Abstract
In man, mutations in different regions of the prion protein (PrP) are associated with infectious neurodegenerative diseases that have remarkably different clinical signs and neuropathological lesions. To explore the roots of this phenomenon, we created a knock-in mouse model carrying the mutation associated with one of these diseases [Creutzfeldt–Jakob disease (CJD)] that was exactly analogous to a previous knock-in model of a different prion disease [fatal familial insomnia (FFI)]. Together with the WT parent, this created an allelic series of three lines, each expressing the same protein with a single amino acid difference, and with all native regulatory elements intact. The previously described FFI mice develop neuronal loss and intense reactive gliosis in the thalamus, as seen in humans with FFI. In contrast, CJD mice had the hallmark features of CJD, spongiosis and proteinase K-resistant PrP aggregates, initially developing in the hippocampus and cerebellum but absent from the thalamus. A molecular transmission barrier protected the mice from any infectious prion agents that might have been present in our mouse facility and allowed us to conclude that the diseases occurred spontaneously. Importantly, both models created agents that caused a transmissible neurodegenerative disease in WT mice. We conclude that single codon differences in a single gene in an otherwise normal genome can cause remarkably different neurodegenerative diseases and are sufficient to create distinct protein-based infectious elements.
Keywords: neurodegeneration, protein aggregation, protein misfolding, transgenic mice
Prion diseases are among the most enigmatic and fascinating subjects in biology from the standpoint of the diseases of protein folding. They involve highly unusual infectious agents (prions) that lack any detectable information-bearing nucleic acid and instead rely on the self-templated misfolding of an otherwise benign protein [i.e., prion protein (PrP)] to encode the disease (1). Prion diseases share several features with other neurodegenerative diseases, such as Alzheimer’s disease (AD) and Parkinson disease (PD). All typically affect people late in life, and all are characterized by the accumulation and aggregation of misfolded proteins (2, 3). Further, the misfolded proteins that precipitate in these diverse diseases are broadly expressed and are not more abundant in the brain areas that are most severely affected by disease. Intensifying interest in the prion diseases, recent evidence has established that the proteins involved in AD and PD (affecting a far larger patient population than the prion diseases) have self-templating properties that have traditionally been thought to be unique to the prion protein (3). The AD and PD proteins, Aβ and α-synuclein, do not produce infectious agents, but their self-templating properties profoundly influence their pathogenicity (4–6). An understanding of underlying mechanisms is urgently needed. The PrP, with the highly distinct pathologies and pathological progressions that are linked to it, provides an important general model for such investigations.
There are several types of human prion diseases, each beginning with pathologic processes in a different brain region and leading to distinct functional deficits: cognition [Creutzfeldt–Jakob disease (CJD)], movement control (Gerstmann–Sträussler–Scheinker syndrome), or sleep and autonomic functions [fatal familial insomnia (FFI)] (7). Prion diseases also afflict animals and include bovine spongiform encephalopathy (BSE) of cattle, scrapie of sheep and goats, and chronic wasting disease (CWD) of deer and elk (1). Importantly, all forms of prion diseases appear to be caused by misfolded PrP.
Historically, prion diseases were studied by injecting infectious material into indicator mice to model prion diseases initiated by the transmission of an exogenous agent (i.e., acquired disease). However, like other more conventional neurodegenerative diseases, most cases of prion diseases in humans are caused by the inheritance of mutations or occur sporadically (i.e., with uncertain causes) (8). Here, our goal was to model genetic forms because different mutations are linked to different diseases.
More than 20 mutations in the prion protein gene (PRNP) are associated with human prion diseases, many with specific pathological changes and clinical signs (7). Despite decades of research, it remains unclear why different mutations lead to different diseases. Because the mutations arise in different people, one possibility is that host-specific factors might cause these different phenotypes. Another is that the mutations simply sensitize carriers to infection by distinct prion variants encountered in the environment. The countervailing hypothesis is that different PRNP mutations induce specific misfolding events that occur in different specific regions of the brain or that occur broadly but affect only specific regions. Difficulties in generating mouse models that develop disease spontaneously have impeded our understanding. The use of randomly integrated transgenes to model familial forms of prion disease in mice has the widely sought advantage of producing higher-than-normal expression levels, which enhance the rate of misfolding and accelerate disease. However, the resulting mice are prone to variable, and typically incorrect, spatial expression patterns. Indeed, this may explain why most mouse models that have been engineered this way are disease-free. Moreover, because mice with very high overexpression of WT PrP can develop diseases (9–11), in transgenic models, it is difficult to distinguish between the effects of the mutation from the effects of overexpression.
The alternative knock-in approach has been used much less commonly because of the greater degree of difficulty involved. Moreover, within the limited lifetime of a mouse, most knock-in models of late-onset neurodegenerative disorders do not develop spontaneous disease. (Notable exceptions include polyglutamine disease models that carry expansion mutations severe enough to cause disease in younger humans, or models that use ectopic loci or nonnative promoters.) In earlier work, only one knock-in line had been generated for a familial PRNP mutation associated with human disease, P101L (12). (Throughout the present report, we use mouse Prnp codon numbering, which is −1 compared with human PRNP as a result of a single amino acid deletion in the N terminus.) This mouse was disease-free and pathology-free, and did not produce infectious material (12). We hypothesized that the P101L mutation might be a poor candidate to cause disease in the short lifespan of the mouse because it is associated with a very slowly progressing neurodegenerative disease in humans. Further, the P101L substitution is located in a region of the protein that is already unstructured and would be expected to have a relatively modest influence on the protein’s stability (13). Therefore, we developed a knock-in mouse strain that carried a mutation in the structured region of the protein, more likely to cause misfolding, and associated with rapidly progressing disease in humans (14).
