Significance
Our paper describes the high-resolution structures of the complete EphA4 receptor tyrosine kinase ectodomain and its complex with ephrin-A5. These structures reveal for the first time how ephrin binding promotes Eph conformational changes allowing for the formation of ordered signaling clusters at the sites of cell–cell contact. In addition, the structural data, combined with structure-based mutagenesis and cell-based assays, reveal a previously undescribed receptor–receptor interaction between the EphA4 ligand-binding and membrane-proximal fibronectin domains, which is functionally important for efficient Eph activation. Based on the data we suggest a mechanism for Eph preclustering and recruitment of unliganded Eph into existing Eph/ephrin signaling clusters.
Keywords: crystallography, phosphorylation, protein, transmembrane
Abstract
Eph receptor tyrosine kinases and their ephrin ligands mediate cell signaling during normal and oncogenic development. Eph signaling is initiated in a multistep process leading to the assembly of higher-order Eph/ephrin clusters that set off bidirectional signaling in interacting cells. Eph and ephrins are divided in two subclasses based on their abilities to bind and activate each other and on sequence conservation. EphA4 is an exception to the general rule because it can be activated by both A- and B-class ephrin ligands. Here we present high-resolution structures of the complete EphA4 ectodomain and its complexes with ephrin-A5. The structures reveal how ligand binding promotes conformational changes in the EphA4 ligand-binding domain allowing the formation of signaling clusters at the sites of cell–cell contact. In addition, the structural data, combined with structure-based mutagenesis, reveal a previously undescribed receptor–receptor interaction between the EphA4 ligand-binding and membrane-proximal fibronectin domains, which is functionally important for efficient receptor activation.
Eph receptors, the largest family of receptor tyrosine kinases, and their ephrin ligands regulate a variety of cell–cell interactions during development and in the adult organism (1–3). Because both receptors and ligands are membrane-bound, their interactions at sites of cell–cell contact initiate unique bidirectional signaling cascades (4). The signaling downstream of the Eph is referred to as “forward,” and the signaling downstream of the ephrins is referred to as “reverse.”
The 16 Eph receptors and 9 ephrins are divided into two subclasses based on sequence homology and binding affinities. Receptor–ligand binding within each subclass is fairly promiscuous, whereas cross-subclass signaling happens rarely (5). The best-known exception to this general rule is EphA4, which has long been known to bind and be activated by both A- and B-class ligands (6, 7). Because of its unique properties, EphA4 is an attractive target for studying the structural details of subclass specificity (8).
Eph and ephrins were originally identified as axon guidance molecules; they have now been implicated in a vast array of cell communication events. Those include bone morphogenesis and homeostasis, immunological and inflammatory host responses, stem cell plasticity, learning and memory, and Alzheimer’s disease (9). However, currently, the most intensely studied function of the Eph/ephrin system is that during development and progression of different cancers. Many A- and B-class receptors were shown to be overexpressed in various tumor types (10–13) and to regulate critical steps of blood vessel formation (vasculogenesis) and remodeling (angiogenesis) and hence tumor growth.
Structural studies on the minimal binding domains of Eph receptors and ephrins revealed important details of receptor–ligand recognition (2, 14). The initial binding force comes from a penetration of a long, hydrophobic ephrin loop into a hydrophobic cavity on the surface of the receptor to form high-affinity 1:1 ligand–receptor heterodimers (15). The ligand-binding cavity of the receptor consists of loops DE, FG, and JK, which were shown to exhibit various amounts of conformational flexibility, depending on the subclass and the individual receptor. Several structures of the EphA4 ligand-binding domain (LBD) alone and in complex with its ligands have addressed its unusually high ligand promiscuity (8, 16–18).
More recently, the crystal structures of the EphA2 ectodomain (ECD) bound to ephrin-A1 and ephrin-A5 suggested a structural basis for the formation of Eph/ephrin signaling clusters at the interfaces of interacting cells (19, 20). Specifically, it was proposed that these signaling clusters nucleate from high-affinity Eph/ephrin heterodimers, which assemble into heterotetramers that further aggregate into higher-order oligomers. To understand the structural basis of EphA4 ligand-induced clustering and explore underlying receptor–receptor and receptor–ligand interactions, we determined and report here the structures of the complete EphA4 ECD, alone and in complexes with its ephrin-A5 ligand.
Results and Discussion
Structure of the EphA4 ECD.
