Significance
There are six human amino-terminal acetyltransferases (NATs) that mediate a growing number of biological processes and are differentiated from one another on the basis of substrate specificity. Interestingly, only one more promiscuous NAT enzyme exists in archaea. The crystal structure of an archaeal NAT from Sulfolobus solfataricus (ssNAT), together with mutagenesis and kinetic analysis, reveal that the active site of ssNAT represents a hybrid of known eukaryotic NAT active sites. We highlight features of this protein that allow it to facilitate catalysis of distinct substrates through different catalytic strategies, which is a unique characteristic of this enzyme. The data presented here have implications for the evolution of eukaryotic NAT enzymes and substrate specificities therein.
Keywords: structural biology, evolutionary biology, enzymology, X-ray crystallography
Abstract
Amino-terminal acetylation is a ubiquitous modification in eukaryotes that is involved in a growing number of biological processes. There are six known eukaryotic amino-terminal acetyltransferases (NATs), which are differentiated from one another on the basis of substrate specificity. To date, two eukaryotic NATs, NatA and NatE, have been structurally characterized, of which NatA will acetylate the α-amino group of a number of nonmethionine amino-terminal residue substrates such as serine; NatE requires a substrate amino-terminal methionine residue for activity. Interestingly, these two NATs use different catalytic strategies to accomplish substrate-specific acetylation. In archaea, where this modification is less prevalent, only one NAT enzyme has been identified. Surprisingly, this enzyme is able to acetylate NatA and NatE substrates and is believed to represent an ancestral NAT variant from which the eukaryotic NAT machinery evolved. To gain insight into the evolution of NAT enzymes, we determined the X-ray crystal structure of an archaeal NAT from Sulfolobus solfataricus (ssNAT). Through the use of mutagenesis and kinetic analysis, we show that the active site of ssNAT represents a hybrid of the NatA and NatE active sites, and we highlight features of this protein that allow it to facilitate catalysis of distinct substrates through different catalytic strategies, which is a unique characteristic of this enzyme. Taken together, the structural and biochemical data presented here have implications for the evolution of eukaryotic NAT enzymes and the substrate specificities therein.
The cotranslational process of amino-terminal acetylation occurs on a majority of eukaryotic proteins and mediates many biological processes, including cellular apoptosis, enzymatic regulation, protein localization, and protein degradation (1–4). In humans, three major amino-terminal acetyltransferase (NAT) complexes, called NatA, NatB, and NatC, are responsible for modifying ∼85% of all proteins that undergo amino-terminal acetylation (5). These complexes are conserved in yeast, exist as obligate heterodimers, and are differentiated from one another on the basis of substrate specificity, which is dictated by the amino-terminal sequence of the substrate protein (5, 6). Each complex consists of a single unique catalytic subunit and an additional unique auxiliary subunit that has been shown to potentiate activity and alter substrate specificity of the enzymatic component, as well as anchor the complex to the ribosome during translation (6–11). Three additional human NAT enzymes, NatD–NatF, have also been identified, but they have a much more limited set of physiological substrates, appear to be independently active, and are not well characterized across eukaryotes (12–14).
The only two eukaryotic NATs that have been structurally characterized are Schizosaccharomyces pombe NatA, consisting of the Naa10p catalytic subunit and Naa15p regulatory subunit, and human NatE, which is the independently active NAA50 enzyme (15, 16). NatA, which harbors the most diversity for substrate selection, is responsible for modifying the majority of all amino-terminally acetylated proteins, which it accomplishes by recognizing proteins with amino-terminal Ala-, Cys-, Gly-, Ser-, Thr-, or Val- residues (5). NatE/NAA50 has only one known biologically relevant substrate that contains the amino-terminal sequence Met-Leu-Gly-Pro, of which only the amino-terminal Met- is absolutely required for catalysis (14). These structures and their accompanying biochemical characterization have provided significant insight into the mechanisms of substrate specificity and catalysis used by NAT enzymes. Interestingly, although NatA and NatE both catalyze α-amino group acetylation, they use unique catalytic strategies and substantially different substrate amino-terminal residue binding pockets to achieve sequence-based substrate specificity.
Although only six amino-terminally acetylated proteins have been identified in Escherichia coli, large-scale proteomics studies of three different archaeal species revealed that ∼14–29% of all proteins isolated are acetylated at their amino terminus (17–20). Analysis of the thermophilic archaea Sulfolobus solfataricus genome resulted in the identification of a protein with 37% sequence identity to human NAA10 that is believed to be the only NAT in this species (21). In agreement with this hypothesis, the same study showed that this protein is able to independently acetylate NatA and NatE substrates, as well as other amino-terminal sequences corresponding to NatB and NatC substrates in vitro (21) Furthermore, no auxiliary subunit homologs could be identified in this genome, suggesting that this archaeal species dedicates just one protein to a task that requires at least nine unique proteins in humans. This ancestral NAT variant, herein referred to as ssNAT, is therefore an ideal candidate for probing the evolution of this family of enzymes and the mechanisms they use to achieve substrate-specific acetylation. Here, we report the X-ray crystal structure of the ssNAT enzyme at 1.98 Å resolution, along with a detailed comparison of ssNAT with the NatA and NatE structures. We combine information from the structure and accompanying alignments to generate a series of ssNAT mutants that we kinetically characterized to dissect the molecular features that promote the acetylation promiscuity observed for this enzyme and to propose evolutionary events that led to a diverse family of substrate-specific monomeric and heterodimeric NATs.
