Abstract
Synergistic microbial communities are ubiquitous in nature and exhibit appealing features, such as sophisticated metabolic capabilities and robustness. This has inspired fast-growing interest in engineering synthetic microbial consortia for biotechnology development. However, there are relatively few reports of their use in real-world applications, and achieving population stability and regulation has proven to be challenging. In this work, we bridge ecology theory with engineering principles to develop robust synthetic fungal-bacterial consortia for efficient biosynthesis of valuable products from lignocellulosic feedstocks. The required biological functions are divided between two specialists: the fungus Trichoderma reesei, which secretes cellulase enzymes to hydrolyze lignocellulosic biomass into soluble saccharides, and the bacterium Escherichia coli, which metabolizes soluble saccharides into desired products. We developed and experimentally validated a comprehensive mathematical model for T. reesei/E. coli consortia, providing insights on key determinants of the system’s performance. To illustrate the bioprocessing potential of this consortium, we demonstrate direct conversion of microcrystalline cellulose and pretreated corn stover to isobutanol. Without costly nutrient supplementation, we achieved titers up to 1.88 g/L and yields up to 62% of theoretical maximum. In addition, we show that cooperator–cheater dynamics within T. reesei/E. coli consortia lead to stable population equilibria and provide a mechanism for tuning composition. Although we offer isobutanol production as a proof-of-concept application, our modular system could be readily adapted for production of many other valuable biochemicals.
Keywords: lignocellulosic biofuel, consolidated bioprocessing, renewable energy
Biosynthesis of fuels and commodity chemicals from lignocellulosic biomass represents a promising and sustainable alternative to present petroleum feedstock platforms. However, high processing costs and inefficient biocatalysts remain key barriers to commercialization (1). Microbial conversion of lignocellulosic biomass requires multiple biological functionalities, including production of saccharifying enzymes (cellulases and hemicellulases), enzymatic hydrolysis of lignocellulose to soluble saccharides, and metabolism of soluble saccharides to desired products. Economic analyses indicate that substantial cost reductions can be achieved by integrating all biologically mediated transformations into a single step, an approach termed consolidated bioprocessing (CBP) (1). The typical CBP strategy entails engineering microbes that incorporate all required functionalities into a single strain (1). However, engineering and optimizing multiple heterologous functionalities in a single microbial strain have proven inherently challenging. Despite intensive research efforts spanning decades, there have been few reports of achieving commercially viable product yields and titers with CBP (1). Additionally, the most notable CBP successes have been in fuel ethanol production (2), whereas yields and titers remain low for desirable next-generation biofuels, such as higher molecular-weight alcohols and hydrocarbons.
In contrast to the CBP “superbug” paradigm, microbes live in synergistic communities in most natural environments in which individual species with specialized roles cooperate to survive and thrive together (3). Natural microbial consortia hold many appealing properties in a bioprocessing context, such as stability, functional robustness, and the ability to perform complex tasks (3, 4). Inspired by the powerful features of natural consortia, there is rapidly growing interest in engineering synthetic consortia for biotechnology applications (3, 4). Examples include cocultures of genetically modified Escherichia coli for cofermentation of hexose and pentose sugars (5–7) or direct conversion of lignocellulosic biomass to advanced biofuels (8), a synthetic Saccharomyces cerevisiae consortium for assembly of extracellular minicellulosomes and direct production of cellulosic ethanol (9), improving CBP with the cellulolytic ethanologen Clostridium phytofermentans via coculture with S. cerevisiae (10), and a coculture of Actinotalea fermentans and genetically engineered S. cerevisiae for converting lignocellulose to methyl halides (11). These examples illustrate a broader trend of using synthetic consortia to compartmentalize pathways into different hosts for individual optimization and/or demonstrate that through division of labor, synthetic consortia can accomplish complex tasks that are difficult to achieve with monocultures. Another broad approach explores the “bottom-up” strategy of programming specific interactions between microbes using synthetic genetic circuits and intercellular communication (3). Such approaches have been used to construct canonical ecological and logic systems for proof-of-concept and fundamental study, but they have been less used for biotechnology applications (3). As a notable exception, Prindle et al. (12) recently developed a microbial arsenic detection system by engineering an oscillatory circuit synchronized across an entire cell population in which the oscillatory period is modulated as a function of arsenic concentration.
In sharp contrast to their natural counterparts, synthetic microbial consortia are often fragile and unstable, limiting their use in real-world applications like industrial bioprocessing. In mixed cultures created by arbitrarily combining different species, population compositions are often unstable and prone to domination by a single species or extinction (13), whereas consortia featuring programmed interactions via synthetic genetic circuits are predisposed to mutational aberration (14). In this work, we apply ecology theory to the design and construction of robust synthetic fungi/bacteria consortia for flexible biosynthesis of valuable products from lignocellulosic feedstocks. The required biological functions are divided between two specialists: a fungal cellulolytic specialist, which secretes cellulase enzymes to hydrolyze lignocellulosic biomass into soluble saccharides, and a fermentation specialist, which metabolizes soluble saccharides into desired products. We developed a comprehensive mathematical model for cellulolytic fungi/bacteria consortia that semimechanistically captures salient features and allows us to elucidate key behaviors and ecological interactions. In parallel, we experimentally implemented the consortium with the cellulolytic fungus Trichoderma reesei and an E. coli strain metabolically engineered to produce isobutanol, a promising next-generation biofuel. We demonstrate direct conversion of microcrystalline cellulose (MCC) and ammonia fiber expansion (AFEX) pretreated corn stover (CS) to isobutanol with the consortium, reaching titers up to 1.88 g/L and yields up to 62% of the theoretical maximum. Although we offer isobutanol production as a proof of concept, our modular design could be readily adapted to the large portfolio of existing metabolically engineered microbial strains to produce a wide variety of valuable biofuels and chemicals.
Results
Design and Theoretical Analysis of a Synthetic T. reesei/E. coli Consortium.
As a proof of concept, we designed a synthetic microbial consortium for direct conversion of lignocellulosic biomass into isobutanol, a promising next-generation biofuel with superior properties (15) (Fig. 1A). Isobutanol can be produced via decarboxylation and subsequent reduction of 2-ketoisovalerate, an endogenous valine biosynthesis intermediate (15). The highest reported isobutanol titers and yields have been achieved using engineered E. coli strains under microaerobic conditions (15, 16); we selected one of these strains, E. coli NV3 pSA55/69, as the isobutanol production specialist (16). To produce isobutanol, the consortium must hydrolyze lignocellulose into soluble saccharides that can be metabolized by E. coli. Natural saprophytic organisms have evolved sophisticated synergistic enzyme systems for efficient hydrolysis of cellulose, which is recalcitrant to degradation (17). We selected the aerobic filamentous fungus T. reesei RUTC30 as the cellulolytic specialist, because it is a prodigious cellulase producer (17), physiologically compatible with E. coli (i.e., environmental conditions, media composition), and not antagonistic toward bacteria (18).
Fig. 1.

