Abstract
Central nervous system tumors carry grave clinical prognoses due to limited effectiveness of surgical resection, radiation, and chemotherapy. Thus, improved strategies for brain tumor visualization and targeted treatment are critically needed. We demonstrate that mouse cerebellar medulloblastoma (MB) can be targeted and illuminated with a fluorescent, engineered cystine knot (knottin) peptide that binds with high affinity to αvβ3, αvβ5, and α5β1 integrin receptors. This integrin-binding knottin peptide, denoted EETI 2.5F, was evaluated as a molecular imaging probe in both orthotopic and genetic models of MB. Following tail vein injection, fluorescence arising from dye-conjugated EETI 2.5F was localized to the tumor compared with the normal surrounding brain tissue, as measured by optical imaging. The imaging signal intensity correlated with tumor volume. Due to its unique ability to bind to α5β1 integrin, EETI 2.5F showed superior in vivo and ex vivo brain tumor imaging contrast compared with other engineered integrin-binding knottin peptides and with c(RGDfK), a well-studied integrin-binding peptidomimetic. Next, EETI 2.5F was fused to an antibody fragment crystallizable (Fc) domain (EETI 2.5F–Fc) to determine if a larger integrin-binding protein could also target intracranial brain tumors. EETI 2.5F–Fc, conjugated to a fluorescent dye, illuminated MB following i.v. injection and was able to distribute throughout the tumor parenchyma. In contrast, brain tumor imaging signals were not detected in mice injected with EETI 2.5F proteins containing a scrambled integrin-binding sequence, demonstrating the importance of target specificity. These results highlight the potential of using EETI 2.5F and EETI 2.5–Fc as targeted molecular probes for brain tumor imaging.
Keywords: protein engineering, miniprotein, tumor targeting, Hedgehog pathway
Despite significant advances in cancer therapy and improved understanding of brain tumor biology (1, 2), the quality of life and overall life expectancy for patients diagnosed with malignant brain tumors have shown marginal improvements (3, 4). Children diagnosed with medulloblastoma (MB), the most common childhood brain tumor, require aggressive chemotherapy and radiation due to the tumor’s marked propensity to recur and metastasize (5, 6). Although MB can be surgically resected, the extent of diseased tissue removal has been shown to correlate with overall survival rates (7–9). Improvements in intraoperative tumor visualization would allow for more refined surgical resection and minimize removal of healthy brain tissue. Thus, there is a critical need for molecular imaging agents that bind specifically and with high affinity to tumor-associated receptors, are cleared rapidly from nontarget tissues, localize to intracranial tumors following distal venous injection, and can be conjugated to fluorescent dyes for optical imaging.
The elevated level of cell surface integrins on tumors and their vasculature has spurred great interest in using these receptors as targets for cancer therapy (10) and molecular imaging (11). Specifically, αvβ3, αvβ5, and α5β1 integrins have been shown to play critical roles in tumor survival, invasion, metastasis, and angiogenesis (12–17) and are present at high levels in a variety of cancers (reviewed in ref. 18). A variety of small molecules, peptides, peptidomimetics, and proteins have been developed that target tumor-associated integrin receptors, and several have advanced to human trials for therapeutic and diagnostic applications (11, 19). As an example, Cilengitide, which is currently in clinical trials for glioblastoma therapy, is a cyclic pentapeptide containing an Arg-Gly-Asp (RGD) motif that binds to αvβ3 and αvβ5 integrins (20). Chemically modified variants of cyclic RGD-containing peptides, developed as noninvasive molecular imaging probes, have been limited due to relatively weak integrin binding affinities or suboptimal pharmacokinetics and tissue distribution, requiring physicochemical modifications to increase tumor contrast (21). Although there are numerous small molecules and peptides that bind αvβ3 and αvβ5 integrins, examples of high-affinity α5β1 integrin-targeting agents have been limited to monoclonal antibodies. Molecular imaging agents with broad specificity toward multiple tumor-associated integrins could potentially target malignant tissue with higher sensitivity and selectivity compared with probes that bind a more limited range of receptor targets (22).
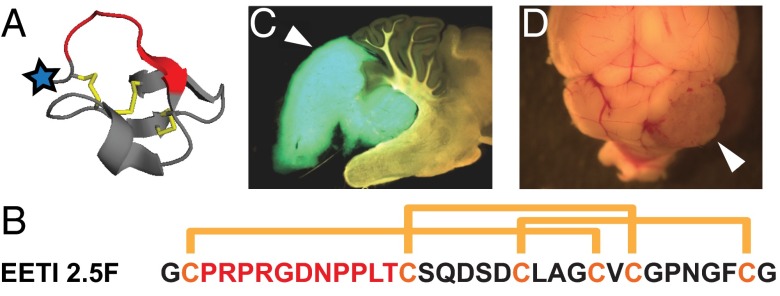
We engineered a small (∼3.5 kDa), conformationally constrained cystine knot peptide, termed EETI 2.5F, that binds with high affinity and unique specificity to αvβ3, αvβ5, and α5β1 integrins (23). The cystine knot structural family, also known as knottins, consists of small polypeptides (30–50 amino acids) linked by at least three interwoven disulfide bonds, creating a rigid molecular “knot” that confers high chemical, thermal, and proteolytic stability (24). EETI 2.5F, which is based on the Ecballium elaterium trypsin inhibitor-II (EETI-II, Fig. 1 A and B), was identified via high-throughput screening of yeast-displayed knottin libraries (23). Radiolabeled versions of EETI 2.5F and other engineered integrin-binding knottin peptides were previously shown to be promising molecular probes for PET imaging in s.c. mouse xenograft models due to their high tumor contrast and low imaging signals in nondiseased tissue, including the liver and kidneys (25, 26).
Fig. 1.
Overview of knottins and mouse medulloblastoma models. (A) Structure of wild-type EETI-II (PDB 2IT7). Blue star indicates location of N-terminal AF680 dye in our studies; red indicates native trypsin-binding loop, which has been replaced by an engineered integrin-binding loop in EETI 2.5F. (B) Sequence and disulfide bond connectivity for EETI 2.5F. Engineered integrin-binding loop sequence shown in red. (C) Ptch+/−;Math1-GFP mice spontaneously develop MBs, which express GFP. (D) Med1-MB cells derived from Ptch+/−;LacZ mice form MBs when surgically implanted into the cerebella of nude mice. White arrowhead indicates a tumor.
