Abstract
Mitochondria are emerging as important players in the transformation process of cells, maintaining the biosynthetic and energetic capacities of cancer cells and serving as one of the primary sites of apoptosis and autophagy regulation. Although several avenues of cancer therapy have focused on mitochondria, progress in developing mitochondria-targeting anticancer drugs nonetheless has been slow, owing to the limited number of known mitochondrial target proteins that link metabolism with autophagy or cell death. Recent studies have demonstrated that two members of the newly discovered family of NEET proteins, NAF-1 (CISD2) and mitoNEET (mNT; CISD1), could play such a role in cancer cells. NAF-1 was shown to be a key player in regulating autophagy, and mNT was proposed to mediate iron and reactive oxygen homeostasis in mitochondria. Here we show that the protein levels of NAF-1 and mNT are elevated in human epithelial breast cancer cells, and that suppressing the level of these proteins using shRNA results in significantly reduced cell proliferation and tumor growth, decreased mitochondrial performance, uncontrolled accumulation of iron and reactive oxygen in mitochondria, and activation of autophagy. Our findings highlight NEET proteins as promising mitochondrial targets for cancer therapy.
Keywords: Cisd1, Cisd2, Miner1, MCF7, MDA-MB-231
Mitochondria play a key role in many human diseases related to their functions in cellular energy production, biosynthesis of essential cellular compounds, and involvement in autophagy and/or apoptosis regulation (1). Contrary to previously held beliefs, recent studies have demonstrated that mitochondria are also key players in the transformation process of cancer cells and may be used as targets for anticancer therapy (2–6). The involvement of mitochondria in cancer cell function could be linked to the enhanced accumulation of iron and reactive oxygen species (ROS) in mitochondria of cancer cells, which is thought to result from the increased metabolic and energetic demands of the transformed phenotype (7). An interesting, recently discovered group of proteins that could link iron and ROS homeostasis with mitochondrial function in cancer cells are NEET proteins (8–15).
NEET proteins [mitoNEET (mNT; CISD1), Nutrient-deprivation autophagy factor-1 (NAF-1; CISD2), and CISD3] are a class of iron-sulfur proteins involved in several human pathologies, including diabetes, cystic fibrosis, Wolfram syndrome 2, neurodegeneration, and muscle atrophy (16–22). mNT and NAF-1 are localized to the outer mitochondrial membrane. NAF-1 is also localized to the endoplasmic reticulum, where it interacts with BCL-2 and Beclin 1 to regulate autophagy and apoptosis (8–13). Deficiency in mNT causes the accumulation of iron and ROS in mitochondria of animal and plant cells, and deficiency in NAF-1 results in decreased mitochondrial function and stability, as well as activation of autophagy in mice and human cells (13–16, 20, 21).
Interestingly, levels of mNT and NAF-1 mRNA are increased significantly in many different human cancer cells (23–25). The involvement of NAF-1 and mNT in mitochondrial metabolism and homeostasis, as well as in autophagy regulation, points to a possible role for these proteins in cell transformation. Thus, we hypothesized that suppressing the expression of these proteins could inhibit cancer proliferation. To address this hypothesis, we studied the regulation and function of NAF-1 and mNT in human epithelial breast cancer cells (MCF-7 and MDA-231) and, in particular, how controlling the expression level of these proteins affects tumor growth.
Here we report that NAF-1 and mNT protein levels accumulate in human epithelial breast cancer cells, and that suppressing the level of these proteins in cancer cells results in significantly reduced cell proliferation and tumor growth, decreased mitochondrial performance, uncontrolled accumulation of iron and ROS in mitochondria, and activation of autophagy. Taken together, our findings indicate that NEET proteins may be promising mitochondrial targets for cancer therapy.
Results
Accumulation of NEET Proteins and Mitochondrial Function in Human Epithelial Breast Cancer Cells.
Protein blot analysis of NAF-1 and mNT in three different human epithelial breast cancer cell lines (MCF-7, MDA-MB-468, and HCC-70, compared with control breast MCF-10a cells) revealed significantly elevated NAF-1 levels in all three lines, and significantly elevated mNT levels in two of these lines (Fig. 1A). These findings are supported by mRNA expression studies of NEET proteins in breast cancer cells (23–25), and suggest a role for NEET proteins in promoting mitochondrial biogenesis and autophagy resistance in cancer cells (23).
Fig. 1.
Protein expression, cellular proliferation and mitochondrial function of mNT and NAF-1 in human epithelial breast cancer cells. (A) Accumulation of mNT and NAF-1 protein in three different human epithelial breast cancer cell lines. (B) Suppression of mNT and NAF-1 protein expression in MCF-7 cells expressing shRNA against mNT (mNT−) or NAF-1 (NAF-1−). (Upper) Protein blot. (Lower) Relative levels of the respective proteins. (C) Suppression of cell growth and proliferation in mNT− or NAF-1− MCF-7 cells. (Inset) Relative change in growth rate for control, mNT(−), and NAF-1(−) at day 6. (D) Mitochondrial function (Left) and glycolytic activity (Right) in control MCF-7 cells and MCF-7 cells expressing shRNA against mNT (mNT−) or NAF-1 (NAF-1−). (E) Spare respiratory capacity (Left) and glycolytic capacity (Right) in control and mNT− or NAF-1− MCF-7 cells. NAF-1 and mNT are seen to accumulate in breast cancer cells, and their suppression via shRNA results in suppressed cell proliferation and reduced mitochondrial function. Figs. S1, S2, and S3 present similar results with MDA-231 cells. *P < 0.05; **P < 0.01.