Design and theoretical analysis of the TrEc consortium. (A) Schematic of the TrEc consortium. Key parameters, identified via sensitivity analysis (in B), are labeled (SI Appendix, section 1 and Table S1). T. reesei produces cellulases (CBHI, cellobiohydrolase I; CBHII, cellobiohydrolase II; EGI, endoglucanase I) that hydrolyze cellulose to soluble oligosaccharides. Oligosaccharides are further hydrolyzed to glucose via cell wall-localized β-glucosidase (BGL). Soluble saccharides serve as growth substrates for the microbes (cellobiose and glucose for T. reesei, glucose only for E. coli). E. coli ferments glucose into isobutanol, which inhibits microbial growth due to toxicity. (B) Global sensitivity analysis of the TrEc consortium model. PRCCs between model parameters and output metrics are shown with hierarchical clustering (Ward’s method, Pearson correlation distance). Parameters are labeled in A. Output metrics are as follows: 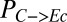 , fraction of substrate carbon consumed by E. coli (grams per total grams); QI, isobutanol productivity (grams per gram of cellulose per hour); Rcel, mean cellulose hydrolysis rate (grams per liter per hour); REc, mean E. coli growth rate (grams per liter per hour); RTr, mean T. reesei growth rate (grams per liter per hour); XEc, E. coli population fraction at fermentation end point (grams per gram of total microbial biomass); and
, fraction of substrate carbon consumed by E. coli (grams per total grams); QI, isobutanol productivity (grams per gram of cellulose per hour); Rcel, mean cellulose hydrolysis rate (grams per liter per hour); REc, mean E. coli growth rate (grams per liter per hour); RTr, mean T. reesei growth rate (grams per liter per hour); XEc, E. coli population fraction at fermentation end point (grams per gram of total microbial biomass); and  , isobutanol yield (grams per gram of cellulose). The most significant PRCCs (
, isobutanol yield (grams per gram of cellulose). The most significant PRCCs ( and
and 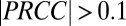 ) are shown here; full results are provided in SI Appendix, Fig. S2. (C) Normalized kernel density estimate (200 × 200 grid, standard bivariate normal distribution kernel) for Rcel vs.
) are shown here; full results are provided in SI Appendix, Fig. S2. (C) Normalized kernel density estimate (200 × 200 grid, standard bivariate normal distribution kernel) for Rcel vs. 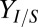 over all sets of parameter and IC values sampled in sensitivity analysis. Individual points are shown in low-density (<1.25) regions. Axes are padded by 4% at each end to ensure visibility of all data. (D) Theoretical analysis of isobutanol production. Parameter values and ICs correspond to the point denoted by the white asterisk in C, with Fa, fraction of substrate bonds accessible to enzymes, modified to 0.011. Numerical solutions were calculated over a range of initial E. coli fraction,
over all sets of parameter and IC values sampled in sensitivity analysis. Individual points are shown in low-density (<1.25) regions. Axes are padded by 4% at each end to ensure visibility of all data. (D) Theoretical analysis of isobutanol production. Parameter values and ICs correspond to the point denoted by the white asterisk in C, with Fa, fraction of substrate bonds accessible to enzymes, modified to 0.011. Numerical solutions were calculated over a range of initial E. coli fraction, 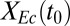 , values. Key fermentation metrics are shown (Rcel,
, values. Key fermentation metrics are shown (Rcel, 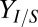 , and QI); the green dashed line denotes the theoretical maximum of
, and QI); the green dashed line denotes the theoretical maximum of  (0.41 g/g). More details are provided in SI Appendix, section 2.
(0.41 g/g). More details are provided in SI Appendix, section 2.
To gain insights into the behavior and ecology of the T. reesei/E. coli (TrEc) consortium, we developed a comprehensive ordinary differential equation (ODE) modeling framework that captures salient features of the system. We derived rate expressions for each of the steps depicted in Fig. 1A (details are provided in SI Appendix, section 1). Parameter values were obtained mainly from the literature, with certain values experimentally measured in-house (SI Appendix, Tables S1 and S3). An important subtlety in the TrEc consortium is that soluble oligosaccharides are hydrolyzed to glucose via cell wall-localized β-glucosidases of T. reesei (19) (Fig. 1A). This leads to a higher glucose concentration at the T. reesei cell surface compared with the bulk medium, which we estimate using a mass transfer analysis (Fig. 1A). Our model contains 50 parameters and includes variables for the concentration of microbial biomass (vegetative and senescent T. reesei mycelium and E. coli), enzymes, isobutanol, soluble oligosaccharides (one to four glucose monomers in size), and each possible cellulose polysaccharide from degree of polymerization (DP) = 5 up to the maximum DP of the substrate. We performed a global sensitivity analysis to dissect this functionally complex model and identify key parameters controlling consortium behavior (Fig. 1B and SI Appendix, section 2). The ODEs were numerically integrated with 1,000 sets of parameter values and initial conditions (ICs) sampled from appropriate statistical distributions (SI Appendix, Table S1), with Latin hypercube (20) selection. For each parameter or IC, partial rank correlation coefficients (PRCCs) (20) were calculated with a set of output metrics. The most significant parameters and ICs (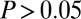 and
and 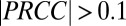 ) are shown in Fig. 1B, with the same parameters/ICs labeled in Fig. 1A (full results are shown in SI Appendix, Fig. S2).
) are shown in Fig. 1B, with the same parameters/ICs labeled in Fig. 1A (full results are shown in SI Appendix, Fig. S2).
Hierarchical clustering reveals that substrate partition between E. coli and T. reesei (quantified as the fraction of substrate carbon consumed by E. coli, 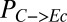 , grams per total gram) is largely related to initial population composition [quantified as initial E. coli fraction,
, grams per total gram) is largely related to initial population composition [quantified as initial E. coli fraction, 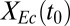 , grams per total gram of microbial biomass] and parameters/ICs associated with growth/substrate uptake kinetics (clusters 1 and 3 in Fig. 1B). Parameters/ICs controlling
, grams per total gram of microbial biomass] and parameters/ICs associated with growth/substrate uptake kinetics (clusters 1 and 3 in Fig. 1B). Parameters/ICs controlling 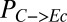 also correlate to final population composition
also correlate to final population composition  and isobutanol production (yield,
and isobutanol production (yield, 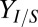 , grams per gram of cellulose; productivity, QI, grams per gram of cellulose per hour), as expected.
, grams per gram of cellulose; productivity, QI, grams per gram of cellulose per hour), as expected. 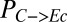 ,
,  , and QI are most strongly correlated with
, and QI are most strongly correlated with  , suggesting that this IC is a key determinant of consortium performance (Fig. 1B; topmost row). The parameters/ICs in cluster 2 primarily control overall rates of enzymatic hydrolysis (and subsequent microbial growth) but have less influence on
, suggesting that this IC is a key determinant of consortium performance (Fig. 1B; topmost row). The parameters/ICs in cluster 2 primarily control overall rates of enzymatic hydrolysis (and subsequent microbial growth) but have less influence on 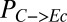 (Fig. 1B); these terms include the fraction of substrate bonds accessible to enzymes, Fa, as well as initial cellulose and microbe concentrations and parameters related to endoglucanase activity. In addition to parameters/ICs, we clustered output metrics. The similarity between cellulose hydrolysis rate (Rcel, grams per liter per hour) and T. reesei growth rate (RTr, grams per liter per hour) reflects the dependence of cellulose hydrolysis on cellulase production by T. reesei, whereas the other cluster contains output metrics related to E. coli.