In the current study we show that EETI 2.5F, conjugated to a near-infrared imaging dye, can specifically target and illuminate brain tumor tissue in mouse models of MB that faithfully recapitulate human disease. In addition, we compared optical imaging signals from integrin-targeting agents of different molecular sizes and binding specificities to provide insight into the mechanism of brain tumor targeting. Our results illustrate some potential limitations of alternative integrin-targeting agents for MB detection and demonstrate the promise of developing knottins as molecular probes for image-guided surgical resection of brain tumors.
Results
Knottin Peptide EETI 2.5F Binds to Integrins Present on Ptch+/− Medulloblastoma Cells.
Ptch+/− mice with or without a Math1-GFP transgene (27) were used as sources of genetically derived MBs for our experiments. In these mouse models, MBs develop due to a mutation in the tumor suppressor Ptch gene, which normally restrains expression of target genes regulated by the Hedgehog (Hh) signaling pathway (28, 29). In humans, similar mutations in the Hh pathway account for about a quarter of MBs (30, 31). When the Math1-GFP transgene is present, aberrantly dividing tumor cells will produce GFP, which is detectable with fluorescent microscopy (Fig. 1C). Because only 10–15% of mice with the Ptch+/− genotype develop MB, we also used a cell transplant tumor model. Cultured Ptch+/−;LacZ MB (Med1-MB) cells were implanted into the cerebella of nude mice to generate a more abundant and predictable source of mice with MB tumors (Fig. 1D).
We used flow cytometry to measure the ability of EETI 2.5F, conjugated to an optical imaging probe, to bind to integrins produced on Med1-MB tumor cells. The near-infrared dye Alexa Fluor 680 (AF680) was coupled to the knottin N terminus using succinimide ester chemistry (Fig. S1 A–C). Binding of AF680-labeled EETI 2.5F knottin (AF680–EETI 2.5F) to integrins on Med1-MB tumor cells was tested and compared with unlabeled EETI 2.5F using a competition binding assay (Fig. S1D). Agouti-related protein (AgRP) 7A, an alternative engineered knottin peptide that binds specifically to αvβ3 integrin (32), was used as the competitor. AgRP 7A has an N-terminal FLAG epitope tag, which allowed detection of its binding by flow cytometry using a fluorescent anti-FLAG antibody. Unlabeled EETI 2.5F and labeled AF680–EETI 2.5F had similar half-maximal inhibitory (IC50) values of 7 ± 2 and 4.4 ± 0.3 nM, respectively (Fig. S1D). AF680–EETI RDG, a knottin peptide containing a scrambled integrin-targeting sequence (23), did not bind to Med1-MB tumor cells. Thus, EETI 2.5F binds to Med1-MB tumor cells with high affinity, and AF680 dye conjugation had negligible effects on integrin-binding interactions.
Tail Vein Injection of Fluorescent EETI 2.5F Illuminates Brain Tumors, Enabling Transcranial in Vivo Imaging.
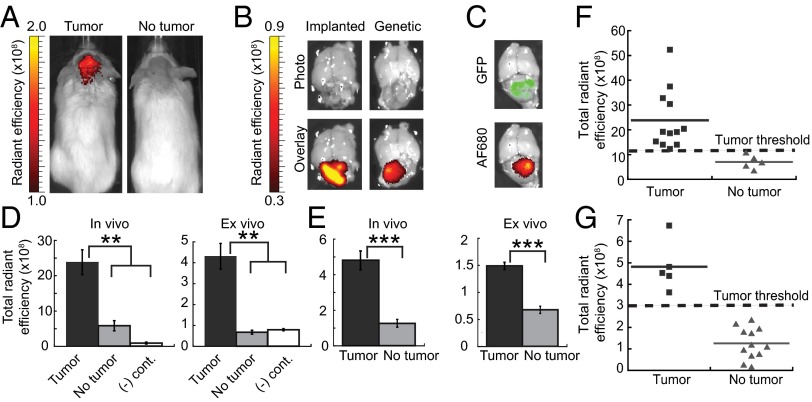
AF680–EETI 2.5F was evaluated as an optical imaging probe in Ptch+/− mice that had become symptomatic from genetically acquired MB and in nude mice with MB arising from orthotopic cerebellar implantation of Med1-MB tumor cells (Fig. 2). Control mice for the genetic model consisted of littermates of the same genetic background that did not develop tumors. In the orthotopic model, control mice underwent the same surgical procedure, with stereotactic injection of saline into the cerebellum instead of tumor cells. These sham surgery mice controlled for the effects of craniotomy, wound healing, and brain penetration. In addition, each mouse contained an intrinsic control of normal cerebellum and other brain parenchyma.
Fig. 2.
AF680–EETI 2.5F illuminates mouse medulloblastoma in vivo and ex vivo. (A) Ptch+/− mouse with a tumor (Left) and mouse with no tumor (Right) imaged 2 h after tail vein injection of AF680–EETI 2.5F. (B) Knottin peptide bound specifically to tumor tissue in orthotopic Med1-MB or Ptch+/− genetic models as confirmed by ex vivo imaging of excised brain tissue. (C) Ptch+/−; Math1-GFP mouse, which produces GFP-labeled tumor cells, showed colocalization of GFP signal and knottin AF680 signal. Quantification of in vivo and ex vivo total fluorescent signal 2 h postinjection discriminated between tumor-bearing and non–tumor-bearing mice for implanted Med1-MB (D) and Ptch+/− (E) tumors. AF680–EETI RDG [(-) cont., negative control] generated low imaging signals (D), confirming the integrin-targeting specificity of AF680–EETI 2.5F. Values are reported as the mean ± SE (**P < 0.01; ***P < 0.001). (F and G) Plots of in vivo transcranial AF680 signals for individual mice. Defined signal thresholds (dashed lines) clearly delineate mice with tumors from mice without tumors for orthotopic Med1-MB (F) and genetic Ptch+/− (G) models.
