Significance
Assembling a large genome faces three challenges: assembly quality, computer memory requirement, and execution time. Our developed assembler, JR-Assembler, uses (a) a strategy that selects good seeds for contig construction, (b) an extension strategy that uses whole sequencing reads to increase the chance to jump over repeats and to expedite extension, and (c) detecting misassemblies by remapping reads to assembled sequences. Compared with current assemblers, JR-Assembler achieves a better overall assembly quality, requires less execution time and requires, with one exception, less memory. The advantages of JR-Assembler in memory usage and execution time will increase slowly as the read length increases. Thus, contrary to the prevailing view, the extension approach seems superior to the de Bruijn graph approach.
Abstract
Assembling a large genome using next generation sequencing reads requires large computer memory and a long execution time. To reduce these requirements, we propose an extension-based assembler, called JR-Assembler, where J and R stand for “jumping” extension and read “remapping.” First, it uses the read count to select good quality reads as seeds. Second, it extends each seed by a whole-read extension process, which expedites the extension process and can jump over short repeats. Third, it uses a dynamic back trimming process to avoid extension termination due to sequencing errors. Fourth, it remaps reads to each assembled sequence, and if an assembly error occurs by the presence of a repeat, it breaks the contig at the repeat boundaries. Fifth, it applies a less stringent extension criterion to connect low-coverage regions. Finally, it merges contigs by unused reads. An extensive comparison of JR-Assembler with current assemblers using datasets from small, medium, and large genomes shows that JR-Assembler achieves a better or comparable overall assembly quality and requires lower memory use and less central processing unit time, especially for large genomes. Finally, a simulation study shows that JR-Assembler achieves a superior performance on memory use and central processing unit time than most current assemblers when the read length is 150 bp or longer, indicating that the advantages of JR-Assembler over current assemblers will increase as the read length increases with advances in next generation sequencing technology.
Short read sequencing (SRS) platforms, such as HiSeq 2500, have much higher throughputs and much lower costs than traditional Sanger sequencing. However, genome assembly is more difficult using SRS data than using Sanger data because the amount of SRS data are huge [≥100 giga base pairs (Gb) per run] and SRS reads (100∼250 bp) are much shorter than Sanger reads (∼1,000 bp).
Most assemblers for Sanger data apply the overlap–layout–consensus (OLC) approach (1). This approach is not suitable for SRS data because the run time quadratically increases with the number of reads. The long run time problem can be alleviated by the indexing technique, as in Extract DE Novo Assembler (Edena) (2). However, Edena and string graph assembler (SGA) (3) are perhaps the only two OLC assemblers for SRS data.
Currently, the two major types of SRS assemblers are extension-based and de Bruijn (or Eulerian) graph-based. Extension-based assemblers [e.g., Short Sequence Assembly by K-mer search and 3′ read Extension (SSAKE)] (4, 5) start from a seed read and extend it by other reads that overlap with the seed. Without constructing a layout graph, this approach saves both computational time and memory. However, it is sensitive to sequencing errors and repeats and usually generates a fragmented assembly. The de Bruijn graph approach is fast at the step of finding overlaps between reads. It decomposes a read into consecutive k-mers, which are represented by nodes connected by edges in a graph. By scanning all reads once, it rapidly finds the overlaps between all reads because overlapping reads have the same k-mer. The resulting graph contains all characteristics of a genomic sequence. The constructed graph, however, has to be reduced using various algorithms and heuristic rules to eliminate sequencing errors and to resolve repeats. This approach is adopted by several leading assemblers for SRS data, including Assembly By Short Sequences (ABySS) (6), ALLPATHS-LG (7), EULER-USR (8), SOAPdenovo (9, 10), and Velvet (11).
The de Bruijn graph approach requires more memory than both the OLC and the extension approach because it needs to save the entire graph in memory for assembly. To handle a large genome such as a 3 Gb genome, ALLPATHS-LG and SOAPdenovo (7, 9, 10) have been carefully engineered, but most other de Bruijn graph assemblers cannot handle large genomes when the memory is limited. Note that the memory requirement increases dramatically as the read length increases, which will occur as the sequencing technology advances. Although correcting errors in sequencing reads can reduce the graph size and increase the assembly quality, the execution time still increases with the total read count and the read length. Therefore, balancing the assembly quality, memory requirement, and execution time is a major challenge for de novo assembly.
In this paper, we propose a unique extension-based assembler for SRS data, called JR-Assembler (J stands for jumping extension and R for read remapping). We revive the extension-based approach because it is straightforward and economical in run time and memory use. A key concept in our approach is that instead of a base-by-base extension, JR-Assembler extends a read by other whole reads—that is, it makes a jump. This approach speeds up extension and can readily jump over small repeats. Second, it includes a dynamic back trimming process, so that the extension can effectively avoid termination by sequencing errors. Finally, to avoid partial repeat resolutions, JR-Assembler remaps reads to each assembled sequence, and if an error is found due to the existence of repeats, it breaks the repeat sequences at their boundaries and reconnects the broken contigs at the stage of contig merging. We conducted performance comparisons of many assemblers using many datasets from genomes of various sizes and using simulation. For large genomes, JR-Assembler was more efficient in memory use and run time than current SRS assemblers, while achieving at least a comparable genome assembly quality. For small- and medium-size genomes, JR-Assembler performed better than most current SRS assemblers. JR-Assembler is freely available at http://jr-assembler.iis.sinica.edu.tw/.
Results
Overview of JR-Assembler.
JR-Assembler runs in five steps: raw read processing, seed selection, seed extension, repeat detection, and contig merging (Fig. 1). First, all reads containing any base “N” or any low-complexity region are filtered out. Second, it selects “good” reads as seeds using the read count—that is, the number of identical reads in the data. Third, JR-Assembler uses a jumping extension, including many whole reads at a time (Fig. 2). Moreover, to deal with sequencing errors at read tails, JR-Assembler uses back trimming to remove low-quality nucleotides at the 3′-end of a read to facilitate extension (Fig. 3). Fourth, when an extension is terminated, JR-Assembler checks whether a misextension was made because of the existence of a repeat. If a misextension occurs, it identifies the boundaries of the repeat and breaks the sequence at the boundaries (Fig. 4). The three steps of seed selection, seed extension, and misextension detection are repeated until no unused seed remains. Finally, JR-Assembler takes care of low-coverage regions by applying a less stringent extension procedure to merge the assembled sequences. JR-Assembler also incorporates a scaffolding program, SSAKE-based Scaffolding of Pre-Assembled Contigs after Extension (SSPACE) (12), for users to construct scaffolds. We will describe each step in detail in Materials and Methods.
Fig. 1.
