghai, People’s Republic of China).
Anti-luciferase siRNA (antisense, 5′-UCGAAGUACU-CAGCGUAAGTT-3′; sense, 5′-CUUACGCUGAGU-ACUUCGATT-3′), negative control siRNA (NC), anti-RhoA siRNA, and FAM-labeled NC siRNA (FAM-siRNA) were synthesized by GenePharma Co, Ltd (Shanghai, People’s Republic of China). Cy5-labeled NC siRNA (Cy5-siRNA) was synthesized by Ribobio Co, Ltd (Guangzhou, People’s Republic of China). The siRNAs were delivered in lyophilized form, and upon delivery, were diluted to obtain a 20 μM solution using RNAse-free water provided by GenePharma Co, Ltd.
Cell culture
SMMC-7721 cells (a human hepatocellular carcinoma cell line) were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and stably transfected with luciferase gene using a pcDNA3.1 vector. The cells were grown as recommended by ATCC and maintained in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum, 25 mM HEPES buffer, 100 U/mL penicillin, and 100 mg/mL streptomycin in a humidified atmosphere of 5% CO2 at 37°C.
Preparation of liposomes containing siRNa
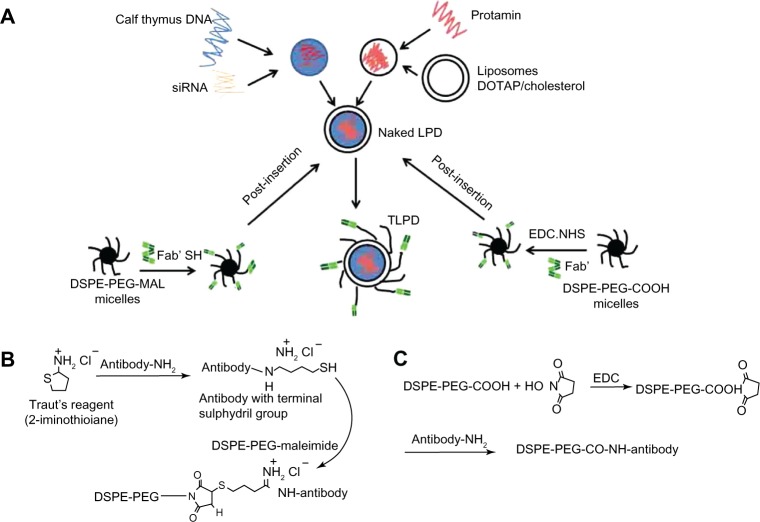
Naked LPDs were prepared as previously described.25 Briefly, cationic liposomes composed of DOTAP and cholesterol (1:1 molar ratio, 10 mM) were prepared using the thin film hydration method and 0.1% (molar ratio) carboxyfluorescein-PE was added in the lipid film to form fluorescent liposomes. Next, 124 μL of cationic liposomes, 15 μL of protamine 2 mg/mL, and 11 μL of diethylpyrocarbonate water were mixed as solution A, and 90 μL of siRNA (24 μg, 20 μM), 2.4 μL of calf thymus DNA (24 μg, 10 mg/mL), and 57.6 μL of diethylpyrocarbonate water were mixed as solution B. Solutions A and B were then mixed quickly to form naked LPD. For DSPE-PEG-MAL conjugated with Fab’, 600 μg of thiolated anti-EGFR Fab’ and a micelle solution containing DSPE-PEG-MAL (10 mg/mL, 28.35 μL) were first incubated at 4°C overnight. For DSPE-PEG-COOH conjugated with Fab’, 67.5 μL of NHS 0.25 M and 67.5 μL of EDC 0.25M were first mixed with 28.4 μL of micelle solution containing DSPE-PEG-COOH 10 mg/mL, dissolved in MES buffer (10 mM MES/150 mM NaCl, pH 5.5), and kept at room temperature for 15 minutes. The mixture was then neutralized with 1 M NaOH to pH 7.5 and incubated with the desired amount of anti-EGFR Fab’ (600 μg) at 4°C with gentle stirring overnight. The following day, the naked LPD was mixed at 50°C for 10 minutes with the antibody-conjugated micelles to form TLPD-FPM or TLPD-FPC (Figure 1A).
Figure 1.
(A) Procedures involved in developing naked LPD and TLPD. (B) Thiolation of antibodies using Traut’s reagent and conjugation of thiolated antibody to maleimide groups on the DSPE-PEG-MAL. (C) Conjugation of DSPE-PEG-COOH with antibody.
Abbreviations: LPD, liposome-polycation-DNA complex; TLPD, targeted liposome-polycation-DNA; DSPE-PEG-MAL, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (ammonium salt); DSPE-PEG-COOH, distearoyl-N-(3-carboxypropionoyl poly(ethylene glycol) succinyl)phosphatidylethanolamine; EDC, 1-ethyl-3-(3-dimethylamino-propyl)carbodiimide; DOTAP, 1,2-dioleoyl-3-trimethylammoniumpropane; NHS, N-hydroxy succinimide; DNA, deoxyribonucleic acid; DSPE-PEG, distearoyl-N-(monomethoxy poly(ethylene glycol) succinyl)phosphatidylethanolamine; Fab’, antigen-binding fragment; SH, hydrosulfide group; NH-, imido-group.
Nontargeted control LPD (NTLPD) was prepared by coupling bovine serum albumin instead of anti-EGFR Fab’ to the liposomes using the same method as for TLPD. The following designations are used: TLPD-FPM (TLPD conjugated with anti-EGFR Fab’ by the post-insertion approach when the linker PEG derivative was DSPE-PEG-MAL) and TLPD-FPC (TLPD conjugated with anti-EGFR Fab’ by the post-insertion approach when the linker PEG derivative was DSPE-PEG-COOH).
Particle size and zeta potential
The mean particle size and zeta potential of the liposomes were determined using the Zetasizer Nano series (Malvern Instruments, Malvern, UK). Prior to measurement, the liposomes were diluted in double-distilled water and measurement was carried out at 25°C. Each sample was measured three times and the mean value was calculated.
Transmission electron microscopy
Transmission electron microscopy was performed on a Hitachi H-600 instrument (Tokyo, Japan) at an acceleration voltage of 75 k V. Briefly, freshly prepared formulations were dropped onto a copper grid coated with a carbon membrane and a short incubation was allowed at room temperature. The grid was then tapped with filter paper to remove the aqueous solution on the surface and air-dried. Negative staining was performed by addition of a drop of 2% phosphotungstic acid to the copper grid with the sample.
siRNA encapsulation efficiency
An Amicon® Ultra-4 centrifugal filter device (100,000 nominal molecular weight limit, Millipore, Bedford, MA, USA) was used to determine the encapsulation efficiency of siRNA by ultrafiltration of the liposomes entrapping FAM-siRNA, as described elsewhere.24,25 siRNA encapsulation efficiency was estimated from the following equation: (Qo − Qn)/Qo × 100%, where Qo and Qn represent the quantity of siRNA originally added and the quantity of unencapsulated siRNA, respectively.
