Abstract
Integration of the DNA copy of the HIV-1 genome into a host chromosome is required for viral replication and is thus an important target for antiviral therapy. The HIV-encoded enzyme integrase (IN) catalyzes two essential steps: 3′ processing of the viral DNA ends, followed by the strand transfer reaction, which inserts the viral DNA into host DNA. Raltegravir binds to IN and blocks the integration of the viral DNA. Using the Rous sarcoma virus-derived vector RCAS, we previously showed that mutations that cause one viral DNA end to be defective for IN-mediated integration led to abnormal integrations in which the provirus had one normal and one aberrant end, accompanied by rearrangements in the host genome. On the basis of these results, we expected that suboptimal concentrations of IN inhibitors, which could block one of the ends of viral integration, would lead to similar aberrant integrations. In contrast to the proviruses from untreated cells, which were all normal, ∼10–15% of the proviruses isolated after treatment with a suboptimal dose of raltegravir were aberrant. The aberrant integrations were similar to those seen in the RCAS experiments. Most of the aberrant proviruses had one normal end and one aberrant end and were accompanied by significant rearrangements in the host genome, including duplications, inversions, deletions and, occasionally, acquisition of sequences from other chromosomes. The rearrangements of the host DNA raise concerns that these aberrant integrations might have unintended consequences in HIV-1–infected patients who are not consistent in following a raltegravir-containing treatment regimen.
Keywords: strand transfer inhibitor, chromosomal rearrangements, integrase inhibitors
HIV-1 integration is a two-step process: first, in the cytoplasm, an integrase (IN) dimer binds to each end of the newly synthesized linear viral DNA and removes two nucleotides from each of the 3′ ends, exposing the conserved CA dinucleotide. The preintegration complex (PIC) is translocated from the cytoplasm into the nucleus, where the viral DNA is integrated into the host genome. In the strand transfer (ST) reaction, the two exposed 3′ hydroxyl groups on the newly processed viral DNA ends attack phosphodiester bonds on the opposite strands of the target DNA at positions that lie across the major groove, 4–6 bp apart. This reaction is carried out by a tetramer of IN; each of the viral DNA ends is associated with an IN dimer. This transesterification reaction generates an intermediate in which the 3′ ends of the viral DNA are covalently joined to the host DNA and there are 4- to 6-bp gaps in the host DNA associated with both of the 5′ ends of the viral DNA. Cellular machinery repairs these gaps, creating a 4- to 6-bp duplication (the size of the duplication varies for different retroviruses) of the host DNA flanking the integrated viral DNA (1–4). HIV-1 integration generates a 5-bp duplication of the host DNA at the integration site (5, 6).
Raltegravir (RAL) and all of the potent IN inhibitors thus far discovered bind to the active site of IN and target the ST reaction. For this reason they are referred to as integrase strand transfer inhibitors (INSTIs) (7–11). The recent crystal structures of the prototype foamy virus (PFV) IN, in complexes with viral DNA and, in some cases, with viral DNA and inhibitors, sheds light on the mechanism of action of the INSTIs. PFV IN is structurally similar to HIV IN, especially in the region around the active site (7, 12–15). RAL and the other known INSTIs have two essential components: a metal binding region that binds the two magnesium ions in the active site of HIV-1 IN, and a modified phenyl ring that displaces the nucleobase of the A at the 3′ end of the viral DNA and stacks on the penultimate C (Fig. S1).
Using the Rous sarcoma virus (RSV)-derived RCAS vector system, we previously showed that when viral DNA was mutated so that one end was not a good substrate for IN, the viral DNA could still be integrated with moderate efficiency. However, most of the resulting proviruses were abnormal (16). The data strongly suggested that the IN mediated the insertion of the “good” end of the viral DNA. However, if the mutation of the other end prevented IN from carrying out the ST reaction, the insertion of the blocked end could still occur with moderate efficiency (approximately 30% of WT), but this insertion was mediated by host enzymes and usually involved rearrangements of the host DNA, which included duplications, deletions, inversions, and the acquisition of sequences from other chromosomes (16).
If the concentration of an INSTI is below optimal levels, only one of the two IN-mediated ST reactions would be blocked. This could have consequences similar to having one end of the viral DNA mutated. Thus, a suboptimal dose of an INSTI could allow one end of the viral DNA to be inserted normally (by IN), and the other (blocked) end would be inserted by host enzymes, leading to aberrant integrations. To test this prediction, we infected cells either in the absence of an INSTI or in the presence of a range of suboptimal concentrations of RAL. We recovered full-length proviruses flanked on both ends by host genomic DNA and determined their structures. A suboptimal concentration of RAL led to aberrant integrations that affected one end and, more rarely, both ends of the viral DNA. The aberrant integrations are accompanied by rearrangement of the host DNA adjacent to the integration site, including large duplications, deletions, inversions and, rarely, the acquisition of sequences from other chromosomes. We also analyzed proviruses from cells treated with higher doses (>IC99) of RAL and found that a majority of these proviruses had not one but two aberrant ends and these aberrant integrations were also accompanied by rearrangements of the host genome.
Results
Types of Viral DNAs Recovered from Cells Infected in the Absence of RAL.