Workflow of JR-Assembler. JR-Assembler runs in five steps. (A) Raw read processing. Input reads containing any N or a low-complexity region are discarded. Then, identical reads are collapsed into unique reads and stored in a table in alphabetical order. (B) Seed selection. The unique reads are ranked by read count (from low to high). Unique reads with medium read counts are selected as seeds for extension. (C) Seed extension. A seed is extended at the 3′ and 5′ directions by jumping extension. (D) Repeat detection. When an extension is terminated, JR-Assembler determines if a misassembly has occurred due to the existence of repeats. If it detects a misassembly, it identifies the boundaries of the repeat and breaks the sequence at each boundary. Steps B, C, and D are repeated until no unused seed remains. (E) Contig merging. After all seeds are used, JR-Assembler merges the contigs that can be connected by unassembled reads.
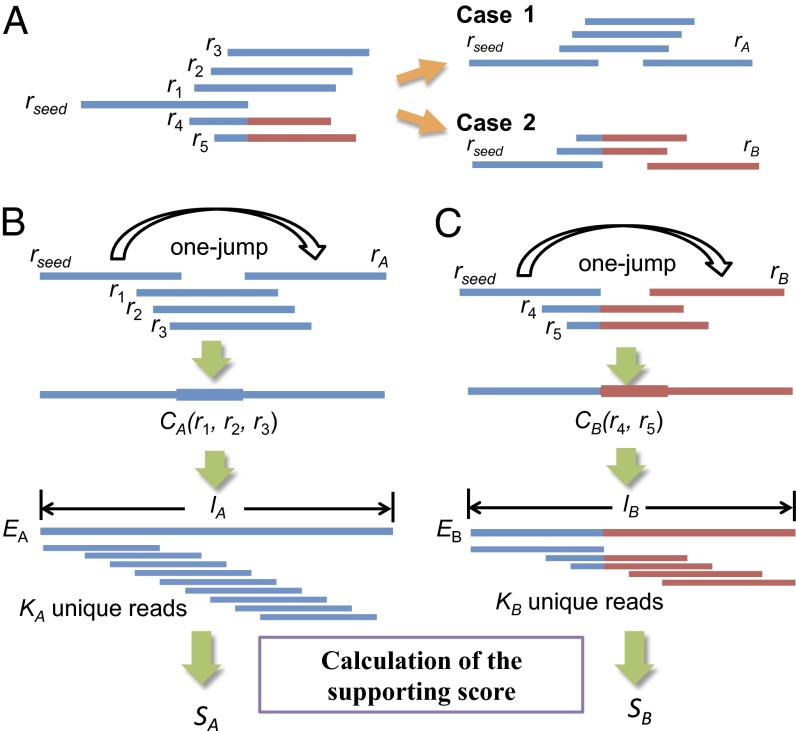
Fig. 2.
An example of extending a seed at the 3′-end. (A) Two extension candidates (cases 1 and 2) of a seed, rseed, are found for a jump extension. Red parts indicate that the corresponding bases are identical between r4 and r5 but cannot be aligned with the corresponding regions of r1, r2, and r3. (B) For case 1 in A, three bridging reads (r1, r2, and r3) of rA are identified, all of which overlap with a common unique read rA at the 3′-end. The consensus sequence of the bridging reads, CA(r1,r2,r3), is used to connect rseed and rA. The extension candidate EA is evaluated by the supporting score SA = KA/(lA – l + 1). (C) For case 2 in A, two bridging reads (r4 and r5) of rB are identified, and the extension candidate EB is built and evaluated by the supporting score SB = KB/(lB – l + 1). JR-Assembler selects the extension candidate with the highest score for extension.
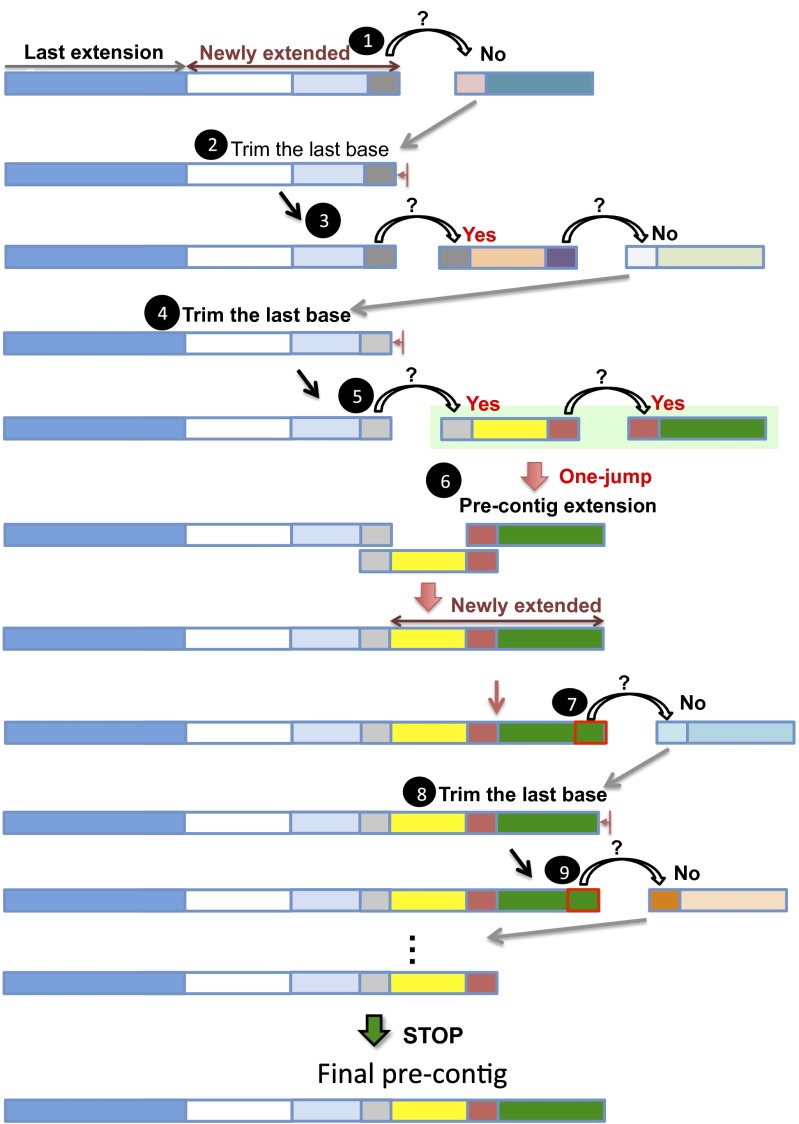
Fig. 3.