AF680–EETI 2.5F, administered by tail vein injection, traversed through the bloodstream and reached intracranial tumor tissue with sufficient affinity and quantity for detection by near-infrared fluorescence imaging through the intact skull and skin (Fig. 2 and Fig. S2). Transcranial imaging signals generated by AF680–EETI 2.5F were confirmed by ex vivo fluorescence imaging of intact brains. The fluorescent signal was emitted specifically by the tumor and not by normal brain tissue from the same mouse, and negligible imaging signals were observed in control mice without tumors (Fig. 2 and Fig. S2). In orthotopically implanted Med1-MB tumors, AF680 fluorescence colocalized with the grossly discernible MBs as visualized under white light (Fig. 2B). For MBs in Ptch+/−;Math1-GFP mice, AF680 fluorescence colocalized with the tumor-specific GFP signal (Fig. 2C). In all experiments, mice with MBs had significantly greater in vivo and ex vivo tumor signals (measured as the total radiant efficiency) compared with non–tumor-bearing controls (Fig. 2 D and E). Stronger ex vivo imaging signals correlated with larger tumor volume (Fig. S3). In the genetic model, we included mice with white, agouti, and black coat colors in the experimental groups and were able to accurately detect the presence of MB regardless of coat color. In mice with or without tumors, rapid renal clearance was observed, resulting in low background levels of fluorescence in nontarget organs (Fig. S2 C and D). Negligible imaging signals were observed with the scrambled control knottin AF680–EETI RDG (Fig. 2D), demonstrating that tumor illumination by AF680–EETI 2.5F was mediated by integrin-binding events rather than nonspecific probe binding or accumulation.
Assessment of transcranial imaging data allowed us to define in vivo tumor detection thresholds in each model that completely distinguished tumor-bearing from non–tumor-bearing mice. The assigned thresholds were as follows: total radiant efficiency > 1.2 × 109 for the orthotopic Med1-MB model (Fig. 2F) and total radiant efficiency > 3.0 × 108 for the Ptch+/− genetic model (Fig. 2G). To further validate the defined tumor detection threshold in the orthotopic Med1-MB model, we used optical imaging to prospectively follow tumor development. Nude mice were imaged with AF680–EETI 2.5F starting 10 days after implantation of Med1-MB cells and approximately weekly thereafter (Fig. S4). A separate dose of AF680–EETI 2.5F was injected 2 h before each imaging experiment. To demonstrate that imaging signals resulted from tumor formation rather than prolonged knottin accumulation over time, we showed that the AF680 signal was no longer detectable in vivo 24 h after knottin injection (Fig. S2). Upon exhibiting MB-related symptoms, mice were immediately euthanized after in vivo imaging, and ex vivo imaging of the resected brain confirmed the presence of a tumor. In sham surgery control mice, minimal imaging signals resulting from localization of AF680–EETI 2.5F at the surgical site decreased twofold over 4 weeks in serial imaging experiments (Fig. S4). For all mice with MB, strong transcranial AF680–EETI 2.5F imaging signals were detected upon the onset of tumor-related symptoms (Fig. S4); in three out of five mice tested, brain tumor imaging signals were present before the presentation of clinical symptoms. The AF680–EETI 2.5F imaging signals were consistently above the defined tumor detection threshold (Fig. 2F and Fig. S4), whereas none of the sham surgery control mice reached this threshold. Collectively, these results demonstrate that signal thresholds can be defined that distinguish between tumor-bearing and non–tumor-bearing mice, providing a useful metric for noninvasive tumor detection and monitoring in animal models.
α5β1 Integrin-Binding Specificity Is Critical for Imaging MB with AF680–EETI 2.5F.
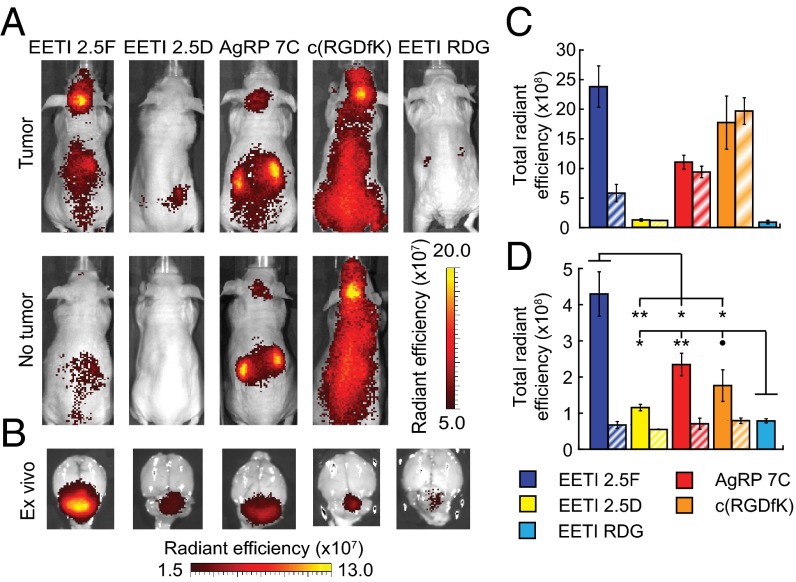
Previous studies of integrin-targeting agents under development as diagnostic agents have provided limited evaluation of the effects of integrin-binding specificity on tumor imaging contrast. To examine the mechanism by which AF680–EETI 2.5F targets intracranial tumors, we performed MB imaging with three additional peptides, each with distinct specificity for various integrins (Fig. 3). As previously described, EETI 2.5F binds with high affinity to αvβ3, αvβ5, and α5β1 integrins. EETI 2.5D is a related engineered knottin peptide that binds to αvβ3 and αvβ5 with high affinity but does not bind α5β1 integrin (23). AgRP 7C, an engineered knottin peptide derived from human AgRP, binds to αvβ3 integrin with high affinity but does not bind to αvβ5 or α5β1 integrins (32). c(RGDfK), a variant of the integrin antagonist Cilengitide, binds specifically to αvβ3 and αvβ5 integrins (33). c(RGDfK) was chosen for comparison because cyclic peptides containing an RGD motif have been evaluated as molecular imaging agents in numerous preclinical and clinical studies, including studies of human brain tumors (34). As with EETI 2.5F, these three peptides were labeled with AF680 such that one dye molecule was attached per peptide. The relative binding affinities of AF680-labeled peptides to αvβ3 integrin on Med1-MB tumor cells were measured, with AF680–c(RGDfK) showing the weakest affinity (IC50 = 550 ± 60 nM) and EETI 2.5F showing the highest affinity (IC50 = 4.4 ± 0.3 nM) (Fig. S5 and Table 1).
Fig. 3.
AF680–EETI 2.5F generates superior in vivo and ex vivo imaging signals compared with other integrin-binding peptides. Mice with tumors arising from cerebellar implantation of Med1-MB cells were imaged with AF680-labeled peptides. Representative in vivo (A) and ex vivo (B) images with similarly sized tumors shown for a panel of integrin-binding peptides with varying specificities. “No tumor” indicates control mice with sham surgeries. The color scale used to illustrate weaker tumor signals in B artificially increases the EETI 2.5F signal beyond the tumor margins. Quantification of in vivo (C) and ex vivo (D) imaging data. Solid bars show mice with tumors, diagonal stripes show control mice with sham surgeries but no tumor implantation. ∙P < 0.1; *P < 0.05; **P < 0.01. Number of mice used is as follows: EETI 2.5F tumor (13), sham (5); EETI 2.5D tumor (6), sham (3); AgRP 7C tumor (7), sham (3); c(RGDfK) tumor (7), sham (3); and EETI RDG tumor (4). All images and data were acquired 2 h postinjection.