(Fig. 1B); these terms include the fraction of substrate bonds accessible to enzymes, Fa, as well as initial cellulose and microbe concentrations and parameters related to endoglucanase activity. In addition to parameters/ICs, we clustered output metrics. The similarity between cellulose hydrolysis rate (Rcel, grams per liter per hour) and T. reesei growth rate (RTr, grams per liter per hour) reflects the dependence of cellulose hydrolysis on cellulase production by T. reesei, whereas the other cluster contains output metrics related to E. coli.
Overall, these results suggest that competition between T. reesei and E. coli for soluble saccharides (via relative population size and growth/uptake kinetics) is a key ecological interaction that drives system behavior. The competitive nature of the TrEc consortium implies an inherent tradeoff between cellulose hydrolysis rate and isobutanol yield. For example, increasing  should increase
should increase  ; however, growth rate and subsequent cellulase synthesis by T. reesei would decrease, leading to lower Rcel. Examining Rcel and
; however, growth rate and subsequent cellulase synthesis by T. reesei would decrease, leading to lower Rcel. Examining Rcel and 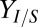 over all sensitivity analysis samplings reveals the feasible region of these two competing processes over the global parameter/IC space (Fig. 1C). The boundaries (corresponding to parameter/IC extrema) represent a production envelope for yield and hydrolysis rate, clearly showing a tradeoff between the two (with
over all sensitivity analysis samplings reveals the feasible region of these two competing processes over the global parameter/IC space (Fig. 1C). The boundaries (corresponding to parameter/IC extrema) represent a production envelope for yield and hydrolysis rate, clearly showing a tradeoff between the two (with  as
as  g/g, the theoretical maximum) (Fig. 1C).
g/g, the theoretical maximum) (Fig. 1C).
We further investigated the interplay between ecological interactions and isobutanol production by examining consortium performance over a range of 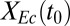 values (Fig. 1D), using an optimistic parameter set (corresponding to the white asterisk in Fig. 1C). As per the expected rate/yield tradeoff,
values (Fig. 1D), using an optimistic parameter set (corresponding to the white asterisk in Fig. 1C). As per the expected rate/yield tradeoff,  increases with
increases with 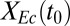 , whereas Rcel decreases. QI is balanced by rate and yield, with a maximum at
, whereas Rcel decreases. QI is balanced by rate and yield, with a maximum at 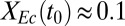 . Interestingly,
. Interestingly, 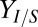 and Rcel are predicted to behave asymptotically as
and Rcel are predicted to behave asymptotically as 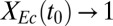 , approaching values of ∼0.34 g/g (84% theoretical) and ∼0.21 g⋅L−1⋅h−1, respectively. We note that this behavior is consistent with the PRCCs for
, approaching values of ∼0.34 g/g (84% theoretical) and ∼0.21 g⋅L−1⋅h−1, respectively. We note that this behavior is consistent with the PRCCs for  (Fig. 1B), because rank transformation linearizes all monotonic relationships (an example is shown in SI Appendix, Fig. S3). The predicted asymptotic effect may be due, in part, to saccharide uptake kinetics (SI Appendix, section 1); increased glucose concentration at the T. reesei cell surface permits higher uptake by T. reesei compared with E. coli at very low glucose concentrations, thus asymptotically limiting substrate flow to E. coli.
(Fig. 1B), because rank transformation linearizes all monotonic relationships (an example is shown in SI Appendix, Fig. S3). The predicted asymptotic effect may be due, in part, to saccharide uptake kinetics (SI Appendix, section 1); increased glucose concentration at the T. reesei cell surface permits higher uptake by T. reesei compared with E. coli at very low glucose concentrations, thus asymptotically limiting substrate flow to E. coli.
Experimental Validation and Investigation of TrEc Consortia.
To study consortium ecology and validate our theoretical framework, we experimentally characterized a monoculture of T. reesei RUTC30 and bicultures of T. reesei RUTC30 with E. coli K12 (reference strain) or E. coli NV3 pSA55/69 (isobutanol production strain) (16), using MCC as a model substrate (Fig. 2 A–C). We used regression to estimate parameter values and fit the model to experimental data, focusing on a subset of key parameters with initial values from the literature or in-house monoculture experiments (SI Appendix, section 2 and Tables S1 and S3). After regression, model predictions for cellulose, microbial biomass, and isobutanol concentrations show reasonable agreement with experimental data, affirming our framework. In addition to microbial biomass and cellulose, we measured enzyme and soluble saccharide concentrations in the RUTC30/K12 biculture (SI Appendix, section 1 and Fig. S4 A and B). Although experimentally observed enzyme activities agree with model predictions, measured glucose concentrations are higher than predicted during the nongrowth phase of the culture (SI Appendix, Fig. S4 A and B).
Fig. 2.

Experimental and model analysis of T. reesei (Tr) RUTC30 monoculture and bicultures of T. reesei RUTC30 with E. coli (Ec) K12 or E. coli NV3 pSA55/69. Error bars are ±SD for n = 3 technical replicates. (A) T. reesei RUTC30 monoculture on 20 g/L MCC. Modeling results are shown as smooth lines, and experimental results are shown as points. (B) RUTC30/K12 biculture 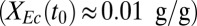 on 10 g/L MCC. (C) RUTC30/NV3 biculture
on 10 g/L MCC. (C) RUTC30/NV3 biculture 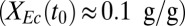 on 20 g/L MCC. E. coli data points are color-coded to indicate population fraction retaining plasmids pSA55/69 (error bars are shown in SI Appendix, Fig. S4C). (D) Fermentation product titers for NV3 monoculture (20 g/L glucose) vs. biculture experiment in C. Error bars are ±SD for n = 2 biological replicates. Acet, acetate; EtOH, ethanol; iButOH, isobutanol; Succ, succinate. (E) Local sensitivity analysis of the RUTC30/NV3 parameter set. The model was integrated with one-at-a-time ±25% perturbations to each parameter/IC (SI Appendix, section 2). Parameter effects were quantified by response coefficients, defined as
on 20 g/L MCC. E. coli data points are color-coded to indicate population fraction retaining plasmids pSA55/69 (error bars are shown in SI Appendix, Fig. S4C). (D) Fermentation product titers for NV3 monoculture (20 g/L glucose) vs. biculture experiment in C. Error bars are ±SD for n = 2 biological replicates. Acet, acetate; EtOH, ethanol; iButOH, isobutanol; Succ, succinate. (E) Local sensitivity analysis of the RUTC30/NV3 parameter set. The model was integrated with one-at-a-time ±25% perturbations to each parameter/IC (SI Appendix, section 2). Parameter effects were quantified by response coefficients, defined as 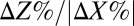 , where
, where 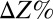 is the percentage change in output Z and
is the percentage change in output Z and  is the percentage change in parameter X. Response coefficients for
is the percentage change in parameter X. Response coefficients for 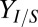 and Rcel are shown for top 10 parameters (ranked by
and Rcel are shown for top 10 parameters (ranked by 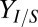 ), with the color code indicating the direction of the parameter/IC perturbation for the plotted response.
), with the color code indicating the direction of the parameter/IC perturbation for the plotted response.