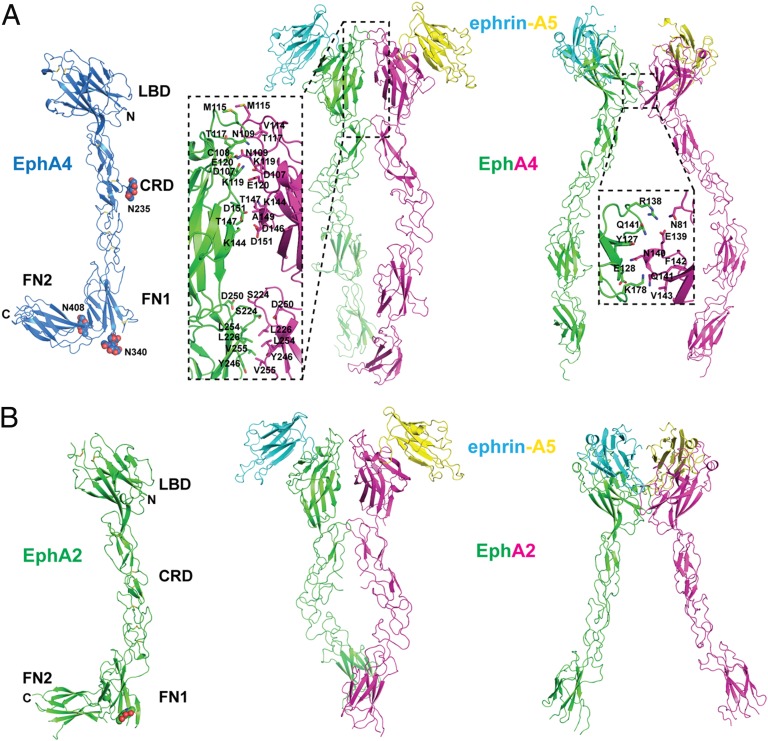
The structure of the complete EphA4 receptor ECD (residues 22–543) (Fig. 1A) was determined by molecular replacement at 2.08-Å resolution and refined to an Rfree of 22.5% (Table S1). It reveals an extended arrangement of tightly packed domains spanning 150 × 50 × 55 Å. The overall structure of unbound EphA4 is similar to that of EphA2 (19) as illustrated in Fig. 1 A and B, and the two ECDs can be superimposed with an rmsd between equivalent Ca positions of ∼2.7 Å. The Eph ECD contains an N-terminal LBD (rmsd between the EphA2 and EphA4 structures for this region is 0.7 Å), followed by a cysteine-rich domain (CRD) (rmsd between the EphA2 and EphA4 CRD is 0.8 Å), two fibronectin type III repeats (FN1 and FN2) (rmsd between the EphA2 and EphA4 is 1.0 Å for FN1 and 0.8 Å for FN2). The Eph CRD, encompassing ∼114 amino acids (residues 201–314) immediately C-terminal to the LBD, can be subdivided into two domains: an N-terminal domain similar to complement control module/short complement regulator domain and a TNF receptor-like cysteine-rich module (19, 20). The LBD, CRD, and FN1 fold into a rigid, rod-like structure with little interdomain flexibility. The connection between FN1 and FN2, on the other hand, is flexible, and these domains are positioned differently in the ligand-bound and unbound structures (Fig. 1A). EphA4 is glycosylated on positions N235, N340, and N408 as predicted, and the carbohydrate was built in the model.
Fig. 1.
Structures of the EphA4-ECD and the EphA4-ECD/ephrin-A5 complexes and comparison with EphA2. (A) (Left) Structure of the unbound EphA4-ECD (blue). The glycosylation moiety at N408 is drawn as spheres. (Center) Structure of the EphA4-ECD (green and magenta) in complex with ephrin-A5 (cyan and yellow). Two EphA4 and two ephrin-A5 molecules are shown, connected via the clustering interface. (Right) Two EphA4 and two ephrin-A5 molecules from the same crystal connected via the heterotetramerization interface. Magnified views of the clustering and heterotetramerization interfaces are shown in the rectangular areas defined by the dashed lines, with the contacting residues drawn in stick and labeled. (B) (Left) Structure of unbound EphA2-ECD (green) with the individual domains indicated and the glycosylation moiety drawn (19). (Center) Structure of the EphA2-ECD (green and magenta) bound to ephrin-A5 (cyan and yellow). Two EphA2 and two ephrin molecules are shown, connected via the clustering interface. (Right) Two EphA2 and two ephrin molecules from the same crystal connected via the heterotetramerization interface. CRD, cysteine-rich domain; FN1, fibronectin 1; FN2, fibronectin 2; LBD, ligand-binding domain.
Structure of EphA4/Ephrin-A5 Complex.
The structure of the EphA4 ECD (residues 22–543) bond to ephrin-A5 (resides 24–196) (Fig. 1A) was determined by molecular replacement at 3.15-Å resolution and refined to an Rfree of 28.5% (Table S1).
High-affinity Eph/ephrin heterodimer.
The structure of the EphA4-ECD/ephrin-A5 complex confirms the model that functional Eph/ephrin signaling clusters assemble from high-affinity Eph/ephrin heterodimers, which aggregate into heterotetramers and higher-order oligomers (2) (Fig. S1). Indeed, the EphA4/ephrin-A5 heterodimer is overall quite similar to the EphA2/ephrin-A1 and EphA2/ephrin-A5 structures (rmsd values of 2.5Å and 4.3Å, respectively) (19, 20). As in all characterized Eph/ephrin complexes, the high-affinity ligand/receptor interface (total buried area of ∼2,360 Å2) centers around the GH loop of ephrin-A5, which is inserted in a channel on the surface of EphA4. This binding is dominated by van der Waals contacts between two largely hydrophobic surfaces. Adjacent to the channel/GH loop interactions, a second, structurally separate, polar contact area encompasses the ephrin-docking site along the upper surface of the receptor. The specific interaction residues in the high-affinity “heterodimerization interface” (24 EphA4-LBD residues and 22 ephrin-A5 residues) are listed in Fig. S2A.
EphA4 clustering.
Upon ephrin binding, Eph/ephin clusters form between interacting cells via extensive Eph/Eph, Eph/ephin, and ephrin/ephrin interactions. The crystal structure of the EphA4/ephrin-A5 complex visualizes these interactions. Specifically, there are two distinct EphA4/ephrin-A5 heterotetrameric formations generated via two distinct EphA4/EphA4 interfaces that, when combined, generate a continuous EphA4/ephrin-A5 assembly (Fig. S1).