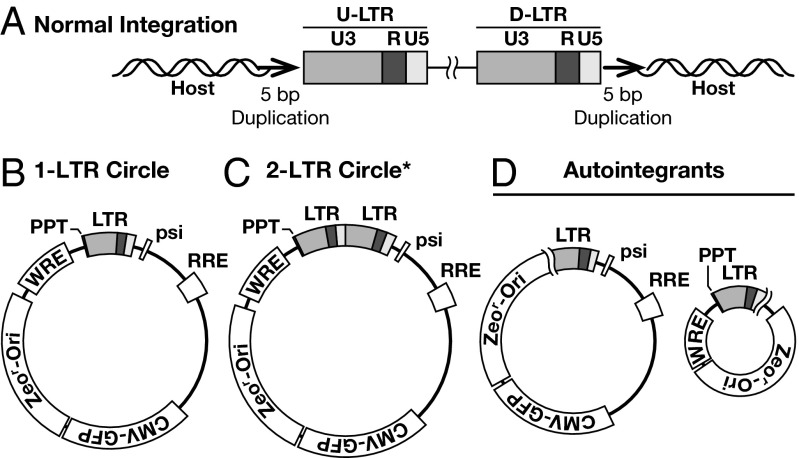
Viral DNAs were recovered from human osteosarcoma (HOS) cells infected with vesicular stomatitis virus G protein (VSV-G) pseudotyped HIV-1 vector that were either treated with different suboptimal concentrations or with high concentrations of RAL, and from parallel untreated control cells. We recovered both integrated and unintegrated viral DNAs (Fig. 1): the integrated viral DNAs were flanked on both ends by the host DNA, and the unintegrated forms of DNA included 1- and 2-LTR circles, autointegrants, and aberrant circles.
Fig. 1.
Viral DNAs recovered from cells infected with HIV-1. Viral LTRs are shown as shaded boxes: the dark gray box represents the U3 region, R is shown by a black box, and U5 is indicated by the light gray box. (A) The structure of normal proviruses with a 5-bp duplication of the host DNA sequences (small bold arrow) flanking the provirus. Unintegrated forms of circular viral DNA include (B) 1-LTR circles formed by the homologous recombination of the LTRs, (C) 2-LTR circles formed by end-joining of the two LTRs, and (D) autointegrants formed by integration of the LTR into different regions of the viral DNA itself. *Although we did not recover any 2-LTR circles in the experiment in which viral DNAs were isolated from untreated cells, these are one of the types of unintegrated viral DNA circles typically found in cells infected with HIV-1, and DNAs of this type were isolated from RAL-treated cells.
All of the proviruses we recovered from cells infected in the absence of RAL (a total of 99) were normal (Table 1) and had features expected for HIV-1 proviruses: two nucleotides were removed from the 3′ ends of the viral DNA, and the insertion of the viral DNA created a 5-bp duplication of the host DNA at the target site (Fig. 1A). The ends of the unintegrated linear form of the vector DNA exactly match the ends of the linear DNA from replication competent HIV-1. Thus, the integration of the vector DNA should be the same as replication competent HIV-1 DNA; this expectation was confirmed in the experiments done in the absence of RAL. The majority of the viral DNAs recovered from untreated cells were derived from integrated proviruses; only 10 of a total of 120 recovered viral DNAs were derived from unintegrated circular forms of viral DNA (Fig. 1 B–D and Table 1). The sequences of the viral DNAs from the remaining 11 samples were uninterpretable. We suspect, but did not prove, that many of these uninterpretable DNAs are aberrant unintegrated circular forms that had lost a significant amount of sequence that corresponds to one or both ends of the linear DNA, making them difficult to sequence with primers that match the viral LTRs.
Table 1.
Recovery of integrated viral DNA
| RAL, nM | Aberrant integration | Normal integration | 1-LTR (aberrant 1-LTR) | 2-LTR (aberrant 2-LTR) | Autointegrants (aberrant circles) | Total no. of samples recovered and sequenced |
| HOS | ||||||
| No drug | 0 | 99 | 7 (0) | 0 (0) | 1 (2) | 120 |
| 2.4 (IC14) | 5 | 34 | 159 (13) | 20 (18) | 13 (53) | 384 |
| 5.1 (IC30) | 4 | 14 | 167 (22) | 46 (22) | 7 (47) | 384 |
| 8.5 (IC50) | 1 | 7 | 83 (11) | 21 (5) | 7 (25) | 192 |
| 10.2 (IC60) | 1 | 7 | 87 (8) | 25 (5) | 5 (30) | 192 |
| 13.5 (IC75) | 1 | 8 | 35 (3) | 1 (2) | 8 (11) | 96 |
| PBMC | ||||||
| No drug | 0 | 85 | 188 (3) | 0 (0) | 4 (23) | 360 |
| 5.1 (IC30) | 5 | 69 | 204 (6) | 2 (3) | 7(28) | 413 |
The concentration of RAL that was used to treat the cells is indicated, and the corresponding inhibitory concentration is indicated in parenthesis. The number of normal and aberrant integrations obtained from the total number of viral DNAs recovered and sequenced is indicated. The number and types of unintegrated circular products recovered are indicated, and the number of aberrant forms of the viral DNAs we recovered are indicated in parenthesis. At all concentrations of RAL used in our experiments, we recovered a few viral DNAs whose sequences were uninterpretable.
Recovery of Viral DNAs from Cells Treated with Suboptimal Doses of RAL.
Viral DNAs were recovered from cells treated with several different concentrations of RAL. In our system, the IC50 for RAL is 8.5 nM. We determined the effects of RAL at concentrations ranging from 2.4 nM to 13.5 nM. In contrast to the viral DNAs recovered from untreated cells, a larger proportion of the viral DNAs recovered from RAL-treated cells were unintegrated circular forms. This is the expected result; RAL interferes with integration, and a larger fraction of the viral DNA is converted into the various circular forms (14). In contrast to what was seen with the proviruses recovered from untreated cells, we recovered both normal and aberrant proviruses from RAL-treated cells. Almost all of the aberrant viral integrations had one normal end and one aberrant end; these aberrant integrations were accompanied by the insertion or deletion of viral DNA sequences (Fig. S2 A and B). The aberrant proviruses could be divided into several different types according to the structure of the host DNA flanking the provirus. In the first type, the integration event created a large duplication of the host DNA at the site of integration (Fig. S2C). In the second type, there was both a duplication and an inversion of the flanking host DNA (Fig. S2D). In the third type, there was a deletion in the host DNA sequence at the site of integration (Fig. S2E), and in the fourth type, a fragment from a different chromosome was inserted next to the aberrant end of the provirus (Fig. S2F). In rare instances, both the ends of the proviral DNA were aberrant; these proviruses were also accompanied by rearrangements of the host genome (Fig. S2G). In experiments done at all of the concentrations of RAL, the sequences of a few of the recovered viral DNAs were uninterpretable.