T. reesei growth and cellulose hydrolysis rates are slower and fermentation times are longer in RUTC30/K12 and RUTC30/NV3 bicultures (Fig. 2 B and C) compared with RUTC30 monoculture (Fig. 2A), supporting theoretically predicted tradeoffs between cellulose hydrolysis rate and substrate flow to E. coli (Fig. 1 C and D). Several key differences were noted for RUTC30/NV3 compared with RUTC30/K12 bicultures, including much lower E. coli concentration, earlier growth cessation, and higher death rates for both organisms (Fig. 2 B and C). Our regression analysis suggests that E. coli NV3 pSA55/69 has much higher glucose-biomass yield (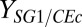 ; grams of glucose per gram of cells) and maintenance (
; grams of glucose per gram of cells) and maintenance (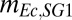 ; grams of glucose per gram of cells per hour) coefficients compared with K12 (SI Appendix, Table S3), thus leading to lower cell concentrations. Early growth cessation of RUTC30 is likely due to toxicity of NV3 fermentation products (i.e., isobutanol as suggested by our model) and perhaps O2 limitation, whereas NV3 growth is predicted to be limited by glucose concentration. Higher death rates for RUTC30/NV3 compared with RUTC30/K12 bicultures may also be related to toxicity; additionally, E. coli NV3 contains a defective rpoS allele (16), which is expected to decrease stationary phase survival.
; grams of glucose per gram of cells per hour) coefficients compared with K12 (SI Appendix, Table S3), thus leading to lower cell concentrations. Early growth cessation of RUTC30 is likely due to toxicity of NV3 fermentation products (i.e., isobutanol as suggested by our model) and perhaps O2 limitation, whereas NV3 growth is predicted to be limited by glucose concentration. Higher death rates for RUTC30/NV3 compared with RUTC30/K12 bicultures may also be related to toxicity; additionally, E. coli NV3 contains a defective rpoS allele (16), which is expected to decrease stationary phase survival.
The RUTC30/NV3 biculture reached an isobutanol titer of 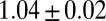 g/L, with
g/L, with 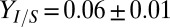 g/g of cellulose hydrolyzed (16 ± 3% theoretical) and
g/g of cellulose hydrolyzed (16 ± 3% theoretical) and  g⋅L−1⋅h−1, thus demonstrating that the TrEc consortium is capable of directly converting cellulose to isobutanol. To establish a performance reference, we characterized an NV3 monoculture under the same growth condition as the biculture but with glucose as the substrate (Fig. 2D). Compared with the RUTC30/NV3 biculture, the NV3 monoculture produced isobutanol with greater selectivity and reached a higher titer (3.5 ± 0.01 g/L; Fig. 2D). Reduced isobutanol selectivity of NV3 under the biculture condition may be due, in part, to loss of isobutanol production plasmids (Fig. 2C). Although isobutanol production by the RUTC30/NV3 biculture is of the same order of magnitude as the NV3 monoculture, our previous theoretical analysis (Fig. 1 C and D) suggests that performance could be substantially improved. Because
g⋅L−1⋅h−1, thus demonstrating that the TrEc consortium is capable of directly converting cellulose to isobutanol. To establish a performance reference, we characterized an NV3 monoculture under the same growth condition as the biculture but with glucose as the substrate (Fig. 2D). Compared with the RUTC30/NV3 biculture, the NV3 monoculture produced isobutanol with greater selectivity and reached a higher titer (3.5 ± 0.01 g/L; Fig. 2D). Reduced isobutanol selectivity of NV3 under the biculture condition may be due, in part, to loss of isobutanol production plasmids (Fig. 2C). Although isobutanol production by the RUTC30/NV3 biculture is of the same order of magnitude as the NV3 monoculture, our previous theoretical analysis (Fig. 1 C and D) suggests that performance could be substantially improved. Because 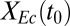 is predicted to be a key determinant of isobutanol production, we conducted biculture studies over a series of NV3/RUTC30 (grams per gram) inoculation ratios (SI Appendix, Fig. S4 E and F). However, increases in
is predicted to be a key determinant of isobutanol production, we conducted biculture studies over a series of NV3/RUTC30 (grams per gram) inoculation ratios (SI Appendix, Fig. S4 E and F). However, increases in  with inoculation ratio were marginal, suggesting that
with inoculation ratio were marginal, suggesting that 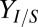 may already be saturated with respect to
may already be saturated with respect to 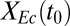 (i.e., as in Fig. 1D).
(i.e., as in Fig. 1D).
To identify parameters/ICs that could be adjusted to improve performance, we conducted a local sensitivity analysis on the RUTC30/NV3 parameter/IC set (Fig. 2E). Sensitivity analysis predicts that increasing maintenance-associated isobutanol yield (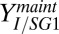 ; grams per gram of glucose consumed by E. coli) or decreasing T. reesei maximum specific growth rate (
; grams per gram of glucose consumed by E. coli) or decreasing T. reesei maximum specific growth rate (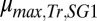 ; reciprocal hours) will produce the largest proportional gains in isobutanol production (Fig. 2E). As expected, parameter/IC changes that increase
; reciprocal hours) will produce the largest proportional gains in isobutanol production (Fig. 2E). As expected, parameter/IC changes that increase 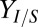 generally decrease Rcel; however, decreasing
generally decrease Rcel; however, decreasing 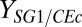 or increasing
or increasing  is predicted to improve
is predicted to improve 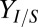 with neutral or positive effects on Rcel (Fig. 2E). These results provide valuable guidance to our future efforts for further improving performance of the TrEc consortium.
with neutral or positive effects on Rcel (Fig. 2E). These results provide valuable guidance to our future efforts for further improving performance of the TrEc consortium.
Isobutanol Production from Corn Stover.
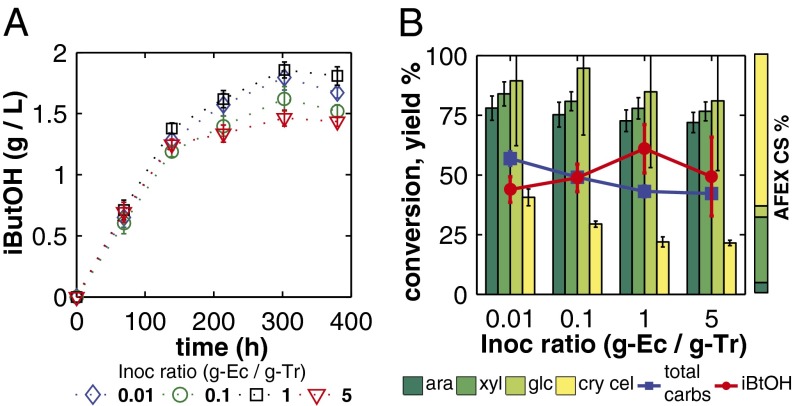
To demonstrate isobutanol production on real lignocellulosic biomass, we conducted RUTC30/NV3 bicultures over a wide range of NV3/RUTC30 inoculation ratios on AFEX (21) pretreated CS. AFEX pretreated CS carbohydrates include both hemicellulose (mixed pentose/hexose polysaccharides) and crystalline cellulose; we characterized conversion (percentage of carbohydrate consumed) of each major carbohydrate, along with titers and overall 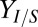 (Fig. 3 A and B). Hemicellulose conversion was high and relatively uniform, whereas crystalline cellulose conversion was lower and tended to decline with inoculation ratio. Isobutanol titer was relatively constant, whereas
(Fig. 3 A and B). Hemicellulose conversion was high and relatively uniform, whereas crystalline cellulose conversion was lower and tended to decline with inoculation ratio. Isobutanol titer was relatively constant, whereas  tended to increase with inoculation ratio until reaching a maximum at an NV3/RUTC30 ratio of 1. Overall, the biculture inoculated at this ratio produced the highest isobutanol titer and yield, at 1.88 g/L and 62% theoretical, respectively. Interestingly, fermentation product distributions differed between MCC and AFEX pretreated CS, with higher relative proportions of isobutanol and succinate in AFEX pretreated CS bicultures compared with MCC (SI Appendix, Fig. S5 C and D).