Gel retardation assay
A gel retardation assay was used to assess the binding affinity of siRNA in liposomes. The liposomes were first solubilized in 5% Triton-X100, then added to independent wells of 2% agarose gel, and finally electrophoresed and visualized with Goldenview™ dye staining. At the same time, untreated liposomes were used as the negative control.
siRNa serum stability
The samples of siRNA solution and the siRNA loaded liposomes were both diluted to the concentration of siRNA about 0.04 μg/μL with diethypyrocarbonate water, and then to 0.02 μg/μL with fetal bovine serum and incubated at 37°C. At different times, after the liposomes were destroyed by 5% Triton-X100, aliquots containing 0.3 μg siRNA of each sample were loaded onto agarose gel for electrophoresis.
In vitro cell viability study
Cell viability was detected using a Cell Counting Kit-8 (CCK-8, Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s protocol. Briefly, SMMC-7721 cells were seeded one day prior to the experiment in a 96-well plate at a density of 3 × 104 cells/well in complete Roswell Park Memorial Institute medium containing 10% fetal bovine serum. Following overnight culture, the cells were treated with the liposomes (the final siRNA concentration was 100 nM or 200 nM) in serum containing medium at 37°C. After a 24-hour incubation period the cells were washed once with phosphate-buffered saline and the medium was exchanged for fresh culture medium. After a total incubation time of 48 hours, 10 μL of CCK-8 solution was added to each well and incubated for 2 hours at 37°C and absorbance was measured using a microplate reader (Bio-Tek, Winooski, VT, USA) at 450 nm/630 nm. Cell viability in each group was calculated as follows: (AE − AB)/(AU − AB) × 100%, where AE, AU, and AB represent the absorbance of experimental samples, untreated samples, and blank controls, respectively.
Confocal microscopic study
Cellular uptake experiments were performed to compare the effect of receptor-mediated endocytosis exerted by anti-EGFR Fab’-conjugated immunoliposomes with two different linkers. Briefly, SMMC-7721 cells were seeded on 22 × 22 mm coverslips at a density of 2.5 × 105/well and treated with 0.1% carboxyfluorescein-PE-labeled liposomes dispersed in serum containing medium, at 37°C for 2 hours, in which the concentration of Cy5 labeled siRNA was 100 nM. The cells were fixed with 4% paraformaldehyde after washing with phosphate-buffered saline twice and incubated with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI, Sigma, Fluka Chemie, Buchs, Israel) for nuclei staining. Immunofluorescence was then observed using a TCS SP5 confocal microscope (Leica Micro-systems, Exton, PA, USA).
In vitro siRNa transfection study
To study the transfection efficiency of the anti-EGFR Fab’-conjugated immunoliposome in vitro, SMMC-7721 cells (6 × 104 per well) were seeded in 48-well plates overnight. The cells were treated with fluorescently labeled liposomes loaded with siRNA (FAM-siRNA, at a concentration of 250 nM or 500 nM) for 24 hours, trypsinized and washed by centrifugation (5 minutes, 1,200× g) in ice-cold phosphate-buffered saline supplemented with 1% fetal bovine serum. The resulting cell pellet was resuspended in ice-cold phosphate-buffered saline supplemented with 1% fetal bovine serum. Fluorescence analysis of a minimum of 10,000 events per sample was performed using a FACScanto fow cytometer (Becton Dickinson, San Jose, CA, USA).
In vitro study of siRNA-mediated gene silencing
To compare the in vitro gene silencing effect of TLPD-FPM with that of TLPD-FPC, SMMC-7721 cells with stable luciferase expression were seeded one day prior to the experiment in 48-well plates. The cells were treated with different concentrations of liposomes containing anti-luciferase or NC siRNA at 37°C for 24 hours. The transfection medium was then changed to fresh culture medium. Following a 48-hour incubation period the cells were washed with phosphate-buffered saline and lysed by incubation in 100 μL of cell lysis buffer at room temperature for 20 minutes. After mixing 5 μL of lysate with 25 μL of substrate from a luciferase assay system kit (Promega, Madison, WI, USA), the luminescence was measured using a 96 microplate luminometer (Glomax™, Promega). The protein concentration in the lysate was determined using a bicinchoninic acid assay (Beyotime Biotechnology, Haimen, People’s Republic of China). Luciferase activity was calculated according to the following equation: (Lis/Cs)/(LiN/CN) × 100%, where Lis and Cs represent luminescence intensity and the protein concentration of the sample, respectively, and LiN and CN represent the luminescence intensity and protein concentration of the untreated control.
In vitro study of RhoA silencing on cell migration
A Boyden chamber assay was employed to evaluate RhoA silencing on migration of SMMC-7721 cells. Briefly, cells (1.25 × 105 cells/well) were seeded overnight in 24-well plates. The cells were then treated with anti-RhoA or NC siRNA-loaded liposomes at a final siRNA concentration of 500 nM and grown for 24 hours. After removal of these complexes, the cells were cultured for an additional 48 hours. Total RNA was extracted from the cells and cDNA was reverse-transcribed from RNA. RhoA silencing was measured by real-time polymerase chain reaction using a 7,500 sequence detection system (Applied Biosystems, Foster City, CA, USA). In addition, SMMC-7721 cells at a density of 1 × 105 were trypsinized and seeded into the upper 8.0 μm pore size membrane inserts in the tissue culture plates. Culture medium was placed in the bottom wells. Following a 48-hour incubation period, the cells were fixed and stained with 1% crystal violet solution. Cells that migrated through the filter were counted in five random fields, and the mean number was calculated.
Statistical analysis
The statistical significance of the data was evaluated using the Student’s t-test. A P-value ≤ 0.05 was considered to be statistically significant.
Results and discussion
Preparation of Pegylated immunoliposomes
The degree of PEGylation is key for efficient gene delivery using PEGylated LPD nanoparticles. On the one hand, PEGylation can protect LPD against nonspecific uptake by the reticuloendothelial system, thus ensuring circulating LPD nanoparticles have enough time to reach the target tissue via the enhanced permeation and retention effect. On the other hand, PEGylation represents a major barrier for endosomal escape.29 In our previous study, the degree of PEGylation of anti-EGFR-Fab’-conjugated LPD was optimized as 7.5 mol%, which was demonstrated to have targeting ability and gene silencing activity in hepatocellular carcinoma cells.24 In the present study, the prepared formulations of TLPD contained 7.5 mol% surface-grafted PEG-phospholipid conjugated with anti-EGFR Fab’ via the post-insertion method. For antibody conjugation, the thiolated anti-EGFR Fab’ was conjugated with DSPE-PEG-MAL by forming a thiol-ether bond between the thiol and maleimide groups (Figure 1B). For conjugation between DSPE-PEG-COOH and anti-EGFR Fab’, the coupling reaction was carried out in the presence of EDC and NHS to form an acyl amino ester, which could subsequently react with the primary amine of the ligand to yield an amide bond (Figure 1C).
