It has been 20 y since the gene responsible for Huntington disease—a fatal neurodegenerative disorder—was identified by an intensive international collaborative effort (1). Although the features of the gene offered few clues to its normal function, the cloning of the IT15 gene immediately provided insight into the mechanism underlying disease: expansion of a CAG trinucleotide repeat within the first exon of the gene, which encodes a polyglutamine stretch in the huntingtin protein, was found to correlate with the onset of Huntington disease and the autosomal dominant mode of inheritance. At the time, this was a relatively novel observation, with the first example of a polyglutamine-encoding CAG repeat disease—spinal and bulbar muscular atrophy (Kennedy disease)—having been uncovered just 2 y prior (2). These diseases were the initial members of a group of at least nine polyglutamine-based neurodegenerative disorders that have been subsequently described (3). Interestingly, many of these diseases exhibit a similar critical polyglutamine length threshold that leads to pathogenesis (4). In the case of Huntington disease, alleles with a repeat size of 40 or greater are completely penetrant, whereas alleles with 36–39 repeats are incompletely penetrant (5), with genetic and environmental factors likely contributing to this variation. Amino-terminal fragments of huntingtin with expanded polyglutamine stretches misfold and form aggregates in vitro and in brains of Huntington patients (6, 7). A great deal of research supports the notion that misfolding/aggregation of huntingtin leads to a “toxic gain-of-function” mechanism at the root of disease pathogenesis, although loss of endogenous huntingtin function (8) and RNA-based toxicity (9) may also play a role in Huntington disease. A caveat of many of the gain-of-function studies is that extremely expanded polyglutamine tracts (often >90 repeats) have often been used to yield robust readouts in disease models, whereas polyglutamine lengths at the disease threshold fail to provide measurable phenotypes (10). The exact cellular function of huntingtin is not clear, but it is required for embryonic development, plays roles in axonal and vesicle transport, and controls production of the neurotrophin BDNF (8). In PNAS, Caron et al. (11) explore the normal biological role of the polyglutamine tract in huntingtin and find that it serves as a flexible hinge permitting interactions between flanking domains.
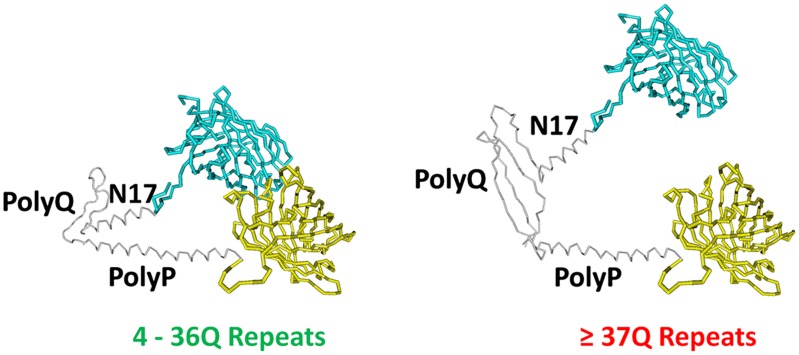
The polyglutamine region in huntingtin lies immediately adjacent to the first 17 amino-terminal residues of the protein, which constitutes a domain required for membrane localization (12, 13), and upstream of a polyproline region that enhances huntingtin interactions with lipid membranes (14). These studies use a Förster resonance energy transfer (FRET)-based conformation sensor to measure the intramolecular interactions between these polyglutamine flanking regions. Efficiency of energy transfer between donor (mCerulean) and acceptor (eYFP) fluorophores localized to the amino and carboxyl termini of huntingtin fragments were measured using fluorescent lifetime imaging microscopy (FLIM) in living cells (Fig. 1), giving a measure of the proximity of these domains. Testing versions of huntingtin-containing repeat lengths within the normal range (17 and 32), the authors find that the polyglutamine stretch acts as a flexible hinge permitting the amino-terminal domain to interact with the downstream polyproline region. Intriguingly, constructs with disease-causing polyglutamine lengths (46 and 138 repeats) reduced FRET efficiency, indicative of decreased flexibility and suggestive of a potential role in pathogenesis of disease. Strikingly, the authors also find that a repeat length at the clinical threshold of 37 was also able to significantly reduce polyglutamine flexibility; this is the first time a sufficiently sensitive readout has been developed to measure a phenotypic change at this repeat length. Interestingly, deletion of the polyglutamine stretch in the sensor eliminated the interaction of the flanking regions, whereas replacement of the polyglutamine stretch with a flexible glycine linker restored these interactions, further supporting the above observations.