The first of these heterotetramers (Fig. 1B, Right) is generated by Eph–Eph interactions that encompass only residues within the LBD. This interface is simply referred to as “heterotetramerization interface” (15, 19). It is relatively small, burying a total of ∼620 Å2, and contains van der Waals contacts, hydrogen bonds, and salt bridges between six Eph residues in each interacting partner as illustrated in Fig. 1A, Right, Inset, and Fig. S2B.
The second EphA4/ephrin-A5 heterotetrameric assembly (Fig. 1B, Center) is generated via Eph–Eph interactions involving the LBD and the CRD. It is referred to as “clustering interface” (19). The LBD region of the clustering interface, which involves the GH loop, beta strand J, and the JK loop, is mostly polar and includes hydrogen bonds and three salt bridges as illustrated in Fig. 1A, Center, Inset, and Fig. S2C. The CRD region of the clustering interface involves a leucine zipper-like assembly with a large number of van der Waals interactions. The hydrophobic zipper is formed by residues L226, Y246, L254, and V255. Overall, the clustering interface buries 1,780 Å2 (Fig. S2C).
The EphA4/ephrin-A5 heterotetramerization and clustering interfaces are analogous to the interfaces observed in the EphA2/ephrin-A5 structures (19), which are shown, for comparison, in Fig. 1B, Right and Center, respectively). There are differences between the heterotetramerization and clustering interfaces observed in the EphA4/ephrin-A5 and the EphA2/ephrin-A5 structures, most notably that the heterotetramerization interface is much smaller in the EphA4/ephrin-A5 structure (∼620 Å2 vs. ∼2,000 Å2 in the EphA2/ephrin-A5 structure) and that it only involves Eph–Eph interaction. Nevertheless, as can be seen in Fig. 1, the overall heterotetramerization and clustering arrangements are similar, and they lead to analogous architectures of the continuous EphA4/ephrin-A5 and EphA2/ephrin-A5 assemblies (Fig. S1).
Eph activation as an ordered multistep process.
The structures reported here, combined with the previously reported EphA2/ephrin-A5 and EphA2/ephrin-A1 structures (19, 20), support a “seeding” mechanism for the assembly of Eph/ephrin clusters: First, upon cell–cell contact, high-affinity 1:1 Eph/ephrin heterodimers are formed. Then, the ligand-bound Eph receptors use two separate interfaces for the assembly of signaling-competent clusters (see Fig. 3). The heterotetramerization interface is responsible for receptor dimerization and formation of 2:2 ligand–receptor heterotetramers (15). Once Eph receptors are in ligand-induced close contact, they can independently use the clustering interface to bind other receptor dimers (or 2:2 Eph/ephrin complexes), thus forming “trimers of receptor dimers.” This process would then go on until large-sized assemblies, consisting of hundreds of receptors, are formed. Fig. S1 shows the packing of the EphA4/ephrin-A5 complexes in the crystals, illustrating a continuous 2D EphA4/ephrin-A5 assembly. The precise size or composition of these assemblies in vivo, however, is still unknown.
Fig. 3.
LBD–FNIII interactions in the unbound EphA4. (A) Two EphA4 molecules (orange and blue) interacting in the crystals of ligand-free EphA4-ECD. The main Eph–Eph interface, which presumably mediates EphA4 preclustering, involves the LBD of one EphA4 molecule interacting with the membrane-proximal FNIII domain (FN2) of a neighboring EphA4 molecule. A second, smaller Eph–Eph interface involves residues in the Eph CRD. The two interface regions are magnified and shown in the rectangular area defined by the dashed line, with the contacting residues shown in stick and labeled. The glycosylation moiety is shown as spheres. (B) (Left) EphA4-LBD (magenta) bound to ephrin-A5 (yellow) in the crystals of the EphA4-ECD/ephrin-A5 complex. (Center) EphA4-LBD (blue) bound to the EphA4-FN2 of a neighboring molecule (orange) in the crystals of unliganded EphA4-ECD. (Right) EphA2-LBD (green) bound to the EphA2-FN2 of a neighboring molecule (red) in the crystals of unliganded EphA2-ECD (19).
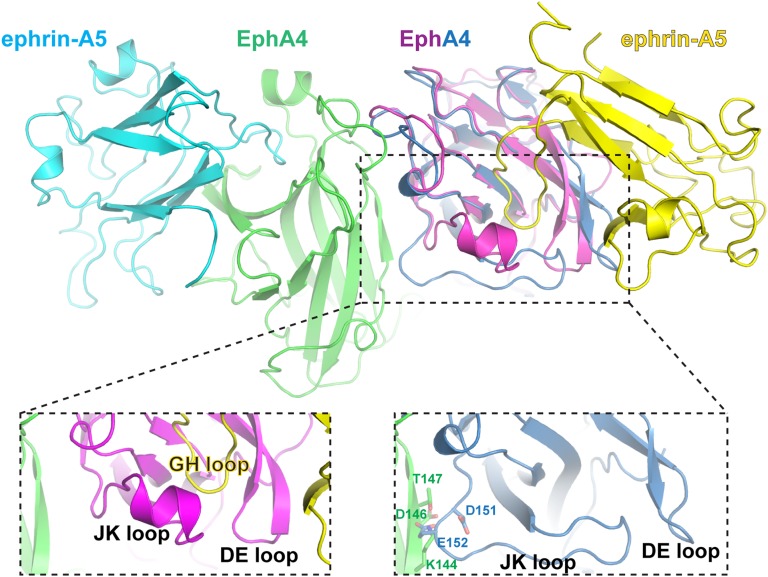
Ephrin Binding Induces Conformational Changes in the EphA4 LBD, Allowing for Formation of Eph/Ephrin Signaling Clusters.