Characterization of liposomes
Particle size and zeta potential
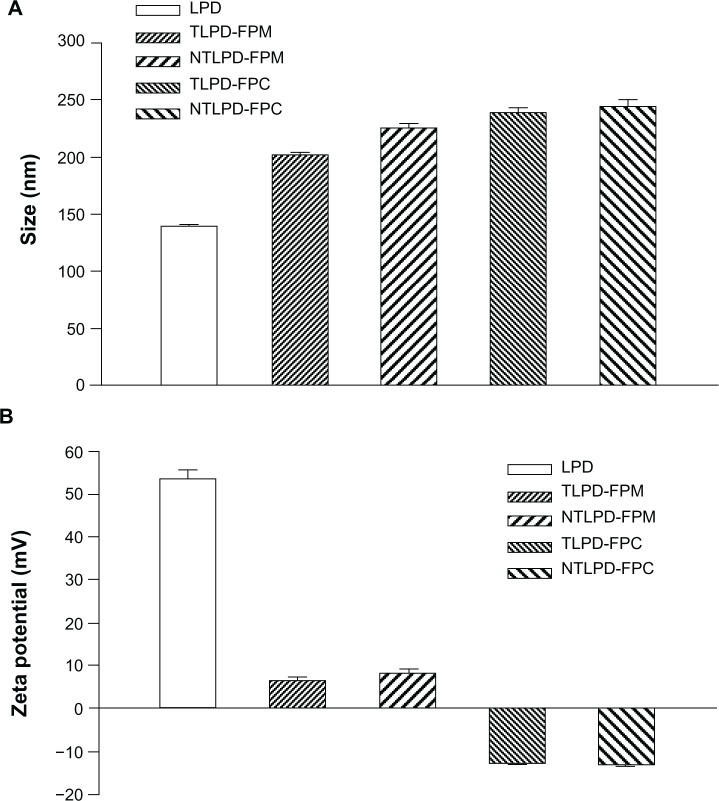
We prepared different formulations of liposomes composed of DOTAP and cholesterol at a molar ratio of 1:1 using the thin film hydration method with two conjugation techniques. The characteristics of the liposomes are shown in Figure 2. Naked LPD had a size of about 130 nm, but the particle sizes of the four TLPD nanoparticle formulations were similar to each other at about 200 nm, indicating that PEG conjugation with Fab’ was able to increase the size of the liposomes. The size of TLPD with DSPE-PEG-COOH as the conjugation linker seemed larger than that of particles with DSPE-PEG-MAL as the conjugation linker. However, statistical analysis showed that there was no significant difference between the two particle groups. Previous studies30,31 have shown that extravasation of PEG-grafted liposomes from the vascular compartment into the tumor interstitium is size-dependent and also dependent on the type of tumor, but mainly limited by the ability to diffuse through the 100–1,200 nm pores in the vessel wall. Thus, a particle size of immunoliposomes at 200 nm is critical for them to pass through the small capillaries and penetrate throughout the tumor.
Figure 2.
Mean size (A) and zeta potential (B) of LPD, TLPD-FCM, NTLPD-FPM, TLPD-FPC, and NTLPD-FPC. Data are expressed as the mean ± standard deviation (n = 3).
Abbreviations: LPD, liposome-polycation-DNA; TLPD, targeted liposome-polycation-DNA; TLPD-FPM, targeted LPD when linker PEG was DSPE-PEG-MAL; TLPD-FPC, targeted LPD when linker PEG was DSPE-PEG-COOH; NTLPD-FPM, nontargeted control LPD when linker PEG was DSPE-PEG-MAL; NTLPD-FPC, nontargeted control LPD when linker PEG was DSPE-PEG-COOH; DSPE-PEG-MAL, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (ammonium salt); DSPE-PEG-COOH, distearoyl-N-(3-carboxypropionoyl poly(ethylene glycol) succinyl)phosphatidylethanolamine; EDC, 1-ethyl-3-(3-dimethylamino-propyl)carbodiimide.
The surface charge on liposomes plays an important role in determining their final destination. When liposomes have a strong anionic charge, they are usually absorbed by scavenger cells, resulting in the injected dose being rapidly eliminated from the blood. A strong positive charge can also create some problems. In our prepared LPD system, the mean zeta potential of naked LPD was 49.4 ± 4.1 mV (n = 4). This highly positively charged nanoparticle formulation tended to aggregate when incubated with high-salt buffer or serum32 and had a strong charge-charge interaction with cells. When the naked LPD was modified by PEG conjugated with antibody, the zeta potential of the formulations decreased rapidly, indicating that PEG derivatives had inserted into LPD. We found that TLPD-FPM and NTLPD-FPM were similar as no more than 8 mV (Figure 2B), but TLPD-FPC and NTLPD-FPC decreased to about −11 mV, probably due to the negative charge of DSPE-PEG-COOH itself. The difference in zeta potential between the two types of LPD may have an influence on electrostatic interactions with targeted cells.
Transmission electron microscopy
Transmission electron microscopy was used to visualize the particle structure of the different post-inserted functionalized PEG derivatives, TLPD-FPM and TLPD-FPC. Figure 3 shows that there is no difference in shape or size distribution between TLPD-FPM and TLPD-FPC, suggesting that the different post-inserted PEG derivatives conjugated with Fab’ have little impact on the structure of the liposomes. Notably, the particles observed by transmission electron microscopy were smaller than those observed by dynamic light scattering (Figure S1). The reason may be that large-sized particles showed significantly greater light scattering compared with small-sized particles at the same concentration.25
Figure 3.