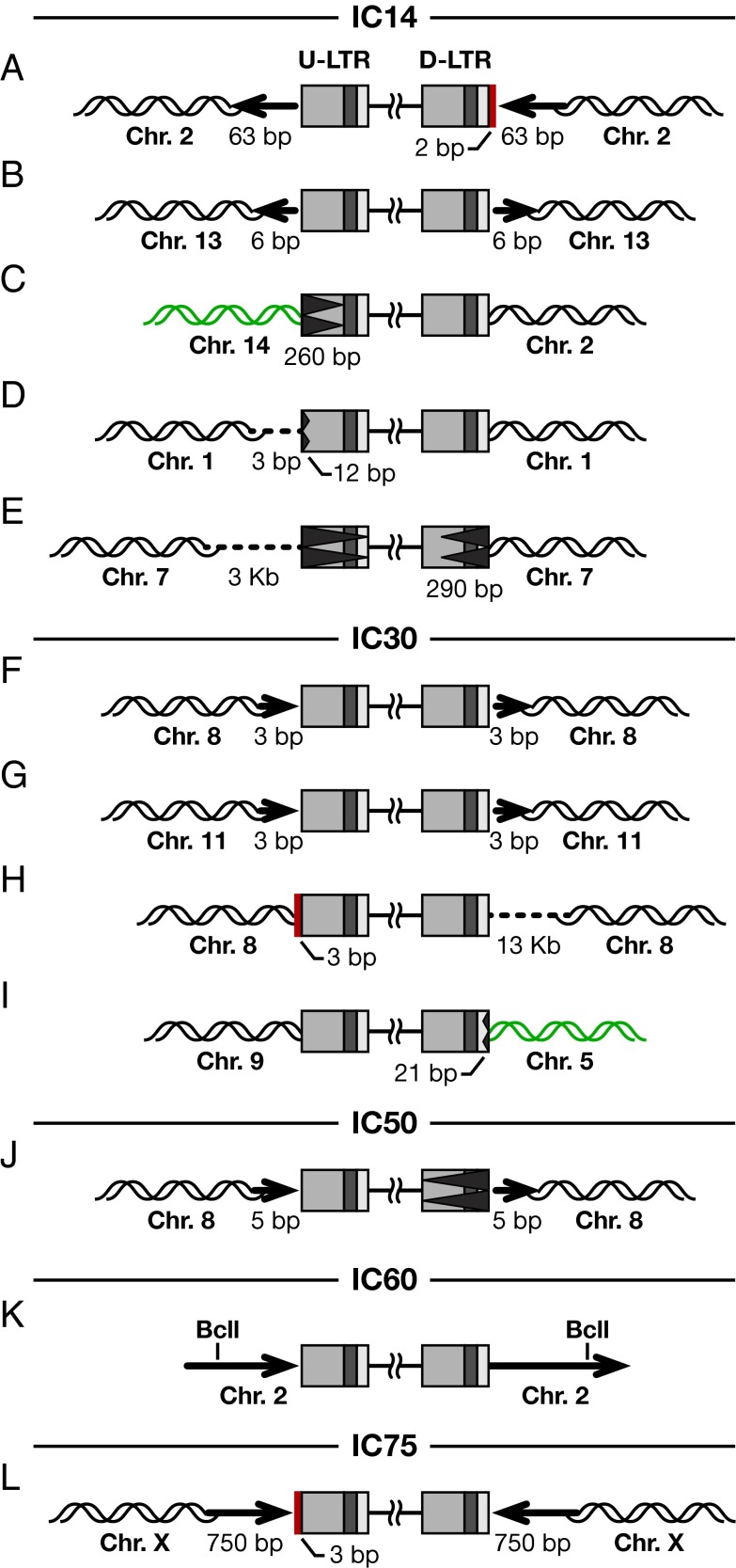
Of 384 viral DNAs recovered from cells treated with 2.4 nM RAL (IC14), we recovered 34 normal and 5 aberrant proviruses (Table 1); the remaining viral DNAs whose sequences we could interpret were unintegrated circular forms. As shown in Fig. 2, with 2.4 nM RAL, four out of the five aberrant proviruses had one normal end and one aberrant end; in the remaining provirus, both ends were aberrant. In one recovered provirus (Fig. 2A), there was an insertion of two extra nucleotides, AG, at the junction between the host and the viral DNA. These two bases did not match either the end of the viral DNA or the flanking host DNA but could have been copied from some other viral or host DNA sequence (Discussion). The host DNA flanking the integration site consisted of a duplication of 63 bp (Fig. 2A). Both ends of the second aberrant provirus seemed to be normal (Fig. 2B); however, the provirus was flanked by a host DNA duplication of 6 bp, and this duplicated host DNA fragment was inverted. Even among the aberrant proviruses, inversion of the host sequences flanking the provirus was relatively rare (a mechanism for generating an inversion is discussed below) (Fig. S3). In the third provirus, 260 bp of the upstream-LTR (U-LTR) (in the provirus, the U-LTR is adjacent to the primer binding site) were deleted, and there was an insertion of sequences from a different host chromosome (chromosome 14) at the viral DNA/host DNA junction (Fig. 2C). The fourth provirus had a 12-bp deletion at the end of the U-LTR and a deletion of 3 bp from the host DNA at the integration site (Fig. 2D). The fifth aberrant provirus had deletions from both ends of the viral DNA (Fig. 2E); the U-LTR was entirely deleted, and 290 bp were deleted from the downstream-LTR (D-LTR) (the D-LTR is adjacent to the polypurine tract). There was also a deletion of 3 kb in the host DNA at the integration site.
Fig. 2.
