Abstract
Huntington disease (HD) is a neurodegenerative disorder caused by a CAG expansion within the huntingtin gene that encodes a polymorphic glutamine tract at the amino terminus of the huntingtin protein. HD is one of nine polyglutamine expansion diseases. The clinical threshold of polyglutamine expansion for HD is near 37 repeats, but the mechanism of this pathogenic length is poorly understood. Using Förster resonance energy transfer, we describe an intramolecular proximity between the N17 domain and the downstream polyproline region that flanks the polyglutamine tract of huntingtin. Our data support the hypothesis that the polyglutamine tract can act as a flexible domain, allowing the flanking domains to come into close spatial proximity. This flexibility is impaired with expanded polyglutamine tracts, and we can detect changes in huntingtin conformation at the pathogenic threshold for HD. Altering the structure of N17, either via phosphomimicry or with small molecules, also affects the proximity between the flanking domains. The structural capacity of N17 to fold back toward distal regions within huntingtin requires an interacting protein, protein kinase C and casein kinase 2 substrate in neurons 1 (PACSIN1). This protein has the ability to bind both N17 and the polyproline region, stabilizing the interaction between these two domains. We also developed an antibody-based FRET assay that can detect conformational changes within endogenous huntingtin in wild-type versus HD fibroblasts. Therefore, we hypothesize that wild-type length polyglutamine tracts within huntingtin can form a flexible domain that is essential for proper functional intramolecular proximity, conformations, and dynamics.
Keywords: polyglutamine diseases, conformational switching, FLIM-FRET, neurodegeneration, fluorescence lifetime imaging microscopy
Huntington disease (HD) is an autosomal dominant, progressive neurodegenerative disorder caused by the expansion of a CAG trinucleotide repeat within the huntingtin (HTT) gene (1). The pathogenic threshold of CAG expansion is ∼37 repeats, with increased repeats leading to an earlier age-onset of HD (2–4). This CAG mutation results in an expanded polyglutamine tract in the amino terminus of the gene’s protein product, huntingtin (1). To date, no phenotypes at the level of huntingtin molecular biology or animal models can be attributed to polyglutamine lengths near the human pathogenic disease threshold (5).
Polyglutamine or glutamine-rich domains are also found in transcription factors such as the Sp-family and cAMP response element-binding protein (CREB) and are defined as protein–protein interaction motifs used to scaffold and regulate transcription by RNA polymerase II (6, 7). In the transducin-like enhancer of split (TLE) corepressor proteins, the glutamine-rich domains are thought to allow dimerization (8). However, the role of polyglutamine in proteins such as huntingtin may be distinct from that of a protein–protein interaction or dimerization domain.
The polyglutamine tract of huntingtin is flanked on the amino-terminal side by the first 17 amino acids, termed N17, an amphipathic alpha-helical targeting domain that can mediate huntingtin localization to membranes (9, 10). The N17 domain can be modified posttranslationally at multiple residues (11–14). Within N17, phosphorylation of two serines at positions 13 and 16 is critical for modulating huntingtin localization during stress (10, 12), N17 structure (12), and the toxicity of mutant huntingtin in cell biological and mouse models of HD (12–15). We have previously demonstrated that mutant huntingtin is hypophosphorylated at serines 13 and 16; this results in an inability to respond to cell stress (12).
On the carboxyl-terminal side of the polyglutamine tract is a region containing two pure proline tracts separated by a leucine-proline–rich intervening region within the human huntingtin protein (16). Similar to the N17 domain, interacting proteins within this polyproline region have been found to affect mutant huntingtin toxicity (16). Protein kinase C and casein kinase 2 substrate in neurons (PACSIN1), or syndapin 1, binds directly to the polyproline domain (17). This interaction is enhanced in the presence of expanded polyglutamine (18). PACSIN1 is a predominantly cytoplasmic neuronal protein that has functions in NMDA receptor recycling (18, 19), actin/microtubule reorganization (20), and neuronal spine formation (21). Both N17 and PACSIN1 are substrates of casein kinase 2 (CK2) (12, 17).
Because both of the flanking regions to the polyglutamine tract are critical in mediating the toxicity of the mutant huntingtin protein in mammalian and yeast models (22), we wanted to determine whether the two domains could be interacting with each other in 3D space using the polyglutamine tract as a flexible region. To test this hypothesis, we developed a Förster resonance energy transfer (FRET) sensor with donor and acceptor fluorophores at the amino and carboxyl-termini of huntingtin fragments, respectively. Using fluorescence lifetime imaging microscopy (FLIM), we quantified the FRET efficiency of live cells expressing huntingtin fragments with multiple polyglutamine lengths. We discovered that the N17 domain folds back to the polyproline region of huntingtin and that this conformation is altered by expanded polyglutamine at the clinical pathogenic threshold for HD of 37 repeats.
We have previously used a chemical biology approach to determine that CK2 can phosphorylate huntingtin within N17. This could be prevented by treatment of cells with CK2 inhibitors (12). Conversely, Iκβ kinase (IKK) inhibitors paradoxically increased phosphorylation of N17 (12). This suggested that the protective effect of serine 13 and 16 phosphomimicry in the bacterial artificial chromosome Huntington disease mouse model (15) may be attainable using small molecule treatments. The lipid ganglioside GM1 treatment of the yeast artificial chromosome 128 mouse model resulted in reversion of the HD motor phenotype to normal and was found to restore the hypophosphorylation of mutant huntingtin at N17 to normal levels (14). Therefore, we wanted to test if there was a direct effect of serines 13 and 16 on the conformation of the amino terminus of huntingtin and whether the conformations of huntingtin could be affected by phosphomutants or true phosphomodulation of N17.