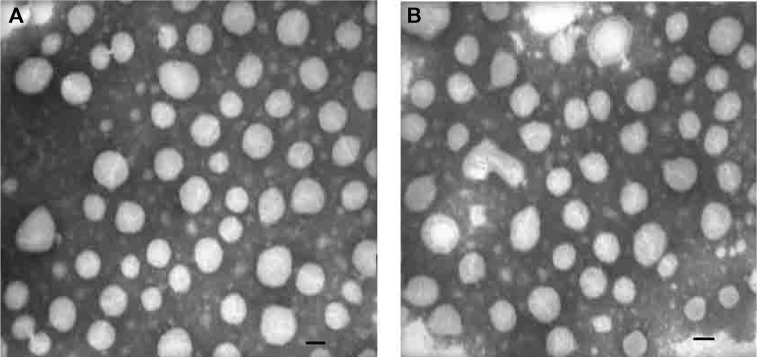
Transmission electron microscopic images of TLPD-FPM (A) and TLPD-FPC (B). Bar indicates 100 nm.
Abbreviations: TLPD-FPM, targeted LPD when linker PEG was DSPE-PEG-MAL; TLPD-FPC, targeted LPD when linker PEG was DSPE-PEG-COOH; LPD, liposome-polycation-DNA complex; DSPE-PEG-MAL, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (ammonium salt); DSPE-PEG-COOH, distearoyl-N-(3-carboxypropionoyl poly(ethylene glycol) succinyl)phosphatidylethanolamine.
Gel retardation assay
The effect of the different conjugation techniques on the siRNA binding affinity of liposomes was evaluated using a gel retardation assay. Figure 4A shows that encapsulated siRNA was completely retarded in the gel wells when the samples were added directly. However, when Triton-X100 destroyed all liposomes of intact naked LPD, TLPD-FPM, NTLPD-FPM, TLPD-FPC, and NTLPD-FPC, siRNA was released from the liposomes, presenting as bright bands. These results indicate that the siRNA binding affinity remains powerful regardless of different Fab’ conjugation technologies, confirming that the siRNA binding affinity would not be affected by these different technologies.
Figure 4.
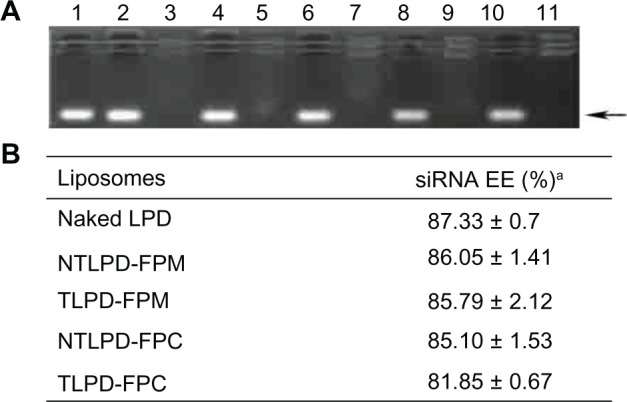
(A) Gel retardation assay and (B) siRNA encapsulation efficiency. In the gel retardation assay, naked LPD (lanes 2 and 3), TLPD-FPM (lanes 4 and 5), NTLPD-FPM (lanes 6 and 7), TLPD-FPC (lanes 8 and 9), NTLPD-FPC (lanes 10 and 11) are shown. The former and latter lanes indicate samples destroyed by Triton-X100 or untreated samples, respectively. Untreated siRNA was run in lane 1. Each lane contains 0.3 μg of siRNA. The arrows indicate free siRNA. aThe data are given as the mean ± standard deviation (n = 3).
Abbreviations: EE, encapsulation efficiency; LPD, liposome-polycation-DNA; TLPD, targeted liposome-polycation-DNA; TLPD-FPM, targeted LPD when linker PEG was DSPE-PEG-MAL; TLPD-FPC, targeted LPD when linker PEG was DSPE-PEG-COOH; NTLPD-FPM, nontargeted control LPD when linker PEG was DSPE-PEG-MAL; NTLPD-FPC, nontargeted control LPD when linker PEG was DSPE-PEG-COOH; siRNA, small interfering RNA; SD, standard deviation; DSPE-PEG-MAL, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (ammonium salt); DSPE-PEG-COOH, distearoyl-N-(3-carboxypropionoyl poly(ethylene glycol) succinyl)phosphatidylethanolamine.
siRNA encapsulation efficiency
Cationic liposomes with their positive charge can absorb negatively charged siRNA to increase encapsulation efficiency.19 LPD as an effective nanovector for systemic siRNA delivery has high encapsulation efficiency, not only because of the positive charge, but because of the thymus DNA used as a carrier in this formulation to improve core compaction. The different Fab’ conjugation technologies used for LPD did not change the particle structure (Figure 3), but the effect on encapsulation efficiency remains unknown. It was found that the siRNA encapsulation efficiency of all liposomes was as high (about 90%) as that of naked LPD (Figure 4B), which is consistent with the result obtained from the gel retardation assay, indicating that there was hardly any impact on siRNA encapsulation efficiency for TLPD or NTLPD using the different Fab’ conjugation technologies.
siRNa degradation in serum
In our previous study, we demonstrated that LPD was an effective carrier that could protect siRNA against the influence of external factors. To observe whether different PEG derivatives can block siRNA degradation in serum, all samples were incubated in an aqueous solution of 50% serum at 37°C, and then agarose gel electrophoresed at designated time points. As shown in Figure 5, after a 6-hour incubation period, naked siRNA was broken down completely (lane 2), while siRNA molecules entrapped inside the PEGylated liposomes remained relatively intact even after 48 hours, irrespective of the method of preparation. These results suggest that both conjugation technologies efficiently protected siRNA against serum, which is consistent with the finding of Kim et al that PEGylated liposomes increased the stability of siRNA against RNase.33 LPD with the two different PEG derivatives also had the same effect of prolonging the residence time of liposomes in serum by providing a steric barrier.
Figure 5.
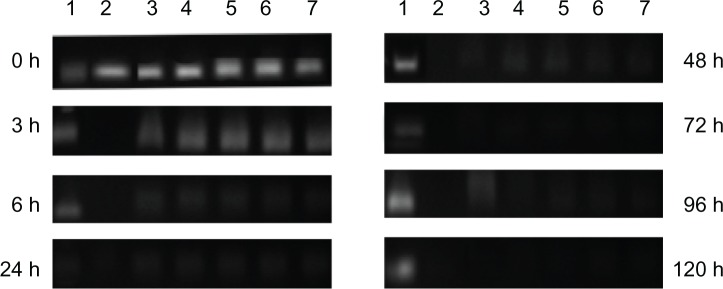
siRNA serum stability. Lane 1, intact siRNA; lane 2, siRNA; lane 3, naked LPD; lane 4, TLPD-FPM; lane 5, NTLPD-FPM; lane 6, TLPD-FPC; and lane 7, NTLPD-FPC. Samples of siRNA solution or liposomes incubated with 50% fetal bovine serum. After the liposomes were destroyed by 5% Triton-X100, these samples containing 0.3 μg siRNA were applied to agarose gel electrophoresis.