Results
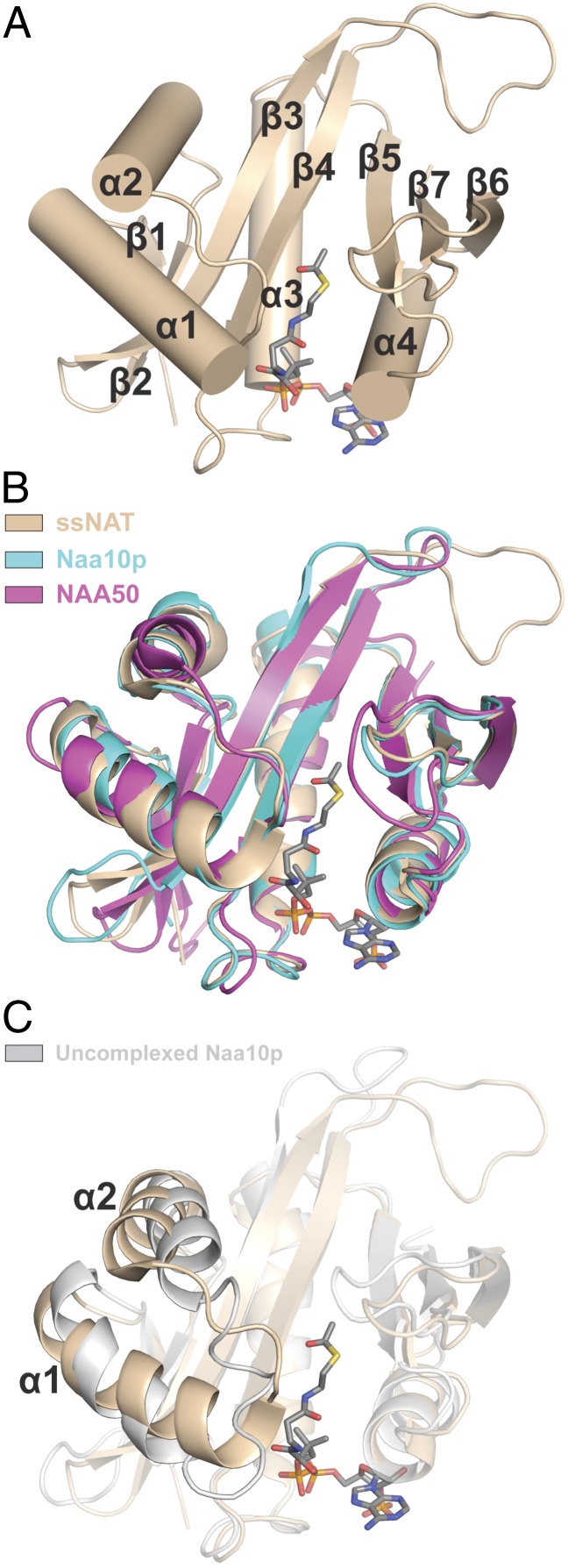
Overall Structure of the Active ssNAT Enzyme.
Crystals of the ssNAT enzyme (residues 50–216) bound to acetyl CoA were obtained using recombinant protein expressed in E. coli and diffracted to 1.98 Å resolution in the space group C2 with one molecule per asymmetric unit. The structure was determined with molecular replacement using ssARD1 [Protein Data Bank (PDB) ID code 2x7B] as a search model. Electron density was observed for residues 60–216 and acetyl CoA, and the structure was refined to a final Rwork/Rfree of 19.9%/24.3% (Fig. S1 and Table S1). The ssNAT structure adopts a general control of amino acid synthesis protein 5 (Gcn5)-related N-acetyltransferase (GNAT) fold with a central acetyl CoA binding core region and flanking N- and C-terminal segments that is highly similar to the active form of the NatA catalytic subunit Naa10p (rmsd = 1.46 Å) and NatE/NAA50 (rmsd = 1.98 Å) (Fig. 1 A and B). ssNAT also contains an extended β3-β4 loop that is the only nonconserved feature in the catalytic core of this enzyme relative to the eukaryotic NATs (Fig. 1B). In addition, an alignment of ssNAT with the uncomplexed form of Naa10p reveals more significant differences, particularly with regard to the position of the α1-loop-α2 segment (Fig. 1C). More specifically, the C-terminal portion of the α1 helix in ssNAT is extended by a single complete helical turn, a conformation that Naa10p can only adopt in the presence of its auxiliary subunit. This helix extension, which is also present in NAA50, seems to be crucial for proper positioning of the α1-α2 loop, a region that has been shown to be essential for substrate-specific acetylation by both eukaryotic NATs discussed here. Notably, the α1-loop-α2 segments in ssNAT, NAA50, and the active form of Naa10p all adopt nearly identical conformations, which is supports a previous report that ssNAT does not require a binding partner for activity; as a result, no auxiliary subunit exists in the S. solfataricus proteome (21).
Fig. 1.
Overall structure of the ssNAT enzyme. (A) The structure of the ssNAT enzyme (tan cartoon representation) bound to acetyl CoA (gray stick representation). Elements of secondary structure are labeled. (B) A 3D alignment of the ssNAT/acetyl CoA complex with NAA50 (pink cartoon representation) and the complexed form of Naa10p (teal cartoon representation). A color-coded legend is also included. (C) A 3D alignment of the ssNAT/acetyl CoA complex with the uncomplexed, inactive Naa10p (silver cartoon representation). Regions of negligible structural variation are shown at 50% transparency. α-helices 1 and 2 are labeled.