Based on previous analysis of EphA2-ECD/ephrin structures (19), it has been suggested that the role of the ligand is to increase the local concentration of the receptors on cell surface above the critical value for efficient clustering. The EphA4/ephrin-A5 structure now reveals that ephrin-A5 binding also directly facilitates the formation of Eph/ephrin clusters by inducing conformational changes in the LBD of EphA4. Specifically, as illustrated in Fig. 2, upon ephrin-A5 binding, the EphA4 JK and DE loops undergo major conformational rearrangements: both the Eph DE beta hairpin and the JK loop bend toward the incoming ephrin GH loop to form an intimate interaction interface which leads to large structural changes in this Eph region, including the formation of a new alpha helix in the JK loop. These rearrangements result in the formation of complementary interaction surfaces in adjacent Eph receptors, facilitating the Eph/Eph interactions at the clustering interface (Fig. 2, Right Inset). Unbound EphA2 would not be able to form a similar clustering interface because of steric overlap of the JK loop (residues D151 and E152) in one receptor with the beta strand J (residues K144, D146, and T147) of its neighbor. As mentioned above, both of these regions are integral parts of the clustering interface in the ligand/receptor complex. The same ligand-induced conformational change in the EphA4 LBD was also observed in the studies reporting the structures of the EphA4-LBD and the EphA4-LBD/ephrin-A2 and ephrin-B2 complexes (8, 16, 18), but because the isolated Eph LBD does not cluster, its significance was not appreciated.
Fig. 2.
Ligand-induced conformational changes in the EphA4 LBD facilitate receptor clustering. The LBDs of two Ephs (green and magenta) bound to two ephrins (yellow and cyan) in the EphA4-ECD/ephrin-A5 complex are shown in a view centered on the LBD region of the heterotetramerization interface (as in Fig. 1A, Center, top of the rectangular Inset). Superimposed on the magenta ephrin-bound EphA4 LBDs is the structure of the LBD of the unbound EphA4 (blue). The superimposed area is magnified and shown in the rectangular area defined by the dashed line. Upon ligand engagement, the JK loop of EphA4 changes conformation (undergoes coil-to-helix transition), and both the JK and DE loops move toward the ephrin-A5 GH loop, frapping the entrance of the pocket for the GH loop. These rearrangements create the complementary interacting surfaces forming the Eph–Eph heterotetramerization interface (Left Inset). Ligand-free EphA4 (Right Inset) cannot participate in similar Eph–Eph heterotetramerization interactions because of a steric clash mediated by the unbound conformation of the EphA4 JK loop. The clashing residues are shown in stick (Right Inset).
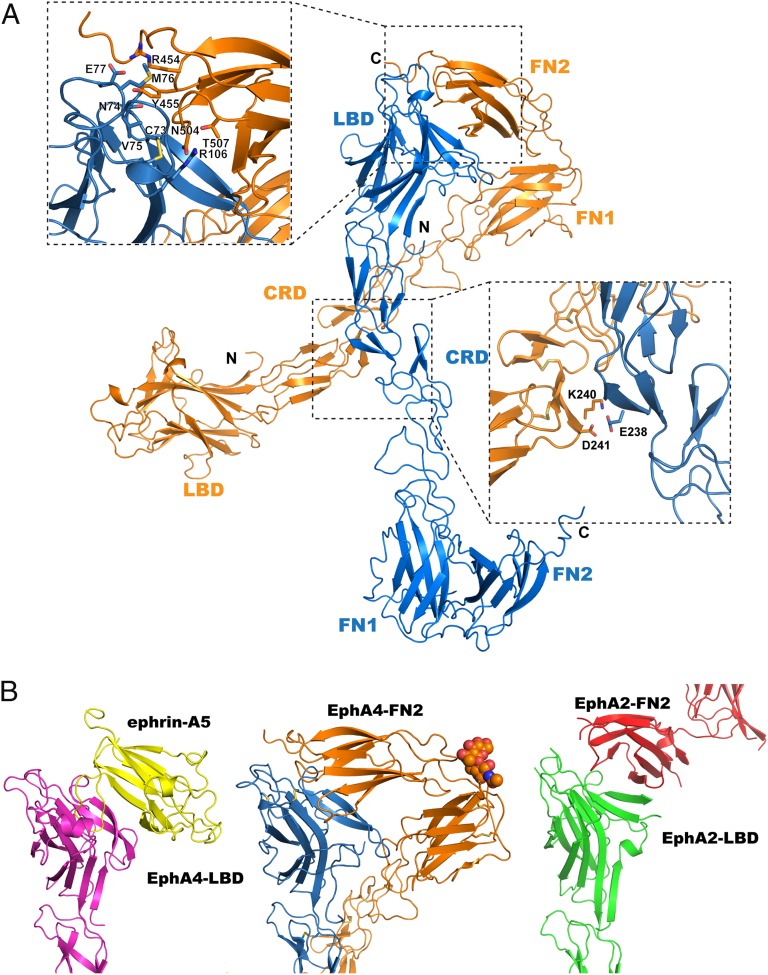
LBD-FNIII Interactions in the Unbound EphA4.