Two knock-in mouse lines were analyzed, one a control without a disease mutation and one carrying the aspartate-to-asparagine substitution associated with FFI, D177N (14). Both mice carried a 2-aa substitution (L108M, V111M) known as the 3F4 epitope, which served several purposes. First, this epitope is encoded by the human gene, making our constructs closer to the human version. Second, this epitope creates a transmission barrier (confirmed in our report; ref. 14) that reduces the possibility of mice becoming infected by exogenous mouse prions. Third, it creates a convenient antigenic tool for distinguishing the protein generated by the knock-in gene from the allelic WT form. Mice with this variant knocked-in to the endogenous Prnp locus (ki-3F4-WT) express a PrP protein with normal structure and stability. Moreover, they have normal brain morphology and behavior and do not produce infectious material. Herein, these mice serve as our WT controls. In contrast, the FFI knock-in (ki-3F4-FFI, hereafter referred to as FFI) express PrP with abnormal structure and stability and develop clinical and neuropathological abnormalities similar to human FFI (14). Importantly, the FFI mice spontaneously generate infectious material that can be serially propagated in mice expressing only WT PrP. This model therefore fulfilled a long-postulated tenet of the prion hypothesis: that the misfolding of PrP is itself sufficient to generate infectious material de novo (14).
However, FFI is a very unusual prion disease. In addition to having distinct clinical manifestations, FFI typically lacks the two most common markers of prion diseases, spongiform degeneration and proteinase K (PK)-resistant PrP (PrPres) (15). Our ability to reproduce its general characteristics in mice established a true model of the disease but raised additional questions: can familial PrP mutations associated with the more classical features of prion disease produce a disease with these features in mice? In addition, can proteins differing by only a single amino acid produce distinct diseases with distinct infectious characteristics? Humans carrying the E199K mutation develop CJD, a disease clinically distinct from FFI, with spongiosis and PrPres (7). Here we create and analyze the distinguishing characteristics of mice carrying a knock-in of this mutation and, in the process, also provide unique characterizations of FFI mice.
Results
Generation of CJD Knock-In Mice.
We used our previous strategy (14) to develop a CJD knock-in line that differs from the WT line by a single glutamate-to-lysine codon substitution (Fig. S1). A major challenge in developing mouse models of neurodegenerative disease is to accelerate disease processes, which typically require at least four decades to develop in humans, so that they occur within the short lifespan of a mouse. Because humans homozygous for the CJD mutation develop disease faster than heterozygotes (16), we focused on homozygous mice.
The PrP protein in the brains of our CJD mice was expressed at approximately the same steady state-level as the WT PrP protein in WT brains, indicating the CJD mutation did not strongly destabilize PrP. Both were expressed at a higher level than the PrP protein of FFI mice, confirming that the FFI mutation is destabilizing (Fig. S1). The glycosylation patterns of the FFI and CJD proteins were different, indicating a differential effect on PrP metabolism or trafficking (Fig. S1). However, all variants, including the WT, were equally sensitive to Peptide-N-Glycosidase F (PNGaseF) and equally resistant to endoglycosidase H, indicating that they all trafficked through the ER and Golgi (Fig. S1). The differences in steady-state levels and glycoform patterns between the two mutant proteins suggest they populate different conformations.
Clinical Abnormalities in CJD Mice.
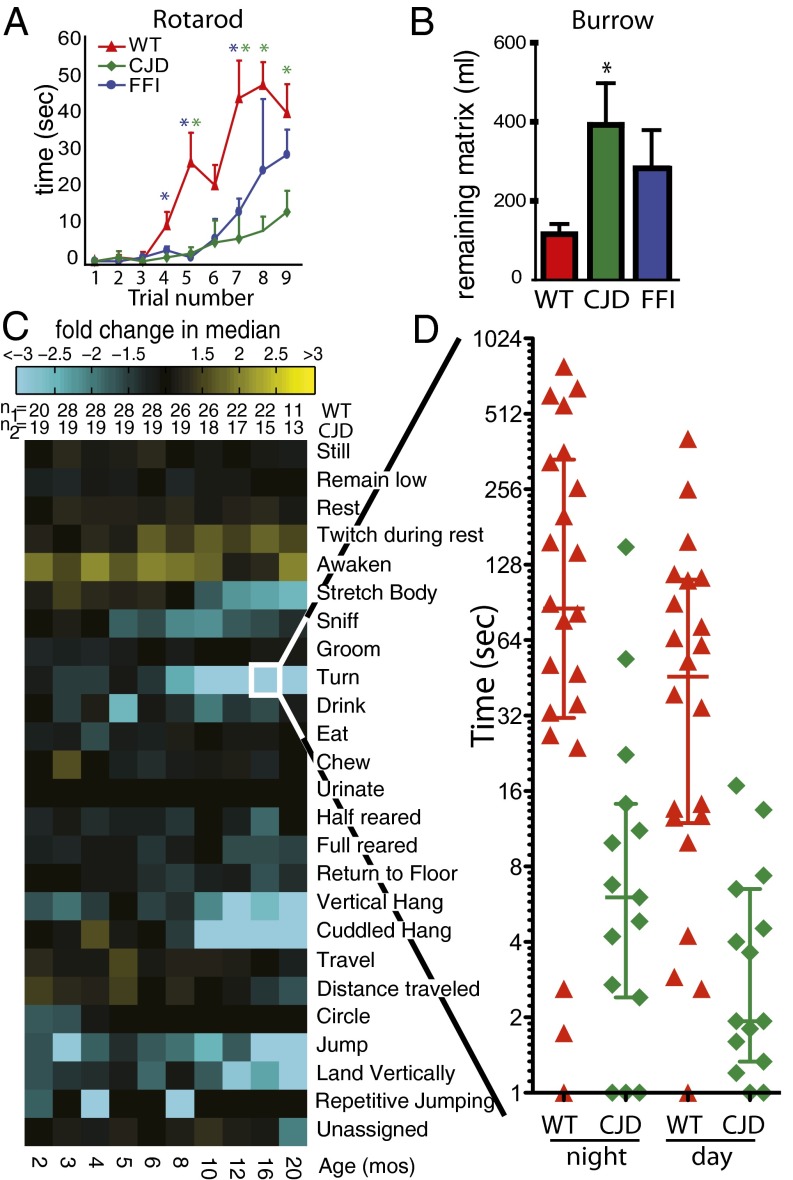
The behaviors of the mice were compared in diverse assays. The rotarod is an instrument that tests their ability to walk on a cylinder rotating at an accelerating pace. Experiments were performed on 18-mo-old mice, three times each day for three consecutive days. Upon the first trial, all mice performed poorly, with median times of less than 5 s (Fig. 1A, trials 1–3). Median rotarod times improved for all lines with repeated trials, but CJD mice lagged far behind WT mice at later time points. FFI mice performed only slightly worse than WT mice (Fig. 1A, trials 7–9).