Implications for the Evolution of NAT Substrate-Acetylation Specificity.
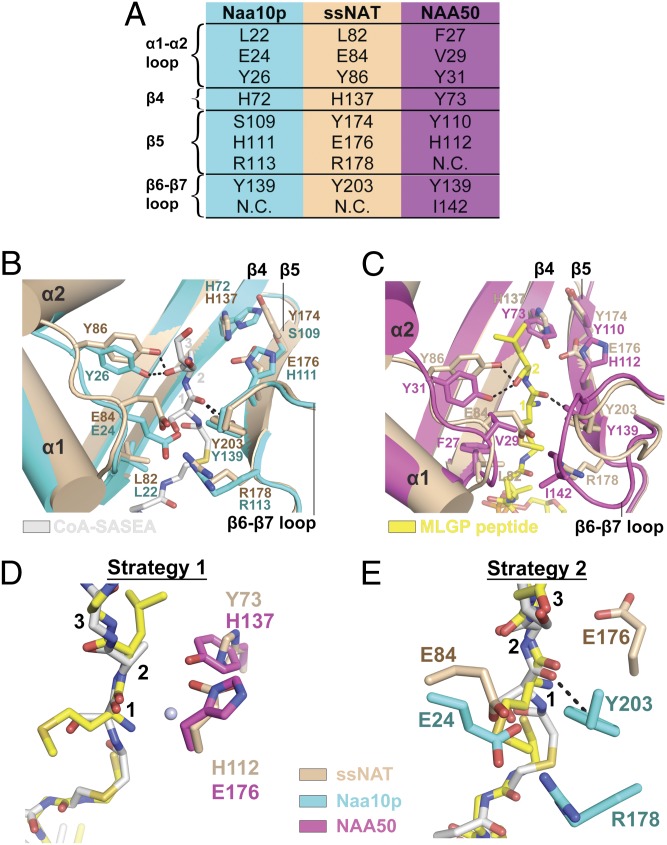
The structure of the NatA complex bound to a bisubstrate peptide-CoA conjugate inhibitor established the position of the substrate amino-terminal residue in the active site of the catalytic subunit (15). An alignment of the active site from this structure with the corresponding region of ssNAT reveals that all key catalytic and cognate substrate binding residues in the active site of Naa10p are conserved in ssNAT (Fig. 2 A and 2B). Specifically, residues L82, E84, and Y86 in the α1-α2 loop of ssNAT align with corresponding residues of Naa10p. Similarly, ssNAT residues R178 and Y203 correspond to Naa10p residues R113 and Y139. In contrast, another alignment of the ssNAT active site with the ternary NAA50⋅CoA⋅substrate peptide fragment structure shows significant differences in the amino-terminal residue binding pockets of the two enzymes (Fig. 2C). In NAA50, residues F27, P28, V29, and I142 align with ssNAT residues L82, E84, Y86, and R178 and form a substantially more sterically open, hydrophobic pocket that is ideal for an amino-terminal Met- substrate. Indeed, it appears that ssNAT-E84 residue would clash with the Met- side chain of the NAA50 substrate peptide (Fig. 2 A and 2D). Therefore, at first glance, it appears that ssNAT is more tailored for binding to NatA vs. NatE substrates.
Fig. 2.
ssNAT active site and catalytic strategies. (A) A list of key secondary structure elements from the NATs discussed here, and active site residues within. Each row represents corresponding residues from each enzyme. N.C. represents a corresponding residue that is not in a position to effect catalysis. (B) A 3D alignment of the ssNAT active site (tan) with the corresponding region in Naa10p (teal). Putative ssNAT substrate binding and catalytic residues are shown in stick format, as are corresponding residues in Naa10p. The bisubstrate inhibitor from Naa10p is shown in silver stick format, and residues from the peptide portion of this inhibitor are labeled with orange numbers. Key secondary structure elements are also labeled. Hydrogen bonds are represented as black dotted lines. (C) A 3D alignment of the ssNAT active site (tan) with the corresponding region in NAA50 (pink). Putative ssNAT substrate binding and catalytic residues are shown in stick format, as are corresponding residues in NAA50. The CoA and substrate peptide ligands from NAA50 are shown in yellow stick format, and residues from the substrate peptide are labeled with yellow numbers. Key secondary structure elements are also labeled. Hydrogen bonds are represented as black dotted lines. (D) ssNAT catalytic residues from catalytic strategy 1 are shown in tan stick format and aligned with catalytic residues from NAA50 (pink stick format). A conserved active site water molecule is shown as a light blue sphere. The bisubstrate inhibitor from Naa10p is shown in silver stick format and the CoA and substrate peptide ligands from NAA50 are shown in yellow stick format. Residues from the peptide portions of these ligands are labeled with black numbers. (E) ssNAT catalytic residues from catalytic strategy 2 are shown in tan stick format, and Naa10p residues are shown in teal stick format. The bisubstrate inhibitor from Naa10p is shown in silver stick format, and the CoA and substrate peptide ligands from NAA50 are shown in yellow stick format. Residues from the peptide portions of these ligands are labeled with black numbers. A hydrogen bond is represented as a black dotted line. A color-coded legend for the panel has been included for clarity.