Table 1.
Affinity, specificity, and imaging contrast of fluorescent integrin-binding peptides
| AF680 peptide | IC50, nM | Integrin specificity | Ex vivo contrast | In vivo contrast |
| EETI 2.5F | 4.4 ± 0.3 | αvβ3, αvβ5, α5β1 | 6.5 ± 0.6 | 6.5 ± 1.5 |
| EETI 2.5D | 38 ± 1 | αvβ3, αvβ5 | 2.7 ± 0.3 | 0.9 ± 0.2 |
| AgRP 7C | 15 ± 9 | αvβ3 | 3.2 ± 0.4 | 1.2 ± 0.1 |
| c(RGDfK) | 550 ± 60 | αvβ3, αvβ5 | 3.3 ± 0.4 | 0.9 ± 0.2 |
| EETI RDG | — | No binding (- cont.) | 1.6 ± 0.2 | n.d. |
Relative binding affinities of AF680-labeled peptides to Med1-MB cells as measured by competition binding assays. Integrin-binding specificities were determined in previous studies (20, 23, 32, 33). Ex vivo contrast is measured as the ratio of tumor signal to normal cerebellum signal in the same mouse. In vivo contrast is measured as the ratio of signal in mice with tumors to signal in mice without tumors (sham surgery controls). IC50 values are reported as mean ± SD. Imaging contrast values are reported as mean ± SE. n.d., not determined; (- cont.), negative control.
We compared in vivo and ex vivo imaging signals generated by these four integrin-targeting peptides and the nonbinding EETI RDG control in the orthotopic Med1-MB model. AF680–EETI 2.5F was a superior brain tumor imaging probe compared with all other integrin-targeting agents tested (Fig. 3 and Table 1). In vivo tumor signals generated from AF680–EETI 2.5D were low, similar to those seen with the AF680–EETI RDG control (Fig. 3 A and C). In comparison, in vivo imaging signals observed for AF680–AgRP 7C and AF680–c(RGDfK) were equivalent to those found in control mice with sham surgeries, indicating nonspecific tissue distribution (Fig. 3 A and C). Only AF680–EETI 2.5F was able to effectively differentiate between mice with or without brain tumors using transcranial imaging, with greater than sixfold higher tumor contrast (Fig. 3 A and C and Table 1). EETI 2.5F was the only peptide tested that binds to α5β1 integrin, suggesting the importance of this receptor for MB localization and detection.
AF680–EETI 2.5F also gave the highest total signal and greatest tumor contrast in ex vivo imaging experiments (Fig. 3 B and D and Table 1). Ex vivo tumor contrast for AF680–EETI 2.5F was 6.5 ± 0.6 (mean ± SE), at least twice what was measured for any other peptide (Table 1). Total ex vivo tumor signal for AF680–EETI 2.5D was above the background level of the AF680–EETI RDG control peptide but was fourfold less than that measured for AF680–EETI 2.5F (Fig. 3 B and D and Table 1). In comparison, AF680–AgRP 7C and AF680–c(RGDfK) exhibited ex vivo imaging signals that were roughly half those measured with AF680–EETI 2.5F. Although α5β1 integrin-binding specificity appeared to be responsible for in vivo MB imaging signals observed with AF680–EETI 2.5F, the ability of other integrin-binding peptides to generate low levels of specific ex vivo MB imaging signals suggests that αvβ3 (and perhaps αvβ5) integrin might also contribute to tumor targeting.
An EETI 2.5F–Fc Fusion Illuminates Intracranial MB and Distributes Throughout Brain Tumor Tissue.
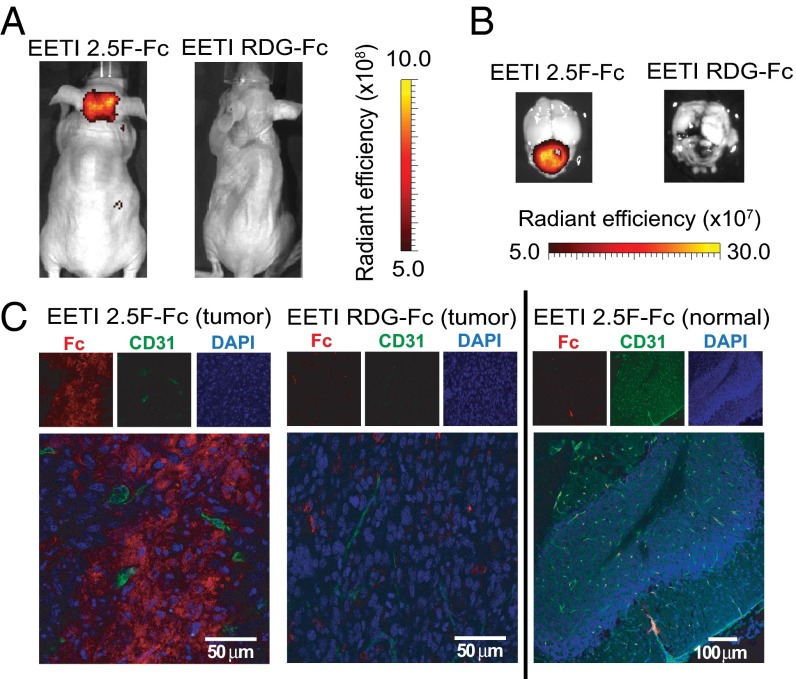
Recent theoretical and experimental studies have examined the relationship between molecular size and affinity needed for effective tumor targeting (35, 36). In particular, small proteins have been shown to accumulate rapidly in tumors, provided that they bind their targets with high affinities (i.e., in the pM to nM range) (37, 38). To study the effects of molecular size on brain tumor targeting and illumination in MB, we fused EETI 2.5F to the fragment crystallizable (Fc) region of mouse IgG2a. The resulting construct, EETI 2.5F–Fc, is a larger, bivalent version of EETI 2.5F (60 kDa versus 3.5 kDa). AF680–EETI 2.5F–Fc exhibited robust targeting of intracranial MB in the orthotopic Med1-MB model, as measured by in vivo and ex vivo optical imaging 2 h postinjection (Fig. 4 A and B). The control construct AF680–EETI RDG–Fc, which contains a scrambled integrin-binding sequence, failed to illuminate intracranial MB, indicating that the tumor-targeting ability of AF680–EETI 2.5F–Fc relies on specific integrin-binding interactions. Postmortem confocal imaging revealed extensive dissemination of AF680–EETI 2.5F–Fc throughout the tumor tissue (Fig. 4C, Left). Costaining for CD31 in the same tissue section confirmed that AF680–EETI 2.5F–Fc was located in both blood vessels and the tumor parenchyma. In contrast, there was minimal distribution of the control protein AF680–EETI RDG–Fc within tumor tissue (Fig. 4C, Center), indicating that tumor localization of AF680–EETI 2.5F–Fc depended upon its integrin-binding properties. Additionally, AF680–EETI 2.5F–Fc staining was not observed in healthy cerebellar tissue adjacent to the tumor (Fig. 4C, Right). These results indicate that small and large integrin-targeting proteins can illuminate MB in vivo and that AF680–EETI 2.5F–Fc can access and bind specifically to brain tumor cells through integrin-mediated interactions.