To translate this conformational sensor to look at endogenous huntingtin, we developed an antibody-based FLIM-FRET assay to measure the conformation of the amino terminus of huntingtin in the context of the full-length protein. This assay confirmed the finding in human patient HD cells that N17 folds back to the vicinity of the polyproline region in endogenous huntingtin.
Results
Huntingtin N17 Folds Back to the Polyproline Region.
To test for an intramolecular interaction between the regions flanking the polyglutamine tract of huntingtin, we developed a conformational sensor using FRET efficiency as readout. We chose a fluorophore pair for FRET that consisted of a mCerulean donor and an enhanced yellow fluorescent protein (eYFP) acceptor. FRET was calculated using fluorescence lifetime imaging microscopy (FLIM) by time-correlated single photon counting (TCSPC) (23, 24). Fluorescence lifetime refers to the amount of time a valence electron from a fluorophore remains in the excited state before returning to ground state and emitting a photon. The lifetime of a fluorophore can be directly affected by FRET, which results in a decrease in the donor fluorophore lifetime. All of the necessary controls were performed to validate the fluorophore lifetime values in our live-cell system (Fig. 1 A–C).
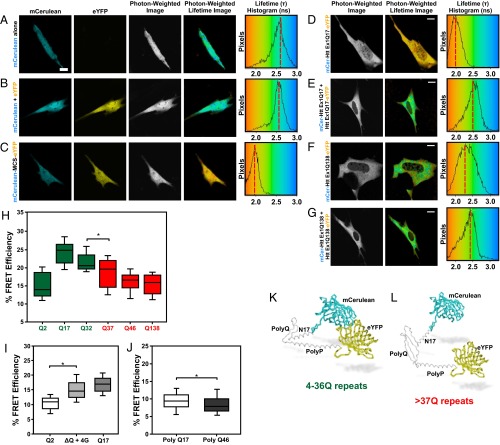
Fig. 1.
Huntingtin exon1 FLIM-FRET sensor. Sample live-cell fluorescence and FLIM images for the (A) mCerulean (mCer) donor-alone control, (B) mCer coexpressed with the YFP acceptor control, and (C) mCer-multiple cloning site-YFP positive control. Sample FLIM images of (D) mCer-huntingtin exon1 Q17-eYFP (mCer-HttEx1 Q17-eYFP) fusion, (E) mCer-HttEx1 Q17 + HttEx1 Q17-eYFP control for intermolecular FRET, (F) mCer-HttEx1 Q138-eYFP fusion, and (G) mCer-HttEx1 Q138 + HttEx1 Q138-eYFP control for intermolecular FRET expressed in STHdhQ7/Q7 cells. (H) FLIM-FRET data using the mCer-HttEx1-eYFP FRET sensor with varying lengths of polyglutamine. Black lines represent the median value, boxes encompass 25% and 75% confidence intervals, and whiskers indicate the 5% and 95% confidence intervals. *P < 0.001. n = 50, four replicates. Box-whisker plot shows data for all trials. (I) FRET efficiency comparing mCer-HttEx1 Q2-YFP and the mCer-HttEx1 Q17-YFP FRET sensor to the mCer-HttEx1 ∆Q + 4 glycines-YFP positive control. *P < 0.001. n = 90, three replicates. (J) FRET efficiency of 17 and 46 glutamines. *P < 0.001. n = 100, three replicates. Models of huntingtin FRET sensor with (K) wild-type and (L) mutant polyglutamine lengths. (Scale bars: 10 µm.)
We constructed a FRET sensor using exon1 fragments of varying polyglutamine lengths with a donor and an acceptor fluorophore at the amino and carboxyl termini of exon1 with a 1:1 ratio of donor:acceptor. At a wild-type length of 17 polyglutamine repeats, we noted a robust lifetime decrease, thus a relative increase in percent FRET efficiency, indicating a close proximity of the donor and acceptor fluorophores (Fig. 1 D and H). This FRET was intramolecular, because coexpression of mCerulean-huntingtin exon1 and huntingtin exon1-eYFP together, on separate plasmids, did not result in a donor lifetime change at these expression levels (Fig. 1 E and G). When a similar sensor with a polyglutamine expansion of 138 repeats was tested, the fluorescence lifetime values were increased relative to the wild-type polyglutamine length, resulting in a reduced FRET efficiency (Fig. 1 F and H). We controlled for expression of huntingtin fragment levels in the assays to avoid the effects of protein aggregation on lifetime values.
Typical pathogenic polyglutamine lengths for midlife onset of HD are between 45–50 repeats (1); however, HD is fully penetrant at any length over 40 repeats. Most HD models use huntingtin genes with very long CAG repeats (>100) to induce a robust phenotype (5). To date, no assay in live cells has noted a phenotype at polyglutamine lengths close to 37 repeats or even typical HD repeat lengths. Using the huntingtin exon1 FRET sensor, we tested the effect of varying glutamine repeat lengths to determine if we could detect a conformational change at the pathogenic threshold for HD. We detected a significant decrease in FRET efficiency between 32 and 37 repeats; however, minimal changes were measured between 37, 46, and 138 repeats (Fig. 1H). To prove that the FRET efficiency changes we detected were not due to simply moving the acceptor probe farther away from the donor, we synthesized and tested an exon1 FRET sensor with only two glutamines. This construct resulted in a lower FRET efficiency relative to all other polyglutamine lengths tested, despite the shorter alpha-carbon backbone (Fig. 1H). To demonstrate that the intramolecular FRET between N17 and the polyproline region was not an artifact of the STHdhQ7/Q7 cell type used, we repeated experiments using the exon1 FRET sensor with wild-type (Q17) and mutant (Q138) polyglutamine tracts in both NIH 3T3 fibroblasts and primary human fibroblasts (Fig. S1 A and B). Consistent with the data generated in the STHdhQ7/Q7 cells, we measured a higher percentage of FRET efficiencies with the wild-type versus the mutant sensor in both fibroblast cell lines.