tended to increase with inoculation ratio until reaching a maximum at an NV3/RUTC30 ratio of 1. Overall, the biculture inoculated at this ratio produced the highest isobutanol titer and yield, at 1.88 g/L and 62% theoretical, respectively. Interestingly, fermentation product distributions differed between MCC and AFEX pretreated CS, with higher relative proportions of isobutanol and succinate in AFEX pretreated CS bicultures compared with MCC (SI Appendix, Fig. S5 C and D).
Fig. 3.
Isobutanol production with RUTC30/NV3 bicultures on 20 g/L AFEX pretreated CS (21). Error bars are ±SD for n = 2 biological replicates. (A) Isobutanol concentration over time. (B) Carbohydrate conversion (percentage consumed) and isobutanol yield. ara, arabinose; cry cel, crystalline cellulose; glc, hemicellulose-derived glucose; Inoc, inoculation; xyl, xylose. For reference, relative proportions of major AFEX pretreated CS carbohydrates are shown to the right; more details are provided in SI Appendix, Table S7.
Exploiting Cooperator–Cheater Dynamics for Stabilizing and Tuning TrEc Consortia.
Microbial consortia with stable and tunable population compositions are highly desirable for bioprocessing applications, because these properties could expand possible process configurations and improve efficiency. For instance, stability permits the use of continuous reactors or repeated batch fermentation, whereas tunability would allow population composition to be optimized for desired performance (e.g., maintain economically optimal balance between rate and yield in TrEc consortia). Ecology theory provides a framework to predict the outcomes and stability of interactions in microbial consortia, which we can exploit to design novel mechanisms for stabilizing and tuning TrEc consortia.
In a game theory context, T. reesei acts as a cooperator and E. coli as a cheater. Cellulase secretion by T. reesei is cooperative because it is metabolically expensive and T. reesei does not have exclusive access to hydrolysis products. E. coli behaves as a cheater by using hydrolysis products without bearing the burden of cellulase production. Game theory suggests that cooperators and cheaters can stably coexist provided that cooperators receive high enough net benefits during their interactions with cheaters, a defining feature of the so-called “snowdrift game” (22). Experimental studies with S. cerevisiae sucrose metabolism demonstrate that cooperator–cheater coexistence is possible, given that fitness benefits are concave and cooperators have privileged access to the products of cooperation (22). In TrEc consortia, these criteria are satisfied by concave growth kinetics (i.e., Monod kinetics) and increased saccharide concentration at the T. reesei cell surface due to localized hydrolysis, respectively. Because equilibrium population composition has been shown to depend on relative cooperation/cheating benefits (22), it may be possible to modulate related parameters to tune composition.
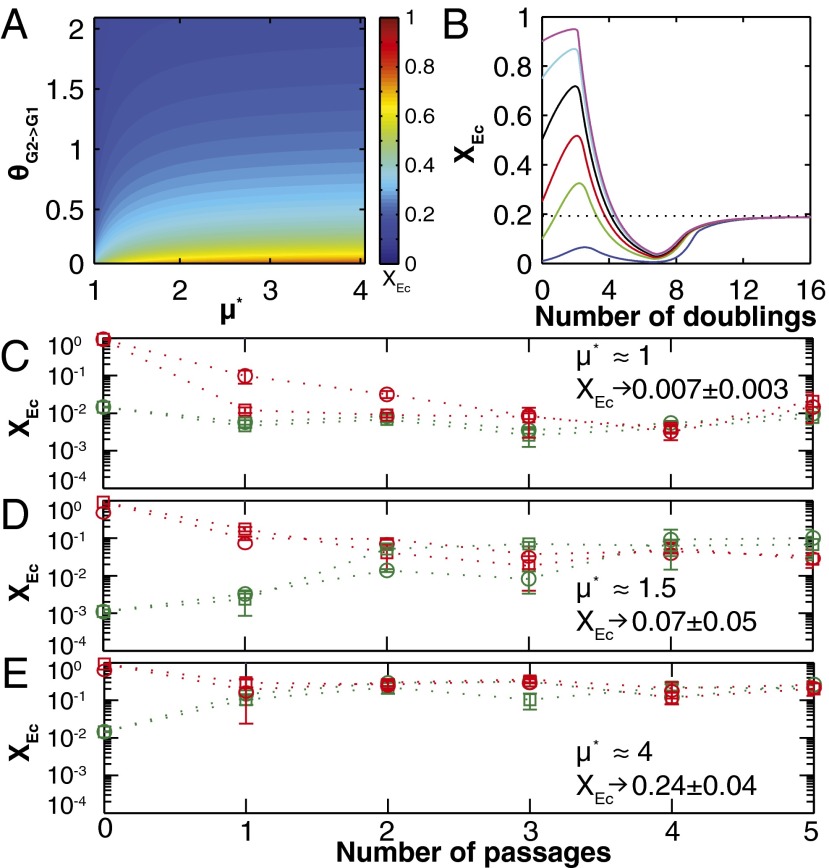
To explore the cooperator–cheater coexistence/tuning concept, we performed a stability analysis on a simplified version of the TrEc consortium model over a design space of potentially adjustable parameters: E. coli cheating benefits, quantified by  (the ratio of the maximum growth rate of E. coli to that of T. reesei), and T. reesei privileged access, quantified by
(the ratio of the maximum growth rate of E. coli to that of T. reesei), and T. reesei privileged access, quantified by 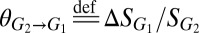 (increase in glucose concentration relative to the bulk medium at the T. reesei cell surface due to cell wall-localized cellobiose hydrolysis; SI Appendix, sections 1 and 2). Our analysis suggests that a wide range of equilibrium population compositions can be achieved by tuning
(increase in glucose concentration relative to the bulk medium at the T. reesei cell surface due to cell wall-localized cellobiose hydrolysis; SI Appendix, sections 1 and 2). Our analysis suggests that a wide range of equilibrium population compositions can be achieved by tuning  and/or
and/or 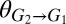 (Fig. 4A). In addition to
(Fig. 4A). In addition to 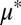 and
and 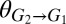 , equilibrium composition is a function of other model parameters, which could also be used for tuning (SI Appendix, section 2, Fig. S6, and Table S4). Carbon flow partition is proportional to population composition, underscoring that population tuning could be used to modulate biosynthesis of desired products (SI Appendix, Fig. S7). In dynamic simulations over the
, equilibrium composition is a function of other model parameters, which could also be used for tuning (SI Appendix, section 2, Fig. S6, and Table S4). Carbon flow partition is proportional to population composition, underscoring that population tuning could be used to modulate biosynthesis of desired products (SI Appendix, Fig. S7). In dynamic simulations over the 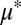 and
and  space, equilibria were reached within 12–60 doubling times regardless of ICs, suggesting that equilibrium states are stable and readily accessible (examples are shown in Fig. 4B and SI Appendix, Fig. S8 A and B). Population trajectories tended to undergo damped oscillations as they converged to equilibrium (possibly due to feedback delay between cellulose hydrolysis, saccharide concentrations, and microbial growth), with equilibration time increasing as
space, equilibria were reached within 12–60 doubling times regardless of ICs, suggesting that equilibrium states are stable and readily accessible (examples are shown in Fig. 4B and SI Appendix, Fig. S8 A and B). Population trajectories tended to undergo damped oscillations as they converged to equilibrium (possibly due to feedback delay between cellulose hydrolysis, saccharide concentrations, and microbial growth), with equilibration time increasing as 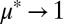 (SI Appendix, Fig. S8 A and B).