Fig. 4.
AF680–EETI 2.5F–Fc illuminates MB tumors in vivo and distributes throughout the tumor tissue. Representative in vivo (A) and ex vivo (B) images of signal from tumor-bearing mice injected with AF680–EETI 2.5F–Fc and AF680–EETI RDG–Fc. Images were taken 2 h after probe injection. (C) Brain tissue was processed for histology 2 h postinjection. AF680–EETI 2.5F–Fc was detected with an anti-Fc antibody (red) in tumor tissue costained with the vasculature marker CD31 (green), revealing widespread distribution within the tumor (Left) that was absent in the adjacent normal cerebellum (Right). In contrast, the AF680–EETI RDG–Fc control showed minimal distribution in tumor tissue (Center).
Discussion
Due to their critical role in mediating tumor survival and progression, integrins have been identified as potential targets for therapeutic intervention and molecular imaging applications (reviewed in refs. 18 and 39). Numerous integrin-binding agents have been developed (11, 20), but their ability to specifically target intracranial brain tumors has been limited. Unlike previously developed molecules which bind only αvβ3, or in some cases αvβ3 and αvβ5, EETI 2.5F is a first-in-class peptide that binds with high affinity (single-digit nM) and specificity to three tumor-associated integrins: αvβ3, αvβ5, and α5β1. Peripheral injection of EETI 2.5F, conjugated to a near-infrared fluorescent dye, illuminated intracranial tumors with significantly higher imaging contrast compared with other integrin-targeting agents. Given its high tumor specificity and unique ability to bind to α5β1 integrins, EETI 2.5F has great potential for brain tumor targeting in applications such as fluorescence-guided surgical resection or tumor-specific pharmacotherapy.
Numerous peptides and peptidomimetics containing an RGD integrin-binding motif have been developed and used as molecular imaging agents. Many of these probes have limitations such as weak target-binding affinity, suboptimal imaging contrast, and undesirable pharmacokinetics or tissue distribution, particularly for the difficult challenge of targeting brain tumors. For example, an RGD-containing cyclic peptide [c(RGDyK)], conjugated to a near-infrared dye, could detect αvβ3 integrin in s.c. mouse xenograft models (40–42), but attempts to image glioblastoma cells injected into the mouse forebrain were unsuccessful (40). Removal of the skull before imaging revealed only minimal contrast between tumor cells and normal brain (43). Several factors appear to have contributed to the enhanced tumor imaging contrast observed with EETI 2.5F compared with other integrin-targeting agents. First, the high integrin-binding affinity of EETI 2.5F generated increased imaging signals compared with molecules with relatively weak integrin-binding affinities, such as RGD-containing cyclic pentapeptides. Additionally, EETI 2.5F binds to a broad range of tumor-associated integrin receptors, including α5β1 integrin, which appears to be a critical factor for MB targeting. Brain tumors other than MB also display αvβ3, αvβ5, or α5β1 integrins; for example, recent evidence suggests that α5β1 integrin plays an important role in drug-resistant and aggressive glioblastoma (13, 16). High levels of at least one of these three integrin subtypes have also been found in a variety of tumors including melanoma; glioma; non–small-cell lung cancer; ovarian cancer; and tumors of the prostate, pancreas, cervix, and breast (18). Thus, EETI 2.5F has potentially wide-reaching applications for tumor targeting beyond MB.
Another benefit of EETI 2.5F is the ability to conjugate a variety of imaging probes directly to the knottin N terminus without disrupting integrin binding (26, 44). In comparison, modifications to some RGD-containing molecules to make them suitable for imaging applications have had limited success. For example, derivatives of Cilengitide containing lysine or cysteine substitutions as chemical handles for prerequisite probe attachment have significantly weaker integrin-binding affinities [ref. 45 and c(RGDfK) in Table 1]. Many RGD-containing molecules have required multimerization and the attachment of carbohydrates or synthetic polymers to improve tumor targeting, pharmacokinetic profiles, and tissue distribution (42, 45). Although such modifications could potentially further enhance the tumor imaging contrast observed with EETI 2.5F, they make manufacturing and clinical development more expensive and challenging. Thus, a significant advantage of EETI 2.5F compared with other integrin-targeted imaging agents is its high tumor-targeting efficiency, rapid blood clearance, and lack of accumulation in nontarget organs and tissue (i.e., kidneys, liver, and normal brain), without the need for additional modifications. In addition to EETI 2.5F, an imaging agent based on chlorotoxin, a naturally occurring knottin peptide derived from the venom of the deathstalker scorpion, has also shown promise for MB detection (46, 47). In future studies, a side-by-side comparison of these two knottin peptides as brain tumor imaging agents would be instructive.