Structure of aberrant proviruses isolated from cells treated with suboptimal concentrations of RAL. (A–L) Proviruses recovered from cells treated with RAL at IC14, IC30, IC50, IC60, and IC75. Viral LTRs are shown as shaded boxes: the dark gray box represents the U3 region, R is shown by the black box, and U5 is indicated by the light gray box. Insertion of additional bases at the viral DNA ends is indicated by a solid red box. Deletion from the viral DNA is indicated by a black jagged end; deletion of the entire LTR is indicated by the jagged end stretching through the LTR. Large duplications of flanking host DNA are indicated by long bold arrows; the direction of the arrowheads indicates the orientation of the flanking host sequences. Acquisition of sequences from a different host chromosome is indicated by green DNA strands. Deletion of sequences from the host chromosome is indicated by a hashed line. In one case (K), there was a BclI restriction enzyme site in the duplicated host sequence. Digestion with BclI and subsequent self-ligation of the digested fragment, during the recovery of the integrated provirus, led to truncation of the flanking host sequences. The numbers of nucelotides that were deleted or inserted are indicated.
When cells were treated with 5.1 nM RAL (IC30), of 384 viral DNAs recovered and sequenced, 14 were normal proviruses and 4 were aberrant proviruses; the other recovered viral DNAs were unintegrated circular forms (Table 1). All four of the aberrant proviruses had one normal and one abnormal end. The first two aberrant proviruses had a shorter than normal host DNA duplication at the integration site, 3 bp rather than 5 bp (Fig. 2 F and G). The third aberrant provirus (Fig. 2H) had an insertion of three extra nucleotides, GTT, at the end of the U-LTR, and there was a large deletion (approximately 13 kb) in the host sequences at the integration site. The presence of the extra GT at the end of the U-LTR suggests that the 3′ end was not cleaved by IN; the two nucleotides removed in the 3′ processing reaction are GT. There was a deletion of 21 bp in the D-LTR of the fourth provirus (Fig. 2I), and, despite the fact that the normal end of this provirus was inserted into chromosome 9, the aberrant end of the provirus was flanked by sequences from chromosome 5.
As the concentration of RAL was increased to 8.5 nM, 10.2 nM, or 13.5 nM (IC50, IC60, and IC75, respectively), fewer proviruses were recovered. However, one aberrant provirus was recovered at each of these suboptimal concentrations (Table 1). The aberrant provirus recovered from cells treated with 8.5 nM RAL had lost the entire D-LTR; however, there was a 5-bp duplication at the host DNA flanking the integrated viral DNA (Fig. 2J). The aberrant provirus recovered from cells treated with 10.2 nM RAL appeared to be flanked by a relatively large direct repeat of host sequences; however, we did not recover the entire duplicated segment because there was a BclI restriction enzyme recognition site within the repeated sequence (Fig. 2K). As shown in Fig. 2L, the aberrant provirus recovered from cells that were treated with 13.5 nM RAL had an insertion of three extra nucleotides, GTA, at the end of the U-LTR, and as indicated earlier, GT matches the viral sequences that are present in an unprocessed viral DNA end. This provirus was flanked by a 750bp DNA duplication of the host sequences at the target site; however, in this case, the duplication was an inverted rather than a direct repeat.
Besides the normal and aberrant proviruses, in the presence of suboptimal doses of RAL, ∼80–90% of the viral DNAs we recovered were unintegrated circular viral DNAs including 1- and 2-LTR circles, and other circular forms, some of which could have been autointegrants (Table 1).
To prove that the results we obtained were not affected by the use of the HOS cell line, we did additional experiments using peripheral blood mononuclear cells (PBMCs) obtained from healthy donors. The results obtained in the experiments done in PBMCs were similar to the results obtained in HOS cells. There were no aberrant integrations in the absence of RAL; however, in the presence of 5.1 nM RAL, ∼7% of the proviruses were aberrant (Table 1). The same types of aberrant integrations were seen in PBMCs and HOS cells.
Joining of Aberrant Viral DNA to the Host Genome Often Involves Microhomology.
We previously showed, in experiments done with viral DNAs that had one end mutated to prevent the modified end from being a good substrate for IN, that the viral–host DNA junctions of the aberrant integration events frequently had short stretches of homology (16). We proposed that these regions of microhomology were the result of the participation of a host DNA polymerase in the generation of the aberrant integrations (Discussion). Because it is likely that the same, or similar, host enzymes are responsible for the aberrant integrations in the experiments done with suboptimal concentrations of RAL, we examined the viral–host DNA junctions for microhomologies. In 67% of the RAL-induced aberrant integrations, there was microhomology of one to seven nucleotides at the virus–host junction (Table 2). Of the 12 aberrant integrations obtained at suboptimal concentrations of RAL, only 4 showed no homology at the virus–host junction. This is higher than the frequency at which microhomologies are seen in normal integrations, where the presence of matching host–viral sequences corresponds to what would be expected from chance (16).
Table 2.
Microhomology at virus–host DNA junctions in aberrant proviruses
| RAL | Provirus | U-LTR | D-LTR |
| IC14 | A | cttcca | tctagca |
| B | cttcca | tctaGCA | |
| C | caaacct | tctagca | |
| D | gagtgaA | tctagca | |
| E | cttcca | tctagca | |
| IC30 | F | cttcca | tctagcA |
| G | cttcCA | tctagca | |
| H | cttcca | tctagCA | |
| I | cttcca | ttttagTTA | |
| IC50 | J | cttcca | tctagcA |
| IC60 | K | cttcca | TCTAGCA |
| IC75 | L | cttcca | tctagca |
Seven nucleotides from the U-LTR and D-LTR virus–host DNA junctions are shown. Boldface capital letters indicate viral sequences showing microhomology with host DNA. The inhibitory concentration of RAL used is shown. The letters A to L correspond to those in Fig. 2.
High Doses of RAL Cause Aberrant Integrations.