Fig. 1.
Clinical abnormalities of CJD mice. (A) Median time (±SEM) mice remained on the rotarod during each trial. Red triangles, green diamonds, and blue dots represent WT, CJD, and FFI data, respectively. Color-coded asterisks are placed above the triangles where WT mice had better scores (P < 0.05) than CJD (green) or FFI (blue) mice, calculated with the nonparametric Mann–Whitney test. Numbers of mice: WT, n = 14; CJD, n = 11; FFI, n = 19. (B) The median (±SEM) amount of burrowing matrix remaining in the burrowing chambers. Compared with WT mice (red), CJD mice (green) removed less (*P < 0.05, nonparametric Mann–Whitney test) but FFI mice (blue) did not. Numbers of mice: WT, n = 14; CJD, n = 9; FFI, n = 9. (C) Phenotypic array representing median differences between WT and CJD mice for specific behaviors, labeled (Right). Yellow tiles depict comparisons for which the CJD mice scored higher than WT; cyan represents the opposite. The brightness corresponds to the magnitude of the difference. The age in months is directly below the array. The number of animals for each comparison is immediately above the array. (D) Scatter plot of the data making up the tile “turn” at 16 mo (framed in white). Error bars depict median and quartile values.
Another assay exploited the strong, instinctive drive of mice to build burrows (17) (Movie S1). We tested their ability to remove a burrowing matrix from a tube placed in a large cage, a task that is performed quickly and efficiently by healthy mice. CJD mice were only half as efficient as WT mice (P < 0.05, Mann–Whitney test; Fig. 1B). The slightly reduced performance of FFI mice was not statistically significant (Fig. 1B).
Finally, we used automated mouse behavioral analysis (AMBA), a computerized system that quantifies the activities of mice in their home cages (18). The key benefits of AMBA are that animals are allowed to roam freely in the absence of interventions by experimenters, with 24 spontaneous activities scored by computer in an unbiased manner (Movie S2). This system enables the detection of behavioral changes before overt neurological abnormalities. More than 1 billion video frames were analyzed in these experiments. A composite of the data are presented in a highly condensed, readily comparable form—as a “phenotypic array”—in Fig. 1C. Representative data corresponding to several individual behaviors over a 24-h period for each mouse tested are shown in Fig. 1D and Fig. S2.
CJD mice had normal amounts of “rest,” a correlate of sleep (Fig. 1C and Fig. S2). In contrast, FFI mice had abnormally high rest (14). “Turn,” a metric frequently scored during body twisting, was very strongly reduced in CJD mice (Fig. 1 C and D and Fig. S2), but this behavior was rarely abnormal in FFI mice (14). “Cuddled hang,” measured as hanging from the ceiling of the cage, was strongly reduced in both mutant lines but more so in CJD mice (Fig. 1C and Fig. S2) . CJD mice in general spent less time doing physically demanding behaviors (“jump,” “rear,” and “cuddled hang”) and more time with resting-related behaviors (“twitch during rest” and “awaken”; Fig. 1C and Fig. S2). The more quiescent behaviors of CJD mice overall represented a general reduction in activity rather than the specific disturbance in sleep we observed in FFI mice (14).
Distinct Histopathological Changes in CJD Mice.
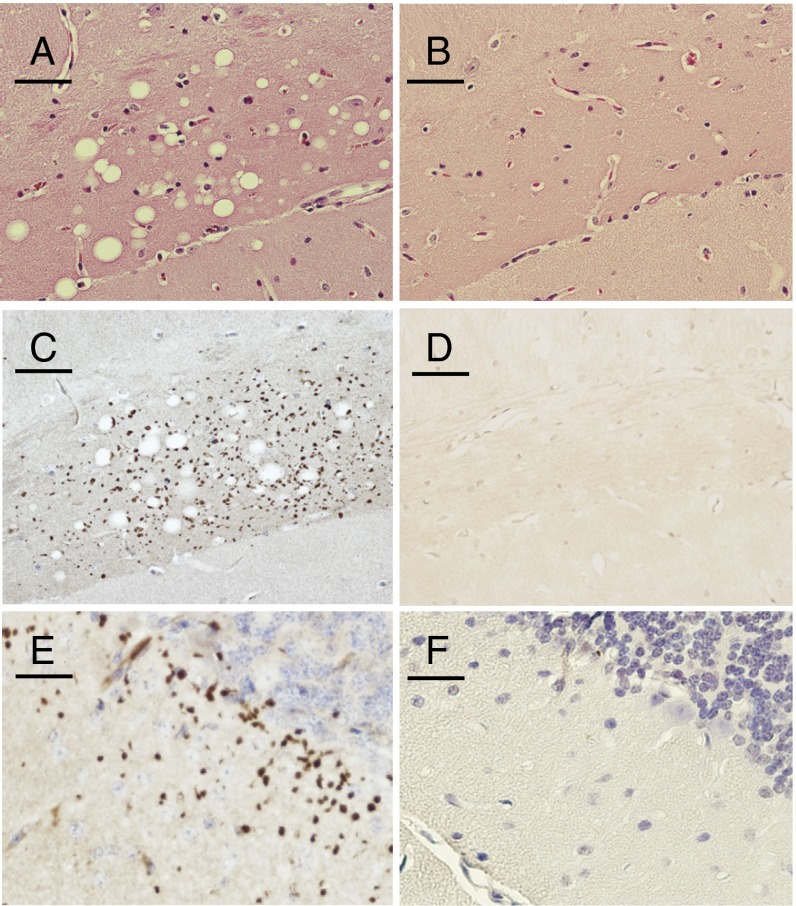
To reveal neuropathological changes, we used a variety of histological techniques. We first examined paraffin-embedded 4-μm-thick sections stained with H&E. CJD mice had prominent spongiform degeneration in the hippocampal CA1 region (Fig. 2A), specifically in the synapse-rich area (i.e., neuropil) of the molecular layer. FFI brains did not (Fig. 2B). In humans, spongiosis is a rare feature that distinguishes most of the prion diseases, including CJD but not FFI, from other neurodegenerative diseases (19). Other notable differences between the mice also characteristically distinguish CJD and FFI diseases in humans. For example, FFI brains had dilated ventricles, atrophied cerebella, and massive neuronal loss in the thalamus (14), but CJD brains did not.