One potential translational application of EETI 2.5F is intraoperative guidance for improved surgical resection of brain tumors. The extent of resection in many primary and metastatic brain tumors has been correlated with survival (7–9, 48) because incomplete removal of tumors can lead to treatment-resistant recurrences. To help demarcate tumor from normal tissue, fluorescent EETI 2.5F could allow a surgeon to more clearly visualize remaining tumor tissue after removal of obvious bulk tumor. Various integrin subtypes have been associated with inflammatory responses (49, 50), raising potential concerns for nonspecific probe binding; however, αvβ3, αvβ5, and α5β1 integrins have not been reported to be present at high levels in inflamed brain tissue. In Europe, 5-aminolevulinic acid (5-ALA) has been used for fluorescence-guided resection of glioblastoma, leading to more complete tumor resection (51). Limitations of 5-ALA imaging include rapid photobleaching, difficult timing and methodology of dosing, and reliance upon nonspecific heme metabolism for probe uptake in tumors. Integrin-targeted imaging of brain tumors with EETI 2.5F could potentially improve upon 5-ALA imaging in three ways: (i) EETI 2.5F binding is not dependent on tumor metabolism; (ii) EETI 2.5F could be readministered as needed throughout a surgical procedure; and (iii) EETI 2.5F could be conjugated to a variety of fluorophores to obtain desired spectroscopic properties and compatibility with different surgical instruments and detection systems while 5-ALA can only be visualized within a limited spectral range.
The blood–brain barrier is considered a major obstacle for efficient drug delivery to the central nervous system due to its limited paracellular permeability as well as its abundance of active efflux transporters (52). It is known that some molecules can access brain tumor tissue through a compromised blood–tumor barrier (53, 54), whereas other molecules exploit active transport–import mechanisms to traverse intact vasculature in the brain (55). The precise mechanism by which engineered integrin-binding knottin peptides access the tumor parenchyma from the bloodstream is currently being investigated. Regardless of whether EETI 2.5F or EETI 2.5F–Fc can cross an intact BBB, experiments with nontargeting control proteins demonstrate that integrin-specific binding is essential for MB localization compared with healthy brain tissue.
In summary, distal i.v. injection of integrin-binding knottin proteins EETI 2.5F and EETI 2.5F–Fc allowed detection of intracranial MB in mouse models. Brain tumor imaging was driven by the ability of these proteins to bind α5β1 integrin. The difficulty in targeting tumor markers in the brain and the great sensitivity and specificity of EETI 2.5F and EETI 2.5F–Fc in delineating tumor versus normal surrounding brain tissue make these engineered knottin proteins particularly promising for clinical applications.
Materials and Methods
Please also see SI Materials and Methods.
Peptide Synthesis and Fluorescent Labeling.
Knottin peptides were prepared as previously described in detail (23, 25). Briefly, the linear peptide sequences were made by solid-phase peptide synthesis on a CS Bio (Menlo Park, CA) instrument using standard 9-fluorenylmethyloxycarbonyl chemistry. Knottin peptides were folded by promoting disulfide bond formation in oxidizing buffer at room temperature with gentle rocking overnight. Folded knottin peptides were purified by reversed-phase HPLC, where they appeared as a sharp peak with a shorter retention time than unfolded or misfolded precursors. The molecular masses of folded knottins were determined by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (Stanford Protein and Nucleic Acid Facility) (Fig. S1C). Folded knottin peptides (2 mg/mL) were incubated with an amine-reactive succinimidyl ester derivative of Alexa Fluor 680 carboxylic acid in 0.1 M sodium bicarbonate, pH 8.0, at a 5:1 dye/peptide molar ratio for 1 h at room temperature and then at 4 °C overnight. The resulting dye-conjugated knottins were purified by RP-HPLC (Fig. S1B), and masses were confirmed by MALDI-TOF mass spectrometry (Fig. S1C). Conjugation of AF680 to commercially available c(RGDfK) (Peptides International) was performed in a similar manner. AF680-labeled peptides were lyophilized and resuspended in PBS, and concentration was determined using UV-visible spectroscopy, measuring absorption of the dye at 679 nm (ε = 184,000 cm−1⋅M−1). AF680-labeled peptides, at a concentration of 15 µM in PBS, were passed through a 0.22-µm filter for animal experiments. Because each peptide has exactly one fluorophore per peptide molecule, fluorescent imaging signals are comparable when administering a standard dose of 1.5 nmol labeled peptide per animal.
Genetic and Transplant Models of Medulloblastoma.
Animal procedures were carried out according to a protocol approved by Stanford University Administrative Panels on Laboratory Animal Care. Ptch+/− mice, with and without the Math1-GFP transgene, were housed and bred in the Stanford University animal facility. For tumor cell implantation, 6-wk-old male nude mice (Charles River Laboratories) were anesthetized, and 4 µL of a suspension of 6 × 106 Med1-MB cells in 50 µL of Dulbecco’s PBS was stereotactically injected in the cerebellar hemisphere.
In Vivo and ex Vivo Tumor Imaging.
Mice were defined as symptomatic from MB when they exhibited altered motor coordination, hunched posture, tilted head, or weight loss. Mice were anesthetized with isofluorane and injected with 1.5 nmol of AF680–peptide in 100 µL of PBS via the tail vein. For Ptch+/− mice, heads were shaved to allow better transmission of excitation light and signal emission. Mice were anesthetized with isofluorane and imaged at 2 h after peptide injection, unless otherwise specified, using an IVIS 200 system (Caliper Life Sciences). The AF680 near-infrared fluorophore was detected using an excitation range of 615–665 nm and monitoring emission signals at 695–770 nm. Background autofluorescence was measured using an excitation range of 580–610 nm and monitoring emission signals at 695–770 nm. In each imaging set, a mouse not injected with fluorescent probe was included to measure the background signal for data processing. For ex vivo imaging, mice were euthanized at 2 h, and whole brains were excised and imaged using the same excitation and emission settings as for in vivo imaging experiments. Brain tissue was fixed overnight in 4% (vol/vol) PFA at 4 °C for histological processing.
Supplementary Material
Acknowledgments
The authors acknowledge Dr. Ervin Epstein for his generous contribution of Med1-MB cultured cells and Dr. Amato Giaccia for the pADD2 mammalian cell expression vector. All in vivo imaging was conducted in the Stanford Small Animal Imaging Facility, with Tim Doyle, Frezghi Habte, and Laura Pisani providing advice on image acquisition and quantitation. The authors also thank Carmen Avram for providing assistance with mouse work and Kathleen Hershberger for assistance with protein expression and purification. This work has been funded in part by a Wallace H. Coulter Foundation Translational Partnership Award, the V Foundation for Cancer Research, the James S. McDonnell Foundation, a Stanford Cancer Institute Developmental Research Award, and the Stanford Center for Children’s Brain Tumors at Lucile Packard Children’s Hospital. The authors acknowledge fellowship support from the following sources: National Science Foundation Graduate Research Fellowship, Stanford Graduate Fellowship (Medtronic Fellow), Siebel Scholars Fellowship, and Gerald J. Lieberman Fellowship (to S.J.M.); California Institute of Regenerative Medicine (M.G.H.G.); and the Stanford Child Health Research Institute, the Lucile Packard Foundation for Children’s Health, and Stanford CTSA UL1 RR025744 (to J.M.B.). M.P.S. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311333110/-/DCSupplemental.