(SI Appendix, Fig. S8 A and B).
Fig. 4.
Cooperator–cheater dynamics in TrEc consortia. (A) Stability analysis of the TrEc model. Equilibrium population compositions 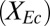 were calculated over a range of plausible values for
were calculated over a range of plausible values for 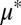 and
and 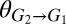 . (B) Sample numerical solutions for
. (B) Sample numerical solutions for 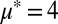 and
and  over a range of
over a range of 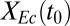 . (C) Experimentally observed dynamics for RUTC30/K12 bicultures at pH 5.3
. (C) Experimentally observed dynamics for RUTC30/K12 bicultures at pH 5.3 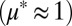 . Cultures were inoculated with high or low
. Cultures were inoculated with high or low  (red or green points/lines, respectively) in duplicate (circles/squares). Data are shown for each culture, and error bars are ±SD for n = 2 technical replicates. (D) RUTC30/NV3 bicultures at pH 6.0
(red or green points/lines, respectively) in duplicate (circles/squares). Data are shown for each culture, and error bars are ±SD for n = 2 technical replicates. (D) RUTC30/NV3 bicultures at pH 6.0 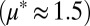 . (E) RUTC30/K12 bicultures at pH 6.0
. (E) RUTC30/K12 bicultures at pH 6.0  .
.
Isobutanol production and toxicity were neglected in this analysis because isobutanol accumulation prevents our model from reaching nontrivial steady states. We extended our simplified model to include isobutanol production/toxicity and performed a numerical stability analysis by simulating serial cultures (details are provided in SI Appendix, section 2). Although simulations converge to stable population equilibria, toxicity effects lead to steady oscillations (driven by fluctuating isobutanol concentrations in the serial culture scheme) and higher equilibrium E. coli population fraction relative to the model without toxicity effects (due to higher isobutanol tolerance of E. coli compared with T. reesei) (sample trajectories are provided in SI Appendix, Fig. S8 C and D).
We experimentally investigated population dynamics and equilibria in TrEc consortia by conducting serial culturing studies. Ten grams per liter of cellobiose (a soluble glucose disaccharide) was used for growth substrate instead of cellulose; this substitution preserves requisite features for cooperator–cheater coexistence (concave fitness benefits and cooperator privileged access) while simplifying measurement of microbial composition. To explore cooperator–cheater tuning, we modulated 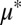 by using strains of E. coli with different intrinsic
by using strains of E. coli with different intrinsic  values (K12 and NV3;
values (K12 and NV3; 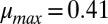 1/h and
1/h and 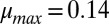 1/h, respectively; SI Appendix, Table S3) or by varying culture pH (T. reesei prefers more acidic pH compared with E. coli;
1/h, respectively; SI Appendix, Table S3) or by varying culture pH (T. reesei prefers more acidic pH compared with E. coli;  vs. pH shown in SI Appendix, Table S6). Experimental results for TrEc bicultures at different
vs. pH shown in SI Appendix, Table S6). Experimental results for TrEc bicultures at different  values are shown in Fig. 4 C–E. As predicted, bicultures inoculated at different initial XEc values converge to stable equilibria (Fig. 4 C–E). Furthermore, steady-state XEc increases with
values are shown in Fig. 4 C–E. As predicted, bicultures inoculated at different initial XEc values converge to stable equilibria (Fig. 4 C–E). Furthermore, steady-state XEc increases with  , whereas equilibration time decreases (Fig. 4 C–E), consistent with theoretical predictions (Fig. 4A and SI Appendix, Fig. S8 A and B). Results with RUTC30/NV3 bicultures (Fig. 4D) demonstrate that cooperator–cheater dynamics are preserved with isobutanol production/toxicity effects, although plasmid maintenance and subsequent isobutanol production were unstable and varied with inoculation fraction (SI Appendix, Fig. S8 E and F). Additionally, our experimental methods do not have sufficient resolution to determine whether the predicted oscillations occur with isobutanol production/toxicity. Overall, these experimental results support theoretical predictions of cooperator–cheater dynamics within TrEc consortia and illustrate the feasibility of tuning population composition via control of relative cooperation/cheating benefits.
, whereas equilibration time decreases (Fig. 4 C–E), consistent with theoretical predictions (Fig. 4A and SI Appendix, Fig. S8 A and B). Results with RUTC30/NV3 bicultures (Fig. 4D) demonstrate that cooperator–cheater dynamics are preserved with isobutanol production/toxicity effects, although plasmid maintenance and subsequent isobutanol production were unstable and varied with inoculation fraction (SI Appendix, Fig. S8 E and F). Additionally, our experimental methods do not have sufficient resolution to determine whether the predicted oscillations occur with isobutanol production/toxicity. Overall, these experimental results support theoretical predictions of cooperator–cheater dynamics within TrEc consortia and illustrate the feasibility of tuning population composition via control of relative cooperation/cheating benefits.
Discussion
Synthetic microbial consortia offer a plethora of potential biotechnology applications. However, our limited understanding of the dynamics and interactions of microbial populations hinders their use in real-world applications, and achieving population stability and regulation has proven challenging. In this work, we developed TrEc bicultures as model systems for stable, tunable, and modular consortia for CBP of lignocellulosic biomass to valuable products. Although the general division of labor we used (saccharolytic and fermentation specialists) has been considered by others (10, 23), our investigation has yielded unique insights into ecological interactions and properties that arise from this design motif. In addition to fundamental interest, these insights suggest strategies to control consortium performance via manipulation of ecological parameters. For instance, we show that cooperator–cheater dynamics within TrEc consortia lead to stable coexistence between T. reesei and E. coli, and that equilibrium population composition can be tuned by modulating relative cooperation/cheating benefits and possibly other ecological parameters. More broadly, the cooperator–cheater concept could serve as a general tool for designing and regulating consortia, and it represents a complementary approach to previous efforts to stabilize/tune consortia via mutualistic interactions (4, 10, 24). Cooperator–cheater tuning could be accomplished via manipulation of culture conditions or genetically programmed cellular behavior, as we have illustrated above.