Abbreviations: LPD, liposome-polycation-DNA; TLPD, targeted liposome-polycation-DNA; TLPD-FPM, targeted LPD when linker PEG was DSPE-PEG-MAL; TLPD-FPC, targeted LPD when linker PEG was DSPE-PEG-COOH; NTLPD-FPM, nontargeted control LPD when linker PEG was DSPE-PEG-MAL; NTLPD-FPC, nontargeted control LPD when linker PEG was DSPE-PEG-COOH; DSPE-PEG-MAL, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (ammonium salt); DSPE-PEG-COOH, distearoyl-N-(3-carboxypropionoyl poly(ethylene glycol) succinyl)phosphatidylethanolamine.
Cell viability
Prior to the siRNA transfection experiment, we investigated the cytotoxicity of TLPD-FPM/FPC and NTLPD-FPM/FPC in SMMC-7721 cells. As shown in Figure 6, there was no significant decrease in viability of TLPD-FPM/FPC-treated cells or NTLPD-FPM/FPC-treated cells after 24 hours of incubation as compared with that in untreated cells, indicating that all the liposome formulations had good biocompatibility with SMMC-7721 cells at concentrations of 100 nM or 200 nM siRNA, and the cytotoxicity of (N)TLPD-FPM was similar to that of (N) TLPD-FPC. Therefore, we can conclude that there is no significant difference in cytotoxicity to SMMC-7721 cells between the two technologies for conjugation of Fabs’, and the prepared TLPD system can serve as a secure gene carrier to cells.
Figure 6.
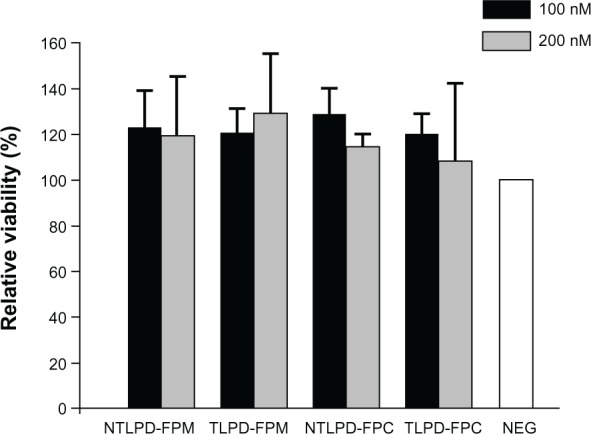
Cell viability of SMMC-7721 cells transfected with NTLPD-FPM, TLPD-FPM, NTLPD-FPC, or TLPD-FPC at an siRNA concentration of 100 nM or 200 nM. The data are shown as the mean ± standard deviation (n = 3).
Abbreviations: TLPD-FPM, targeted LPD when linker PEG was DSPE-PEG-MAL; TLPD-FPC, targeted LPD when linker PEG was DSPE-PEG-COOH; NTLPD-FPM, nontargeted control LPD when linker PEG was DSPE-PEG-MAL; NTLPD-FPC, nontargeted control LPD when linker PEG was DSPE-PEG-COOH; DSPE-PEG-MAL, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (ammonium salt); DSPE-PEG-COOH, distearoyl-N-(3-carboxypropionoyl poly(ethylene glycol) succinyl)phosphatidylethanolamine; NEG, negative.
In vitro cellular uptake of liposome/siRNa complexes
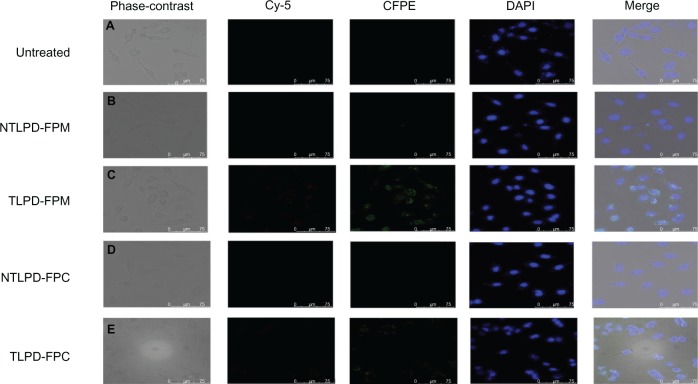
Cellular internalization of siRNA-encapsulated nanoparticles was evaluated by confocal microscopy after culture of nanoparticles with SMMC-7721 cells. As shown in Figure 7, the uptake of green fluorescently labeled carboxyfluorescein-PE and red fluorescently labeled siRNA was much greater in cells treated with TLPD-FPM than that in TLPD-FPC. Further, for both liposomes containing different conjugation linkers, the fluorescence signal in the cells treated with nontargeted liposomes (Figure 7B and D) was much weaker than that of cells treated with LPD-PEG-targeted liposomes (Figure 7C and E). Taken together, these data indicate that the targeted TLPD-FPM liposome was superior to the targeted TLPD-FPC liposome in efficient delivery of siRNA to tumor cells and that intracellular delivery was ligand-dependent. The results are in good agreement with the transfection efficiency data shown in Figure 8.
Figure 7.
Internalization of the nanoparticles was evaluated by confocal microscopy. SMMC-7721 cells were treated with different formulations of liposomes as follows: (A) row, untreated; (B) row, NTLPD-FPM; (C) row, TLPD-FPM; (D) row, NTLPD-FPC; (E) row, TLPD-FPC. Scale bar 75 μm.
Abbreviations: LPD, liposome-polycation-DNA; TLPD, targeted liposome-polycation-DNA; TLPD-FPM, targeted LPD when linker PEG was DSPE-PEG-MAL; TLPD-FPC, targeted LPD when linker PEG was DSPE-PEG-COOH; NTLPD-FPM, nontargeted control LPD when linker PEG was DSPE-PEG-MAL; NTLPD-FPC, nontargeted control LPD when linker PEG was DSPE-PEG-COOH; DSPE-PEG-MAL, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (ammonium salt); DSPE-PEG-COOH, distearoyl-N-(3-carboxypropionoyl poly(ethylene glycol) succinyl)phosphatidylethanolamine; CFPE, carboxyfluorescein-PE; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride.
Figure 8.