The herein reported structure of the unbound EphA4 ECD reveals a previously undescribed interaction between the EphA4 LBD and the FNIII region of a neighboring EphA4 molecule (Fig. 3). The primary binding is between two surface loops of the N-terminal EphA4 FNIII repeat (FN2) and the CD, DE, EF, GH, and JK loops and D, E, G, and M strands of the EphA4 LBD. It is noteworthy that this LBD surface partially overlaps with the surface that binds the ephrin ligands, and Fig. 3B illustrates the similarities between the EphA4-LBD/ephrin-A5 and EphA4-LBD/EphA4-FN2 binding. A second adjacent Eph–Eph contact area involves CRD residues forming a salt bridge (Fig. 3A, Lower Inset). The total binding interface is large (2,460 Å2) and includes multiple van der Waals contacts, hydrogen bonds, and salt bridges (all interacting residues are listed in Fig. S2D), indicating that it is biologically relevant. Moreover, an Eph LBD-FN2 interaction is also present in the structure of the unbound EphA2 ECD (19), but the interface, as illustrated in Fig. 3B, is much smaller (980 Å2 total buried area), and it was not discussed in the corresponding publication.
We used structure-based mutagenesis, in combination with in vitro binding assays and a cell-based EphA4 activation assay, to evaluate the significance of this Eph LBD-FNIII interface. Specifically, we generated mutations in FN2 (R454A/Y455A, or M2) and the CRD (E238A, or M3) designed to weaken the Eph/Eph interface by abolishing hydrogen bonds and salt bridges as well as a mutation (N504D/T507D, or M1) designed to strengthen the Eph/Eph interface by introducing two additional salt bridges. Mutations of these surface-exposed EphA4 residues would not affect ligand binding because these regions are neither part of the Eph/ephrin heterodimerization interface nor part of the Eph/Eph heterotetramerization and clustering interfaces (see above).
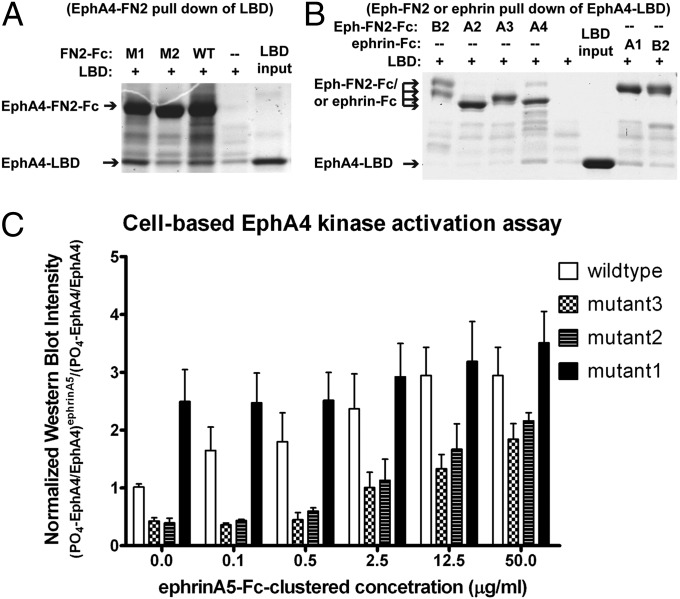
Pull-down protein-binding assays shown in Fig. 4B documented that the M2 mutation decreases LBD binding to the mutated FN2, whereas the M1 mutation increases it (M3 was not tested in this assay because the mutation is outside the LBD).
Fig. 4.
Mutations in the EphA4 preclustering interface functionally affect forward signaling. The effects of various mutations in the EphA4 preclustering interface were evaluated in pull-down and kinase activation assays. Mutants M3 and M2 (E238A and R454A/Y455A, respectively) weaken the interface, whereas mutant M1 (N504D/T507D) strengthens the interface. (A) Pull-down assay between LBD and mutant FN2 of EphA4: The Fc-tagged EphA4 FN2 domains with mutations that were designed to either enhance (M1) or weaken (M2) the preclustering interface were used to pull down the EphA4 LBD. WT EphA4 FN2 was used as positive control. The M1 FN2 binds to the LBD better than WT, whereas the M2 FN2 binding is weaker than WT. (B) EphA4 LBD binding to FN2 domains of different Ephs and to ephrins: The EphA4-LBD was tested in a pull-down assay with Fc-tagged FN2 of different Eph family members as labeled above lanes 1–4. EphA4 LBD binds only the EphA4 FN2. Fc-tagged ephrin-A1 and ephrin-B2 were used as positive control (lanes 7–8), confirming that ephrin-A1 binds EphA4 better than ephrin-B2. (C) Cell-based EphA4 kinase activation assay: HEK293 cells stably expressing WT or mutant full-length EphA4 were treated with increasing concentrations of ephrin-A5-Fc for 10 min and harvested. The activated (phosphorylated) EphA4 in the cell lysate was immunoprecipitated with an anti-phosphotyrosine antibody and was then detected with an anti-EphA4 antibody in Western blots. The Western blot films were scanned, and the signal intensity was quantified and plotted. The amount of phosphorylated EphA4 was normalized for the total EphA4 amount in the total cell lysates. All intensity readings were calculated as the ratio to the basal (ligand-independent, resting) EphA4 phosphorylation level. The experiments were performed in triplicate, and the error bars are shown in the plot.
The mutations were then introduced into full-length EphA4, and the engineered proteins were transfected into HEK293 cells. EphA4 activation was measured by quantifying the phosphorylated fraction of EphA4 following stimulation with ephrin-A5. Specifically, varying amounts of preclustered ephrin-A5-Fc (using anti-Fc mAb; Materials and Methods) was added to HEK293 cells, transfected with either WT EphA4 or the EphA4 mutants, 30 min before lysis. Anti-phosphotyrosine immunoprecipitates from lysates were analyzed by Western blotting with an anti-EphA4 mAb. The overall EphA4 expression levels were evaluated by probing the total cell lysates with anti-EphA4. The phosphorylation levels (corrected for EphA4 expression level) for WT and mutant EphA4 are shown in Fig. 4 as a function of the concentration of ephrin-A5. The results document that the FN2–LBD interaction is functionally relevant, regulating the level of EphA4 activation following ephrin stimulation. Mutations that weaken the unliganded Eph–Eph interactions decrease EphA4 activation, whereas mutations that strengthen the FN2–LBD interface increase EphA4 activation. Indeed, the M1 Eph mutant exhibits greatly elevated basal phosphorylation levels even in the absence of ligand stimulation. This suggests that the FN2–LBD interaction mediates ligand-independent Eph association or “preclustering,” thus presumably facilitating fast and efficient receptor activation upon contact with ligand-expressing cells.