Our results also reveal that population composition and growth/substrate uptake kinetics control carbon flow partition between T. reesei and E. coli, which, in turn, determines the tradeoff between cellulose hydrolysis rate and product yields. Theoretical analysis suggests that rate/yield tradeoff is continuous over global parameter/IC space, with  as
as 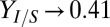 g/g, the theoretical maximum (Fig. 1C). However, for a fixed parameter set (i.e., a specific consortium), only limited ranges of rate/yield may be accessible (e.g., Figs. 1D and 3B). We identified parameters that could be adjusted to optimize and expand possible rates/yields. For instance, product yield/titer can potentially be improved by increasing the competitiveness of E. coli relative to T. reesei for glucose (e.g., decreasing
g/g, the theoretical maximum (Fig. 1C). However, for a fixed parameter set (i.e., a specific consortium), only limited ranges of rate/yield may be accessible (e.g., Figs. 1D and 3B). We identified parameters that could be adjusted to optimize and expand possible rates/yields. For instance, product yield/titer can potentially be improved by increasing the competitiveness of E. coli relative to T. reesei for glucose (e.g., decreasing 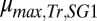 or
or 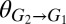 ), whereas volumetric productivity can be improved by simultaneously increasing the growth rates of T. reesei and E. coli.
), whereas volumetric productivity can be improved by simultaneously increasing the growth rates of T. reesei and E. coli.
As a proof-of-concept CBP application, we demonstrated direct conversion of MCC and AFEX pretreated CS to isobutanol, reaching titers up to 1.88 g/L and yields up to 62% of theoretical maximum (Fig. 3 A and B). To the best of our knowledge, these are the highest titers/yields reported to date for CBP production of advanced biofuels, and they were achieved without detailed optimization or nutrient supplementation beyond minimal salts. However, further analysis of this system is needed to evaluate economic feasibility and to determine performance targets for titer, yield, and productivity, especially because aerobic bioprocessing incurs higher costs (due to agitation/aeration) compared with more preferable anaerobic operations.
Our investigation suggests that reduced isobutanol production by E. coli NV3 pSA55/66 under biculture conditions is a key limitation. NV3 monocultures can reach isobutanol titers of 13.6 g/L on rich glucose media (16). However, on minimal media (more representative for commercial production), titers are only  g/L (Fig. 2D). Even controlling for media effects, isobutanol yield is ∼50% lower for NV3 in biculture vs. monoculture (0.123 and 0.25 g/g of glucose consumed, respectively; Fig. 2D). Biculture yield was determined via model regression to experimental data, with an upper bound estimate of
g/L (Fig. 2D). Even controlling for media effects, isobutanol yield is ∼50% lower for NV3 in biculture vs. monoculture (0.123 and 0.25 g/g of glucose consumed, respectively; Fig. 2D). Biculture yield was determined via model regression to experimental data, with an upper bound estimate of 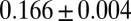 g/g (via mass balance of fermentation products; details are provided in SI Appendix, section 3 and Table S8). Sensitivity analysis suggests that NV3 isobutanol yield (i.e.,
g/g (via mass balance of fermentation products; details are provided in SI Appendix, section 3 and Table S8). Sensitivity analysis suggests that NV3 isobutanol yield (i.e., 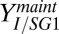 ) is the most limiting factor for isobutanol production (Fig. 2E). Reduced isobutanol yield in biculture may be due to plasmid loss (SI Appendix, Fig. S4C); however, metabolic changes caused by unidentified interactions between E. coli and T. reesei or very low growth rates in the biculture environment are also potential factors. Additional study of E. coli metabolism under biculture conditions and chromosomal integration of plasmid genes may be beneficial for improving yield. Further development of TrEc bicultures for isobutanol production will require other improvements, including investigating/improving isobutanol production from pentoses and improving tolerance of T. reesei/E. coli to isobutanol and pretreatment inhibitors. Finally, to be useful for process design, our modeling framework should be expanded to include hemicellulose, and further refinements are needed to improve predictions for enzyme and soluble saccharide concentrations.
) is the most limiting factor for isobutanol production (Fig. 2E). Reduced isobutanol yield in biculture may be due to plasmid loss (SI Appendix, Fig. S4C); however, metabolic changes caused by unidentified interactions between E. coli and T. reesei or very low growth rates in the biculture environment are also potential factors. Additional study of E. coli metabolism under biculture conditions and chromosomal integration of plasmid genes may be beneficial for improving yield. Further development of TrEc bicultures for isobutanol production will require other improvements, including investigating/improving isobutanol production from pentoses and improving tolerance of T. reesei/E. coli to isobutanol and pretreatment inhibitors. Finally, to be useful for process design, our modeling framework should be expanded to include hemicellulose, and further refinements are needed to improve predictions for enzyme and soluble saccharide concentrations.
Although we offer isobutanol production as a proof-of-concept application, our modular system could be readily adapted to the large existing portfolio of E. coli strains metabolically engineered to produce fuels and other valuable biochemicals. The TrEc consortium is an aerobic system; however, this constraint is not overly prohibitive because E. coli can be engineered to produce anaerobic metabolites under aerobic conditions (25). In a broader context, the TrEc consortium represents a versatile CBP platform that could be deployed with other microbial cellulolytic and fermentation specialists under various conditions. Because many cellulolytic microbes have cell wall-localized (e.g., complexed) cellulase systems (17), cooperator–cheater dynamics could be preserved; additionally, using cellulolytic species with different levels of privileged access to hydrolysis products represents a possible means of tuning. Although many challenges remain, this study provides tools and a useful foundation for understanding and engineering stable, tunable, and modular consortia for CBP.
Materials and Methods
Model and Simulations.
Detailed descriptions of model derivation, parameter values, numerical solutions, regression, sensitivity analysis, and stability analysis are included in SI Appendix, sections 1 and 2.
Strains and Media.
T. reesei RUTC30 was used as the cellulolytic specialist. E. coli K12 MG1655 was used as a reference strain for the fermentation specialist. Isobutanol production experiments were conducted with E. coli NV3 (16) with plasmids pSA55 (ColE1 ori, AmpR, PLlacO1:: kivd-ADH2) and pSA69 (p15A ori, KanR, PLlacO1::alsS-ilvCD) (15). Media and antibiotics are described in SI Appendix, section 3.
Culture Conditions and Analysis.
All cultures were conducted in Trichoderma minimal media (TMM) (pH 6) with 10- or 20-g/L carbon source (MCC, AFEX pretreated CS, or glucose). Batch cultures were conducted in a BioFlo 3000 bioreactor (New Brunswick Scientific) with a culture volume of 3 L. The bioreactor was maintained at pH 6, 30 °C, 200-rpm agitation (dual six blade Rushton impellers, 75-mm diameter), and 6.0-L/min (2 volumes air per volume culture per min) airflow. E. coli NV3 pSA55/69 monocultures and RUTC30/NV3 bicultures were conducted in 2-L (400-mL culture volume) or 250-mL (50-mL culture volume) screw-cap flasks (to prevent isobutanol evaporation) at 30 °C with agitation. NV3 cultures were supplemented with 100 μg/mL ampicillin, 30 μg/mL kanamycin, and 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cultures were sampled periodically and analyzed as indicated for total dry mass, carbohydrate composition, E. coli cell counts, soluble saccharides, isobutanol, total protein, β-glucosidase activity, exoglucanase activity, and endoglucanase activity (details are provided in SI Appendix, section 3).