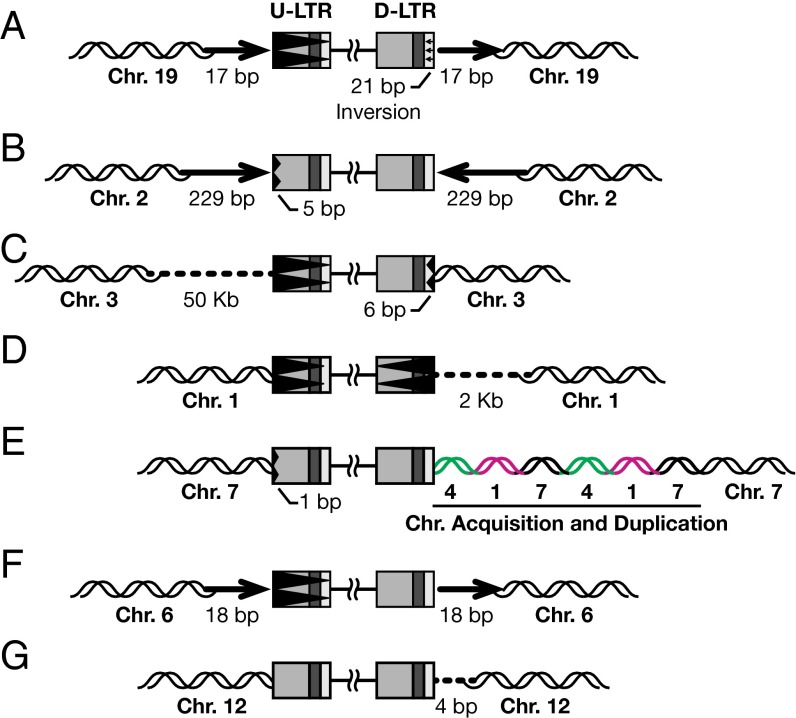
We tested whether high doses (2 µM, >IC99) of RAL would completely abolish integration, or whether there would be some residual integration events. Of a total of 1,100 viral DNA sequences analyzed, 7 were integrated into the host DNA. Although the total number of integrated proviruses was very low, all of the proviruses we recovered were aberrant. As shown in Fig. 3, and unlike our results with suboptimal doses of RAL, both the viral DNA ends of more than half of the proviruses recovered from cells treated with a high concentration of RAL were aberrant. In the first recovered provirus, the U-LTR was entirely deleted, and there was a 21-bp inversion in the U5 region of the D-LTR and a 17-bp DNA duplication of the host DNA at the target site (Fig. 3A). The second provirus had a 5-bp deletion in the U3 region of the U-LTR that was associated with the duplication and inversion of a 229-bp fragment of the host DNA (Fig. 3B). The third and fourth proviruses each had deletions in both of the viral DNA ends and a deletion of host sequences flanking the proviruses (Fig. 3 C and D). The fifth provirus had a small deletion in the U3 region of the U-LTR, and the host DNA flanking the D-LTR had short fragments from two other chromosomes inserted and duplicated as shown in Fig. 3E. The U-LTR was entirely deleted in the sixth provirus, and the flanking host DNA had an 18-bp duplication (Fig. 3F). In the seventh provirus, there was a deletion in the host DNA at the target site (Fig. 3G). These results indicate that viral DNA can still be inserted into the host genome even at a concentration of RAL far above the IC50. Not surprisingly, these aberrant integrations were accompanied by deletions, duplications, or other rearrangements of the host genome.
Fig. 3.
Structures of the aberrant proviruses isolated from cells treated with a high concentration of RAL. (A-G) Proviruses recovered from cells treated with 2 µM RAL. Viral LTRs are shown as shaded boxes. The aberrations in viral and host DNA sequences are indicated according to the scheme described in Fig. 2. In one case (E) DNA was acquired from two different chromosomes; this is indicated by green and pink DNA strands. The numbers of nucleotides that were deleted or inserted are indicated.
Discussion
When the concentration of RAL is suboptimal, the IN-mediated integration of one end of some of the viral DNAs is blocked by the drug, and the blocked end is inserted by host enzymes. In contrast to IN-mediated integrations, the host-mediated insertions often involved microhomology of 1–7 bp at the host–virus DNA junction, suggesting the involvement of a host DNA polymerase. A host polymerase could use the 3′ end of the viral DNA to copy a variety of nearby host or viral DNA sequences. These viral DNA primed copying events would explain the presence of extra viral sequences and/or sequences from other chromosomes at the aberrant integration sites. Ultimately, to create an integrated provirus, the copying/insertion reaction would have to involve the same chromosome into which the “good” viral end was already integrated. Copying a segment of the chromosome into which the “good” end was integrated would create a perfectly matched DNA segment, which would be a substrate for homologous recombination. Depending on whether this final copying/insertion of the viral DNA occurs to the right or left of the initial IN-mediated insertion, a deletion or duplication of the host genome would be generated (Fig. S3 A, B, and D).
The fact that all of the normal integrations that we have recovered have the normal 5-bp duplication of the host DNA and the IN inhibitors preferentially block the ST reaction suggests that the duplications smaller than 5 bp are also the result of a host enzyme-mediated insertion of one of the viral DNA ends. We believe that this is also the most likely explanation for the structure of the provirus in Fig. 2 F and G, and in Fig. 2J, where the entire D-LTR is missing but there is a 5-bp duplication of the host DNA at the integration site. This conjecture is supported by the fact that there was microhomology at the virus–host DNA junction sequences of all three of the aberrant proviruses flanked by short host DNA repeats (Fig. 2 and Table 2). Although the number of proviruses flanked by short repeats is low, the presence of microhomology at the junctions in all three cases supports the inference that these are a result of host-mediated insertions.
Two proviruses recovered from cells infected with virus in the presence of suboptimal doses of RAL were flanked by an inverted repeat of the host DNA (Fig. 2 B and L). This could happen if, in the initial host-mediated DNA copying reaction, the 3′ viral DNA end copied a DNA segment from the same strand of host DNA in which the original (IN-mediated) insertion occurred. Insertion of this segment into the host DNA, in a subsequent copying/recombination step, would create an inversion (Fig. S3C).