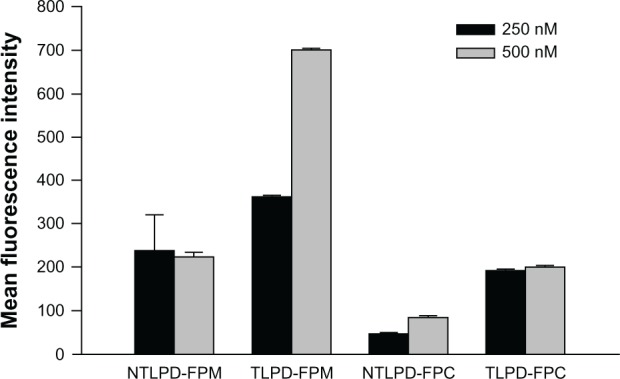
Cellular uptake of SMMC-7721 cells in vitro. After treatment of the cells at a FAM-siRNA concentration of 250 nM or 500 nM for 24 hours, mean fluorescence intensity was analyzed by flow cytometry. The data are shown as the mean ± standard deviation (n = 3).
Abbreviations: TLPD-FPM, targeted LPD when linker PEG was DSPE-PEG-MAL; TLPD-FPC, targeted LPD when linker PEG was DSPE-PEG-COOH; NTLPD-FPM, nontargeted control LPD when linker PEG was DSPE-PEG-MAL; NTLPD-FPC, nontargeted control LPD when linker PEG was DSPE-PEG-COOH; DSPE-PEG-MAL, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (ammonium salt); DSPE-PEG-COOH, distearoyl-N-(3-carboxypropionoyl poly(ethylene glycol) succinyl)phosphatidylethanolamine.
In vitro transfection of liposome/siRNa complexes
To investigate the transfection efficiency of the liposomes, SMMC-7721 cells with high EGFR expression were treated with TLPD at a concentration of 250 nM or 500 nM FAM-siRNA for 24 hours. As shown in Figure 8, TLPD-FPM significantly increased the transfection efficiency of SMMC-7721 cells compared with NTLPD-FPM. The transfection efficiency of TLPD-FPC was also higher than that of NTLPD-FPC in 250 nM or 500 nM siRNA, indicating that both immunoliposomes possess targetability. Compared with TLPD-FPC, TLPD-FPM showed higher transfection efficiency of about 1.88-fold and 3.46-fold in 250 nM and 500 nM siRNA, respectively, that could be attributed to their charges, ie, TLPD-FPM or NTLPD-FPM with a positive charge and TLPD-FPC or NTLPD-FPC with a negative charge. These findings are consistent with the results reported by Morille et al16 that nonviral vectors with a positive charge could bind to a negatively charged cell membrane through electrostatic interactions, leading to higher transfection efficiency than vectors with a negative charge.
In vitro gene silencing
In vitro gene silencing was studied using SMMC-7721 cells stably transduced with the firefly luciferase gene. As shown in Figure 9, the silencing effect in cells treated with targeted liposomes was much higher than that in cells treated with nontargeted liposomes, indicating that Fab’ conjugation to liposomes exerted its targeting effect through receptor-mediated endocytosis. In addition, more than 50% silencing was achieved in targeted liposomes containing DSPE-PEG-MAL at a 250 nM siRNA concentration, which was higher than that of targeted liposomes containing DSPE-PEG-COOH. When the siRNA concentration was increased to 500 nM, the silencing efficiency for TLPD-FPM reached about 80% versus 32% for TLPD-FPC. On the basis of these data, we can conclude that gene silencing activity is highly correlated with intracellular siRNA uptake (Figure 7) and transfection efficiency (Figure 8). As TLPD-FPM showed higher intracellular siRNA uptake and more efficient in gene transfection than TLPD-FPC, TLPD-FPM is more efficiently in silencing reporter gene in hepatocellular carcinoma cells.
Figure 9.
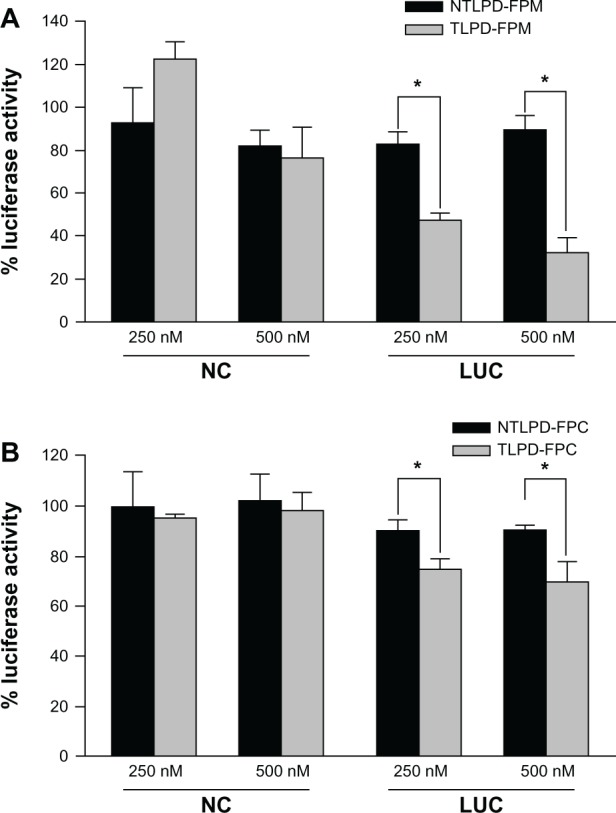
In vitro gene silencing. The percent luciferase activity of SMMC-7721 after transfection with (A) TLPD-FPM, NTLPD-FPM and (B) TLPD-FPC, NTLPD-FPC at a concentration of 250 nM or 500 nM siRNA. The data are shown as the mean ± standard deviation (n = 3). *P < 0.05.
Abbreviations: TLPD-FPM, targeted LPD when linker PEG was DSPE-PEG-MAL; TLPD-FPC, targeted LPD when linker PEG was DSPE-PEG-COOH; NTLPD-FPM, nontargeted control LPD when linker PEG was DSPE-PEG-MAL; NTLPD-FPC, nontargeted control LPD when linker PEG was DSPE-PEG-COOH; LUC, luciferase; NC, negative control siRNA; DSPE-PEG-MAL, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (ammonium salt); DSPE-PEG-COOH, distearoyl-N-(3-carboxypropionoyl poly(ethylene glycol) succinyl)phosphatidylethanolamine.