Procedure of back trimming. When an extension cannot proceed, JR-Assembler trims one base from the 3′-end of the previous extended sequence and then checks whether a jump extension can be made (step 1). If it cannot find a read to jump, the last base is trimmed (step 2) and a new jump is again tried (step 3). The trimming and checking processes are repeated until a jump extension can be made (step 5). After an extension is made, a new precontig is formed (step 6) and the extension process is continued (step 7). In Fig. 3, no extension (steps 7–9) was successful after step 6, so the final precontig was the precontig formed in step 6.
Fig. 4.
Repeat detection and resolution. (A) P is a precontig covered by a set of full-length supporting reads Sw and two sets of partial-length supporting reads S1 and S2. Blue parts in S1 and S2 indicate the regions that can be aligned with the corresponding regions in P, and the parts in red indicate that they cannot be aligned with P. Two possible genomic arrangements, B and C, can explain the precontig P. (B) The precontig P is fully consistent with the genomic region (P1–P2–P3). (C) The precontig P is only partially consistent with two genomic regions (P1–P2–Q3 and Q1–P2–P3), although each of the two local regions is supported by a subset of full-length reads. (D) Because the true genomic arrangement is uncertain because of the repeat P2, JR-Assembler identifies the repeat boundaries (Xp1 and Xp2) by finding a unique read rb that satisfies the following conditions: (i) rb is extendable, (ii) over 90% of prefix or suffix bases match to P perfectly, and (iii) the remaining bases contain at least one mismatch to P. (E) JR-Assembler breaks P into three separated contigs: P1, P2, and P3.
Performance Comparison for Assembling the Escherichia coli Genome.
We used the SRS data (10.3 million PE 101 bp reads, 450× coverage) of the E. coli genome, which is small, so we could efficiently scan a large parameter space to optimize each assembly for a fair comparison (Table S1). We compared JR-Assembler with ABySS (6), Edena (2), SOAPdenovo (9), Taipan (13), and Velvet (11). For each assembler, we aligned the contigs to the reference genome (retrieved from GenBank with accession no. NC_000913) using BLAST-like alignment tool (BLAT) (14) and checked misassemblies (see Table 1 for methods). For all methods compared, the misassembled contigs were broken at the junctions of misassemblies. The resulting pieces were also included in the calculation of genome coverage.
Table 1.
Assembly statistics of the E. coli dataset by different assemblers
| Assembler | No. of contigs* | Total†, Mb | Max‡ | Mean§ | N50¶ | Misassembled contigs|| | Time, min | Memory, GB |
| JR-Assembler | 192 | 4.53 | 237,952 | 23,571 | 48,673 | 0 | 23 | 5.1 |
| Edena | 413 | 4.53 | 63,737 | 10,963 | 22,264 | 0 | 172 | 13.7 |
| Taipan | 345 | 4.53 | 135,114 | 13,124 | 25,506 | 0 | 11 | 11.8 |
| Velvet | 216 | 4.54 | 138,645 | 21,019 | 43,998 | 3 (38k) | 26 | 19.0 |
| ABySS | 249 | 4.54 | 138,115 | 18,223 | 36,565 | 0 | 75 | 22.0 |
| SOAPdenovo | 285 | 4.53 | 117,147 | 15,904 | 31,136 | 0 | 7 | 19.8 |
Contigs of length <300 bp were not counted.
“Total” refers to the total number of bases in the contigs.
“Max” and “Mean” refer to the length of the longest contig and the mean length of contigs, respectively.
N50 is the size of the smallest contig such that 50% of the assembled bases are in the contigs of size equal to or larger than the N50 value.
A contig is misassembled if it cannot be aligned in full-length to the reference genome.
The proportion of the reference genome covered by the aligned contigs.
Table 1 shows the assembly statistics by the six assemblers. JR-Assembler was the best in most assembly metrics, including the number of contigs, the maximal contig length, the N50 length, the number of misassemblies, and the memory use. JR-Assembler used only ∼5.1 gigabytes (GB) of memory, whereas the others required 11–22 GB. The run times of JR-Assembler, Taipan, and Velvet were all under 30 min, whereas SOAPdenovo only required 7 min because it ran in multiple threads; the other assemblers all ran in a single thread (Velvet had not then provided the multithread function yet when the experiment was conducted). The genome coverages by Taipan, Velvet, and ABySS contigs were slightly higher than that generated by JR-Assembler. However, Velvet made three misassemblies.
We found that JR-Assembler could handle well low-coverage regions in the E. coli genome assembly. We remapped the processed reads to the E. coli reference genome and calculated the coverage at each base. A low-coverage region was defined as a region in which the coverage at each base was in the bottom 1% of the ranked coverages. In total, 386 low-coverage regions were identified. JR-Assembler had a higher ratio (89.1%) of contigs in low-coverage regions than Velvet (87.8%) and SOAPdenovo (79.5%). Two examples are shown in Fig. S1.
Performance Comparison for Assembling Small and Medium Genomes.
The first dataset consists of a bacterial genome [Streptomyces roseosporus, genome size 7.7 mega base pairs (Mb)] and three fungal genomes (Neurospora crassa, Plasmodium falciparum, and Saprolegnia parasitica, genome sizes 37.1, 22.9, and 53.1 Mb, respectively). We assembled the datasets of S. roseosporus and the three fungal genomes by the same six assemblers as above. No assembler ranked highest among all metrics (Table 2). Thus, we proposed a ranking approach to evaluate the overall performance for an assembler. For each assembly metric and dataset, we marked the top two values (in bold italic in Table 2). We then counted the number of marked values as the voting score for an assembler. The scores for JR-Assembler, Velvet, SOAPdenovo, ABySS, and Edena were 22, 15, 9, 8, and 3, respectively. Thus, JR-Assembler had the best overall performance. We did not show the results by Taipan because it produced no output on the four datasets.
Table 2.