Because we consistently measured the highest FRET efficiencies for huntingtin fragments with wild-type polyglutamine lengths, this suggested that the polyglutamine tract may exist as a flexible “hinge” to allow the N17 domain to loop back and come into close spatial proximity with the polyproline region (Fig. 1 H, K, and L). To test this hypothesis, we designed a synthetic exon1 fragment where we substituted the polyglutamine tract with a four glycine linker (25). We detected a similar change in FRET efficiency with the exon1 ∆Q + 4 glycine construct relative to the exon1 Q17 FRET sensor, suggesting that wild-type polyglutamine tracts can behave similarly to a flexible glycine linker (Fig. 1I). Conversely, we hypothesized that the mutant polyglutamine expansion results in a diminished flexibility, which induces a conformational change in the amino terminus of huntingtin. To test the importance of the flanking sequences on the flexibility of the polyglutamine tract, we inserted pure polyglutamine tracts with either 17 or 46 repeats into the FRET sensor. For both constructs, we measured lower FRET efficiencies relative to the exon1 constructs. There was only a modest difference between the wild-type and mutant pure polyglutamine constructs, indicating that just polyglutamine was not sufficient to properly place the amino and carboxyl termini in close proximity in the absence of flanking sequences of huntingtin (Fig. 1J). Our next step was to analyze if we could detect polyglutamine-dependent conformational changes in larger huntingtin fragments.
We then constructed FRET sensors with increasing fragment lengths of huntingtin in both the wild-type and mutant context (Fig. S2). FRET efficiency of the huntingtin 1–117 (exon1+2) sensor was higher than that of the exon1 sensor. This indicated that N17 was likely in closer proximity to a distal region of huntingtin beyond the polyproline region or that additional amino acids beyond this domain were important to stabilize this conformation. As with the huntingtin exon1 sensor, polyglutamine expansion of huntingtin 1–117 resulted in a similar drop of FRET efficiency. Placement of the FRET acceptor at amino acids 171, 220, or 465 in huntingtin resulted in a drop in FRET efficiency, with no significant difference between polyglutamine lengths.
N17 Serine Phosphomimetics Affect Huntingtin Conformation.
To test the importance of serines 13 and 16 on the conformation of huntingtin exon1 in live cells, we generated constructs with N17 phosphomimetic mutants in the context of the FRET sensor (Fig. S3). S13AS16A mutations in either the Q17 (Fig. S3A) or Q142 (Fig. S3C) context of exon1 had little effect on the FRET efficiency of the sensor relative to wild-type N17 constructs (Fig. 2A). However, S13ES16E substitutions in both the wild-type (Fig. S3B) and mutant (Fig. S3D) context significantly reduced FRET efficiency (Fig. 2A). Previously, we demonstrated that phosphorylation and phosphomimicry at serine residues 13 and 16 disrupted the structure of N17 (12), whereas a proline substitution at methionine 8 (M8P) completely abolished N17 structure (10). In the context of the FRET sensor, the M8P mutations with either Q17 (Fig. S3E) or Q150 (Fig. S3F) both resulted in a significant decrease in FRET efficiency (Fig. 2A). The result with the M8P mutant was consistent with the control experiment of just 17 or 46 glutamines in the FRET sensor (Fig. 1J). Thus, we conclude that the structure of N17 contributes to the overall conformation of the amino terminus of huntingtin as well as the proximity of N17 to the polyproline region. The caveat of this data is that phosphomimicry is not true phosphorylation. To address this, we used a chemical biology approach with kinase inhibitors that we previously described as modulating the phosphorylation of huntingtin in N17 (12).
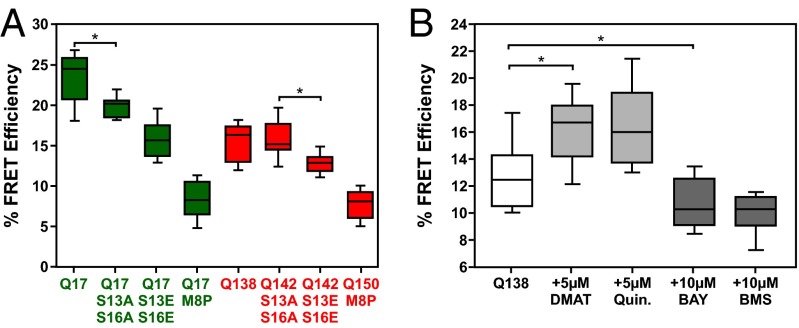
Fig. 2.
N17 phospho-mimicry mutants and kinase inhibitors can affect the conformation of huntingtin exon1. (A) Percent FRET efficiency for mCer-HttEx1 Q17 or Q142-eYFP with serines 13 and 16 mutated to alanines or glutamic acids, mCer-HttEx1 Q17 or Q150-eYFP with the M8P mutation. *P < 0.001. n = 40, four replicates. (B) Percent FRET efficiency for the mCer-HttEx1 Q138-eYFP sensor following no treatment, treatment with CK2 inhibitors (DMAT, Quinalizarin), or treatment with IKK inhibitors (Bay 11-7082, BMS-345541). *P < 0.001. n = 30, three replicates.
Kinase Inhibitors Directly Affect the Conformation of Amino Terminus of Huntingtin.