Similar LBD–FN2 interactions have not been reported for B-class ephrins, and the potential LBD–FN2 interaction as observed in the EphA2 structure is significantly weaker than the one in EphA4. Thus, this strong, unliganded Eph–Eph interaction might be unique to EphA4 and might be responsible, at least in part, for the ligand promiscuity of the receptor—EphA4 is more preclustered than other Ephs and easier to activate.
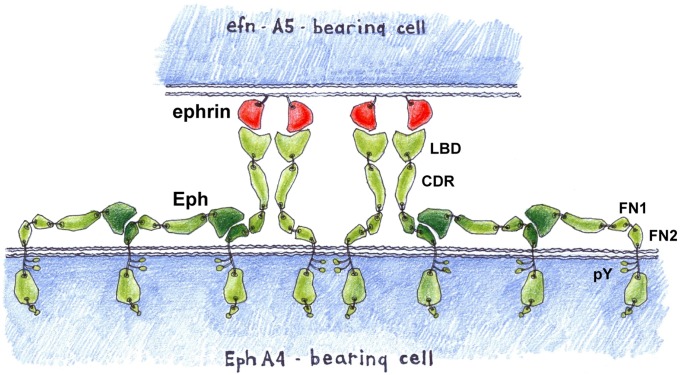
Eph signaling clusters often span cell surface areas larger than the areas of immediate Eph/ephrin contact (21, 22). In addition, it has been shown that Ephs that are not ligand-bound can sometimes be recruited to the Eph/ephrin clusters (21, 23, 24). The observed LBD–FNIII interactions provide a structural basis for the observed functional participation of unliganded Ephs in the Eph/ephrin signaling assemblies, and Fig. 5 presents a schematic of two interacting cells where homotypic Eph LBD–FNIII interactions increase the size of the signaling site on the Eph-expressing cell by recruiting unliganded Ephs to the Eph/ephrin complexes.
Fig. 5.
The homotypic, head-to-tail Eph–Eph (LBD–FN2) interactions observed in the crystals of unbound EphA4-ECD presumably represent Eph preclustering interactions and could also explain the observation that Ephs form clusters well beyond the area of immediate ephrin contact (22). Provided is a schematic representation of two interacting cells where the Eph clusters on the Eph-expressing cells are larger than the Eph/ephrin contact area, mediated by interactions between the LBDs of ephrin-free Ephs and the FNIII regions of their neighbors. The Eph molecules are colored in green, and the ephrins are in red. Phosphorylated intracellular tyrosines are shown as small circles.
The observed head-to-tail homotypic interactions between EphA4 molecules represent yet another example of the subtle ways Eph/ephrin signaling is controlled. Such interactions might work in concert with in cis Eph/ephrin interactions that have been reported in some cell types coexpressing Eph and ephrins (25, 26) to regulate, for example, Eph receptor phosphorylation (27) and pathway selection by spinal motor axons (28).
Conclusion
The herein reported structures of the complete EphA4 ECD and its complex with ephrin-A5 reveal several important characteristics of Eph signaling: (i) They confirm the general mechanism of assembly of Eph/ephrin signaling clusters, suggested from the EphA2 ECD/ephrin structures, which uses three types of Eph/ephrin interfaces: a high-affinity Eph/ephrin heterodimerization interface and lower-affinity heterotetramerization and clustering interfaces. (ii) They provide direct visualization of how ephrin binding induces conformational changes in the Eph LBD facilitating the clustering interactions required for formation of the expansive Eph/ephrin signaling centers at the sites of cell–cell contact. (iii) They reveal a previously undescribed homotypic interaction between unliganded neighboring Eph molecules which could facilitate both Eph preclustering before ephrin binding and an increase in the size of the signaling clusters via recruitment of unliganded Ephs, thus promoting efficient and potent signaling initiation.
Materials and Methods
Cloning and Mutagenesis.
Human EphA4 ECD (residues 22–543) and human ephrin-A5 (residues 24–169) were cloned into a modified pAcGP67 baculovirus expression vector (BD Bioscience), with GP67 secretion signal and human Fc fragment as C-terminal tag. Recombinant baculovirus was generated by cotransfecting the expression plasmid along with linearized BaculoGold DNA (Pharmingen) into SF9 cells. The human EphA4 full-length protein and the mutants were cloned into a pcDNA3.1 + hygromycin vector (Invitrogen) for stable expression in a HEK293 cell line. Mutations were introduced by site-directed mutagenesis (Stratagene) and were sequenced.
Protein Expression and Crystallization.