Fig. 1.
A model of the huntingtin FRET sensor used to detect polyglutamine flexibility. A “normal” polyglutamine (polyQ) tract (4–36 repeats) is flexible and permits intramolecular interactions between the amino terminus of huntingtin (N17) and a carboxyl-terminal polyproline (polyP) stretch, bringing together the mCerulean and eYFP fluorophores, which can be quantified using FRET-FLIM approaches. Expanded disease-causing polyglutamine lengths of 37 repeats and greater disrupt this interaction. Artwork courtesy of Ray Truant (McMaster University) and modified from Caron et al. (11).
As two serine residues at positions 13 and 16 in the amino-terminal domain of huntingtin are known to be phosphorylated and modulate huntingtin localization and structure (13, 15), the authors used genetic and pharmacological approaches to investigate whether phospho-status of these residues modulates polyglutamine flexibility. The authors find that genetically mimicking phosphorylation by substituting these serines with glutamic acid leads to a significant reduction in interactions between the polyglutamine-flanking domains. Furthermore, treatment with Iκβ kinase inhibitors, which promotes phosphorylation of the amino-terminal serines, yielded a robust reduction in FRET efficiency, indicative of a conformational change similar to phospho-mimicry. Provocatively, increased phosphorylation of these serine residues improves symptoms in a mouse model of Huntington disease (16).
The nature of the FRET sensor developed for this study required the constructs being expressed at higher than physiological levels in the cell lines investigated, potentially introducing caveats into the experiments. To address this concern, Caron et al. developed an antibody-based FRET-FLIM assay, which exploited two fluorescently tagged antibodies known to specifically target the huntingtin amino-terminal and polyproline domains, respectively. This sensor permits the authors to investigate huntingtin conformations in cells derived from Huntington disease transgenic knock-in mice and a Huntington disease patient where huntingtin is expressed at endogenous levels. In both cases, the authors validate their previous results, finding that FRET efficiency dropped significantly in cells expressing expanded, disease-causing lengths of polyglutamine compared with normal controls.
Previous work has found that protein kinase C and casein kinase 2 substrate in neurons 1 (PACSIN1) interacts with the polyproline region in huntingtin, an interaction that is promoted with expanded polyglutamine (17, 18). PACSIN1 is an endocytic adaptor that is required for NMDA receptor recycling, including receptor subtypes containing GluN3A subunits (19). Implementing biochemical and microscopic approaches, Caron et al. find that PACSIN1 also interacts with the amino terminus of huntingtin, suggesting that PACSIN1 may help stabilize the interactions between these two polyglutamine flanking domains. This was confirmed using RNA interference to reduce expression of PACSIN1 in cells containing the huntingtin FRET sensors with either normal or expanded polyglutamine. The authors find that reduced PACSIN1 levels decreased FRET efficiency of the normal
This study by Caron et al. is an important step forward in the understanding of huntingtin and how polyglutamine expansion contributes to Huntington disease pathogenesis.
huntingtin sensor, but not in the case of expanded polyglutamine, supporting the notion that PACSIN1 acts as a bridge to stabilize these domains. A recent study has highlighted the importance of the PACSIN1–huntingtin interaction (20). This work finds that neurons expressing mutant huntingtin exhibit increased surface levels of GluN3A-containing NMDA receptors, which is likely due to sequestration of PACSIN1 by mutant huntingtin. Provocatively, overexpression of GluN3A in the striatum of WT mice recapitulated the striatal synapse loss observed in Huntington disease, whereas targeted deletion of GluN3A ameliorated phenotypes in a mouse model of the disease.