To study the effects of phosphomodulation on the conformation of the amino terminus of huntingtin, we used kinase inhibitors known to either inhibit or promote phosphorylation at serine residues 13 and 16 of N17 (12). N17 phosphorylation can be inhibited by CK2 inhibitors 2-Dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole (DMAT) and 1,2,5,8-tetrahydroxyanthraquinone (quinalizarin). Alternatively, N17 phosphorylation can be promoted by treatments with IKK inhibitors, the ATP analog Bay 11-7082 [(E)-3-(4-methylphenylsulfonyl)-2- propenenitrile], and the allosteric inhibitor Bristol Myers Squibb (BMS)-345541 [N-(1,8-Dimethylimidazo[1,2-a]quinoxalin-4-yl)-1,2-ethanediamine hydrochloride]. These compounds were tested on STHdhQ7/Q7 cells expressing the polyglutamine expanded (Q138) huntingtin FRET sensor (Fig. 2B). Unlike alanine substitutions, CK2 inhibition by DMAT and quinalizarin caused an increase in FRET efficiency by affecting the conformation of mutant huntingtin exon1. This indicates that the hydroxyl-side group of the serine is important for the contribution of N17 to huntingtin conformation. IKK inhibition by Bay 11-7082 and BMS-345541, leading to hyperphosphorylation of N17 (12), resulted in reduced FRET efficiency to a greater extent than phosphomimicry (Fig. 2 B versus A). From these data, we concluded that the promotion of huntingtin N17 phosphorylation by IKK inhibitors showed similar effects to phosphomimicry on the conformation of soluble mutant huntingtin.
PACSIN1 Interacts with both the N17 and Polyproline Domains of Huntingtin.
PACSIN1 has previously been reported to interact with the proline-rich intervening region of the polyproline domain of huntingtin in a polyglutamine length-dependent manner (18). Using coimmunoprecipitation, we were able to purify PACSIN1 using the N17 domain alone (Fig. S4A). Importantly, although both N17 and PACSIN1 are substrates of CK2, treatment of cells with the DMAT before coimmunoprecipitation had little effect on the amount of PACSIN1 recovered using N17. This indicated that the phosphorylation state of N17 or PACSIN1 does not affect the interaction between the two proteins.
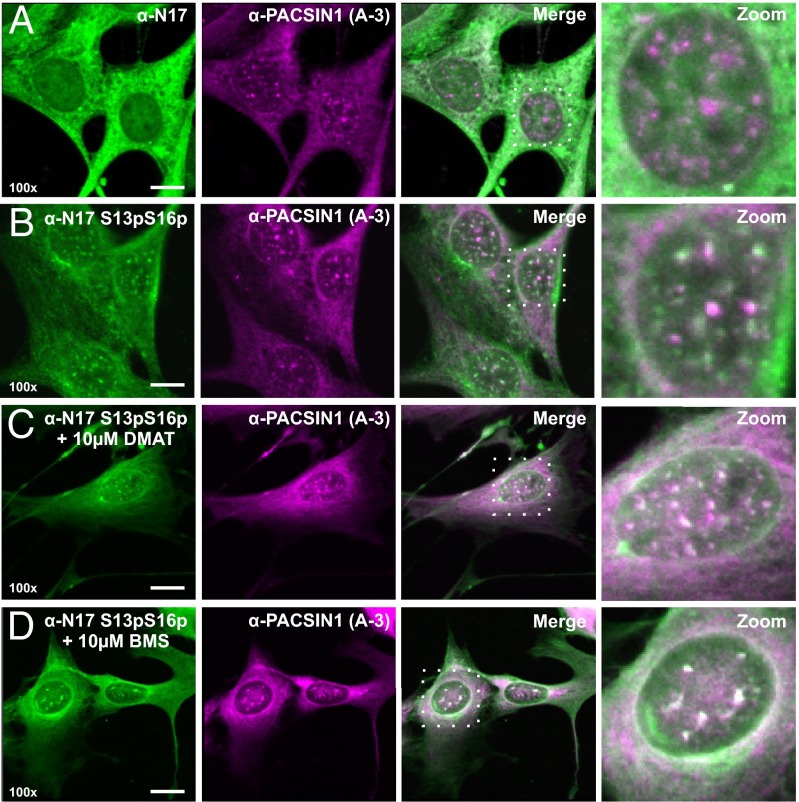
Coimmunofluorescence with antibodies recognizing PACSIN1 and N17 demonstrated that these two proteins show a high degree of signal overlap in the cytoplasm (Fig. 3A). When coimmunofluorescence was done using antibodies raised toward PACSIN1 and N17 S13pS16p, we observed no signal overlap in the cytoplasm; instead, both proteins were strongly colocalized to nuclear puncta (Fig. 3B). When we treated cells with compounds that are known to decrease (DMAT; Fig. 3C) or increase (BMS-345541; Fig. 3D) the phosphorylation of huntingtin at residues S13 and S16 (12), we noted a profound effect on the larger morphology and reduced number of the puncta present in the nucleus.
Fig. 3.
PACSIN1 interacts with N17 and facilitates the proximity between N17 and the polyproline region of huntingtin. Coimmunofluorescence images of STHdhQ7/Q7 cells taken at 100× magnification using an anti-PACSIN1 antibody (A-3) and (A) N17 or (B) N17 S13pS16p, respectively. Immunofluorescence against PACSIN1 and N17 S13pS16p was also done on STHdhQ7/Q7 cells following 16 h treatments with either (C) 10 μM DMAT or (D) 10 μM BMS-345541. (Scale bars: 10 µm.)