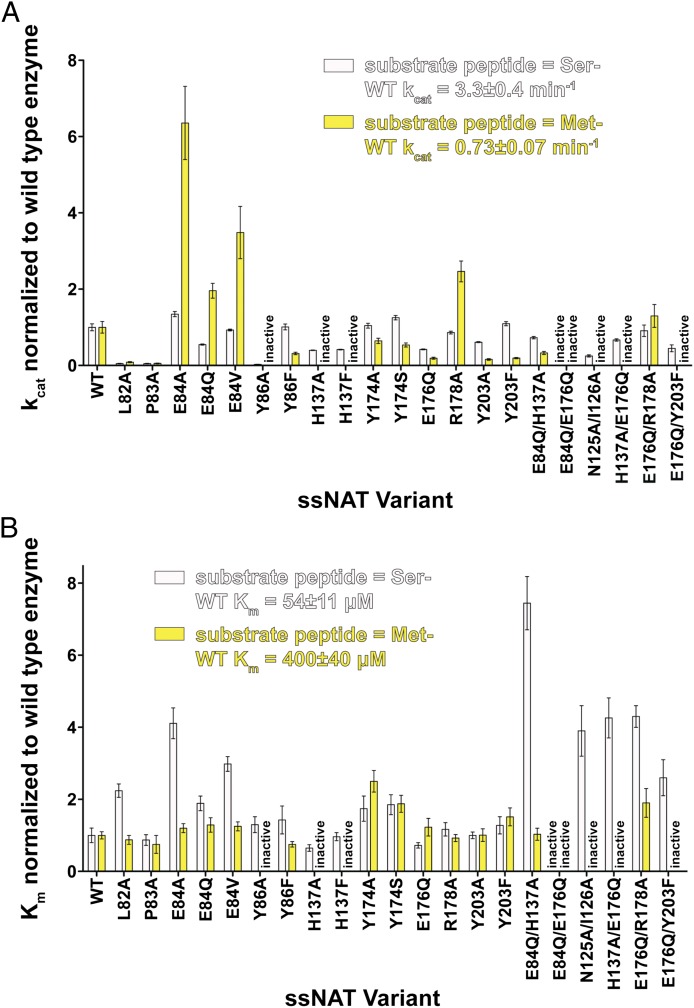
To directly establish the ability of ssNAT to process NatA and NatE substrates, we generated catalytic parameters for the enzyme against amino-terminal Ser- and Met- containing peptides (Tables S2 and S3). Consistent with the structural comparison, we observed that ssNAT has a preference for the NatA/Naa10p over the NatE/NAA50 substrate; specifically, ssNAT has a Km for the Ser- amino-terminal Naa10p substrate that is 7.4-fold lower than its Km for the NAA50 Met- amino-terminal substrate, and the kcat toward the Ser- amino-terminal substrate is 4.5-fold higher than the kcat toward the Met- amino-terminal substrate (Fig. 3). Although this enzyme displays more favorable catalytic parameters with the Ser- substrate, its ability to modify amino-terminal Met- substrates is in stark contrast to NatA, which shows only background acetylation levels with similar peptides (15) To directly test the importance of the ssNAT-E84 side chain in N-terminal residue processing, we prepared ssNAT-E84A/V mutants and assayed their ability to process Ser- and Met- amino-terminal substrate peptides. As expected, we found that these mutants were more accommodating toward the Met substrates, a conclusion that is supported by increases in the kcat of the enzyme by 4.6- and 2.5-fold, respectively, toward the Met- amino-terminal substrate, whereas the kcat toward the Ser- amino-terminal substrate was unaffected. These mutants also resulted in an increased Km for the Ser- amino-terminal substrate (4.1- and 3.5-fold, respectively) with no effect on the Km for the Met- amino-terminal substrate. Furthermore, an alanine point mutation at R178, which also contributes to the hydrophilicity of the binding pocket and forms an ionic interaction with E84, exhibits a 2.5-fold increase in kcat toward the Met- amino-terminal substrate with no effect on the kcat toward the Ser- amino-terminal substrate. These results strongly support our previous hypothesis that NAA50, which has a valine (V29) in the position of ssNAT-E84, uses a more open hydrophobic binding pocket to achieve Met- amino-terminal substrate binding specificity.
Fig. 3.
Catalytic parameters for wild-type and mutant ssNAT enzymes. (A) kcat values for the wild-type (WT) ssNAT and accompanying mutants toward both the Ser- amino-terminal substrate peptide (silver bars) and the Met- amino-terminal substrate peptide (yellow bars). All values have been normalized to the kcat value calculated for the WT enzyme. (B) Km values for the WT ssNAT and accompanying mutants toward both the Ser- amino-terminal substrate peptide and the Met- amino-terminal substrate peptide. All values have been normalized to the Km value calculated for the WT enzyme. Errors represent SDs (n = 3).
The Y86 and Y203 residues in ssNAT are conserved in both Naa10p and NAA50 and form hydrogen bonds with substrate peptide backbone carbonyl groups in each of the previously reported structures (Fig. 2 B and 2C). Although these residues do form van der Waals interactions with the amino-terminal residue of the substrate peptide in each structure, it seems that their primary function is to anchor the substrate peptide to the enzyme in a catalytically competent position, and it is unlikely that they play a role in substrate specificity. We found that the ssNAT-Y86F and ssNAT-Y203F mutants do not affect the Km in the context of the Met- amino-terminal substrate but do show decreases of 3.2- and 5.2-fold in kcat, respectively. The same mutations show no effect on catalysis or substrate binding relative to wild-type enzyme in the context of the Ser- amino-terminal substrate. Thus, the substrate peptide backbone interactions are more critical for productive binding of the Met- amino-terminal substrate, likely because the ssNAT amino-terminal residue binding pocket favors binding of the Ser- side chain over the Met- side chain.
ssNAT Combines Unique Features from NatA and NatE to Use Two Independent Catalytic Strategies.