Cell migration after RhoA silencing
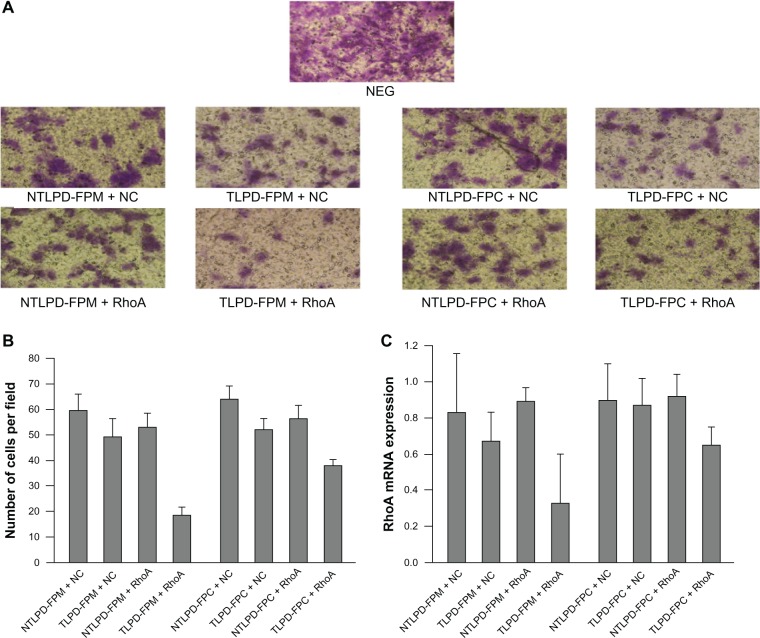
As a small Rho GTPase, RhoA has been shown to play a key role in regulation of tumor growth, migration, and response to therapy.34 When the cell surface receptor is simulated, RhoA becomes activated and regulates tumor growth, migration, and gene expression through Rho kinases and ROCK.35 RhoA has been used as a therapeutic target to demonstrate the potential of TLPD-FPM and TLPD-FPC in cancer gene therapy. In contrast with untreated cells, transfection of SMMC-7721 cells with TLPD-FPM and TLPD-FPC containing anti-RhoA siRNA reduced RhoA messenger RNA expression by 70% and 30%, respectively (Figure 10C) and appeared to suppress cell migration compared with NTLPD-FPM and NTLPD-FPC containing anti-RhoA siRNA (Figure 10A and B). As expected, targeted or untargeted LPD of the two different conjugation technologies entrapping NC siRNA had no RhoA gene silencing activity and could not inhibit SMMC-7721 cell migration. In summary, anti-RhoA siRNA formulated in targeted liposomes could effectively downregulate RhoA messenger RNA expression and reduce cell migration, and the effect of targeted TLPD-FPM liposome was much higher than that of the targeted TLPD-FPC liposome.
Figure 10.
RhoA silencing of SMMC-7721 cell migration. The cells were treated with anti-RhoA or NC siRNA encapsulated in liposomes at a final siRNA concentration of 500 nM. (A) After 48 hours of transfection, cell migration induced by RhoA was assessed. Original magnification ×200. (B) The number of cells per field was calculated from five random fields. (C) After 72 hours of transfection, total RNA was extracted from the cells and RhoA expression was demonstrated by real-time polymerase chain reaction. The data are shown as the mean ± standard deviation (n = 3).
Abbreviations: TLPD-FPM, targeted LPD when linker PEG was DSPE-PEG-MAL; TLPD-FPC, targeted LPD when linker PEG was DSPE-PEG-COOH; NTLPD-FPM, nontargeted control LPD when linker PEG was DSPE-PEG-MAL; NTLPD-FPC, nontargeted control LPD when linker PEG was DSPE-PEG-COOH; DSPE-PEG-MAL, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (ammonium salt); DSPE-PEG-COOH, distearoyl-N-(3-carboxypropionoyl poly(ethylene glycol) succinyl)phosphatidylethanolamine; NC, negative control siRNA; NEG, negative.
Conclusion
In the present study, we prepared different formulations of liposomes with two Fab’ conjugation technologies using the post-insertion method and made a series of comparisons between them using a variety of analytical techniques in vitro. It was found that liposomes prepared by the two conjugation technologies possessed high siRNA encapsulation efficiency, superior serum stability, and little immunotoxicity, and both could be used as siRNA carriers. In addition, targeted liposomes showed a better effect than untargeted liposomes in transfection efficiency and gene silencing. However, compared with TLPD-FPC, TLPD-FPM showed significantly enhanced EGFR targeting efficiency and achieved superior gene silencing activity in vitro. These findings suggest that when the targeting LPD is modified by anti-EGFR Fab’ conjugation with DSPE-PEG-MAL or DSPE-PEG-COOH, the former would have significant superiority in siRNA delivery in comparison with the latter. The data obtained from this study may provide new information about selection of linkers in preparing targeted drug carriers.
Supplementary figure
Size distribution of TLPD-FPM and TLPD-FPC as measured by dynamic light scattering.
Abbreviations: TLPD-FPM, targeted LPD when linker PEG was DSPE-PEG-MAL; TLPD-FPC, targeted LPD when linker PEG was DSPE-PEG-COOH; DSPE-PEG-MAL, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (ammonium salt); DSPE-PEG-COOH, distearoyl-N-(3-carboxypropionoyl poly(ethylene glycol) succinyl)phosphatidylethanolamine.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (81101141), the Shanghai Commission of Science and Technology (12nm0501000, 13NM1401503) and the Ministry of Science and Technology of China (973 and 863 program projects).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Lee RJ, Huang L. Folate-targeted, anionic liposome-entrapped polylysine-condensed DNA for tumor cell-specific gene transfer. J Biol Chem. 1996;271(14):8481–8487. doi: 10.1074/jbc.271.14.8481. [DOI] [PubMed] [Google Scholar]
- 3.Tseng YC, Mozumdar S, Huang L. Lipid-based systemic delivery of siRNA. Adv Drug Deliv Rev. 2009;61(9):721–731. doi: 10.1016/j.addr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6(6):443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leung RK, Whittaker PA. RNA interference: from gene silencing to gene-specific therapeutics. Pharmacol Ther. 2005;107(2):222–239. doi: 10.1016/j.pharmthera.2005.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim WJ, Kim SW. Efficient siRNA delivery with non-viral polymeric vehicles. Pharm Res. 2009;26(3):657–666. doi: 10.1007/s11095-008-9774-1. [DOI] [PubMed] [Google Scholar]
- 7.Schaffert D, Wagner E. Gene therapy progress and prospects: synthetic polymer-based systems. Gene Ther. 2008;15(16):1131–1138. doi: 10.1038/gt.2008.105. [DOI] [PubMed] [Google Scholar]
- 8.Kaufmann J, Ahrens K, Santel A. RNA interference for therapy in the vascular endothelium. Microvasc Res. 2010;80(2):286–293. doi: 10.1016/j.mvr.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Bedi D, Musacchio T, Fagbohun OA, et al. Delivery of siRNA into breast cancer cells via phage fusion protein-targeted liposomes. Nanomedicine. 2011;7(3):315–323. doi: 10.1016/j.nano.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Res. 2005;65(19):8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 11.Song E, Zhu PC, Lee SK, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat Biotechnol. 2005;23(6):709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 12.Kawakami S, Hashida M. Targeted delivery systems of small interfering RNA by systemic administration. Drug Metab Pharmacokinet. 2007;22(3):142–151. doi: 10.2133/dmpk.22.142. [DOI] [PubMed] [Google Scholar]