PACSIN1 interacts with both the N17 and polyproline domains that flank the polyglutamine tract of huntingtin, which we hypothesized may stabilize the positioning of these two domains in close 3D proximity. To determine the involvement of PACSIN1 between these domains, we tested our huntingtin exon1 FRET sensor following treatment of the cells with a mixture of three small interfering RNAs (siRNA) directed to mouse PACSIN1. Both a Western blot and immunofluorescence were performed to confirm the efficacy of the knock-down (Fig. S4 B–D). In cells expressing the wild-type huntingtin FRET sensor (Q17), treatment with PACSIN1 siRNAs significantly reduced FRET efficiency, whereas this was not seen with the mutant polyglutamine expanded FRET sensor (Q138) (Fig. S4E). Our data indicate that PACSIN1 may facilitate the proximity of N17 and polyproline by acting as a scaffold to bridge these two domains.
Measuring the Conformation of the Amino Terminus Within the Context of Full-Length, Endogenous Huntingtin.
Making protein fusions with fluorophores at both termini can alter the folding dynamics and conformations of huntingtin fragments. We were also limited by the need to overexpress the FRET sensor to superphysiological concentrations to collect enough photons to generate accurate fluorophore lifetime decay curves. Thus, we wanted to determine if we could measure the conformation of the amino terminus of huntingtin within the context of the full-length protein at endogenous expression levels, based on evidence of a N17-polyproline proximity learned from the exon1 FRET sensor. We conjugated several primary antibodies raised to epitopes known to fall between residues 1 and 117 of huntingtin. We chose to use both the 2B7 and the 4C9 monoclonal antibodies that target residues 8–13 of the N17 domain and residues 61–71 of the polyproline region of huntingtin, respectively (26). As an optimal pair for FLIM-FRET, we chose to conjugate the antibodies with either alexa488 or alexa546 dyes.
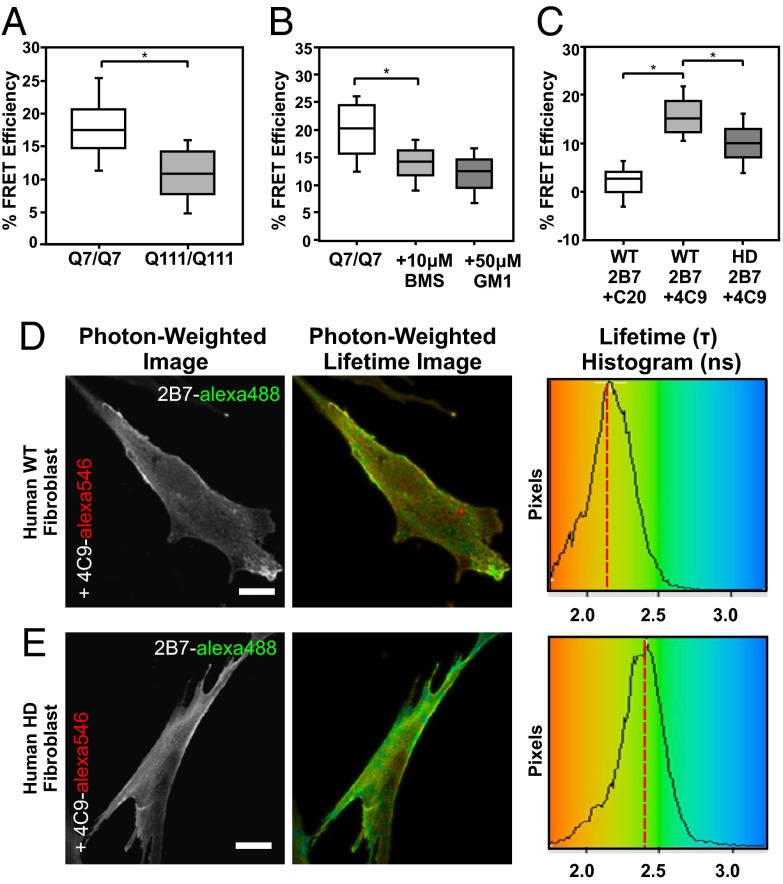
When we compared FRET efficiency values between STHdhQ7/Q7 and STHdhQ111/Q111 lines both stained with 2B7-alexa488 and 4C9-alexa546 conjugates, we measured a significantly lower percent FRET efficiency in the mutant compared with the wild-type cells (Fig. 4A). As a negative control, we used an acceptor conjugated to an antibody that recognized an epitope at the carboxyl terminus of huntingtin (C-20) and saw no significant change in the donor lifetime. This was due to the acceptor conjugate now being too far away from the donor for FRET. Thus, using this antibody-based assay on full-length, endogenous huntingtin, we were able to recapitulate our earlier findings with the huntingtin exon1 FRET sensor (Fig. S5).
Fig. 4.
An antibody-based FLIM-FRET assay to measure the conformations of the amino terminus of full-length huntingtin. (A) Percent FRET efficiency for the amino terminus of huntingtin in STHdhQ7/Q7 versus STHdhQ111/Q111 cells following immunofluorescence with conjugated primary antibodies 2B7 and 4C9. *P < 0.001. n = 150, five replicates. (B) Percent FRET efficiency for the amino terminus of huntingtin in STHdhQ7/Q7 following treatment with 10 μM BMS-345541 or 50 μM ganglioside GM1. *P < 0.001. n = 100, 3 replicate trials. (C) FRET efficiency for the amino terminus of huntingtin in wild-type versus HD fibroblasts with conjugated primary antibodies 2B7/C20 or 2B7/4C9. FLIM images of 2B7-alexa488 and 4C9-alexa546 conjugates in fixed (D) wild-type or (E) HD patient fibroblasts. *P < 0.001. n = 100, three replicates. (Scale bars: 10 µm.)