Assembly statistics of S. roseosporus, N. crassa, S. parasitica, and P. falciparum
| Species | Assembler | No. of contigs | Total, Mb | Max | Mean | N50 | Time, min | Memory, GB |
| S. roseosporus | JR-Assembler | 1,189* | 7.68* | 40,501 | 6,461* | 11,374* | 99* | 26.1* |
| ABySS | 1,127* | 7.73* | 55,078* | 6,859* | 12,499* | 325 | 67.0 | |
| Velvet | 1,192 | 7.49 | 61,423* | 6,286 | 11,075 | 166 | 59.5 | |
| SOAPdenovo | 2,453 | 7.65 | 24,303 | 3,120 | 4,691 | 19* | 34.1* | |
| N. crassa | JR-Assembler | 12,244* | 38.61 | 58,672* | 3,153* | 6,074 | 76* | 14.9* |
| ABySS | 13,420 | 38.05 | 45,381 | 2,835 | 6,350* | 191 | 31.9 | |
| Velvet | 10,187* | 36.11 | 45,599* | 3,544* | 6,781* | 155 | 30.5 | |
| SOAPdenovo | 16,261 | 40.25* | 31,423 | 2,475 | 5,029 | 30* | 19.7* | |
| Edena | 17,083 | 39.95* | 42,952 | 2,338 | 4,534 | 659 | 20.4 | |
| S. parasitica | JR-Assembler | 40,587* | 46.09* | 119,543* | 1,135* | 1,510* | 172 | 14.1* |
| ABySS | 52,087* | 38.26 | 94,931* | 734 | 740 | 145 | 24.1 | |
| Velvet | 53,736 | 47.38* | 91,073 | 881* | 1,021* | 116* | 22.0 | |
| SOAPdenovo | 66,456 | 45.59 | 30,400 | 686 | 712 | 18* | 32.5 | |
| Edena | 62,357 | 44.13 | 41,473 | 707 | 746 | 610 | 19.5* | |
| P. falciparum | JR-Assembler | 13,352* | 11.02 | 7,939* | 825* | 975* | 133 | 9.0* |
| ABySS | 16,658 | 11.80 | 7,934 | 708 | 826 | 134 | 26.5 | |
| Velvet | 16,423* | 11.91* | 7,940* | 725* | 848* | 60* | 18.2 | |
| SOAPdenovo | 17,424 | 11.93* | 7,939* | 684 | 786 | 7* | 21.5 | |
| Edena | 16,531 | 11.76 | 7,936 | 711 | 831 | 359 | 13.4* |
The assembly metrics here follow those in Table 1.
The top two best values of each assembly metrics are marked in bold italic.
The other dataset is the one used in the Genome Assembly Gold-standard Evaluations (GAGE) assembly comparison (15) and consists of two bacterial genomes (Staphylococcus aureus and Rhodobacter sphaeroides, genome sizes of 2.9 and 4.6 Mb, respectively) and human chromosome 14 (88.3 Mb). In the GAGE study (15), all reads were error-corrected before assembly by ABySS, ALLPATHS-LG, Bambus2, Celera Assembler with the Best Overlap Graph (CABOG), Maryland Super-Reads Celera Assembler (MSR-CA), SGA, SOAPdenovo, and Velvet. For a fair comparison, we also obtained these corrected datasets for use in JR-Assembler. Because the two bacterial genomes and human chromosome 14 had been assembled using Sanger data, each of them could be used as a reference assembly. We used the assembly evaluation script provided by GAGE to assess various assembly metrics. Briefly, the GAGE script aligns contigs to the reference genome and calculates the corrected N50 length by breaking contigs at misassembled sites.
Table 3 shows the assembly metrics for JR-Assembler and eight others; the statistics for these eight assemblers were taken from the GAGE study (15). For the S. aureus dataset, ALLPATHS-LG and JR-Assembler achieved the longest corrected N50 lengths (66.2 and 66.1 Mb, respectively). SOAPdenovo achieved a corrected N50 length of 62.7 Mb, but made more misassemblies. Moreover, its ratio of the corrected N50 length to the uncorrected length (0.22) was much lower than those of ALLPATHS-LG (0.68) and JR-Assembler (0.65). For the R. sphaeroides dataset, JR-Assembler achieved the largest corrected N50 length and a high ratio (0.80) of the corrected N50 length to the uncorrected length.
Table 3.
Evaluation of JR-Assembler by GAGE
| Dataset | Contig statistics | JR-Assembler | ABySS | ALLPATHS-LG | Bambus2 | CABOG | MSR-CA | SGA | SOAPdenovo | Velvet |
| S. aureus | Contig No. | 58 | 302 | 60 | 109 | NA† | 94 | 252 | 107 | 162 |
| N50 | 101.4 | 29.2 | 96.7 | 50.2 | NA† | 59.2 | 4 | 288.2 | 48.4 | |
| N50 corr.* | 66.1 | 24.8 | 66.2 | 16.7 | NA† | 48.2 | 4 | 62.7 | 41.5 | |
| R. sphaeroides | Contig No. | 319 | 1,915 | 204 | 177 | 322 | 395 | 3,067 | 204 | 583 |
| N50 | 48 | 5.9 | 42.5 | 93.2 | 20.2 | 22.1 | 4.5 | 131.7 | 15.7 | |
| N50 corr.* | 38.3 | 4.2 | 34.4 | 12.8 | 17.9 | 19.1 | 2.9 | 14.3 | 14.5 | |
| Human chromosome 14 | Contig No. | 4,893 | 51,924 | 4,529 | 13,592 | 3,361 | 30,103 | 56,939 | 22,689 | 45,564 |
| N50 | 36.2 | 2 | 36.5 | 5.9 | 45.3 | 4.9 | 2.7 | 14.7 | 2.3 | |
| N50 corr.* | 18.2 | 2 | 21 | 4.3 | 23.7 | 4.3 | 2.7 | 7.4 | 2.1 |
The assembly result of JR-Assembler is compared with the statistics of the assemblers reported in ref. 15.
N50 corr., the N50 values of correction were computed after correcting contigs by breaking them at each misjoint or each indel longer than five bases.
NA, could not run, incompatible read lengths in one library. All N50 and N50 corr. values are all in kb.
For the human chromosome 14 dataset, CABOG, ALLPATHS-LG, and JR-Assembler were the top three assemblers in terms of the corrected N50 length (23.7, 21.0, and 18.2 Mb, respectively), whereas the corrected N50 length of the other assemblers were less than 10 Mb. We inspected the contigs that were connected by CABOG and ALLPATHS-LG but were broken by JR-Assembler. We found that 85% of the gaps between these JR-Assembler contigs contained low-complexity regions (all <200 bp), as detected by DustMasker (16).
Performance Comparison for Assembling Larger Genomes.
We used four datasets. The mouse and human datasets had 4.4 and 3.2 billion 101 bp reads, respectively. We compared our assemblies of these two genomes with those obtained using SOAPdenovo and ALLPATH-LG (7). We also obtained datasets of two other vertebrates, Metriaclima zebra (fish; 1.4 Gb) and Sorex araneus (shrew; 2.9 Gb); they had 1.27 and 4.23 billion 101 bp reads, respectively.
We compared JR-Assembler with ALLPATHS-LG, SOAPdenovo, and Velvet, which are three leading assemblers that can assemble vertebrate-sized genomes (∼3 Gb). We first used the base correction tool in the SOAPdenovo package to correct the errors in mouse and human reads, using default parameter values. Because many characters at the first fifth base in the data were Ns and the last 10 bases were often of low quality, we trimmed the first five bases and the last 10 bases from each read. We then assembled the processed reads by JR-Assembler using its default parameter values. For the mouse dataset, 2.24 Gb were assembled by JR-Assembler, 2.28 Gb by ALLPATHS-LG, and 2.26 Gb by SOAPdenovo; for the last two assemblers, we downloaded their assembled results and counted the total unambiguous bases of each assembly. The N50 length for JR-Assembler was 16 kilobases (kb), the same as generated by the ALLPATHS-LG and SOAPdenovo assemblies (7). For the human dataset, ∼2.25 Gb were assembled by JR-Assembler, 2.61 Gb by ALLPATHS-LG, and 2.38 Gb by SOAPdenovo. The N50 length (16 kb) of JR-Assembler was lower than that of ALLPATHS-LG (24 kb), but higher than that by SOAPdenovo (5.5 kb), as provided by ref. 7.