References
- 1.Huse JT, Holland EC. Targeting brain cancer: Advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10(5):319–331. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- 2.Gibson P, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468(7327):1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollack IF. Multidisciplinary management of childhood brain tumors: A review of outcomes, recent advances, and challenges. J Neurosurg Pediatr. 2011;8(2):135–148. doi: 10.3171/2011.5.PEDS1178. [DOI] [PubMed] [Google Scholar]
- 4.Saury JM, Emanuelson I. Cognitive consequences of the treatment of medulloblastoma among children. Pediatr Neurol. 2011;44(1):21–30. doi: 10.1016/j.pediatrneurol.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Paulino AC, et al. Local control after craniospinal irradiation, intensity-modulated radiotherapy boost, and chemotherapy in childhood medulloblastoma. Cancer. 2011;117(3):635–641. doi: 10.1002/cncr.25601. [DOI] [PubMed] [Google Scholar]
- 6.Wu X, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482(7386):529–533. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albert FK, Forsting M, Sartor K, Adams HP, Kunze S. Early postoperative magnetic resonance imaging after resection of malignant glioma: Objective evaluation of residual tumor and its influence on regrowth and prognosis. Neurosurgery. 1994;34(1):45–61. doi: 10.1097/00006123-199401000-00008. [DOI] [PubMed] [Google Scholar]
- 8.Devaux BC, O’Fallon JR, Kelly PJ. Resection, biopsy, and survival in malignant glial neoplasms. A retrospective study of clinical parameters, therapy, and outcome. J Neurosurg. 1993;78(5):767–775. doi: 10.3171/jns.1993.78.5.0767. [DOI] [PubMed] [Google Scholar]
- 9.Lacroix M, et al. A multivariate analysis of 416 patients with glioblastoma multiforme: Prognosis, extent of resection, and survival. J Neurosurg. 2001;95(2):190–198. doi: 10.3171/jns.2001.95.2.0190. [DOI] [PubMed] [Google Scholar]
- 10.Cox D, Brennan M, Moran N. Integrins as therapeutic targets: Lessons and opportunities. Nat Rev Drug Discov. 2010;9(10):804–820. doi: 10.1038/nrd3266. [DOI] [PubMed] [Google Scholar]
- 11.Gaertner FC, Schwaiger M, Beer AJ. Molecular imaging of αvß3 expression in cancer patients. Q J Nucl Med Mol Imaging. 2010;54(3):309–326. [PubMed] [Google Scholar]
- 12.Brooks PC, et al. Integrin alpha v beta 3 antagonists promote tumor regression by inducing apoptosis of angiogenic blood vessels. Cell. 1994;79(7):1157–1164. doi: 10.1016/0092-8674(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 13.DeLay M, et al. Microarray analysis verifies two distinct phenotypes of glioblastomas resistant to antiangiogenic therapy. Clin Cancer Res. 2012;18(10):2930–2942. doi: 10.1158/1078-0432.CCR-11-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedlander M, et al. Definition of two angiogenic pathways by distinct alpha v integrins. Science. 1995;270(5241):1500–1502. doi: 10.1126/science.270.5241.1500. [DOI] [PubMed] [Google Scholar]
- 15.Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285(5430):1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 16.Janouskova H, et al. Integrin α5β1 plays a critical role in resistance to temozolomide by interfering with the p53 pathway in high-grade glioma. Cancer Res. 2012;72(14):3463–3470. doi: 10.1158/0008-5472.CAN-11-4199. [DOI] [PubMed] [Google Scholar]
- 17.Kim S, Bell K, Mousa SA, Varner JA. Regulation of angiogenesis in vivo by ligation of integrin alpha5beta1 with the central cell-binding domain of fibronectin. Am J Pathol. 2000;156(4):1345–1362. doi: 10.1016/s0002-9440(10)65005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desgrosellier JS, Cheresh DA. Integrins in cancer: Biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10(1):9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodman SL, Picard M. Integrins as therapeutic targets. Trends Pharmacol Sci. 2012;33(7):405–412. doi: 10.1016/j.tips.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Dechantsreiter MA, et al. N-methylated cyclic RGD peptides as highly active and selective alpha(V)beta(3) integrin antagonists. J Med Chem. 1999;42(16):3033–3040. doi: 10.1021/jm970832g. [DOI] [PubMed] [Google Scholar]
- 21.Haubner R, Beer AJ, Wang H, Chen X. Positron emission tomography tracers for imaging angiogenesis. Eur J Nucl Med Mol Imaging. 2010;37(Suppl 1):S86–S103. doi: 10.1007/s00259-010-1503-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z, Wang F. Dual-targeted molecular probes for cancer imaging. Curr Pharm Biotechnol. 2010;11(6):610–619. doi: 10.2174/138920110792246546. [DOI] [PubMed] [Google Scholar]
- 23.Kimura RH, Levin AM, Cochran FV, Cochran JR. Engineered cystine knot peptides that bind alphavbeta3, alphavbeta5, and alpha5beta1 integrins with low-nanomolar affinity. Proteins. 2009;77(2):359–369. doi: 10.1002/prot.22441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolmar H. Natural and engineered cystine knot miniproteins for diagnostic and therapeutic applications. Curr Pharm Des. 2011;17(38):4329–4336. doi: 10.2174/138161211798999465. [DOI] [PubMed] [Google Scholar]
- 25.Kimura RH, Cheng Z, Gambhir SS, Cochran JR. Engineered knottin peptides: a new class of agents for imaging integrin expression in living subjects. Cancer Res. 2009;69(6):2435–2442. doi: 10.1158/0008-5472.CAN-08-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore SJ, Leung CL, Cochran JR. Knottins: Disulfide-bonded therapeutic and diagnostic peptides. Drug Discov Today Technol. 2011;9(1):e3–e11. doi: 10.1016/j.ddtec.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Lumpkin EA, et al. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr Patterns. 2003;3(4):389–395. doi: 10.1016/s1567-133x(03)00089-9. [DOI] [PubMed] [Google Scholar]