If the copying/insertion of the aberrant 3′ viral end involves host sequences to the right of the original insertion (Fig. S3D), it would lead to the deletion of the host sequences. We recovered three aberrant proviruses that had deletions of the host chromosome at the site of integration, fewer than those with duplications. This is consistent with the results obtained in the experiments done with the RCAS vector system and suggests that the host-mediated insertion has a preference for one side of the original (IN-mediated) insertion (16). In the case of the proviruses in Fig. 2 C and I, the proviruses had acquired fragments from a different chromosome. If, as we propose, the insertions involve a copying reaction carried out by a host DNA polymerase, copying sequences from a nearby chromosome is an obvious possibility.
RAL has a much smaller effect on the 3′ processing of the viral DNA ends than on the insertion of viral DNA (17). As might have been expected, in some of the aberrant proviruses, both the ends of the viral DNA seem to have been processed normally, with two nucleotides removed from each 3′ end. However, in the proviruses shown in Fig. 2 H and L, the nucleotides GT are present at the aberrant end of the viral DNA. These match the sequence found at the ends of unprocessed viral DNA; however, these two proviruses also have a third inserted nucleotide, either a T or an A, respectively, and these additional nucleotides could have been derived by copying either host or viral DNA. The two additional nucleotides found at the end of the viral DNA in the provirus shown in Fig. 2A do not match either the viral or the adjacent host DNA sequences. This insertion could be a result of a host polymerase copying these two nucleotides from some portion of either the viral or the host DNA before copying sequences from the host genome at the final integration site. A copying/recombination model can also be used to explain the structures of the aberrant unintegrated circles that we recovered in the presence of suboptimal doses of RAL (Fig. S4).
Our data also show that the aberrations seen in the viral and the host DNAs are created as a result of using suboptimal doses of RAL and are not merely rare aberrant integrations that are uncovered owing to the inhibition of normal integrations. The suboptimal doses of RAL reduced the titer of the virus by 25–85%. Approximately 10–15% of the integrations recovered were aberrant. So, if the aberrant integrations were uncovered by RAL treatment, ∼5–10% of the integrations in our untreated samples would have been aberrant. However, the fact that we did not find any aberrant integrations in a sample of 99 proviruses obtained from infections in untreated cells shows that the aberrant integrations seen in the presence of RAL were the result of the action of the drug.
We also showed that high doses of RAL lead to rare aberrant integration events. Approximately 1% of the viral DNAs that we recovered from infections done in the presence of high doses of RAL were integrated into the host genome, and all of them were aberrant. In more than half the proviruses, both ends were aberrant, and these aberrant integrations were accompanied by rearrangements in the host genome. These inserted proviruses are similar to the aberrant proviruses that arise in the absence of functional IN (16, 18, 19). Recently, Ebina et al. (20) also reported that HIV-1 integrations occurred at a reduced efficiency using either an IN-deficient virus or a wild-type virus in the presence of high doses of IN inhibitors. They showed that either high doses of the drug or the active site mutations in IN seemed to change the integration site preference and caused deletions or insertions of nucleotides at the LTR–genomic DNA junction. However, this report did not describe the structure of the proviruses, nor did it describe the impact of the drug on the structure of adjacent host sequences.
Because all of the INSTIs bind to the PIC and prevent the integration in a similar way, we expect that any drug in this class will have similar effects. In patients for whom their drug therapy completely blocks viral replication, it is unlikely that treatment with an INSTI would lead to a significant number of aberrant integrations. However, if a patient who is taking an INSTI is not compliant, he/she will experience periodic suboptimal doses of the drug. Noncompliance could lead to the development of drug resistance. The half-life of RAL has been estimated to be approximately 8 h in patients (21), and we show that a range of suboptimal doses of RAL can lead to aberrant integrations. This suggests that there might be a 1- to 2-d window of suboptimal dosing that could lead to aberrant integrations in a patient who stops a therapeutic regimen that contains RAL. If the virus becomes resistant to the INSTI, and the patient stays on an RAL-containing therapeutic regimen, it is likely that the concentration of the drug will be suboptimal. Because one or both of these two situations occur in infected patients, there are reasons to be concerned. In many cases, malignancies that have arisen in HIV-infected patients are known to be associated with infections by herpes viruses that are normally controlled by the host’s immune system (22–24). However, non–AIDS-defining malignancies have been on the increase since the availability of combination antiretroviral therapy, and the underlying cause of these additional malignancies is poorly understood (25–27). Although the activation of protooncogenes due to normal HIV-1 integration seems to be extremely rare, which is one of the reasons lentiviral vectors are being tested for gene therapy, there is a report suggesting that HIV integration can cause oncogene activation (28). The fact that the treatment with INSTIs leads to aberrant insertions that are accompanied by rearrangements of the host genome could increase the potential for the insertional activation of oncogenes and/or the inactivation of genes encoding tumor suppressors. We suggest, when malignancies arise in patients treated with INSTIs, that DNA from these malignancies should be analyzed to determine whether aberrant integrations may have contributed to the development of the malignancies.
Materials and Methods
Plasmid Construction.
A four-vector system was used to generate the virus stocks. A ClaI-MluI shuttle cassette that contains sequences that permit the replication of circular forms of viral DNA as plasmids in Escherichia coli was derived from pHIV-SH (29). The cassette was shortened by removing the Pol II promoter. The final cassette contained a zeocin resistance gene with an upstream EM-7 Promoter (EM-zeo), a lac operator sequence, and a ColE1 origin of replication (oriE). A coding region for enhanced GFP (eGFP), under the control of the cytomegalovirus promoter (CMV), was inserted as a NotI-ClaI fragment immediately upstream of the plasmid-recovery cassette. The plasmid used to produce the viral RNA, pSICO-LZF, was derived from pSICO-XBX by inserting the shuttle cassette and the eGFP coding region as a NotI-MluI fragment (30). The viral RNA was expressed from a chimeric 5′ LTR with a CMV promoter in place of U3, and the vector also contained an HIV Psi packaging/Rev Response element (RRE)/RNA export signal (31). The four-vector system included, in addition to pSICO-LZF, a plasmid expressing HIV-1 Gag and Gag-Pol: pMDL-SH.IN+; a plasmid that expresses REV: pRSV-REV; and a plasmid that expresses VSV-G: pCMV-VSV-G. pMDL-SH.IN+ was derived from pMDLg/pRRE by replacing the gag and pol genes with the equivalent sequences from pHIV-SH (29). Both the pRSV-REV and the parental pMDLg/pRRE expression plasmids were obtained from Didier Trono (École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) through Addgene, Inc. (31).