In terms of memory use, JR-Assembler used 418 and 433 GB of memory for the human and mouse datasets, respectively. The exact memory use by ALLPATHS-LG and SOAPdenovo were not documented in ref. 7, but were smaller than 512 GB. About the execution time, JR-Assembler took 1.5 central processing unit (CPU) wk to complete each genome starting from base correction to assembly completion. ALLPATHS-LG took 3 and 3.5 wk to complete the assemblies of the human and mouse genomes, respectively, whereas SOAPdenovo only took ∼3 d for the mouse genome (Dell R815, 48 processors, 512 GB), as documented in ref. 7. For ALLPATHS-LG and SOAPdenovo, the time recorded was the run time rather than the CPU time. As both ALLPATHS-LG and SOAPdenovo run in multiple threads, the sum of the CPU times for all threads should be longer than the run time.
For almost all current assemblers, assembling large genomes consumes high amounts of CPU time and memory. Therefore, for the datasets of M. zebra and S. araneus, we only compared JR-Assembler with SOAPdenovo and Velvet. As these two genomes have no reference sequence available, we only considered the memory requirements by these assemblers. In all three assemblers, we trimmed the original raw reads to 76 bp to remove the poor quality bases at read ends and filtered out the reads with one or more Ns. In the assembly of the M. zebra genome, 1.27 billion raw 101 bp reads were used. The total numbers of assembled bases by JR-Assembler and SOAPdenovo were 667.8 and 649.8 Mb, and the N50 lengths were 4.5 and 2.2 kb, respectively. In terms of memory use, JR-Assembler required only 160 GB, whereas SOAPdenovo required 340 GB. Velvet did not output any result due to a memory allocation error during assembly.
In the assembly of the S. araneus genome, 4.23 billion raw 101 bp reads were used. Running on a server with 1 TB of memory, only JR-Assembler finished the assembly (it used 562 GB) and produced an assembly with a total of 2.15 Gb assembled bases and an N50 length of 3.6 kb. Both SOAPdenovo and Velvet ran out of memory (1 TB). Therefore, JR-Assembler handles a much larger genome than either SOAPdenovo or Velvet.
Memory Use and CPU Time for Assembling Longer Sequencing Reads.
As the SRS technology advances, reads will increase in length. Therefore, we studied whether JR-Assembler will use memory better than other assemblers as read length increases. We obtained the Caenorhabditis elegans (nematode worm) genome from the University of California Santa Cruz (UCSC) Genome Bioinformatics Site (http://genome.ucsc.edu/) and simulated four SRS datasets with read lengths of 100, 150, 200, and 250 bp, respectively. We simulated 100 million reads for each dataset and assumed a uniform distribution of reads across each genome. The error rate was set to 0.1% for the first half of the read bases and quadratically increased from 0.1% to 0.2% for the second half of the read bases. Due to the special input library requirement by ALLPATHS-LG, we set 80% of them as fragment pair-end (PE) reads and 20% of them as short-jump PE reads. For each read length (l), the fragment PE reads were generated by randomly selected sequences of length 1.8 × l from the C. elegans genome, and for each PE pair, read 1 and read 2 were extracted from the 5′- and 3′-ends of the selected sequence, respectively. For the short-jump PE reads, we randomly selected 3 kb sequences from the genome and each read 1 and read 2 were also retrieved from the 5′- and 3′-ends of the selected 3 kb sequence, respectively. Here, we used the PE data for the comparison because ALLPATHS-LG did not provide a standalone program only for contig assembly. Thus, for a fair comparison, all of the compared assemblers were executed with the default scaffolding functions, and JR-Assembler used SSPACE (12) to scaffold the assembled contigs.
We recorded the maximum memory requirements and total CPU times of JR-Assembler, ALLPATHS-LG, SOAPdenovo, SOAPdenovo2, and Velvet; the k-mer sizes of the last three assemblers were both set to 70% of the read length, and the library lengths were separately set 1.8 × l and 3,000 bp, respectively, where l is the read length. For all four read lengths, JR-Assembler required less memory use than ALLPATHS-LG, SOAPdenovo, and Velvet but larger than SOAPdenovo2 (Fig. 5). When the read length increased to 200 bp, SOAPdenovo and Velvet required more than 256 GB, thus requiring a server with >256 GB of memory. For the cases of 200 and 250 bp, ALLPATHS-LG required 126.9 and 159.9 GB, whereas JR-Assembler required only 59.8 and 71.3 GB, respectively. The memory uses of SOAPdenovo2 were lower than those of the other assemblers tested (Fig. S2A).
Fig. 5.
Memory use and total CPU time of assemblers under various read lengths. Four datasets each of 100 million reads (read length 100, 150, 200, and 250 bp, respectively) are simulated from the C. elegans genome. Each dataset was assembled by JR-Assembler, ALLPATH-LG, SOAPdenovo, SOAPdenovo2, and Velvet, separately. The required memory use is in GB and the CPU time is in hours. With 200 bp reads, the memory uses by SOAPdenovo and Velvet exceeded the random access memory (RAM) of the server (256 GB). Thus, these two assemblers had no results for 200 and 250 bp. The horizontal and vertical axes represent the required memory (GB) and total CPU time (hour in a log10 scale), respectively. The performance data points of JR-Assembler, ALLPATHS-LG, SOAPdenovo, and Velvet are colored by blue, read, light green, dark green, and deep purple, respectively. The labels near each data point indicate the read length.
In terms of CPU time, ALLPATHS-LG required ∼135.2, 281.4, 445.8, and 560.7 h and SOAPdenovo2 required ∼12.6, 25.9, 51.4, and 116.1 h to complete the cases of l = 100, 150, 200, and 250 bp, respectively (Fig. 5). In contrast, JR-Assembler only required 2.6, 3.1, 3.6, and 4.4 h to complete for these cases (Fig. 5, Fig. S2B), indicating that JR-Assembler is potentially greater than 100 times and 10 times faster than ALLPATHS-LG and SOAPdenovo2, respectively, when the read length is 200 bp or longer. Fig. 5 shows that JR-Assembler required less memory use than ALLPATHS-LG, Velvet, and SOAPdenovo and a shorter CPU time than the other four assemblers. Thus, JR-Assembler is likely to further outcompete other assemblers in memory use (except for SOAPdenovo2) and CPU time as SRS reads are sequenced in greater length.