- 28.Hahn H, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85(6):841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 29.Johnson RL, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272(5268):1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 30.Cho YJ, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29(11):1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Northcott PA, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29(11):1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silverman AP, Levin AM, Lahti JL, Cochran JR. Engineered cystine-knot peptides that bind alpha(v)beta(3) integrin with antibody-like affinities. J Mol Biol. 2009;385(4):1064–1075. doi: 10.1016/j.jmb.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haubner R, et al. Structural and functional aspects of RGD-containing cyclic pentapeptides as highly potent and selective integrin alphavbeta3 antagonists. J Am Chem Soc. 1996;118(32):7461–7472. [Google Scholar]
- 34.Schnell O, et al. Imaging of integrin alpha(v)beta(3) expression in patients with malignant glioma by [18F] Galacto-RGD positron emission tomography. Neuro-oncol. 2009;11(6):861–870. doi: 10.1215/15228517-2009-024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt MM, Wittrup KD. A modeling analysis of the effects of molecular size and binding affinity on tumor targeting. Mol Cancer Ther. 2009;8(10):2861–2871. doi: 10.1158/1535-7163.MCT-09-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wittrup KD, Thurber GM, Schmidt MM, Rhoden JJ. Practical theoretic guidance for the design of tumor-targeting agents. Methods Enzymol. 2012;503:255–268. doi: 10.1016/B978-0-12-396962-0.00010-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orlova A, et al. Tumor imaging using a picomolar affinity HER2 binding affibody molecule. Cancer Res. 2006;66(8):4339–4348. doi: 10.1158/0008-5472.CAN-05-3521. [DOI] [PubMed] [Google Scholar]
- 38.Zahnd C, et al. Efficient tumor targeting with high-affinity designed ankyrin repeat proteins: Effects of affinity and molecular size. Cancer Res. 2010;70(4):1595–1605. doi: 10.1158/0008-5472.CAN-09-2724. [DOI] [PubMed] [Google Scholar]
- 39.Beer AJ, Schwaiger M. Imaging of integrin alphavbeta3 expression. Cancer Metastasis Rev. 2008;27(4):631–644. doi: 10.1007/s10555-008-9158-3. [DOI] [PubMed] [Google Scholar]
- 40.Chen X, Conti PS, Moats RA. In vivo near-infrared fluorescence imaging of integrin alphavbeta3 in brain tumor xenografts. Cancer Res. 2004;64(21):8009–8014. doi: 10.1158/0008-5472.CAN-04-1956. [DOI] [PubMed] [Google Scholar]
- 41.Wang W, et al. Near-infrared optical imaging of integrin alphavbeta3 in human tumor xenografts. Mol Imaging. 2004;3(4):343–351. doi: 10.1162/15353500200404148. [DOI] [PubMed] [Google Scholar]
- 42.Cheng Z, Wu Y, Xiong Z, Gambhir SS, Chen X. Near-infrared fluorescent RGD peptides for optical imaging of integrin alphavbeta3 expression in living mice. Bioconjug Chem. 2005;16(6):1433–1441. doi: 10.1021/bc0501698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsu AR, et al. In vivo near-infrared fluorescence imaging of integrin alphavbeta3 in an orthotopic glioblastoma model. Mol Imaging Biol. 2006;8(6):315–323. doi: 10.1007/s11307-006-0059-y. [DOI] [PubMed] [Google Scholar]
- 44.Kimura RH, Miao Z, Cheng Z, Gambhir SS, Cochran JR. A dual-labeled knottin peptide for PET and near-infrared fluorescence imaging of integrin expression in living subjects. Bioconjug Chem. 2010;21(3):436–444. doi: 10.1021/bc9003102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mas-Moruno C, Rechenmacher F, Kessler H. Cilengitide: The first anti-angiogenic small molecule drug candidate design, synthesis and clinical evaluation. Anticancer Agents Med Chem. 2010;10(10):753–768. doi: 10.2174/187152010794728639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stroud MR, Hansen SJ, Olson JM. In vivo bio-imaging using chlorotoxin-based conjugates. Curr Pharm Des. 2011;17(38):4362–4371. doi: 10.2174/138161211798999375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veiseh M, et al. Tumor paint: A chlorotoxin:Cy5.5 bioconjugate for intraoperative visualization of cancer foci. Cancer Res. 2007;67(14):6882–6888. doi: 10.1158/0008-5472.CAN-06-3948. [DOI] [PubMed] [Google Scholar]
- 48.Stummer W, et al. Fluorescence-guided resection of glioblastoma multiforme by using 5-aminolevulinic acid-induced porphyrins: A prospective study in 52 consecutive patients. J Neurosurg. 2000;93(6):1003–1013. doi: 10.3171/jns.2000.93.6.1003. [DOI] [PubMed] [Google Scholar]
- 49.Constantin G. Chemokine signaling and integrin activation in lymphocyte migration into the inflamed brain. J Neuroimmunol. 2008;198(1-2):20–26. doi: 10.1016/j.jneuroim.2008.04.023. [DOI] [PubMed] [Google Scholar]
- 50.Milner R, Campbell IL. Cytokines regulate microglial adhesion to laminin and astrocyte extracellular matrix via protein kinase C-dependent activation of the alpha6beta1 integrin. J Neurosci. 2002;22(5):1562–1572. doi: 10.1523/JNEUROSCI.22-05-01562.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stummer W, et al. ALA-Glioma Study Group Fluorescence-guided surgery with 5-aminolevulinic acid for resection of malignant glioma: A randomised controlled multicentre phase III trial. Lancet Oncol. 2006;7(5):392–401. doi: 10.1016/S1470-2045(06)70665-9. [DOI] [PubMed] [Google Scholar]
- 52.Deeken JF, Löscher W. The blood-brain barrier and cancer: Transporters, treatment, and Trojan horses. Clin Cancer Res. 2007;13(6):1663–1674. doi: 10.1158/1078-0432.CCR-06-2854. [DOI] [PubMed] [Google Scholar]
- 53.Gerstner ER, Fine RL. Increased permeability of the blood-brain barrier to chemotherapy in metastatic brain tumors: Establishing a treatment paradigm. J Clin Oncol. 2007;25(16):2306–2312. doi: 10.1200/JCO.2006.10.0677. [DOI] [PubMed] [Google Scholar]
- 54.Lockman PR, et al. Heterogeneous blood-tumor barrier permeability determines drug efficacy in experimental brain metastases of breast cancer. Clin Cancer Res. 2010;16(23):5664–5678. doi: 10.1158/1078-0432.CCR-10-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gabathuler R. Approaches to transport therapeutic drugs across the blood-brain barrier to treat brain diseases. Neurobiol Dis. 2010;37(1):48–57. doi: 10.1016/j.nbd.2009.07.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.