Hi5 cells were infected with passage 3 baculoviruses (for EphA4 or ephrin-A5 expression) at a multiplicity of infection of 10. The medium containing the secreted fusion proteins was collected 72 h postinfection and loaded onto a Protein A-Sepharose affinity column. Recombinant proteins were eluted with low-pH buffer containing 150 mM NaCl and 100 mM glycine (pH 3.0). The Fc tag was cleaved by thrombin proteolysis and removed by Protein A-Sepharose. Both proteins were further purified by gel filtration chromatography. EphA4 was concentrated to 10 mg/mL in Hepes-buffered saline (HBS) and crystallized by hanging-drop vapor diffusion at room temperature against a well solution of 22% PEG 400, 0.1 M 2-(N-morpholino)ethanesulfonic acid (Mes) buffer pH 6.0, 3% DMSO. For production of the EphA4/ephrin-A5 complex, purified EphA4 and ephrin-A5 were mixed in a 1:2 molar ratio and incubated on ice for 1 h. The complex was purified on a SD200 gel filtration column (GE Healthcare) and concentrated to 10 mg/mL in HBS. The complex was crystallized by sitting-drop vapor diffusion at room temperature against a well solution of 2.0 M sodium acetate, 0.1 M sodium acetate buffer pH 4.5. For data collection, the crystals were frozen in a cryobuffer containing an additional 25% (vol/vol) glycerol. X-ray diffraction data were collected at beamline 24ID-C (Northeastern Collaborative Access Team, Advanced Photon Source) and processed with HKL2000 (29). Both structures were determined by molecular replacement with EphA2 [Protein Data Bank (PDB) ID is 3FL7] (19) and EphA4/ephrin-B2 complex (PDB ID is 2WO3) (16) as search templates, using the Phaser program (30) in the Phenix suite (31). Subsequent refinement proceeded with iterative rounds of model adjustments, using the programs Coot (32) and PhenixRefine (31). Crystallographic details and statistics are listed in Table S1.
Cell Manipulations and Transfections.
HEK293 was grown in DMEM (Invitrogen) supplemented with 10% (vol/vol) FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin. Cells were consistently transfected at 80–90% confluence in six-well plates using Lipofectamine 2000 (Invitrogen).
Cell-Based EphA4 Kinase Activation Assay.
For the EphA4 activation assays, HEK293 cells were stably transfected with full-length EphA4 or Eph mutants with alterations in the preclustering interface (M1, N504D/T507D; M2, R454A/Y455A; and M3, E238A). Clones with similar expression levels, tested by anti-EphA4 antibody in Western blots, were chosen for the assay. EphA4-expressing cells were then challenged with ephrin-A5-Fc, which was clustered using anti-human IgG antibody at 25 nM concentration. After 10 min of incubation with ligand, cells were washed with PBS and harvested. Total cell lysate was prepared by lysing cell pellets in a buffer containing 20 mM Hepes (pH 7.4), 150 mM NaCl, 1% (wt/vol) Nonidet P-40, and 1 mM EDTA. Activated receptor was immunoprecipitated with anti-phosphotyrosine antibody (Upstate Biotechnology) and Protein A-Sepharose beads, resolved on SDS/PAGE, and blotted onto PVDF. Membranes were then blotted with anti-EphA4 antibody (R&D Biotechnology). The Western blot films were scanned and quantified by spot densitometry using ImageQuantTL software (GE Healthcare Biosciences). All intensity readings were normalized for the EphA4 amount in total cell lysate and calculated as the ratio to the WT basal activity (ligand-independent phosphorylation level). The experiments were done in triplicate, and the error bars are shown in Fig. 4.
Pull-Down Experiments.
Pull-down experiments were performed essentially as described earlier (5). Briefly, 5 μg of recombinant EphA4 LBD were incubated with the Fc-tagged recombinant ephrins or with isolated Eph FN2 domains at room temperature for 30 min in 500 μL binding buffer containing 20 mM Hepes (pH 8.2), 150 mM KCl, and 2 mM MgCl2. Protein A-Sepharose Fast Flow beads (GE Healthcare) were washed with the binding buffer, added to the reaction mixture, and mixed at room temperature for 30 min. The beads were then harvested by centrifugation and washed twice with 500 μL of the binding buffer, and the bound proteins were separated on a 10–20% polyacrylamide gel.
Illustrations.
Figures were prepared using Adobe Illustrator (Adobe). All molecular representations were produced with PyMOL (33).
Supplementary Material
Acknowledgments
We thank Dr. Rüdiger Klein for protein expression constructs and advice, Momchil Kolev for technical support, Marina Himanen for hand-drawn illustrations, and K. R. Rajashankar for help with data collection. This research was supported by National Institutes of Health Grant R01NS038486 (to D.B.N.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4M4P and 4M4R).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311000110/-/DCSupplemental.