Fig. 2.
Neuropathologic processes of CJD mice. Sections (5 µm thick) from paraffin-embedded hippocampal (A–D) or cerebellar (E and F) areas stained with H&E (A and B) or with PrP antibody 3F4 following PK digestion (C–F). CJD sections are shown (A,C, and E) with corresponding FFI sections (B, D, and F). (Scale bars: 0.05 mm.)
A hallmark of most neurodegenerative diseases is an increase in the size and/or number of astrocytes, a pathological state known as reactive gliosis. This was present in CJD hippocampi and deep cerebellar white matter (Fig. S3), but was notably absent from the thalamus. In contrast, FFI brains had severe reactive gliosis in the thalamus, and in the deep cerebellar white matter, but not in the hippocampus (14).
Finally, we used a silver staining procedure to detect disintegrating neurons (20) (SI Discussion). CJD brains stained similarly to control brains, but FFI brains had intense silver staining in their thalami (Fig. S3). Despite the presence of spongiosis, neuronal loss is not obvious in CJD mice, and systematic stereology experiments would be required to resolve this issue. Thus, as for behavioral tests, histopathologic analysis revealed very different diseases in these mouse models.
Distinct Aggregated Forms in CJD Mice.
In most human prion diseases, including the genetic forms of CJD, misfolded PrP accumulates into an aggregated state that renders the C-terminal core, but not the N terminus, resistant to digestion by PK (7, 21). This distinct form of the disease-associated protein is termed PrPres. CJD brains developed numerous punctate PrPres aggregates in the hippocampus, the same area that was subject to spongiform degeneration (Fig. 2C). The only other regions with abundant PrPres aggregates were the cerebellar molecular layer (Fig. 2E), small patches of the olfactory bulb, and a synapse-rich region of the retina (Fig. S4), which also develops PrP aggregates in human CJD (22). In humans, PrPres is generally much more difficult to detect in FFI than in CJD. Mirroring this contrast in our mice, PrPres deposits were not detected in FFI brains with the use of conventional protocols (14) (Fig. 2 D and F).
In CJD brain homogenates, however, PrPres was more difficult to detect by Western blotting than we might have expected from experiments with humans (Fig. S5). However, the PrPres detected in tissue sections and the spongiform pathologic process were found in many (i.e., 6 of 7) CJD mice by 12 mo of age, and were found in all (N = 17) by 16 mo. A systematic study of serial sections that used antibodies directed at the N and C terminus revealed that the PrP aggregates consisted of full-length PrP. As in human CJD, the N terminus could be digested by PK (Fig. S5).
Next, ultrathin sections were examined by EM. Surprisingly, the PrPres aggregates in CJD brains that were observed by light microscopy were not detected by EM. Although it seems counterintuitive, this distinction has been reported by others (19). In sharp contrast, PrPres aggregates were not detected in FFI brains by using standard procedures. We did, however, find numerous fibrillar deposits in FFI brains by EM, specifically in the thalamus (Fig. S5). [This region of the brain is particularly affected by FFI pathologic processes, although it was negative for many amyloid stains (14).] Standard epitope retrieval procedures are too harsh to be useful for EM. However, mild fixation permitted modest labeling of these deposits with PrP antibodies (Fig. S5). Similar deposits were never observed in CJD mice, which we found perplexing (SI Discussion). The requirement for two very different methods to detect PrP aggregates in these two disease models indicates that the mutations, carried by the same protein and expressed from the same genomic locus, created different types of misfolding. These observations complement aforementioned differences in stability and glycosylation, confirming that the two mutant forms of PrP populate distinct conformational states in the mouse brain.
Additional Changes in CJD Brains.
To identify additional differences between our mutant mice we used some additional imaging technologies. First, we used an activity-dependent MRI technique that detected, in living mice, changes in brain regions that appeared normal with conventional histological methods (23). This approach revealed structural changes in the cerebellum and ventricles and a reduced MRI signal (likely a result of reduced neural activity) in the thalamus of FFI mice (14). In contrast to FFI brains, MRI did not reveal any gross structural changes in the CJD brains (Fig. S6).