Discussion
Selection of Datasets and Assemblers for Comparison.
To have a comprehensive performance comparison, we considered genomes of a wide size range and compared assemblers using different assembly algorithms. Edena is the only SRS assembler using the OLC approach. ALLPATHS-LG, SOAPdenovo, Velvet, and ABySS use the popular de Bruijn graph approach. Taipan applies a hybrid strategy that combines the extension-based and graph-based approaches. Our JR-Assembler is a unique extension-based assembler. To make a fair comparison among these approaches, we optimized the parameter values for each assembler. For the genome assemblies studied by Gnerre et al. (7), using ALLPATHS-LG and SOAPdenovo, we directly cited their results.
Seed Selection.
In an extension-based assembler, a good seed should not contain any sequencing errors and should not be selected from a repeat region. A read containing sequencing errors usually has a very low read count (17). On the other hand, a read from a repeat region usually has a high read count because identical reads from other repeat loci are counted as well. Thus, in seed selection we avoided reads with a very low or a very high read count.
We had also tried two other seed selection strategies. One strategy selected seeds randomly and the other selected reads in a descending order of read count. We used the E. coli dataset and selected the same number of seeds to evaluate each strategy. We defined a good seed as a read that matches uniquely and perfectly to the reference sequence. We found that the proportion of good seeds among the seeds selected was 84% for our proposed approach, but only 52% for the descending read count and 20% for the random approach.
Efficiency in Repeat Resolution.
The jumping extension strategy in JR-Assembler not only speeds up assembly, but also efficiently resolves repeats smaller than the read length. Because JR-Assembler does extension by using whole reads, a repeat completely embedded in reads should be unnoticed during extension. On the other hand, a repeat buried in reads cannot be easily resolved by a de Bruijn graph-based assembler if the k-mer is shorter than the repeat size.
To support the above argument, we simulated reads of length 100 bp from human chromosome 22 using a window of 100 bp, sliding one base at a time. For simplicity, we assumed no sequencing errors. We assembled the simulated reads by JR-Assembler (overlap range set as 30∼35 bp), Velvet, and SOAPdenovo (k-mer size set as 51, 61, 71, 81, and 91 bp). When the k-mer size was 51 bp (half of the read length), the N50 length of Velvet and SOAPdenovo contigs were both <5 kb (Fig. S3), only 10% of their best N50 lengths (45.0 and 47.2 kb, respectively), which were reached only when the k-mer size was set to 91 bp, which is close to the read length (i.e., 100 bp). In contrast, JR-Assembler achieved the longest N50 length (49.6 kb) using a relatively short overlapping range, 30∼35 bp. Thus, users do not need to scan a wide range of parameters to find the optimal assembly by JR-Assembler.
One may argue that this should not be considered a disadvantage of SOAPdenovo and Velvet if it can be avoided by setting a large k-mer size. However, a large k-mer length increases the probability that a k-mer contains a sequencing error(s), becoming a “false” k-mer. As a result, the graph complexity increases and pruning the false graph nodes becomes more difficult. JR-Assembler’s advantage in repeat resolution will become even greater as SRS reads become longer. Unless sequencing errors are also decreased to a very low level, a de Bruijn graph assembler will not benefit from longer reads as much as JR-Assembler.
Scaffolding.
Here, we discuss whether the longer contigs assembled by JR-Assembler also result in longer scaffolds. Using the PE libraries of small and medium genomes as mentioned above, we scaffolded the contigs assembled by JR-Assembler using SSPACE (12). On average, the number of scaffolds was 46% of the number of contigs, and the scaffold N50 length was 2.5-fold of the contig N50 length. We also used the built-in scaffolding tools of SOAPdenovo and Velvet to do scaffolding. By our voting system, the voting scores of JR-Assembler+SSPACE, Velvet, and SOAPdenovo were 22, 19, and 8, respectively (Table S2). Thus, JR-Assembler+SSPACE has a better overall performance in terms of scaffolding than Velvet and SOAPdenovo.
Assembly of Reads with Low-Quality Bases at the 3′-End.
When input reads have too many sequencing errors at the 3′end, JR-Assembler may not be able to find correct overlapping reads for selected seeds to do the extension and may thus generate a set of very short contigs. One way to deal with this situation is to trim input reads at the 3′-end into shorter but better quality reads. However, how does a user know that input reads should be trimmed? For this purpose, JR-Assembler calculates the proportion of the seeds that can be extended or can be used by other extensions among the total seeds and then outputs the value in the final step for the user to judge. If the portion is very low, it indicates that most seeds cannot be extended because of low coverage or low quality. For the low-coverage problem, the user may need to obtain more data to increase the coverage. For the low-quality problem, the user can trim some 3′-end bases to get a better quality of input data. We have implemented the trimming process in JR-Assembler. However, how many bases need to be trimmed depends on output results. One approach, for example, may trim different lengths of 3′-end bases and check whether the seed use proportion is above a suggested threshold.
Sequencing Strategies for JR-Assembler.
Similar to the strategies recommended by ALLPATHS-LG (7), we recommend that for the Illumina technology one should use an overlapping paired-end library with a suitable insert size to generate PE raw reads for contig assembly and use several mate-pair libraries with different insert lengths to generate long-distant jumping reads for scaffolding. The average genome coverage should be at least ≥100∼150× or higher. For generating overlapping paired-end reads, we provide a simple formula to calculate the insert size for constructing a paired-end library:
 |
For example, if the read length is l =150 bp, the maximum overlap length is m = 40 bp, and the number of bases to be trimmed at the 3′-end is 30 bp, then the recommended insert size is 200 bp.
For a small or medium genome (≤50 Mb), we suggest to use MiSeq to sequence the genome because MiSeq produces longer reads than Hi-Seq200, so that it has a better chance to jump over repeats. On the other hand, for a large genome (∼1 Gb or larger) one should use HiSeq2000 to generate enough reads for genome assembly.
Future Development.
The current version of JR-Assembler only allows the input reads of the same length because the initial program kernel was designed for a fixed-length data structure. However, for a large genome, users usually have several read sets with different read lengths and qualities. These different read sets can be from the same platform or from different platforms, such as Roche 454, Illumina Hiseq2000, or MiSeq. For the former case, we have started to remodel the kernel to deal with this problem. For the latter, it will be a challenge to develop a new program to assemble the read sets simultaneously as the sequencing quality, error distribution, and throughputs are quite different from these platforms.
Concluding Remarks.