Cooperator–Cheater Coexistence Experiments.
Duplicate bicultures of T. reesei RUTC30 with E. coli K12 or NV3 pSA55/69 were inoculated at 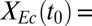 0.9, 0.01, or 0.001 (grams per gram) in 10 g/L cellobiose TMM buffered with maleate-NaOH at the indicated pH, with antibiotics/IPTG added for RUTC30/NV3 bicultures. Cultures were incubated at 30 °C with agitation until saturated and then inoculated 1:50 (by volume) into fresh media. This was repeated for five to six passages; at each passage, cultures were analyzed as indicated for total dry mass, E. coli cell counts, and isobutanol (details are provided in SI Appendix, section 3).
0.9, 0.01, or 0.001 (grams per gram) in 10 g/L cellobiose TMM buffered with maleate-NaOH at the indicated pH, with antibiotics/IPTG added for RUTC30/NV3 bicultures. Cultures were incubated at 30 °C with agitation until saturated and then inoculated 1:50 (by volume) into fresh media. This was repeated for five to six passages; at each passage, cultures were analyzed as indicated for total dry mass, E. coli cell counts, and isobutanol (details are provided in SI Appendix, section 3).
Supplementary Material
Acknowledgments
We thank Bruce Dale (Michigan State University) for providing us with AFEX pretreated biomass, Robert Ziff (University of Michigan) for proofreading our supporting information, Michael Nelson (University of Michigan) for providing assistance with HPLC, Paul Kulhman for helpful discussions about game theory, Shreyas Hirekhan for assisting with experimental work, and Edgar Meyhofer and Erdogan Gulari (University of Michigan) for generously allowing us to use their laboratory facilities. This work was supported by the National Science Foundation (Grants EEC 0926926 and CBET 1055227), the Department of Energy (DOE) Great Lakes Bioenergy Research Center (DOE Office of Science Biological and Environmental Research Grant DE-FC02-07ER64494), and the University of Michigan Office of the Vice President for Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218447110/-/DCSupplemental.
References
- 1.Olson DG, McBride JE, Shaw AJ, Lynd LR. Recent progress in consolidated bioprocessing. Curr Opin Biotechnol. 2012;23(3):396–405. doi: 10.1016/j.copbio.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 2.Argyros DA, et al. High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes. Appl Environ Microbiol. 2011;77(23):8288–8294. doi: 10.1128/AEM.00646-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mee MT, Wang HH. Engineering ecosystems and synthetic ecologies. Mol Biosyst. 2012;8(10):2470–2483. doi: 10.1039/c2mb25133g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuroff TR, Curtis WR. Developing symbiotic consortia for lignocellulosic biofuel production. Appl Microbiol Biotechnol. 2012;93(4):1423–1435. doi: 10.1007/s00253-011-3762-9. [DOI] [PubMed] [Google Scholar]
- 5.Trinh CT, Unrean P, Srienc F. Minimal Escherichia coli cell for the most efficient production of ethanol from hexoses and pentoses. Appl Environ Microbiol. 2008;74(12):3634–3643. doi: 10.1128/AEM.02708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eiteman MA, Lee SA, Altman R, Altman E. A substrate-selective co-fermentation strategy with Escherichia coli produces lactate by simultaneously consuming xylose and glucose. Biotechnol Bioeng. 2009;102(3):822–827. doi: 10.1002/bit.22103. [DOI] [PubMed] [Google Scholar]
- 7.Xia T, Eiteman MA, Altman E. Simultaneous utilization of glucose, xylose and arabinose in the presence of acetate by a consortium of Escherichia coli strains. Microb Cell Fact. 2012;11:77. doi: 10.1186/1475-2859-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bokinsky G, et al. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. Proc Natl Acad Sci USA. 2011;108(50):19949–19954. doi: 10.1073/pnas.1106958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goyal G, Tsai SL, Madan B, DaSilva NA, Chen W. Simultaneous cell growth and ethanol production from cellulose by an engineered yeast consortium displaying a functional mini-cellulosome. Microb Cell Fact. 2011;10:89. doi: 10.1186/1475-2859-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuroff T, Xiques SB, Curtis W. Consortia-mediated bioprocessing of cellulose to ethanol with a symbiotic Clostridium phytofermentans/yeast co-culture. Biotechnol Biofuels. 2013;6(1):59. doi: 10.1186/1754-6834-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bayer TS, et al. Synthesis of methyl halides from biomass using engineered microbes. J Am Chem Soc. 2009;131(18):6508–6515. doi: 10.1021/ja809461u. [DOI] [PubMed] [Google Scholar]
- 12.Prindle A, et al. A sensing array of radically coupled genetic ‘biopixels’. Nature. 2012;481(7379):39–44. doi: 10.1038/nature10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci USA. 2008;105(47):18188–18193. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arkin AP, Fletcher DA. Fast, cheap and somewhat in control. Genome Biol. 2006;7(8):114. doi: 10.1186/gb-2006-7-8-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451(7174):86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 16.Smith KM, Liao JC. An evolutionary strategy for isobutanol production strain development in Escherichia coli. Metab Eng. 2011;13(6):674–681. doi: 10.1016/j.ymben.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66(3):506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seidl V, et al. The Hypocrea jecorina (Trichoderma reesei) hypercellulolytic mutant RUT C30 lacks a 85 kb (29 gene-encoding) region of the wild-type genome. BMC Genomics. 2008;9:327. doi: 10.1186/1471-2164-9-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubicek CP. Release of carboxymethyl-cellulase and β-glucosidase from cell walls of Trichoderma reesei. Appl Microbiol Biotechnol. 1981;13(4):226–231. [Google Scholar]
- 20.Marino S, Hogue IB, Ray CJ, Kirschner DE. A methodology for performing global uncertainty and sensitivity analysis in systems biology. J Theor Biol. 2008;254(1):178–196. doi: 10.1016/j.jtbi.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teymouri F, Laureano-Pérez L, Alizadeh H, Dale BE. Ammonia fiber explosion treatment of corn stover. Appl Biochem Biotechnol. 2004;113-116:951–963. doi: 10.1385/abab:115:1-3:0951. [DOI] [PubMed] [Google Scholar]
- 22.Gore J, Youk H, van Oudenaarden A. Snowdrift game dynamics and facultative cheating in yeast. Nature. 2009;459(7244):253–256. doi: 10.1038/nature07921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hahn-Hägerdal B, Häggström M. Production of ethanol from cellulose, Solka Floc BW-200, in a fedbatch mixed culture of Trichoderma reesei, C 30, and Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 1985;22(3):187–189. [Google Scholar]
- 24.Kerner A, Park J, Williams A, Lin XN. A programmable Escherichia coli consortium via tunable symbiosis. PLoS ONE. 2012;7(3):e34032. doi: 10.1371/journal.pone.0034032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Portnoy VA, Herrgård MJ, Palsson BO. Aerobic fermentation of D-glucose by an evolved cytochrome oxidase-deficient Escherichia coli strain. Appl Environ Microbiol. 2008;74(24):7561–7569. doi: 10.1128/AEM.00880-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.