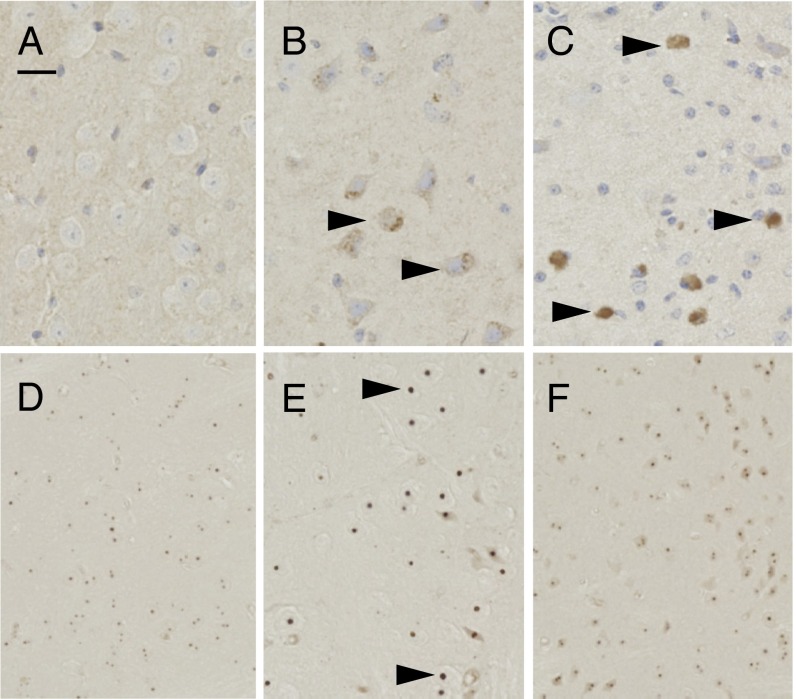
To determine if additional regions were degenerating, we examined markers of early stages of programmed cell death (PCD). In this highly conserved biological process, cells deliberately activate signaling cascades, which, when past a series of checkpoints, cause them to die. Endonuclease G (EndoG) is a mitochondrial protein that increases in total levels during disease and translocates to a perinuclear location, and eventually to the nucleus, to facilitate PCD (24). As expected, it was absent from WT brains. Surprisingly, given the normal results from other tests for pathologic conditions, EndoG was present specifically in the thalamus of CJD brains, although less intense than in FFI brains (Fig. 3 A–C). Another PCD protein is apoptosis inducing factor, which migrates from mitochondria to the cytosol, and eventually into the nucleus to trigger cell death (25). Although WT brains were negative, perinuclear deposits were detected in the thalami of FFI mice (Fig. S7), and, surprisingly, only in the thalami of CJD mice as well (Fig. S7). Thus, despite appearing normal with other tests of pathology, the CJD thalamus contained signs of PCD, whereas the hippocampus and cerebellum did not (SI Discussion).
Fig. 3.
Activation of cell cycle and death markers. Thalamus (A–C) and striatum (D–F) of paraffin-embedded sections from 18-mo-old WT (A and D), CJD (B and E), and FFI (C and F) mice, stained with antibodies against EndoG (A–C) and H2A.X (D–F). Arrowheads in B and C mark abnormal protein staining. Arrowheads in E mark abnormally large immunopositive nuclear foci. (Scale bar: 0.05 mm.)
Most neurons in adult mammalian brains are postmitotic, but, during neurodegenerative disease, a small number of neurons reexpress cell cycle markers (26). We therefore probed brain sections for changes in several cell cycle-related proteins. Although most proteins we examined were not abnormally expressed (Methods), two were. Proliferating cell nuclear antigen (PCNA), a protein associated with mitotically active cells, is found in neuronal nuclei of neurodegenerative diseased brains (27, 28). It was intensely present in small, glial-sized nuclei of all brain sections, as expected. PCNA was also found in neuronal nuclei in the FFI thalamus, but not the CJD thalamus (Fig. S7). Interestingly, in CJD and FFI brains, neuronal nuclei in the superior colliculus were PCNA-positive (Fig. S7).
H2A.X is a histone protein that becomes phosphorylated in response to dsDNA breaks that occur during normal cell division. Adult neurons are nondividing, and H2A.X is associated with the degenerative process (27, 28). Tiny punctate nuclear deposits were present throughout WT brains (Fig. 3D). Abnormally large nuclear puncta were also present in striatal neurons of CJD but not FFI brains (Fig. 3 E and F).
Disease in CJD Mice Is Transmissible.
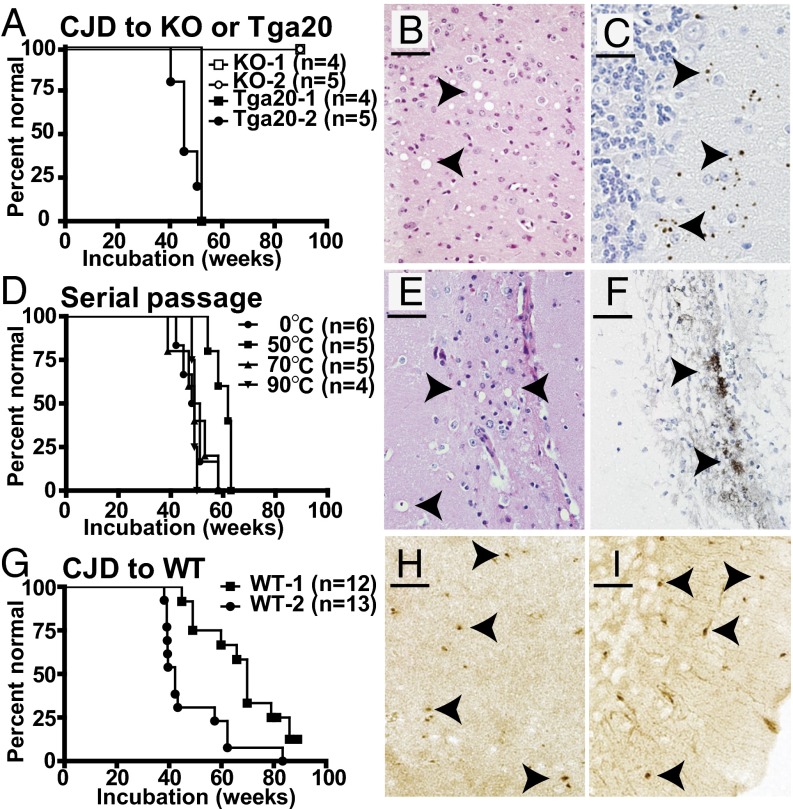
Because human E200K carriers can live into the ninth decade of life before disease onset (7), we were not surprised that many CJD mice lived as long as 2 y without being terminally ill, but some did not. To determine if CJD mice developed spontaneous prion infectivity, brain homogenates from mice that were terminally ill at 14 or 21 mo of age were injected intracranially into several strains of indicator mice. They were examined weekly by an individual highly experienced with detecting neurological illness in mice but blinded to the design of the experiments. Importantly, when injected with brain homogenates from aged WT mice, none of the indicator mice developed behavioral or neuropathological abnormalities (14, 29). KO mice that do not express PrP were challenged with CJD homogenates and remained healthy (Fig. 4A), which was expected because expression of PrP is required for susceptibility to prion infectivity (30).
Fig. 4.