The two most important features of JR-Assembler are the jumping extension and the read remapping approach, which make memory use efficient and ensure a good assembly quality. Thus, JR-Assembler can handle SRS data of genomes of any size. For Illumina reads, the overall performance of JR-Assembler is better than or comparable to those of the current SRS assemblers we compared. Moreover, JR-Assembler requires less memory use and a shorter CPU time than ALLPATHS-LG, SOAPdenovo, and Velvet, especially when the read length is longer than 150 bp. As the sequencing technology advances, read length will increase, and we expect JR-Assembler to further outperform competing assemblers in terms of memory use and CPU time.
In this study, we have compared JR-Assembler with many current assemblers but have not included some others because of practical considerations. For example, SGA is a recent de novo assembly tool that is highly memory efficient (3). However, in a case study of the C. elegans genome (only ∼100 Mb), SGA’s overall performance was not as good as Velvet and SOAPdenovo, and it required 41 CPU h, whereas Velvet and SOAPdenovo only required several hours (3). As SGA is expected to take a very long CPU time for a large genome, we decided not to include it in our comparison study. The version of SOAPdenovo used in our comparison was SOAPdenovo1.05. SOAPdenovo2 includes improvements and new features, including the new contig and scaffold construction improvements (11).
Materials and Methods
Major Steps of JR-Assembler.
Step 1. Raw read processing.
JR-Assembler filters out the raw reads that contain any N, which is noninformative, or any low-complexity region, which may lead to false positive overlaps with other reads. As an option, DustMasker (16) may first be used to convert the low-complexity parts of reads into Ns, and such reads are then removed by JR-Assembler. Then the reads that are exactly the same—that is, the identical reads—are collapsed into one unique read and the read count c—that is, the number of identical reads—is recorded. As it is time consuming to identify unique reads by directly comparing the whole raw reads one by one, JR-Assembler builds a binary search tree to represent all unique reads in alphabetical order. For each raw read, the reverse-complement is also used for building the binary tree. A unique read table R is then constructed by traversing the binary search tree as follows: traverse the left subtree, visit the root, and then traverse the right subtree. The R table contains a set of tuples (r, c), where r represents a unique read and c is its read count. Because all unique reads in R have been stored in alphabetical order, the time complexity for searching a unique read in R is only O(log2N), where N is the size of R.
Step 2. Seed selection.
In JR-Assembler, each extension requires a unique read, called a seed, to initiate the extension. Ideally, a good seed should be a unique read without sequencing error and should not come from a repeat region. A seed with sequencing errors may result in a short contig or a misassembly. On the other hand, a seed from a repeat region may quickly terminate the subsequent extension when the repeat boundary is encountered. Thus, selecting a set of good seeds is essential for fast and good quality assembly by JR-Assembler. The current version of JR-Assembler uses the read count as a criterion to select seeds (Discussion). First, all unique reads are sorted by their read counts (from low to high). Then, those read counts ranked in between 1% and 25% are selected as the seeds for extension.
Step 3. Seed extension.
We first define some terms. Let l be the length of each read, and x and y be two unique reads in the unique read table R. We say that x and y overlap if the suffix t bases of x is identical to the prefix t bases of y, where n ≤ t ≤ m (m and n are, respectively, the allowed maximal and minimal numbers of overlapping bases) and 0 < n ≤ m ≤ l. More explicitly, we say that the 3′-end of x overlaps with the 5′-end of y.
Given a seed, JR-Assembler first extends it at the 3′-end and then at the 5′-end. To extend a seed rseed at the 3′-end, JR-Assembler searches all unassembled unique reads for extendable reads. A read is extendable for rseed if its 5′-end overlaps with the 3′-end of rseed and its 3′-end overlaps with one or more unique reads—for example, rA and rB in Fig. 2A. The extendable reads are also called bridging reads because they connect two unique reads. For example, in case 1 in Fig. 2A, three bridging reads r1, r2, and r3 connect rseed and rA, and in case 2 two bridging reads r4 and r5 connect rseed and rB. The process of connecting rseed and a unique read is termed a jump in this study. In Fig. 2B, to fill the gap between rseed and rA, JR-Assembler constructs a consensus sequence CA(r1,r2,r3) from the bridging reads. After a jump, an extension candidate EA is built by concatenating rseed, CA(r4,r5), and rA (Fig. 2B). The extension candidate EA is evaluated by a supporting score SA, defined as SA(EA) = KA/(lA – l +1), where lA is the length of EA and KA is the number of unique reads in R that can be remapped onto EA (Fig. 2B). If the score is less than the cutoff (0.3 by default)—that is, without enough read support—the extension is neglected. When rseed comes from a repeat, there can be more than one extension candidate (e.g., Fig. 2 B and C). In this case, JR-Assembler selects the candidate with the highest score for extension and deals with the repeat problem later. All unique reads that can be remapped onto this extension are then labeled as assembled. Assembled unique reads will not be used in the remaining extensions. The process is repeated until the 3′- and 5′-ends cannot be extended anymore.
Often an extension cannot proceed because of sequencing errors at the read tail. We propose a back trimming procedure to solve this problem (Fig. 3). JR-Assembler trims one base from the end of the last extended sequence and checks whether a jump is possible from the trimmed sequence. If not, the trimming continues until a jump is possible or until the previous extension is reached (Fig. 3). When a jump is made, the extension and back trimming procedures resume until no jump can be made further. Once the extension at the 3′-end is done, JR-Assembler starts to extend the 5′-end of the initial seed by the same two procedures. After the extensions at both ends are completed, we call the final extended sequence a “precontig.”
Step 4. Repeat detection.
If a precontig contains a misassembly due to a repeat, there should be reads that can be partially mapped to this precontig with the remaining parts entirely or partially mapped to another precontig(s). As shown in Fig. 4, P is a precontig covered by a set of full-length supporting reads, Sw, and two sets of partial-length supporting reads, S1 and S2, in which only partial sequences of the reads can be mapped onto P. Two possible scenarios of genome arrangement can explain P (Fig. 4 B and C). In Fig. 4B, the precontig P is correct in one region (P1–P2–P3), whereas both S1 and S2 support the other region (Q1–P2–Q3). Because all reads for assembling P2 cannot be used again after building P1–P2–P3, Q1–P2–Q3 cannot be rebuilt directly. In the second case (Fig. 4C), the precontig P is incorrect in both regions (P1–P2–Q3 and Q1–P2–P3), but each subregion (e.g., P1–P2 or P2–P3) is supported by a subset of full-length reads. Both ambiguous cases (Fig. 4 B and C) are due to the repeat P2.
To avoid ambiguity, JR-Assembler breaks the precontig P at each boundary of the repeat P2 if there exists a read rb that satisfies the following three conditions: (a) rb is extendable; (b) over 90% of prefix or suffix bases—that is, at least read length×0.9 bases—of rb match perfectly to P; and (c) the remaining bases have at least one mismatch to P. If one or more such reads are found (Fig. 4D), JR-Assembler breaks P at the first/last mismatching base according to the matching region at the 5′/3′-end, and outputs the separated contigs (P1, P2, and P3 in Fig. 4E). These broken contigs may later be merged into scaffolds at the scaffolding step.