NAA50 uses a two-base mechanism to carry out substrate α-amino group deprotonation and facilitate catalysis (16). These two bases, NAA50-Y73 and NAA50-H112, are on the β3 and β4 strands, respectively, and are representative of the traditional general base positions in GNAT enzymes (Fig. 2C) (22). In Naa10p, the NAA50-Y73 and NAA50-H112 residues are replaced by Naa10p-H72 and Naa10p-H111, respectively, of which Naa10p-H72 is flipped out of the active site and is not in a position to act as a general base (Fig. 2B). It is therefore not surprising that neither of these residues is crucial for catalysis by Naa10p (15). The corresponding active site positions in ssNAT are occupied by ssNAT-H137 and ssNAT-E176, both of which have side chains directed into the active site that align well with NAA50-Y73 and NAA50-H112. To assess the roles of ssNAT-H137 and ssNAT-E176 in catalysis, we used site-directed mutagenesis to make single and double mutants at these sites and kinetically characterized the mutant enzymes. The ssNAT-H137A/F/Q mutations were all found to inactivate the enzyme toward the Met- substrate, whereas the ssNAT-E176Q mutant shows a 5.2-fold reduction in kcat toward this substrate with a negligible effect on Km. Although it is possible that ssNAT-H137 is also involved in productive substrate binding, the ssNAT enzyme seems to use multiple catalytic residues, similar to the NAA50 mechanism (herein referred to as strategy 1), to deprotonate the α-amino group of the Met- amino-terminal substrate (Fig. 2D). Notably, all human NATs that acetylate amino-terminal methionine substrates have a potential general base residue in at least one of these positions (Fig. S2). Remarkably, however, we found that the three mutations discussed here all have less than a 2-fold effect on the kcat of the enzyme toward the Ser- amino-terminal substrate, and the ssNAT-H137A/E176Q is the only mutant that has an effect on Km, which increases by 4.3-fold. This finding suggests there is an alternate catalytic strategy within the enzyme that is able to facilitate the acetylation reaction in the presence of the Ser- amino-terminal substrate.
Structural and mutational analysis of Naa10p in the context of the NatA complex suggests that Naa10p-E24 is an essential catalytic residue, as evidenced by a >100-fold reduction in catalysis observed for the E24Q mutant, with no effect on Km (15). In addition, this study revealed that Naa10p-R113 and Naa10p-Y139 are also in position to participate in the catalytic mechanism, and point mutations at these sites decrease the rate of catalysis relative to the wild-type enzyme. Interestingly, all of these residues are conserved in the ssNAT enzyme, where ssNAT residues E84, R178, and Y203 correspond to Naa10p residues E24, R113, and Y139. We found that the sssNAT-E84Q mutant is only 1.8-fold less active toward the Ser- substrate relative to the wild-type enzyme. This result suggests that ssNAT-E84 on its own is not functionally identical to Naa10p-E24 for the Ser- amino terminal substrate. We then addressed the possibility that ssNAT-E84 might cooperate with ssNAT-E176 or ssNAT-H137 for Ser- substrate acetylation and found that the ssNAT-E84Q/E176Q double mutant had no detectable activity toward the Ser- amino terminal substrate, whereas the ssNAT-E84Q/H137A double mutant had only a minor effect on catalysis (1.5-fold decrease in kcat) but showed a 7.4-fold increase in Km. We also assessed the effect of combining the ssNAT-E176Q mutant with mutants of either ssNAT-R178 or ssNAT-Y203 and found that the ssNAT-E176Q/R178A and ssNAT-E176Q/Y203F mutants had a 2-fold or less decrease in kcat toward the Ser- amino-terminal substrate. Taking these results together reveals that residues ssNAT-E84 and ssNAT-E176 cooperate as essential catalytic residues to facilitate Ser- substrate acetylation by ssNAT through an alternative catalytic strategy (strategy 2; Fig. 2E). The observation that the ssNAT-E84Q/E176Q mutant is also unable to acetylate the Met- substrate reflects the importance of ssNAT-E176 for Met-substrate acetylation in the context of an amino-terminal binding pocket with an altered electrostatic environment, which potentially effects the position of the α-amino group within the active site.
A Nonconserved Loop Region Potentiates ssNAT Activity.