Transmission of disease from CJD brains. Kaplan–Meier curves of onset of illness caused by injection of CJD homogenates (A and G) or serial passage of CJD through Tga20 mice (D) in Tga20 (A and D) or WT (G) indicator mice. Sections from CJD-infected Tga20 brain (B and C) or serial passage through Tga20 mice (E and F), stained with H&E (B and E) or stained for PrPres with SAF84 antibody following digestion with 20 μg/mL PK (C and F). (H and I) WT sections infected with CJD homogenates stained for PrPres with SAF84 following digestion with 5 μg/mL PK: (H) hippocampus, (I) cerebellum. Arrowheads in B and E mark spongiform “holes.” Arrowheads mark dark brown punctate (C, H, and I) and coarse (F) PrPres aggregates. (Scale bars: B, 25 µm; C, E, F, H, and I, 50 µm.)
Our test for infectivity first used an indicator mouse strain (known as Tga20) that overexpresses PrP and is especially sensitive to prion infection (14, 29). At 1 y after injection with CJD homogenates, Tga20 mice became terminally ill with reduced body condition (observed as abnormally loose skin), kyphosis, and a highly unusual gait (Fig. 4A and Movie S3). In contrast, Tga20 mice injected with FFI homogenates developed ataxia, a highly unusual paroxysmal hind limb twitch (14), and, later, a persistent scratching activity (i.e., pruritus). CJD inocula induced spongiosis in many areas, including the thalamus (Fig. 4B), and punctate PrPres deposits particularly in the cerebellum (Fig. 4C). In contrast, FFI inocula induced neuronal loss and reactive gliosis in the thalamus and a striking dilation of the lateral ventricles, but not spongiosis or PrPres (14).
Two gold standards of infectious prion diseases are the serial transmission of the infectious agent and its resistance to physical treatments that kill conventional microorganisms, such as high temperatures. Brain homogenate from a Tga20 mouse that had died of the aforementioned CJD infection was split into four aliquots and incubated at 0, 50, 70, or 90 °C for 30 min. Each aliquot was then injected into four to six Tga20 mice. Importantly, all four aliquots induced disease with similar incubation periods of 50 to 60 wk (Fig. 4C). Histopathologic analysis revealed that their brains developed the spongiosis and coarse PrPres aggregates characteristic of infectious CJD in humans (Fig. 4D).
Mice that express WT PrP at endogenous levels provide an even more rigorous indicator of infectious prions. Although they are slower to develop acquired disease, they never develop any PrP-related pathologic conditions, even late in life. We took this principle a step further by using our knock-in WT mice carrying the 3F4 epitope, which makes them resistant to infection by conventional mouse prions (14). Remarkably, CJD brain homogenates induced neurological signs in WT mice (Fig. 4G), including ataxia and kyphosis, similar to the signs induced by FFI brain homogenates, but without a decline in body condition (14). The most prominent neuropathological changes were small amounts of spongiosis and small, punctate PrPres aggregates in the hippocampus and cerebellum (Fig. 4 H and I), which were not induced by FFI inocula (14). Because the WT mice were desensitized to mouse scrapie and did not develop spontaneous disease (14), the diseases they developed following injection with CJD homogenates must have been induced by prion infectivity that had been spontaneously generated in the original knock-in mice. Therefore, although the CJD and FFI mutations appear to modify PrP’s structure differently, they are both sufficient to induce the spontaneous production of prion infectivity.
Discussion
We have created a knock-in mouse model of a familial prion disease, CJD, that causes a late-onset neurodegenerative disease and also generates a spontaneous infectious prion agent. Mouse models rarely recapitulate human disease pathologic conditions without gross overexpression of the disease-associated protein. However, the CJD knock-in mice we created here, by simply changing a single amino acid in the endogenous ORF, develop many hallmarks of the human diseases to which they correspond. Their disease phenotypes, and the nature of the infectious agents they produce, are very different from those of the FFI mice we created earlier (summary in Fig. S8), which instead develop several neurodegenerative characteristics of human FFI. Notably, consistent with in vitro biophysical studies by others (13), the two mutant PrP proteins had different stabilities in the brain. Given that the mice were created by knock-in manipulations in the same ES cell line, and were crossed to the same genetic background, their distinct disease phenotypes can be attributed to the distinct physical properties of the single amino acid substitutions they carry.
Why did two other transgenic lines carrying the same E199K CJD mutation not develop disease or infectivity (31, 32)? One possibility is that our mice expressed a mouse/human PrP chimera, whereas the other mice expressed the mutation in the context of mouse or human PrP (31, 32). In our experiments, the WT version of this mouse/human chimera had a stability and trafficking pattern in mouse brain that was indistinguishable from that of the endogenous mouse protein. However, in the course of the 12 mo required to establish disease, a subtle difference in the stability of the chimeric protein could contribute to the phenotypic distinctions resulting from the additional disease-causing mutations. Another possibility is that knock-in mutations are required to produce spontaneous infectivity because they uniquely recapitulate the complex regulation to which the endogenous PrP gene is subject. Randomly integrated transgenes have the advantage of being far less labor-intensive to engineer. However, they are prone to variable expression levels and patterns, depending on copy number and genome integration site. Indeed, one of the most popular vectors for modeling neurodegenerative diseases, a Prnp promoter fragment lacking an intron that contains regulatory information, is highly prone to integration effects (23, 33, 34). Even very large vectors such as those created by BACs can have variable spatial expression patterns (35). This likely explains why some lines overexpressing WT PrP become sick (9–11). It also highlights why, in the rare past cases in which spontaneous infectivity has been generated by extremely high overexpression levels in transgenic mice, it has been difficult to determine how much the mutation contributed to the result (36).