Step 5. Contig merging in low-read coverage regions.
In the seed extension step, a precontig cannot be further extended if the supporting score—that is, read coverage—is low. To extend such contigs over the regions of low-read coverage, JR-Assembler relaxes the requirement of extendable reads—that is, two or more reads bridging rseed and rA as in Fig. 2A. In this case, it connects Ca and Cb without considering the minimum supporting score because there exists a set of unassembled reads that overlap at least m bases one by one from Ca to Cb. JR-Assembler uses a breadth-first search algorithm to find a path of overlapping reads connecting Ca and Cb. To avoid false connections, it only connects two contigs when the path of reads is unique. To consider contig orientation, JR-Assembler generates the reverse complements of all contigs and examines their connectivity in this step. Although it is an all-against-all approach, this step is not time-consuming in most cases, because the total numbers of unused reads and precontigs are usually much smaller than the total read count.
Performance Comparisons, Datasets, and Running Environments.
We compared JR-Assembler with ABySS 1.2.6 (www.bcgsc.ca/platform/bioinfo/software/abyss) (6), Edena 2.1.1 (www.genomic.ch/edena.php) (2), SOAPdenovo 1.0.5, SOAPdenovo2 (http://soap.genomics.org.cn/soapdenovo.html) (9, 10), Taipan 1.0 (http://taipan.sourceforge.net/) (13), Velvet 1.0.19 (www.ebi.ac.uk/∼zerbino/velvet/) (11), and ALLPATHS-LG 44683 (ftp://ftp.broadinstitute.org/pub/crd/ALLPATHS/Release-LG/) (7).
The SRS datasets of E. coli, S. roseosporus, N. crassa, P. falciparum, and S. parasitica genomes were downloaded from the National Center for Biotechnology Information (NCBI) Short Read Archive (SRA) (www.ncbi.nlm.nih.gov/sra) with the accession nos. SRX016044, SRX026747, SRX030834, SRX022535, and SRX016057 and SRX016059, respectively. Another dataset of two genomes (S. aureus and R. sphaeroides) and human chromosome 14 were downloaded from http://gage.cbcb.umd.edu/data/. The datasets of the whole human and mouse genomes were also downloaded from NCBI SRA; the accession numbers are listed in table S2 in ref. 7. The two datasets of M. zebra and S. araneus were downloaded from the same NCBI site with the accession nos. SRP004788 and SRP005678. The C. elegans genomic sequences were downloaded from the UCSC Genome Bioinformatics Site (http://hgdownload.soe.ucsc.edu/goldenPath/ce10).
The large genome datasets that included the genomes of human, mouse, M. zebra, and S. araneus were assembled on an IBM ×3850 X5 server, which has four 2.00 GHz eight-core Intel Xeon E7-4820 processors and 1 TB of RAM. All other datasets were assembled on a DELL PowerEdge R910 sever with two 2.67 GHz six-core Intel Xeon X7542 processors and 256 GB of RAM. Both servers run on the Linux operating system.
Parameters of Kernel JR-Assembler Program.
The kernel program of JR-Assembler has four parameters: the maximum overlap length (m), the minimum overlap length (n), the minimum remapping ratio (Rmin), and the maximum number of threads (Tno). On the basis of a comprehensive test of genome sizes from 4.5 Mb to 3 Gb and read lengths from 76 bp to 250 bp, as shown in the article, we suggest that only the values of m and n may need to be adjusted for different read lengths, whereas Rmin can be set as a constant (=0.3) for all cases. We recommend the following: if the input read length is 76 bp, set n = 30 and m = 35, and if the input read length is 100 bp or longer, set n = 40 and m = 45. Tno is dependent on the server running environment and is determined by the user.
Supplementary Material
Acknowledgments
We thank Drs. Mei-Yeh Lu, Wen-Lang Fan, Robert Friedman, Toni Gabaldon, Anton Nekrutenko, and Jason Tsai for comments on the manuscript. This work was supported by Academia Sinica and by National Science Council (Taiwan) Grants NSC99-2218-E-006-012, NSC99-3113-B-001-001, NSC 99-2321-B-001-041-MY2, and NSC 99-2621-B-001-003-MY3.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314090110/-/DCSupplemental.
References
- 1.Pop M. Genome assembly reborn: Recent computational challenges. Brief Bioinform. 2009;10(4):354–366. doi: 10.1093/bib/bbp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hernandez D, François P, Farinelli L, Osterås M, Schrenzel J. De novo bacterial genome sequencing: Millions of very short reads assembled on a desktop computer. Genome Res. 2008;18(5):802–809. doi: 10.1101/gr.072033.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simpson JT, Durbin R. Efficient de novo assembly of large genomes using compressed data structures. Genome Res. 2012;22(3):549–556. doi: 10.1101/gr.126953.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warren RL, Sutton GG, Jones SJ, Holt RA. Assembling millions of short DNA sequences using SSAKE. Bioinformatics. 2007;23(4):500–501. doi: 10.1093/bioinformatics/btl629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeck WR, et al. Extending assembly of short DNA sequences to handle error. Bioinformatics. 2007;23(21):2942–2944. doi: 10.1093/bioinformatics/btm451. [DOI] [PubMed] [Google Scholar]
- 6.Simpson JT, et al. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009;19(6):1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gnerre S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci USA. 2011;108(4):1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaisson MJ, Brinza D, Pevzner PA. De novo fragment assembly with short mate-paired reads: Does the read length matter? Genome Res. 2009;19(2):336–346. doi: 10.1101/gr.079053.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20(2):265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo R, et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1(1):18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27(4):578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt B, Sinha R, Beresford-Smith B, Puglisi SJ. A fast hybrid short read fragment assembly algorithm. Bioinformatics. 2009;25(17):2279–2280. doi: 10.1093/bioinformatics/btp374. [DOI] [PubMed] [Google Scholar]
- 14.Kent WJ. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salzberg SL, et al. GAGE: A critical evaluation of genome assemblies and assembly algorithms. Genome Res. 2012;22(3):557–567. doi: 10.1101/gr.131383.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgulis A, Gertz EM, Schäffer AA, Agarwala R. A fast and symmetric DUST implementation to mask low-complexity DNA sequences. J Comput Biol. 2006;13(5):1028–1040. doi: 10.1089/cmb.2006.13.1028. [DOI] [PubMed] [Google Scholar]
- 17.Kelley DR, Schatz MC, Salzberg SL. Quake: Quality-aware detection and correction of sequencing errors. Genome Biol. 2010;11(11):R116. doi: 10.1186/gb-2010-11-11-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.