This study by Caron et al. (11) is an important step forward in the understanding of huntingtin and how polyglutamine expansion contributes to Huntington disease pathogenesis. The knowledge that the polyglutamine region permits conformational changes and intramolecular interactions within huntingtin, which are stabilized by PACSIN1, is likely applicable to other polyglutamine proteins and may have ramifications for other polyglutamine disorders. Particularly relevant here is that chemical approaches (e.g., Iκβ kinase inhibition) may be able to normalize polyglutamine flexibility of mutant huntingtin and may thus have therapeutic relevance. Furthermore, the development of a sensor that detects conformational changes of huntingtin at the clinical threshold will become an important tool for further functional and therapeutic exploration and, due to its simplicity, can be readily scaled up for high content screening. The biological necessity for polyglutamine flexibility in huntingtin is an outstanding question, but undoubtedly future studies aimed at elucidating its functional relevance will not only inform the basic biology of this protein but will also shed light on its role in disease pathogenesis and may ultimately contribute to therapeutic intervention for this devastating disorder.
Footnotes
The author declares no conflict of interest.
See companion article on page 14610.
References
- 1.The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72(6):971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352(6330):77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 3.Orr HT. Polyglutamine neurodegeneration: Expanded glutamines enhance native functions. Curr Opin Genet Dev. 2012;22(3):251–255. doi: 10.1016/j.gde.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11(11):786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bates GP. History of genetic disease: The molecular genetics of Huntington disease - a history. Nat Rev Genet. 2005;6(10):766–773. doi: 10.1038/nrg1686. [DOI] [PubMed] [Google Scholar]
- 6.Scherzinger E, et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90(3):549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 7.DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277(5334):1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 8.Zuccato C, Valenza M, Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol Rev. 2010;90(3):905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 9.Fiszer A, Krzyzosiak WJ. RNA toxicity in polyglutamine disorders: Concepts, models, and progress of research. J Mol Med (Berl) 2013;91(6):683–691. doi: 10.1007/s00109-013-1016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menalled LB, Chesselet MF. Mouse models of Huntington’s disease. Trends Pharmacol Sci. 2002;23(1):32–39. doi: 10.1016/s0165-6147(00)01884-8. [DOI] [PubMed] [Google Scholar]
- 11.Caron NS, Desmond CR, Xia J, Truant R. Polyglutamine domain flexibility mediates the proximity between flanking sequences in huntingtin. Proc Natl Acad Sci USA. 2013;110:14610–14615. doi: 10.1073/pnas.1301342110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rockabrand E, et al. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum Mol Genet. 2007;16(1):61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- 13.Atwal RS, et al. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet. 2007;16(21):2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- 14.Burke KA, Kauffman KJ, Umbaugh CS, Frey SL, Legleiter J. The interaction of polyglutamine peptides with lipid membranes is regulated by flanking sequences associated with huntingtin. J Biol Chem. 2013;288(21):14993–15005. doi: 10.1074/jbc.M112.446237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atwal RS, et al. Kinase inhibitors modulate huntingtin cell localization and toxicity. Nat Chem Biol. 2011;7(7):453–460. doi: 10.1038/nchembio.582. [DOI] [PubMed] [Google Scholar]
- 16.Di Pardo A, et al. Ganglioside GM1 induces phosphorylation of mutant huntingtin and restores normal motor behavior in Huntington disease mice. Proc Natl Acad Sci USA. 2012;109(9):3528–3533. doi: 10.1073/pnas.1114502109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Modregger J, DiProspero NA, Charles V, Tagle DA, Plomann M. PACSIN 1 interacts with huntingtin and is absent from synaptic varicosities in presymptomatic Huntington’s disease brains. Hum Mol Genet. 2002;11(21):2547–2558. doi: 10.1093/hmg/11.21.2547. [DOI] [PubMed] [Google Scholar]
- 18.Plomann M, et al. PACSIN, a brain protein that is upregulated upon differentiation into neuronal cells. Eur J Biochem. 1998;256(1):201–211. doi: 10.1046/j.1432-1327.1998.2560201.x. [DOI] [PubMed] [Google Scholar]
- 19.Pérez-Otaño I, et al. Endocytosis and synaptic removal of NR3A-containing NMDA receptors by PACSIN1/syndapin1. Nat Neurosci. 2006;9(5):611–621. doi: 10.1038/nn1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marco S, et al. Suppressing aberrant GluN3A expression rescues synaptic and behavioral impairments in Huntington’s disease models [published online ahead of print July 14, 2013] Nat Med. 2013 doi: 10.1038/nm.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]