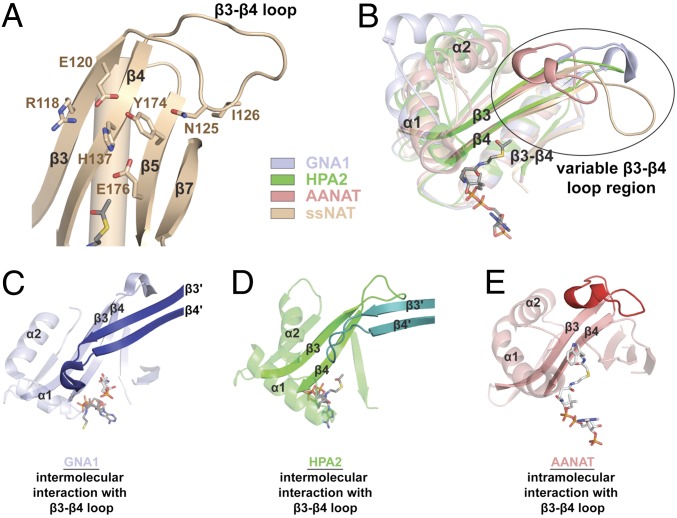
Despite a nearly identical active site landscape between ssNAT and Naa10p, Naa10p is not able to facilitate catalysis via catalytic strategy 1. The active site alignment shows that ssNAT-Y174 is one of the only residues in the active site that is not conserved in Naa10p (Fig. 2B). Significantly, this residue appears to play a role in positioning ssNAT-H137 for Met- amino-terminal substrate catalysis by forcing this general base residue into the active site. The residue corresponding to ssNAT-Y174 in Naa10p, Naa10p-S109, lacks the bulky side chain required to force the corresponding base residue, Naa10p-H72, into the active site of the enzyme. Consequently, the Naa10p-H72 side chain is able to flip out of the active site into a catalytically incompetent position. Contrary to this hypothesis, the ssNAT-Y174S point mutant, which mimics the Naa10p active site, has no effect on catalysis and implies that ssNAT-H137 is held in position by other residues in the local environment. Further inspection of the alignment of ssNAT with the Naa10p and NAA50 structures (Fig. 1B) reveals that the loop segment connecting the β3 and β4 strands of the ssNAT core region is significantly more extended than the corresponding loops of Naa10p and NAA50 and points toward the ssNAT active site (Fig. 4A). In particular, residues ssNAT-N125 and ssNAT-I126, at the tip of the β3-β4 loop, appear to make intramolecular interactions with the β5 and β7 strands. To determine whether the β3-β4 loop region participates in catalysis in some way, we prepared the ssNAT-N125A/I126A mutant, which we found is unable to acetylate the Met- amino-terminal substrate and exhibits a 4.0-fold reduction in kcat toward the Ser- amino terminal substrate, as well as a 3.9-fold increase in Km relative to the wild-type enzyme. On the basis of this observation, we conclude that the extended β3-β4 loop region of ssNAT likely has a stabilizing effect on the 3D fold of the active site and is able to prime the active site of the enzyme for catalysis of the Met- substrate and, to a lesser extent, the Ser- amino terminal substrate.
Fig. 4.
The β3-β4 loop region in ssNAT and other GNAT enzymes. (A) A zoom view of the β3-loop-β4 region of ssNAT. Key residues (shown in tan stick format) and secondary structure elements are labeled. acetyl CoA is represented in gray stick format. (B) An alignment of GNA1 (light blue cartoon representation), HPA2 (neon green cartoon representation), AANAT (pink cartoon representation), and ssNAT/acetyl CoA (tan cartoon representation and gray stick representation, respectively) showing conserved secondary structure elements at 50% transparency. The β3-loop-β4 motifs from these enzymes are shown with no transparency. The variable region has been encircled and labeled. Key secondary structure elements are also labeled. (C) GNA1 (light blue cartoon, 50% transparency) bound to CoA (gray stick representation) and the GlcNAc6P substrate (silver stick representation) with key secondary structure elements labeled, and the β3-loop-β4 motif is shown with no transparency. Secondary structure elements from the β3-loop-β4 motif of the second molecule in the dimer (dark blue cartoon representation) are also labeled with a prime symbol. (D) HPA2 (neon green cartoon, 50% transparency) bound to acetyl CoA (gray stick representation) with key secondary structure elements labeled and the β3-loop-β4 motif shown with no transparency. Secondary structure elements from the β3-loop-β4 motif of the second molecule in the dimer (dark green cartoon representation) are also labeled with a prime symbol. (E) AANAT (light red cartoon, 50% transparency) bound to a bisubstrate analog CoA-S-acetyltryptamine (silver stick representation), with key secondary structure elements labeled, and the β3-loop-β4 motif shown with no transparency. The β3-β4 loop is colored in dark red.
Interestingly, there are several GNAT enzymes that capitalize on the variability of the β3-β4 loop region for functional gain (Fig. 4B). In glucosamine-6-phosphate acetyltransferase (GNA1) and the histone acetyltransferase HPA2, this loop mediates biologically relevant dimerization and facilitates substrate binding by contributing to the active site architecture of the dimer partner (Fig. 4 C and D), whereas in serotonin N-acetyltransferase (AANAT), this region is responsible for orienting the arylalkylamine substrate in the active site through an intramolecular mechanism (Fig. 4E) (23–25).
The Length of the α1 Helix has Implications for Obligate Heterodimerization of Certain NAT Enzymes.