Because prion diseases are infamous for their ability to spread between individuals, they are assumed to easily spread within an individual brain. Thus, it was surprising that the thalamus was primarily affected in FFI mice and the hippocampus was primarily affected in CJD mice, as these regions are adjacent. It was also surprising that, in CJD brains, spongiosis and PrPres intensity increased as the disease progressed, but there was little spreading into neighboring areas. Although they are not likely to be naturally transmissible (3, 37), several other neurodegenerative diseases can be induced by intracerebral injection of diseased brain (4, 6, 38), and some have features reminiscent of prion strains and transmission barriers (5). This has led to renewed interests in spreading mechanisms for many neurodegenerative diseases, whereby a pathologic state is initiated in one area and then spreads to others in a prion-like fashion (2, 39). Because the different phenotypes in our mice can be attributed to the single amino acid substitutions they contain, they provide a case study for the effects that protein conformations alone can have on pathological progressions. Further, they add support to the hypothesis that familial prion diseases arise from misfolding events induced in the endogenous protein, independent of exogenous agents.
Our results also have implications for the origin of prion diseases in the animals we consume. For example, a cow in Alabama with BSE was found to carry the same mutation as our CJD mice (40). If this mutation had simply sensitized carriers to exogenous agents, the environment in which that cow lived was contaminated and the BSE surveillance program was ineffective. However, our work establishes that this mutation can cause disease spontaneously. More broadly, our work has implications for the origin of the BSE epidemic during the 1980s, which caused the “new variant” CJD crisis in humans in the 1990s (40). It is commonly assumed that this epidemic was initiated by an animal that acquired prion disease from the consumption of scrapie-infected sheep brains (41). Our work supports an alternative explanation: that the process started in a cow carrying a PrP mutation that spontaneously gave rise to infectious material and was then spread by the inclusion of cow brain in protein-rich feed (40). Similarly it has been assumed that CWD, a rapidly spreading prion disease of free-roaming deer, moose, and elk in North America (42), was initiated by the accidental exposure of deer to sheep prion contamination. However, it may well have arisen directly and spontaneously in deer as a result of a mutation in PrP, especially in light of the spontaneous development of a severe infectious prion disease in transgenic mice expressing a CWD-related epitope (29). In any case, PrP is unusually vulnerable to misfolding events that generate conformations with a toxic gain of function: the protein is rather small (only 254 aa) yet more than 20 different PrP mutations cause neurodegenerative disease in humans. Our work establishes that spontaneous mutations in the endogenous PrP gene can readily produce not only disease, but also infectious agents.
Methods
Most methods were described previously (14, 18, 23). Animal experiment protocols were approved by the Committee on Animal Care, Massachusetts Institute of Technology. Details of Western blots, antibodies used, measurements of disease, and EM are provided in SI Methods.
Supplementary Material
Acknowledgments
The authors thank M. Duquette and A. Topolszki for excellent technical assistance and J. Ma, A. Steele, and several other laboratory members for critical evaluation of this manuscript. This work was funded by a National Institutes of Health National Research Service Award Fellowship (to W.S.J.) and US Department of Defense funding (A.J. and S.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312006110/-/DCSupplemental.
References
- 1.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95(23):13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cushman M, Johnson BS, King OD, Gitler AD, Shorter J. Prion-like disorders: Blurring the divide between transmissibility and infectivity. J Cell Sci. 2010;123(Pt 8):1191–1201. doi: 10.1242/jcs.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguzzi A, Rajendran L. The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron. 2009;64(6):783–790. doi: 10.1016/j.neuron.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 4.Kane MD, et al. Evidence for seeding of beta -amyloid by intracerebral infusion of Alzheimer brain extracts in beta -amyloid precursor protein-transgenic mice. J Neurosci. 2000;20(10):3606–3611. doi: 10.1523/JNEUROSCI.20-10-03606.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer-Luehmann M, et al. Exogenous induction of cerebral beta-amyloidogenesis is governed by agent and host. Science. 2006;313(5794):1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- 6.Hansen C, et al. α-Synuclein propagates from mouse brain to grafted dopaminergic neurons and seeds aggregation in cultured human cells. J Clin Invest. 2011;121(2):715–725. doi: 10.1172/JCI43366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gambetti P, Kong Q, Zou W, Parchi P, Chen SG. Sporadic and familial CJD: classification and characterisation. Br Med Bull. 2003;66:213–239. doi: 10.1093/bmb/66.1.213. [DOI] [PubMed] [Google Scholar]
- 8.Wadsworth JD, Hill AF, Beck JA, Collinge J. Molecular and clinical classification of human prion disease. Br Med Bull. 2003;66:241–254. doi: 10.1093/bmb/66.1.241. [DOI] [PubMed] [Google Scholar]
- 9.Westaway D, et al. Degeneration of skeletal muscle, peripheral nerves, and the central nervous system in transgenic mice overexpressing wild-type prion proteins. Cell. 1994;76(1):117–129. doi: 10.1016/0092-8674(94)90177-5. [DOI] [PubMed] [Google Scholar]
- 10.Chiesa R, Piccardo P, Biasini E, Ghetti B, Harris DA. Aggregated, wild-type prion protein causes neurological dysfunction and synaptic abnormalities. J Neurosci. 2008;28(49):13258–13267. doi: 10.1523/JNEUROSCI.3109-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watts JC, et al. Spontaneous generation of rapidly transmissible prions in transgenic mice expressing wild-type bank vole prion protein. Proc Natl Acad Sci USA. 2012;109(9):3498–3503. doi: 10.1073/pnas.1121556109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manson JC, et al. A single amino acid alteration (101L) introduced into murine PrP dramatically alters incubation time of transmissible spongiform encephalopathy. EMBO J. 1999;18(23):6855–6864. doi: 10.1093/emboj/18.23.6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Apetri AC, Surewicz K, Surewicz WK. The effect of disease-associated mutations on the folding pathway of human prion protein. J Biol Chem. 2004;279(17):18008–18014. doi: 10.1074/jbc.M313581200. [DOI] [PubMed] [Google Scholar]