Cells, Transfection, and Infection.
Human embryonic kidney 293T and HOS cells were maintained in DMEM (Cellgro) supplemented with 5% (vol/vol) FBS, 5% (vol/vol) newborn calf serum, 100 U/mL penicillin G, and 100 µg/mL streptomycin (Quality Biological). Recombinant virus stocks were generated by calcium phosphate-mediated cotransfection of 293T cells seeded at 1.5 × 106 cells in 100-mm culture plates with the plasmids that make up the four-vector system. Thirteen micrograms of pSICO-LZF (or pSICO-LZR), 12 µg pMDL-SH.IN+, 5 µg pRSV-REV, and 4 µg pCMV-VSV-G were used per plate. Six hours after transfection, cells were gently washed three times with PBS, and fresh media was added. Virus-containing supernatants were harvested 48 h after transfection, clarified by centrifugation at 1620 × g for 10 min, and incubated at 37 °C with 500 U of DNase I (Invitrogen) per 50 mL virus in 5 mM MgCl2 to remove any residual vector DNA carried over from the transfection. HOS cells were seeded at 7.5 × 105 per 100-mm plate the day before infection. PBMCs from healthy human donors were seeded at a minimum of 5 × 106 cells per 25-cm2 cell culture flask (Corning) and maintained in RPMI media supplemented with 10% FBS, 100 U/mL penicillin G, 100 µg/mL streptomycin, 2 mM L-glutamine, and 50 U IL-2. The PBMCs were activated with 5 µg/mL phytohemagglutinin-P (PHA-P) for 48–72 h. The cells were then spun down and resuspended in fresh media containing IL-2. Drug-free cells were infected with 50 ng/mL p24 recombinant virus in the presence of 4 µg/mL of polybrene (Sigma-Aldrich). The cells that were treated with various suboptimal doses of RAL (a gift from Daria Hazuda, Merck Research Laboratories, West Point, PA) were preincubated with the drug for 3 h at 37 °C (PBMCs were treated with 5.1 nM RAL) and then infected with 50 ng/mL p24 recombinant virus in the presence of 4 µg/mL of polybrene. The appropriate concentration of RAL was maintained during the infection. After 24 h of incubation with the virus, the virus-containing media was removed and replaced with fresh media (without virus) in the drug-free cells, whereas fresh media (without virus) containing the appropriate concentrations of RAL was added to the drug-treated cells. The cells were harvested 6 d after infection, and DNA was extracted using the QIAmp DNA Blood Kit from Qiagen.
Recovery of Integrated Retroviral DNA.
Genomic DNAs (100 µg) isolated from the infected HOS cells or PBMCs were treated with DpnI for 2 h at 37 °C to eliminate any remaining plasmid DNA that might have been carried over from the transfection. The DpnI-digested DNA was heated at 80 °C for 20 min and then ethanol precipitated. The DNA was digested overnight with BclI at 50 °C, extracted with phenol/chloroform, and ethanol precipitated. DNA was resuspended in 890 µL of nuclease-free water and was self-ligated overnight at 16 °C in the presence of T4 DNA Ligase and ligation buffer (NEB) in a final volume of 1 mL. The mix was ethanol precipitated and resuspended in 100 µL of nuclease-free water.
DNA (up to 700 ng) was introduced into ElectroMAX DH10B E. coli cells by electroporation using the BTX Electroporation System at 186 Ω and 2.5-kV resistance. The bacterial cells were allowed to recover in 500 µL supra optimal broth with catabolite repression (SOC) media for 3 h at 37 °C with shaking, and plated on L-broth plates containing 100 µg/mL zeocin. The next day, colonies were picked and grown overnight in L-broth containing 100 µg/mL zeocin. DNA was purified using the Qiagen BIO ROBOT Universal System.
Sequencing of the Integrated Viral DNA.
Recovered plasmids were directly sequenced using the primers LTR-FOR (5′ GACTTACAAGGCAGCTGTAG), which hybridizes to the vector in a region that just precedes the PPT, and pSICO REV (5′ GCCTCTTGCCGTGCGCGCTTC), which hybridizes near the PBS, between the LTR and gag. All sequencing was performed by Macrogen (Rockville, MD). In some cases the proviruses had sustained such large deletions that it was necessary to use additional primers to do additional sequencing to determine the end of the provirus. Human and viral sequences were analyzed by BLAST.
Supplementary Material
Acknowledgments
We thank Dr. Daria Hazuda, who generously provided the RAL used in the experiments; Dr. Eric Freed, Ms. Sherimay Ablan and Dr. Angelica Martins for generously providing the PBMCs, IL-2, and PHA-P; Dr. Barry Johnson for helping with the HIV-1 intasome model in Fig. S1; Ms. Andrea Ferris for constructing the pSICO-LZF plasmid and providing cells for the experiments; Mr. Richard Frederickson for help with figures; and Ms. Teresa Burdette for help with the manuscript. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.E. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305066110/-/DCSupplemental.