To assess the effects of phosphorylation on the conformation of the amino terminus within the context of the full-length huntingtin protein, we used two compounds known to promote phosphorylation at serine residues 13 and 16 of N17: BMS-345541 and ganglioside GM1 (12, 14). Treatment of STHdhQ7/Q7 cells with either BMS-345541 or GM1 for 16 h caused a significant reduction of the percent FRET efficiency relative to the nontreated control (Fig. 4B). These data were consistent with our earlier findings using the exon1 FRET sensor that phosphorylation of N17 affects the conformation of the amino terminus of huntingtin.
Next, we wanted to test our antibody-based FLIM-FRET assay on human HD patient cells, rather than models of HD. We chose to use untransformed human fibroblasts from a 54 y old unaffected female and a 51 y old male with HD. Using 2B7 and 4C9 conjugates in these cells, we were able to detect a robust conformational difference of huntingtin in these HD patient fibroblast samples relative to those from the unaffected individual (Fig. 4 D and E). This conformational difference was measured as a significant decrease in the overall FRET efficiency in the HD versus the wild-type fibroblasts (Fig. 4C). Thus, using this assay, we measured a difference in huntingtin conformation in fibroblasts between normal and HD individuals.
Discussion
A long-standing question in HD research has been why clinical CAG repeat lengths beyond 37 repeats result in pathogenesis, whereas even a few repeats below this number do not (27). Animal models of HD require very long polyglutamine tracts to elicit any obvious phenotypic changes; however, using our huntingtin exon1 FRET sensor, we have been able to detect a conformational switch between 32 and 37 glutamine repeats in live cells.
Huntingtin is one of nine polyglutamine expansion disease proteins, where the biological role of normal polyglutamine tracts in these proteins is not understood (28). In transcription factors such as the Sp-family, CREB, and TLE, glutamine-rich domains are thought to mediate protein–protein interactions and/or dimerization (6–8). Our data demonstrate that polyglutamine or glutamine-rich regions may also represent a domain of required flexibility to mediate interactions between flanking protein–protein interaction domains.
All mammalian species have a huntingtin protein that contains at least four glutamines in their polyglutamine tract. In vertebrate species that contain only two glutamines, the proline-rich region is absent (29). The reduced FRET efficiency observed with the synthetic huntingtin exon1 Q2 FRET construct suggests that the polyglutamine tract may be a critical conformational hinge in the amino terminus, allowing N17 to interact with regions downstream. This hinge hypothesis is consistent with the X-ray crystal structures that have been solved with wild-type exon 1 (30) and molecular dynamics simulations (31).
We hypothesize that the decrease of FRET efficiency with mutant fragments is due to a gain of structure in the polyglutamine tract, leading to a reduced flexibility of the hinge region. In vitro, expanded polyglutamine tracts can adopt a rigid β-sheet structure (32). Therefore, our FLIM-FRET data are consistent with the rusty hinge hypothesis (33), where the expansion of polyglutamine leads to reduced flexibility of the tract, resulting in multiple inflexible conformations of the amino terminus. We hypothesize, as have others, that the normal polyglutamine tract in huntingtin is disordered (34). We suggest that polyglutamine tract in proteins may contribute to a phenomenon of stochastic protein conformations and interactions termed “fuzzy complexes” (35). A caveat of using FLIM-FRET to measure conformations of huntingtin is that the data do not tell us exactly what the conformations of the tested proteins are, only the differences between the tested constructs.
The effects of CK2 and IKK inhibitors on skewing the conformation of soluble huntingtin indicate that a protective conformation of mutant huntingtin may be pharmacologically induced by correcting the hypophosphorylation seen in mutant huntingtin (12). Treatment of cells with IKK inhibitors caused an increase in phosphorylation at serines 13 and 16 of N17, which we measured as a decrease in the percent FRET efficiency and interpreted as a change in conformation of the huntingtin. We hypothesized that this decrease in FRET efficiency was either due to a loss of the alpha-helical structure of N17 or due to a change in the interaction network with N17 as a result of phosphorylation. However, because the readout for FLIM is either an increase or a decrease in FRET efficiency values relative to a control, the decrease in FRET efficiency seen following promoting phosphorylation of N17 cannot be compared with the decrease measured when the sensor has an expanded polyglutamine tract.
IKK is a critical regulator of neuroinflammation which has been implicated in the pathogenesis of HD (36). IKK inhibitors form a class of compounds in development for disease therapy (37). In terms of potential therapy for HD, the inhibition of IKK may have two complementary effects; promoting the phosphorylation of N17 and reducing mutant huntingtin induced neuroinflammation. The challenges for the future will be to develop compounds that increase or stabilize the phosphorylation of N17 and can effectively cross the blood–brain barrier.
The biology of PACSIN1 overlaps heavily with known pathways affected in Huntington disease, namely the recycling of NMDA receptors (19) and the formation of neuronal spines (21). PACSIN1 is known to directly interact with human huntingtin within the polyproline region (18), and we have additionally shown that PACSIN1 interacts with the N17 domain. This suggests that PACSIN1 may stabilize or modulate the conformation of the amino terminus of huntingtin. Both N17 and PACSIN1 are substrates for CK2 (12, 17). Therefore, the effect of the CK2 inhibitors on the conformation of the exon1 sensor may not only be due to posttranslational modification of N17 but also to that of PACSIN1. As a corollary, both the N17 and polyproline regions of huntingtin exon1 are also known to interact with actin, either directly with N17 (38) or indirectly through the actin-binding protein, profilin, with the polyproline domain (39). This may indicate the consequence of proper huntingtin conformation and conformational switching on the regulation of actin dynamics during stress (40). The role of PACSIN1 in the nucleus is not known, but the properties of Fes/CIP4 bin-amphiphysin-rvs domains (41) suggest that its role at nuclear puncta may be to either induce or recognize the structure of chromatin.