- 14.Jackson WS, et al. Spontaneous generation of prion infectivity in fatal familial insomnia knockin mice. Neuron. 2009;63(4):438–450. doi: 10.1016/j.neuron.2009.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown P, et al. Intracerebral distribution of infectious amyloid protein in spongiform encephalopathy. Ann Neurol. 1995;38(2):245–253. doi: 10.1002/ana.410380218. [DOI] [PubMed] [Google Scholar]
- 16.Simon ES, et al. Creutzfeldt-Jakob disease profile in patients homozygous for the PRNP E200K mutation. Ann Neurol. 2000;47(2):257–260. [PubMed] [Google Scholar]
- 17.Guenther K, Deacon RM, Perry VH, Rawlins JN. Early behavioural changes in scrapie-affected mice and the influence of dapsone. Eur J Neurosci. 2001;14(2):401–409. doi: 10.1046/j.0953-816x.2001.01645.x. [DOI] [PubMed] [Google Scholar]
- 18.Steele AD, Jackson WS, King OD, Lindquist S. The power of automated high-resolution behavior analysis revealed by its application to mouse models of Huntington’s and prion diseases. Proc Natl Acad Sci USA. 2007;104(6):1983–1988. doi: 10.1073/pnas.0610779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeffrey M, McGovern G, Sisó S, González L. Cellular and sub-cellular pathology of animal prion diseases: Relationship between morphological changes, accumulation of abnormal prion protein and clinical disease. Acta Neuropathol. 2011;121(1):113–134. doi: 10.1007/s00401-010-0700-3. [DOI] [PubMed] [Google Scholar]
- 20.Switzer RC., 3rd Application of silver degeneration stains for neurotoxicity testing. Toxicol Pathol. 2000;28(1):70–83. doi: 10.1177/019262330002800109. [DOI] [PubMed] [Google Scholar]
- 21.Ghoshal N, et al. Codistribution of amyloid beta plaques and spongiform degeneration in familial Creutzfeldt-Jakob disease with the E200K-129M haplotype. Arch Neurol. 2009;66(10):1240–1246. doi: 10.1001/archneurol.2009.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Head MW, et al. Prion protein accumulation in eyes of patients with sporadic and variant Creutzfeldt-Jakob disease. Invest Ophthalmol Vis Sci. 2003;44(1):342–346. doi: 10.1167/iovs.01-1273. [DOI] [PubMed] [Google Scholar]
- 23.Faas H, et al. Context-dependent perturbation of neural systems in transgenic mice expressing a cytosolic prion protein. Neuroimage. 2010;49(3):2607–2617. doi: 10.1016/j.neuroimage.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee BI, Lee DJ, Cho KJ, Kim GW. Early nuclear translocation of endonuclease G and subsequent DNA fragmentation after transient focal cerebral ischemia in mice. Neurosci Lett. 2005;386(1):23–27. doi: 10.1016/j.neulet.2005.05.058. [DOI] [PubMed] [Google Scholar]
- 25.Modjtahedi N, Giordanetto F, Madeo F, Kroemer G. Apoptosis-inducing factor: vital and lethal. Trends Cell Biol. 2006;16(5):264–272. doi: 10.1016/j.tcb.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 26.Khurana V, Feany MB. Connecting cell-cycle activation to neurodegeneration in Drosophila. Biochim Biophys Acta. 2007;1772(4):446–456. doi: 10.1016/j.bbadis.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khurana V, et al. TOR-mediated cell-cycle activation causes neurodegeneration in a Drosophila tauopathy model. Curr Biol. 2006;16(3):230–241. doi: 10.1016/j.cub.2005.12.042. [DOI] [PubMed] [Google Scholar]
- 28.Kim D, et al. Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron. 2008;60(5):803–817. doi: 10.1016/j.neuron.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sigurdson CJ, et al. De novo generation of a transmissible spongiform encephalopathy by mouse transgenesis. Proc Natl Acad Sci USA. 2009;106(1):304–309. doi: 10.1073/pnas.0810680105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Büeler H, et al. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73(7):1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 31.Asante EA, et al. Absence of spontaneous disease and comparative prion susceptibility of transgenic mice expressing mutant human prion proteins. J Gen Virol. 2009;90(pt 3):546–558. doi: 10.1099/vir.0.007930-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Telling GC, et al. Interactions between wild-type and mutant prion proteins modulate neurodegeneration in transgenic mice. Genes Dev. 1996;10(14):1736–1750. doi: 10.1101/gad.10.14.1736. [DOI] [PubMed] [Google Scholar]
- 33.Borchelt DR, et al. A vector for expressing foreign genes in the brains and hearts of transgenic mice. Genet Anal. 1996;13(6):159–163. doi: 10.1016/s1050-3862(96)00167-2. [DOI] [PubMed] [Google Scholar]
- 34.Karapetyan YE, et al. Prion strain discrimination based on rapid in vivo amplification and analysis by the cell panel assay. PLoS ONE. 2009;4(5):e5730. doi: 10.1371/journal.pone.0005730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425(6961):917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 36.Friedman-Levi Y, et al. Fatal prion disease in a mouse model of genetic E200K Creutzfeldt-Jakob disease. PLoS Pathog. 2011;7(11):e1002350. doi: 10.1371/journal.ppat.1002350. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Irwin DJ, et al. Evaluation of potential infectivity of Alzheimer and Parkinson disease proteins in recipients of cadaver-derived human growth hormone. JAMA Neurol. 2013;70(4):462–468. doi: 10.1001/jamaneurol.2013.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luk KC, et al. Intracerebral inoculation of pathological α-synuclein initiates a rapidly progressive neurodegenerative α-synucleinopathy in mice. J Exp Med. 2012;209(5):975–986. doi: 10.1084/jem.20112457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frost B, Diamond MI. Prion-like mechanisms in neurodegenerative diseases. Nat Rev Neurosci. 2010;11(3):155–159. doi: 10.1038/nrn2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richt JA, Hall SM. BSE case associated with prion protein gene mutation. PLoS Pathog. 2008;4(9):e1000156. doi: 10.1371/journal.ppat.1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watts JC, Balachandran A, Westaway D. The expanding universe of prion diseases. PLoS Pathog. 2006;2(3):e26. doi: 10.1371/journal.ppat.0020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams ES, Miller MW. Chronic wasting disease in deer and elk in North America. Rev Sci Tech. 2002;21(2):305–316. doi: 10.20506/rst.21.2.1340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.