References
- 1.Brown PO, Bowerman B, Varmus HE, Bishop JM. Retroviral integration: Structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci USA. 1989;86(8):2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cherepanov P, Maertens GN, Hare S. Structural insights into the retroviral DNA integration apparatus. Curr Opin Struct Biol. 2011;21(2):249–256. doi: 10.1016/j.sbi.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Delelis O, Carayon K, Saïb A, Deprez E, Mouscadet JF. Integrase and integration: Biochemical activities of HIV-1 integrase. Retrovirology. 2008;5:114. doi: 10.1186/1742-4690-5-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li M, Mizuuchi M, Burke TR, Jr, Craigie R. Retroviral DNA integration: Reaction pathway and critical intermediates. EMBO J. 2006;25(6):1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffin JM, Hughes SH, Varmus HE. The interactions of retroviruses and their hosts. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1997. [PubMed] [Google Scholar]
- 6.Flint SJ, Enquist LW, Krug RM, Racaniello VR, Skalka AM. Principles of Virology: Molecular Biology, Pathogenesis and Control. Washington, DC: ASM Press; 2000. [Google Scholar]
- 7.Arts EJ, Hazuda DJ. HIV-1 antiretroviral drug therapy. Cold Spring Harb Perspect Med. 2012;2(4):a007161. doi: 10.1101/cshperspect.a007161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazuda D, Iwamoto M, Wenning L. Emerging pharmacology: Inhibitors of human immunodeficiency virus integration. Annu Rev Pharmacol Toxicol. 2009;49:377–394. doi: 10.1146/annurev.pharmtox.011008.145553. [DOI] [PubMed] [Google Scholar]
- 9.Hicks C, Gulick RM. Raltegravir: The first HIV type 1 integrase inhibitor. Clin Infect Dis. 2009;48(7):931–939. doi: 10.1086/597290. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen BY, et al. Raltegravir: The first HIV-1 integrase strand transfer inhibitor in the HIV armamentarium. Ann N Y Acad Sci. 2011;1222:83–89. doi: 10.1111/j.1749-6632.2011.05972.x. [DOI] [PubMed] [Google Scholar]
- 11.Summa V, et al. Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J Med Chem. 2008;51(18):5843–5855. doi: 10.1021/jm800245z. [DOI] [PubMed] [Google Scholar]
- 12.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010a;464(7286):232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hare S, et al. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc Natl Acad Sci USA. 2010b;107(46):20057–20062. doi: 10.1073/pnas.1010246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hazuda DJ. Resistance to inhibitors of the human immunodeficiency virus type 1 integration. Braz J Infect Dis. 2010;14(5):513–518. [PubMed] [Google Scholar]
- 15.Xue W, Liu H, Yao X. Molecular mechanism of HIV-1 integrase-vDNA interactions and strand transfer inhibitor action: A molecular modeling perspective. J Comput Chem. 2012;33(5):527–536. doi: 10.1002/jcc.22887. [DOI] [PubMed] [Google Scholar]
- 16.Oh J, Chang KW, Hughes SH. Mutations in the U5 sequences adjacent to the primer binding site do not affect tRNA cleavage by rous sarcoma virus RNase H but do cause aberrant integrations in vivo. J Virol. 2006;80(1):451–459. doi: 10.1128/JVI.80.1.451-459.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hazuda DJ, et al. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science. 2000;287(5453):646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 18.Gaur M, Leavitt AD. Mutations in the human immunodeficiency virus type 1 integrase D,D(35)E motif do not eliminate provirus formation. J Virol. 1998;72(6):4678–4685. doi: 10.1128/jvi.72.6.4678-4685.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreau K, Torne-Celer C, Faure C, Verdier G, Ronfort C. In vivo retroviral integration: Fidelity to size of the host DNA duplication might be reduced when integration occurs near sequences homologous to LTR ends. Virology. 2000;278(1):133–136. doi: 10.1006/viro.2000.0641. [DOI] [PubMed] [Google Scholar]
- 20.Ebina H, et al. Integrase-independent HIV-1 infection is augmented under conditions of DNA damage and produces a viral reservoir. Virology. 2012;427(1):44–50. doi: 10.1016/j.virol.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, et al. Pharmacokinetic modeling of plasma and intracellular concentrations of raltegravir in healthy volunteers. Antimicrob Agents Chemother. 2011;55(9):4090–4095. doi: 10.1128/AAC.00593-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carbone A, Cesarman E, Spina M, Gloghini A, Schulz TF. HIV-associated lymphomas and gamma-herpesviruses. Blood. 2009;113(6):1213–1224. doi: 10.1182/blood-2008-09-180315. [DOI] [PubMed] [Google Scholar]
- 23.Chang Y, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266(5192):1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 24.Schulz TF. KSHV/HHV8-associated lymphoproliferations in the AIDS setting. Eur J Cancer. 2001;37(10):1217–1226. doi: 10.1016/s0959-8049(01)00115-0. [DOI] [PubMed] [Google Scholar]
- 25.Deeken JF, et al. The rising challenge of non-AIDS-defining cancers in HIV-infected patients. Clin Infect Dis. 2012;55(9):1228–1235. doi: 10.1093/cid/cis613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiels MS, et al. Cancer burden in the HIV-infected population in the United States. J Natl Cancer Inst. 2011;103(9):753–762. doi: 10.1093/jnci/djr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spano JP, et al. AIDS-related malignancies: State of the art and therapeutic challenges. J Clin Oncol. 2008;26(29):4834–4842. doi: 10.1200/JCO.2008.16.8252. [DOI] [PubMed] [Google Scholar]
- 28.Mack KD, et al. HIV insertions within and proximal to host cell genes are a common finding in tissues containing high levels of HIV DNA and macrophage-associated p24 antigen expression. J Acquir Immune Defic Syndr. 2003;33(3):308–320. doi: 10.1097/00126334-200307010-00004. [DOI] [PubMed] [Google Scholar]
- 29.Oh J, McWilliams MJ, Julias JG, Hughes SH. Mutations in the U5 region adjacent to the primer binding site affect tRNA cleavage by human immunodeficiency virus type 1 reverse transcriptase in vivo. J Virol. 2008;82(2):719–727. doi: 10.1128/JVI.02611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ventura A, et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci USA. 2004;101(28):10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abram ME, Ferris AL, Shao W, Alvord WG, Hughes SH. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. J Virol. 2010;84(19):9864–9878. doi: 10.1128/JVI.00915-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.