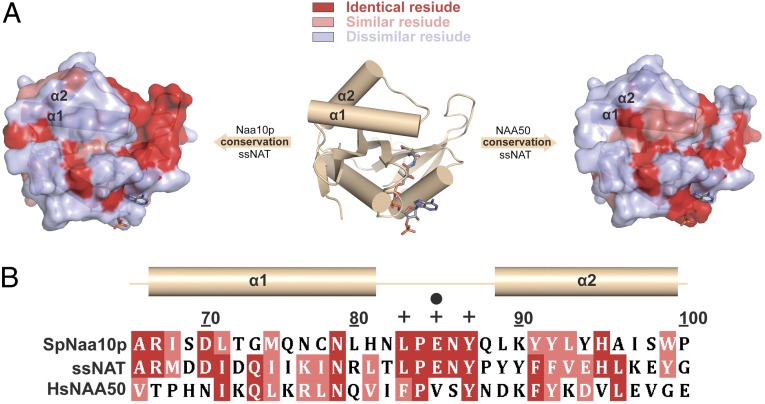
The C-terminal portion of the α1 helix is the only region of ssNAT that is sequentially more similar to NAA50 than to Naa10p (Fig. 5A). Interestingly, as shown in Fig. 1 B and C, the length of this helix is critical for the placement of the α1-α2 loop, which harbors several residues that facilitate substrate binding and/or catalysis in all three NATs discussed here. The independently active ssNAT and NAA50 enzymes have similar α-helix-stabilizing residues in the C-terminal portion of the α1 helix (Fig. 5 A and B), which is able to undergo an additional helical twist relative to monomeric Naa10p (Fig. 1C). As a result, the α1-α2 loop in both ssNAT and NAA50 is able to adopt a conformation that is suitable for catalysis in the absence of an auxiliary subunit. In Naa10p, these helix-stabilizing residues are not conserved; as a consequence, the length of the α1 helix and position of the α1-α2 loop cannot support catalysis by the enzyme in the absence of auxiliary subunit binding. It is therefore tempting to speculate that the length of this helix in the context of the monomeric enzyme may dictate whether or not a NAT enzyme needs a binding partner for catalytic activation.
Fig. 5.
Conservation between the ssNAT α1-loop-α2 motif and the corresponding regions of Naa10p and NAA50. (A) A surface conservation map for ssNAT with Naa10p and NAA50 centered on the α1 helix. Regions in which aligned residues are identical are shown in dark red, regions in which aligned residues are highly similar are shown in light red, and regions in which aligned residues are not similar are shown in light blue. Key secondary structure elements are labeled. (B) A sequence alignment of the α1-loop-α2 motif in ssNAT with the corresponding regions of Naa10p and NAA50. Secondary structure elements observed in the 3-dimensional structure are shown just above the sequence. Residue numbers correspond to ssNAT. Catalytic and substrate binding residues from ssNAT are denoted with ● and ✚ symbols, respectively, above the residue in the sequence.
Discussion
Because S. solfataricus only contains one NAT protein to acetylate substrates with different N-terminal residues, it appears this enzyme has a “hybridized” strategy to acetylate many varied substrates. In particular, we found that the ssNAT contains an active site that has integrated essential features of the NatA and NatE active sites and, as a result, can facilitate acetylation of Met- and non-Met-containing amino-terminal substrate peptides. We show that although ssNAT has higher sequence similarity to Naa10p over NAA50 (34% vs. 24% sequence identity, respectively), and correspondingly has a preference for Ser- over Met- amino-terminal substrates, it still is able to accommodate a Met- amino-terminal substrate. Our results indicate that for the Met- amino-terminal substrate, ssNAT uses H137 in particular, as well as E176 as catalytic residues, and it uses E84 and E176 as catalytic residues for the Ser- amino-terminal peptide characterized here, as well as likely for other NatA-type substrates that it has previously been shown to acetylate (21). We also show that ssNAT uses a structural feature that is not conserved in the eukaryotic NATs, the β3-β4 loop, to stabilize the position of key residues involved in substrate acetylation, and Met- amino-terminal substrate acetylation in particular.
The presence of two different catalytic strategies that facilitate the reaction of a single enzyme toward different substrates is a very rare feature that is unique to this enzyme. One explanation for the existence of the two catalytic strategies is that ssNAT represents a snapshot in the evolution of the eukaryotic NATs, which subsequently evolved into separate enzymes with increased specificity toward different substrates. We show that mutations in the α1-α2 substrate-binding loop can have a significant effect on substrate specificity and provide evidence for the existence of substrate-specific catalytic residues. In addition, we highlight the functional significance of the nonconserved β3-β4 loop region and the importance of the α1 helix length in NAT activity. As a consequence, we propose that evolutionary events that include mutations targeting these features led to a diverse family of monomeric and heterodimeric eukaryotic NATs with different substrate specificities and catalytic strategies.
Materials and Methods
ssNAT protein was expressed in E. coli, and subsequent protein purification and crystallization were carried out as described in SI Materials and Methods. Procedures detailing X-ray diffraction data collection and structure determination also can be found in SI Materials and Methods. Acetyltransferase assays were performed as previously described (16). Protocols describing acetyltransferase assays for both substrates and calculation of catalytic parameters can be found in the SI Materials and Methods. The coordinates of the ssNAT/acetyl CoA structure has been deposited in the PDB under PDB ID code 4LX9. The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grant GM060293 (to R.M.) and NIH Training Grant GM071339 (to G.L.). We acknowledge the use of the Wistar Proteomics Core facility for the work reported here, which is supported in part by NIH Grant CA010815.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4LX9 (ssNAT/AcCoA structure)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310365110/-/DCSupplemental.