Materials and Methods
Tissue Culture.
Immortalized mouse striatal STHdhQ7/Q7 and STHdhQ111/Q111 cell lines were cultured as previously described (10). Primary human fibroblasts from either a 54-y-old unaffected female (Coriell, catalog no. GM02149) or a 51-y-old male with HD (Coriell, catalog no. GM01061) were cultured as per Coriell guidelines.
Plasmid Constructs.
Detailed plasmid construction and DNA primers are described in SI Materials and Methods.
Transfection.
Transfection of SThdh cells was done using TurboFect in vitro reagent (Fermentas, catalog no. R0531) as previously described (42). Transfection of human fibroblasts was done using the Lonza Amaxa 4D-Nucleofector X Kit L (Lonza, catalog no. V4XC-3024) according to the instructions provided.
Antibody Conjugation.
The 2B7 (Novartis), 4C9 (Novartis), and C20 (Santa Cruz) antibodies were all conjugated using Alexa Fluor 488 (invitrogen, catalog no. A20181) or 546 (Invitrogen, catalog no. A20183) monoclonal antibody labeling kit.
Immunofluorescence and Microscopy.
Methods are described in SI Materials and Methods.
SiRNA Treatment.
STHdhQ7/Q7cells were treated with three PACSIN1 siRNAs (Santa Cruz, catalog no. SC-36172) as previously described (40).
Small Molecule and Kinase Inhibitor Treatments.
All compounds were used from sources and at concentrations optimized previously (12, 14). Ganglioside GM1 from bovine brain was sourced from Merck Millipore (catalog no. 345724).
FLIM analysis and subsequent statistical analysis were completed as previously described (40, 42).
Supplementary Material
Acknowledgments
We thank Dr. Andreas Weiss (Istituto Ricerche di Biologia Moleculaire Promidis Srl) for providing us with monoclonal antibodies for use with the full-length huntingtin FLIM-FRET assay. This work is supported by Canadian Institutes of Health Research Grant MOP-119391 and a grant from the Krembil Family Foundation of Toronto with the Huntington Society of Canada.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Commentary on page 14516.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301342110/-/DCSupplemental.
References
- 1.Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72(6):971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Duyao M, et al. Trinucleotide repeat length instability and age of onset in Huntington’s disease. Nat Genet. 1993;4(4):387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- 3.Snell RG, et al. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington’s disease. Nat Genet. 1993;4(4):393–397. doi: 10.1038/ng0893-393. [DOI] [PubMed] [Google Scholar]
- 4.Stine OC, et al. Correlation between the onset age of Huntington’s disease and length of the trinucleotide repeat in IT-15. Hum Mol Genet. 1993;2(10):1547–1549. doi: 10.1093/hmg/2.10.1547. [DOI] [PubMed] [Google Scholar]
- 5.Menalled LB, Chesselet MF. Mouse models of Huntington’s disease. Trends Pharmacol Sci. 2002;23(1):32–39. doi: 10.1016/s0165-6147(00)01884-8. [DOI] [PubMed] [Google Scholar]
- 6.Johannessen M, Delghandi MP, Moens U. What turns CREB on? Cell Signal. 2004;16(11):1211–1227. doi: 10.1016/j.cellsig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Suske G. The Sp-family of transcription factors. Gene. 1999;238(2):291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 8.Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249(1–2):1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- 9.Rockabrand E, et al. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum Mol Genet. 2007;16(1):61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- 10.Atwal RS, et al. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum Mol Genet. 2007;16(21):2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- 11.Aiken CT, et al. Phosphorylation of threonine 3: Implications for Huntingtin aggregation and neurotoxicity. J Biol Chem. 2009;284(43):29,427–29,436. doi: 10.1074/jbc.M109.013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atwal RS, et al. Kinase inhibitors modulate huntingtin cell localization and toxicity. Nat Chem Biol. 2011;7(7):453–460. doi: 10.1038/nchembio.582. [DOI] [PubMed] [Google Scholar]
- 13.Thompson LM, et al. IKK phosphorylates huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187(7):1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Pardo A, et al. Ganglioside GM1 induces phosphorylation of mutant huntingtin and restores normal motor behavior in Huntington disease mice. Proc Natl Acad Sci USA. 2012;109(9):3528–3533. doi: 10.1073/pnas.1114502109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu X, et al. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64(6):828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dehay B, Bertolotti A. Critical role of the proline-rich region in huntingtin for aggregation and cytotoxicity in yeast. J Biol Chem. 2006;281(47):35,608–35,615. doi: 10.1074/jbc.M605558200. [DOI] [PubMed] [Google Scholar]
- 17.Plomann M, et al. PACSIN, a brain protein that is upregulated upon differentiation into neuronal cells. Eur J Biochem. 1998;256(1):201–211. doi: 10.1046/j.1432-1327.1998.2560201.x. [DOI] [PubMed] [Google Scholar]
- 18.Modregger J, DiProspero NA, Charles V, Tagle DA, Plomann M. PACSIN 1 interacts with huntingtin and is absent from synaptic varicosities in presymptomatic Huntington’s disease brains. Hum Mol Genet. 2002;11(21):2547–2558. doi: 10.1093/hmg/11.21.2547. [DOI] [PubMed] [Google Scholar]