- 13.Muratovska A, Eccles MR. Conjugate for efficient delivery of short interfering RNA (siRNA) into mammalian cells. FEBS Lett. 2004;558(1–3):63–68. doi: 10.1016/S0014-5793(03)01505-9. [DOI] [PubMed] [Google Scholar]
- 14.McNamara JO, 2nd, Andrechek ER, Wang Y, et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24(8):1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 15.Gao J, Sun J, Li H, et al. Lyophilized HER2-specific PEGylated immunoliposomes for active siRNA gene silencing. Biomaterials. 2010;31(9):2655–2664. doi: 10.1016/j.biomaterials.2009.11.112. [DOI] [PubMed] [Google Scholar]
- 16.Morille M, Passirani C, Vonarbourg A, Clavreul A, Benoit JP. Progress in developing cationic vectors for non-viral systemic gene therapy against cancer. Biomaterials. 2008;29(24–25):3477–3496. doi: 10.1016/j.biomaterials.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 17.Buyens K, De Smedt SC, Braeckmans K, et al. Liposome based systems for systemic siRNA delivery: stability in blood sets the requirements for optimal carrier design. J Control Release. 2012;158(3):362–370. doi: 10.1016/j.jconrel.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Lonez C, Vandenbranden M, Ruysschaert JM. Cationic liposomal lipids: from gene carriers to cell signaling. Prog Lipid Res. 2008;47(5):340–347. doi: 10.1016/j.plipres.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Rao NM. Cationic lipid-mediated nucleic acid delivery: beyond being cationic. Chem Phys Lipids. 2010;163(3):245–252. doi: 10.1016/j.chemphyslip.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Puri A, Kramer-Marek G, Campbell-Massa R, et al. HER2-specific affibody-conjugated thermosensitive liposomes (Affisomes) for improved delivery of anticancer agents. J Liposome Res. 2008;18(4):293–307. doi: 10.1080/08982100802457377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sapra P, Tyagi P, Allen TM. Ligand-targeted liposomes for cancer treatment. Curr Drug Deliv. 2005;2(4):369–381. doi: 10.2174/156720105774370159. [DOI] [PubMed] [Google Scholar]
- 22.Li SD, Chono S, Huang L. Efficient gene silencing in metastatic tumor by siRNA formulated in surface-modified nanoparticles. J Control Release. 2008;126(1):77–84. doi: 10.1016/j.jconrel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li SD, Chen YC, Hackett MJ, Huang L. Tumor-targeted delivery of siRNA by self-assembled nanoparticles. Mol Ther. 2008;16(1):163–169. doi: 10.1038/sj.mt.6300323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao J, Yu Y, Zhang Y, et al. EGFR-specific PEGylated immunoliposomes for active siRNA delivery in hepatocellular carcinoma. Biomaterials. 2012;33(1):270–282. doi: 10.1016/j.biomaterials.2011.09.035. [DOI] [PubMed] [Google Scholar]
- 25.Gao J, Liu W, Xia Y, et al. The promotion of siRNA delivery to breast cancer overexpressing epidermal growth factor receptor through anti-EGFR antibody conjugation by immunoliposomes. Biomaterials. 2011;32(13):3459–3570. doi: 10.1016/j.biomaterials.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 26.Manjappa AS, Chaudhari KR, Venkataraju MP, et al. Antibody derivatization and conjugation strategies: application in preparation of stealth immunoliposome to target chemotherapeutics to tumor. J Control Release. 2011;150(1):2–22. doi: 10.1016/j.jconrel.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Maruyama K, Takizawa T, Yuda T, Kennel SJ, Huang L, Iwatsuru M. Targetability of novel immunoliposomes modified with amphipathic poly(ethylene glycol)s conjugated at their distal terminals to monoclonal antibodies. Biochim Biophys Acta. 1995;1234(1):74–80. doi: 10.1016/0005-2736(94)00263-o. [DOI] [PubMed] [Google Scholar]
- 28.Gao J, Kou G, Wang H, et al. PE38 KDEL-loaded anti-HER2 nanoparticles inhibit breast tumor progression with reduced toxicity and immunogenicity. Breast Cancer Res Treat. 2009;115(1):29–41. doi: 10.1007/s10549-008-0043-0. [DOI] [PubMed] [Google Scholar]
- 29.Dominska M, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci. 2010;123(Pt 8):1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 30.Moreira JN, Gaspar R, Allen TM. Targeting stealth liposomes in a murine model of human small cell lung cancer. Biochim Biophys Acta. 2001;1515(2):167–176. doi: 10.1016/s0005-2736(01)00411-4. [DOI] [PubMed] [Google Scholar]
- 31.Yang T, Choi MK, Cui FD, et al. Preparation and evaluation of paclitaxel-loaded PEGylated immunoliposome. J Control Release. 2007;120(3):169–177. doi: 10.1016/j.jconrel.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 32.Li SD, Huang L. Targeted delivery of antisense oligodeoxynucleotide and small interference RNA into lung cancer cells. Mol Pharm. 2006;3(5):579–588. doi: 10.1021/mp060039w. [DOI] [PubMed] [Google Scholar]
- 33.Kim HK, Davaa E, Myung CS, Park JS. Enhanced siRNA delivery using cationic liposomes with new polyarginine-conjugated PEG-lipid. Int J Pharm. 2010;392(1–2):141–147. doi: 10.1016/j.ijpharm.2010.03.047. [DOI] [PubMed] [Google Scholar]
- 34.Schmitz AA, Govek EE, Böttner B, Van Aelst L. Rho GTPases: signaling, migration, and invasion. Exp Cell Res. 2000;261(1):1–12. doi: 10.1006/excr.2000.5049. [DOI] [PubMed] [Google Scholar]
- 35.Chang YW, Bean RR, Jakobi R. Targeting RhoA/Rho kinase and p21-activated kinase signaling to prevent cancer development and progression. Recent Pat Anticancer Drug Discov. 2009;4(2):110–124. doi: 10.2174/157489209788452830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Size distribution of TLPD-FPM and TLPD-FPC as measured by dynamic light scattering.
Abbreviations: TLPD-FPM, targeted LPD when linker PEG was DSPE-PEG-MAL; TLPD-FPC, targeted LPD when linker PEG was DSPE-PEG-COOH; DSPE-PEG-MAL, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol)-2000] (ammonium salt); DSPE-PEG-COOH, distearoyl-N-(3-carboxypropionoyl poly(ethylene glycol) succinyl)phosphatidylethanolamine.