References
- 1.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327(5968):973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yi CH, et al. Metabolic regulation of protein N-alpha-acetylation by Bcl-xL promotes cell survival. Cell. 2011;146(4):607–620. doi: 10.1016/j.cell.2011.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forte GM, Pool MR, Stirling CJ. N-terminal acetylation inhibits protein targeting to the endoplasmic reticulum. PLoS Biol. 2011;9(5):e1001073. doi: 10.1371/journal.pbio.1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334(6056):674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnesen T, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci USA. 2009;106(20):8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polevoda B, Sherman F. Composition and function of the eukaryotic N-terminal acetyltransferase subunits. Biochem Biophys Res Commun. 2003;308(1):1–11. doi: 10.1016/s0006-291x(03)01316-0. [DOI] [PubMed] [Google Scholar]
- 7.Gautschi M, et al. The yeast N(alpha)-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol Cell Biol. 2003;23(20):7403–7414. doi: 10.1128/MCB.23.20.7403-7414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polevoda B, Brown S, Cardillo TS, Rigby S, Sherman F. Yeast N(alpha)-terminal acetyltransferases are associated with ribosomes. J Cell Biochem. 2008;103(2):492–508. doi: 10.1002/jcb.21418. [DOI] [PubMed] [Google Scholar]
- 9.Polevoda B, Sherman F. NatC Nalpha-terminal acetyltransferase of yeast contains three subunits, Mak3p, Mak10p, and Mak31p. J Biol Chem. 2001;276(23):20154–20159. doi: 10.1074/jbc.M011440200. [DOI] [PubMed] [Google Scholar]
- 10.Polevoda B, Cardillo TS, Doyle TC, Bedi GS, Sherman F. Nat3p and Mdm20p are required for function of yeast NatB Nalpha-terminal acetyltransferase and of actin and tropomyosin. J Biol Chem. 2003;278(33):30686–30697. doi: 10.1074/jbc.M304690200. [DOI] [PubMed] [Google Scholar]
- 11.Park EC, Szostak JW. ARD1 and NAT1 proteins form a complex that has N-terminal acetyltransferase activity. EMBO J. 1992;11(6):2087–2093. doi: 10.1002/j.1460-2075.1992.tb05267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song OK, Wang X, Waterborg JH, Sternglanz R. An Nalpha-acetyltransferase responsible for acetylation of the N-terminal residues of histones H4 and H2A. J Biol Chem. 2003;278(40):38109–38112. doi: 10.1074/jbc.C300355200. [DOI] [PubMed] [Google Scholar]
- 13.Van Damme P, et al. NatF contributes to an evolutionary shift in protein N-terminal acetylation and is important for normal chromosome segregation. PLoS Genet. 2011;7(7):e1002169. doi: 10.1371/journal.pgen.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evjenth R, et al. Human Naa50p (Nat5/San) displays both protein N alpha- and N epsilon-acetyltransferase activity. J Biol Chem. 2009;284(45):31122–31129. doi: 10.1074/jbc.M109.001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liszczak G, et al. Molecular basis for amino-terminal acetylation by the heterodimeric NatA complex. Nat Struct Mol Biol, in press. [DOI] [PMC free article] [PubMed]
- 16.Liszczak G, Arnesen T, Marmorstein R. Structure of a ternary Naa50p (NAT5/SAN) N-terminal acetyltransferase complex reveals the molecular basis for substrate-specific acetylation. J Biol Chem. 2011;286(42):37002–37010. doi: 10.1074/jbc.M111.282863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirkland PA, Humbard MA, Daniels CJ, Maupin-Furlow JA. Shotgun proteomics of the haloarchaeon Haloferax volcanii. J Proteome Res. 2008;7(11):5033–5039. doi: 10.1021/pr800517a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soppa J. Protein acetylation in archaea, bacteria, and eukaryotes. Archaea. 2010;2010:pii: 820681. doi: 10.1155/2010/820681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aivaliotis M, et al. Large-scale identification of N-terminal peptides in the halophilic archaea Halobacterium salinarum and Natronomonas pharaonis. J Proteome Res. 2007;6(6):2195–2204. doi: 10.1021/pr0700347. [DOI] [PubMed] [Google Scholar]
- 20.Falb M, et al. Archaeal N-terminal protein maturation commonly involves N-terminal acetylation: A large-scale proteomics survey. J Mol Biol. 2006;362(5):915–924. doi: 10.1016/j.jmb.2006.07.086. [DOI] [PubMed] [Google Scholar]
- 21.Mackay DT, Botting CH, Taylor GL, White MF. An acetylase with relaxed specificity catalyses protein N-terminal acetylation in Sulfolobus solfataricus. Mol Microbiol. 2007;64(6):1540–1548. doi: 10.1111/j.1365-2958.2007.05752.x. [DOI] [PubMed] [Google Scholar]
- 22.Marmorstein R. Structure and function of histone acetyltransferases. Cell Mol Life Sci. 2001;58(5-6):693–703. doi: 10.1007/PL00000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angus-Hill ML, Dutnall RN, Tafrov ST, Sternglanz R, Ramakrishnan V. Crystal structure of the histone acetyltransferase Hpa2: A tetrameric member of the Gcn5-related N-acetyltransferase superfamily. J Mol Biol. 1999;294(5):1311–1325. doi: 10.1006/jmbi.1999.3338. [DOI] [PubMed] [Google Scholar]
- 24.Peneff C, Mengin-Lecreulx D, Bourne Y. The crystal structures of Apo and complexed Saccharomyces cerevisiae GNA1 shed light on the catalytic mechanism of an amino-sugar N-acetyltransferase. J Biol Chem. 2001;276(19):16328–16334. doi: 10.1074/jbc.M009988200. [DOI] [PubMed] [Google Scholar]
- 25.Hickman AB, Namboodiri MA, Klein DC, Dyda F. The structural basis of ordered substrate binding by serotonin N-acetyltransferase: Enzyme complex at 1.8 A resolution with a bisubstrate analog. Cell. 1999;97(3):361–369. doi: 10.1016/s0092-8674(00)80745-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.