- 19.Pérez-Otaño I, et al. Endocytosis and synaptic removal of NR3A-containing NMDA receptors by PACSIN1/syndapin1. Nat Neurosci. 2006;9(5):611–621. doi: 10.1038/nn1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimm-Günter EM, Milbrandt M, Merkl B, Paulsson M, Plomann M. PACSIN proteins bind tubulin and promote microtubule assembly. Exp Cell Res. 2008;314(10):1991–2003. doi: 10.1016/j.yexcr.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Schael S, et al. Casein kinase 2 phosphorylation of protein kinase C and casein kinase 2 substrate in neurons (PACSIN) 1 protein regulates neuronal spine formation. J Biol Chem. 2013;288(13):9303–9312. doi: 10.1074/jbc.M113.461293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci USA. 2006;103(29):11,045–11,050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizzo MA, Springer G, Segawa K, Zipfel WR, Piston DW. Optimization of pairings and detection conditions for measurement of FRET between cyan and yellow fluorescent proteins. Microsc Microanal. 2006;12(3):238–254. doi: 10.1017/S1431927606060235. [DOI] [PubMed] [Google Scholar]
- 24.Duncan RR, Bergmann A, Cousin MA, Apps DK, Shipston MJ. Multi-dimensional time-correlated single photon counting (TCSPC) fluorescence lifetime imaging microscopy (FLIM) to detect FRET in cells. J Microsc. 2004;215(1):1–12. doi: 10.1111/j.0022-2720.2004.01343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evers TH, van Dongen EM, Faesen AC, Meijer EW, Merkx M. Quantitative understanding of the energy transfer between fluorescent proteins connected via flexible peptide linkers. Biochemistry. 2006;45(44):13,183–13,192. doi: 10.1021/bi061288t. [DOI] [PubMed] [Google Scholar]
- 26.Weiss A, et al. Single-step detection of mutant huntingtin in animal and human tissues: A bioassay for Huntington’s disease. Anal Biochem. 2009;395(1):8–15. doi: 10.1016/j.ab.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Trottier Y, et al. Polyglutamine expansion as a pathological epitope in Huntington’s disease and four dominant cerebellar ataxias. Nature. 1995;378(6555):403–406. doi: 10.1038/378403a0. [DOI] [PubMed] [Google Scholar]
- 28.Truant R, et al. 2006. Canadian Association of Neurosciences review: Polyglutamine expansion neurodegenerative diseases. Can J Neurol Sci 33(3):278–291.
- 29.Candiani S, Pestarino M, Cattaneo E, Tartari M. Characterization, developmental expression and evolutionary features of the huntingtin gene in the amphioxus Branchiostoma floridae. BMC Dev Biol. 2007;7:127. doi: 10.1186/1471-213X-7-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim MW, Chelliah Y, Kim SW, Otwinowski Z, Bezprozvanny I. Secondary structure of huntingtin amino-terminal region. Structure. 2009;17(9):1205–1212. doi: 10.1016/j.str.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dishinger JF, et al. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat Cell Biol. 2010;12(7):703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagai Y, et al. A toxic monomeric conformer of the polyglutamine protein. Nat Struct Mol Biol. 2007;14(4):332–340. doi: 10.1038/nsmb1215. [DOI] [PubMed] [Google Scholar]
- 33.Truant R, Atwal RS, Desmond C, Munsie L, Tran T. Huntington's Disease: Revisiting the aggregation hypothesis in polyglutamine neurodegenerative diseases. FEBS. 2008;275(17):4252–4262. doi: 10.1111/j.1742-4658.2008.06561.x. [DOI] [PubMed] [Google Scholar]
- 34.Housman D. Gain of glutamines, gain of function? Nat Genet. 1995;10(1):3–4. doi: 10.1038/ng0595-3. [DOI] [PubMed] [Google Scholar]
- 35.Fuxreiter M, Tompa P. Fuzzy complexes: A more stochastic view of protein function. Adv Exp Med Biol. 2012;725:1–14. doi: 10.1007/978-1-4614-0659-4_1. [DOI] [PubMed] [Google Scholar]
- 36.Khoshnan A, Patterson PH. The role of IκB kinase complex in the neurobiology of Huntington’s disease. Neurobiol Dis. 2011;43(2):305–311. doi: 10.1016/j.nbd.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee DF, Hung MC. 2008. Advances in targeting IKK and IKK-related kinases for cancer therapy. Clin Cancer Res 14(18):5656–5662.
- 38.Angeli S, Shao J, Diamond MI. F-actin binding regions on the androgen receptor and huntingtin increase aggregation and alter aggregate characteristics. PLoS ONE. 2010;5(2):e9053. doi: 10.1371/journal.pone.0009053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burnett BG, Andrews J, Ranganathan S, Fischbeck KH, Di Prospero NA. Expression of expanded polyglutamine targets profilin for degradation and alters actin dynamics. Neurobiol Dis. 2008;30(3):365–374. doi: 10.1016/j.nbd.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munsie L, et al. Mutant huntingtin causes defective actin remodeling during stress: Defining a new role for transglutaminase 2 in neurodegenerative disease. Hum Mol Genet. 2011;20(10):1937–1951. doi: 10.1093/hmg/ddr075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halbach A, et al. PACSIN 1 forms tetramers via its N-terminal F-BAR domain. FEBS J. 2007;274(3):773–782. doi: 10.1111/j.1742-4658.2006.05622.x. [DOI] [PubMed] [Google Scholar]
- 42.Caron NS, Munsie LN, Keillor JW, Truant R. Using FLIM-FRET to measure conformational changes of transglutaminase type 2 in live cells. PLoS ONE. 2012;7(8):e44159. doi: 10.1371/journal.pone.0044159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.