References
- 1.Flanagan JG, Vanderhaeghen P. The ephrins and Eph receptors in neural development. Annu Rev Neurosci. 1998;21:309–345. doi: 10.1146/annurev.neuro.21.1.309. [DOI] [PubMed] [Google Scholar]
- 2.Himanen JP, Saha N, Nikolov DB. Cell-cell signaling via Eph receptors and ephrins. Curr Opin Cell Biol. 2007;19(5):534–542. doi: 10.1016/j.ceb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein R. Excitatory Eph receptors and adhesive ephrin ligands. Curr Opin Cell Biol. 2001;13(2):196–203. doi: 10.1016/s0955-0674(00)00197-6. [DOI] [PubMed] [Google Scholar]
- 4.Cowan CA, Henkemeyer M. The SH2/SH3 adaptor Grb4 transduces B-ephrin reverse signals. Nature. 2001;413(6852):174–179. doi: 10.1038/35093123. [DOI] [PubMed] [Google Scholar]
- 5.Himanen JP, et al. Repelling class discrimination: Ephrin-A5 binds to and activates EphB2 receptor signaling. Nat Neurosci. 2004;7(5):501–509. doi: 10.1038/nn1237. [DOI] [PubMed] [Google Scholar]
- 6.Smith A, Robinson V, Patel K, Wilkinson DG. The EphA4 and EphB1 receptor tyrosine kinases and ephrin-B2 ligand regulate targeted migration of branchial neural crest cells. Curr Biol. 1997;7(8):561–570. doi: 10.1016/s0960-9822(06)00255-7. [DOI] [PubMed] [Google Scholar]
- 7.Egea J, et al. Regulation of EphA 4 kinase activity is required for a subset of axon guidance decisions suggesting a key role for receptor clustering in Eph function. Neuron. 2005;47(4):515–528. doi: 10.1016/j.neuron.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 8.Singla N, et al. Crystal structure of the ligand-binding domain of the promiscuous EphA4 receptor reveals two distinct conformations. Biochem Biophys Res Commun. 2010;399(4):555–559. doi: 10.1016/j.bbrc.2010.07.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasquale EB. Eph-ephrin bidirectional signaling in physiology and disease. Cell. 2008;133(1):38–52. doi: 10.1016/j.cell.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Hafner C, et al. Differential gene expression of Eph receptors and ephrins in benign human tissues and cancers. Clin Chem. 2004;50(3):490–499. doi: 10.1373/clinchem.2003.026849. [DOI] [PubMed] [Google Scholar]
- 11.Noblitt LW, Bangari DS, Shukla S, Mohammed S, Mittal SK. Immunocompetent mouse model of breast cancer for preclinical testing of EphA2-targeted therapy. Cancer Gene Ther. 2005;12(1):46–53. doi: 10.1038/sj.cgt.7700763. [DOI] [PubMed] [Google Scholar]
- 12.Robinson D, He F, Pretlow T, Kung HJ. A tyrosine kinase profile of prostate carcinoma. Proc Natl Acad Sci USA. 1996;93(12):5958–5962. doi: 10.1073/pnas.93.12.5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sjöblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 14.Himanen JP, et al. Ligand recognition by A-class Eph receptors: Crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex. EMBO Rep. 2009;10(7):722–728. doi: 10.1038/embor.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Himanen JP, et al. Crystal structure of an Eph receptor-ephrin complex. Nature. 2001;414(6866):933–938. doi: 10.1038/414933a. [DOI] [PubMed] [Google Scholar]
- 16.Bowden TA, et al. Structural plasticity of eph receptor A4 facilitates cross-class ephrin signaling. Structure. 2009;17(10):1386–1397. doi: 10.1016/j.str.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin H, Shi J, Noberini R, Pasquale EB, Song J. Crystal structure and NMR binding reveal that two small molecule antagonists target the high affinity ephrin-binding channel of the EphA4 receptor. J Biol Chem. 2008;283(43):29473–29484. doi: 10.1074/jbc.M804114200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin H, et al. Structural characterization of the EphA4-Ephrin-B2 complex reveals new features enabling Eph-ephrin binding promiscuity. J Biol Chem. 2010;285(1):644–654. doi: 10.1074/jbc.M109.064824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Himanen JP, et al. Architecture of Eph receptor clusters. Proc Natl Acad Sci USA. 2010;107(24):10860–10865. doi: 10.1073/pnas.1004148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seiradake E, Harlos K, Sutton G, Aricescu AR, Jones EY. An extracellular steric seeding mechanism for Eph-ephrin signaling platform assembly. Nat Struct Mol Biol. 2010;17(4):398–402. doi: 10.1038/nsmb.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wimmer-Kleikamp SH, Janes PW, Squire A, Bastiaens PI, Lackmann M. Recruitment of Eph receptors into signaling clusters does not require ephrin contact. J Cell Biol. 2004;164(5):661–666. doi: 10.1083/jcb.200312001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janes PW, Nievergall E, Lackmann M. Concepts and consequences of Eph receptor clustering. Semin Cell Dev Biol. 2012;23(1):43–50. doi: 10.1016/j.semcdb.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Janes PW, et al. Eph receptor function is modulated by heterooligomerization of A and B type Eph receptors. J Cell Biol. 2011;195(6):1033–1045. doi: 10.1083/jcb.201104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noren NK, Yang NY, Silldorff M, Mutyala R, Pasquale EB. Ephrin-independent regulation of cell substrate adhesion by the EphB4 receptor. Biochem J. 2009;422(3):433–442. doi: 10.1042/BJ20090014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marquardt T, et al. Coexpressed EphA receptors and ephrin-A ligands mediate opposing actions on growth cone navigation from distinct membrane domains. Cell. 2005;121(1):127–139. doi: 10.1016/j.cell.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 26.Yin Y, et al. EphA receptor tyrosine kinases interact with co-expressed ephrin-A ligands in cis. Neurosci Res. 2004;48(3):285–296. doi: 10.1016/j.neures.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 27.Carvalho RF, et al. Silencing of EphA3 through a cis interaction with ephrinA5. Nat Neurosci. 2006;9(3):322–330. doi: 10.1038/nn1655. [DOI] [PubMed] [Google Scholar]
- 28.Kao TJ, Kania A. Ephrin-mediated cis-attenuation of Eph receptor signaling is essential for spinal motor axon guidance. Neuron. 2011;71(1):76–91. doi: 10.1016/j.neuron.2011.05.031. [DOI] [PubMed] [Google Scholar]
- 29. Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. [DOI] [PubMed]
- 30.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Emsley P, Cowtan K (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1):2126–2132. [DOI] [PubMed]
- 33.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific LLC; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.