Suppression of mNT (mNT−) or NAF-1 (NAF-1−) protein levels using shRNA in MCF-7 and MDA-231 cells (Fig. 1B and Fig. S1) caused a significant decrease in cell proliferation (Fig. 1C and Figs. S2 and S3). Suppression of mNT or NAF-1 in MCF-7 cells also resulted in diminished spare respiratory capacity of mitochondria and enhanced glycolytic activity, as calculated based on the oxygen consumption rate (OCR), which is an indicator of mitochondrial respiration, and the acid efflux rate (ECAR), which is predominantly a measure of lactic acid formed during glycolytic energy metabolism (Fig. 1 D and E). In contrast, overexpression of mNT or NAF-1 led to an increased spare respiratory capacity of mitochondria and decreased glycolytic activity (Fig. S4). These results demonstrate a physiological role for mNT and NAF-1 in promoting mitochondrial function in cancer cells.
Accumulation of Iron and ROS in Mitochondria of Human Epithelial Breast Cells with Suppressed Expression of NEET Proteins.
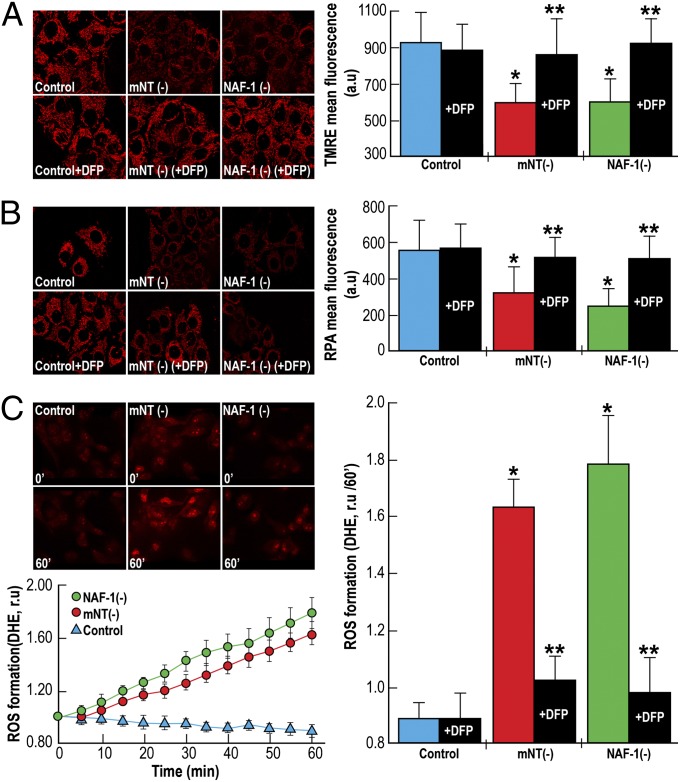
NAF-1 and mNT function as iron-cluster transfer proteins, and mNT is responsible for iron homeostasis in mitochondria (10, 11, 14, 15, 18, 19). Maintaining a balanced iron level in mitochondria is necessary to maintain mitochondrial membrane potential and regulation of mitochondrial ROS levels. To examine the mechanism of mNT and NAF-1 function in cancer cells, we measured mitochondrial membrane potential, mitochondrial iron accumulation, and mitochondrial ROS levels in epithelial human breast cancer cells with suppressed levels of NAF-1 or mNT. Suppression of mNT or NAF-1 expression in breast cancer cells resulted in decreased mitochondrial membrane potential (Fig. 2A and Fig. S5), as indicated by decreased fluorescence of tetramethylrhodamine ethyl ester (TMRE), a positively charged red-orange dye that readily accumulates in active mitochondria owing to their relative negative charge; increased mitochondrial iron levels (Fig. 2B and Fig. S5), as evidenced by the iron-derived quenching of rhodamine B-[(1,10-phenanthrolin-5-yl aminocarbonyl] benzyl ester (RPA) fluorescence in mitochondria; and increased mitochondrial ROS accumulation (Fig. 2C), as evidenced by the increased fluorescence of dehydroetidium (DHE) on interaction with superoxide radicals. The overaccumulation of iron and ROS, along with the decrease in mitochondrial membrane potential, in cells with suppressed mNT or NAF-1 were blocked by the addition of the iron chelator deferiprone (DFP) (Fig. 2 A–C and Fig. S5), demonstrating that the accumulation of ROS and the decrease in mitochondrial membrane potential in breast cancer cells with disrupted levels of mNT or NAF-1 is a result of impaired iron homeostasis caused by the deficiency in NEET proteins.
Fig. 2.
Decreased membrane potential and overaccumulation of iron and ROS in mitochondria of cells with suppressed expression of mNT or NAF-1. (A) Images (Left) and a quantitative graph (Right) showing decreased mitochondrial membrane potential in cells with suppressed expression of mNT (mNT−) or NAF-1 (NAF-1−). Pretreatment of cells with the iron chelator DFP (50 µM) prevents the decrease in mitochondrial membrane potential (Right). (B) Images (Left) and a quantitative graph (Right) showing overaccumulation of iron in mitochondria of mNT− or NAF-1− cells. Pretreatment of cells with the iron chelator DFP (100 µM) prevents mitochondrial iron accumulation (Right). (C) Images (Upper Left), a time course (Lower Left), and a quantitative graph (Right) showing the accumulation of ROS in mitochondria of mNT− or NAF-1− cells. Pretreatment of cells with the iron chelator DFP (100 µM) prevents mitochondrial ROS accumulation (Right). The function of mNT and NAF-1 is seen to be required for maintaining mitochondrial membrane potential, as well as mitochondrial iron and ROS homeostasis in human epithelial breast cancer cells. Fig. S5 presents similar results with MDA-231 cells. *P < 0.05, **P < 0.01.
Activation of Autophagy in Human Epithelial Breast Cells with Suppressed Expression of NEET Proteins.
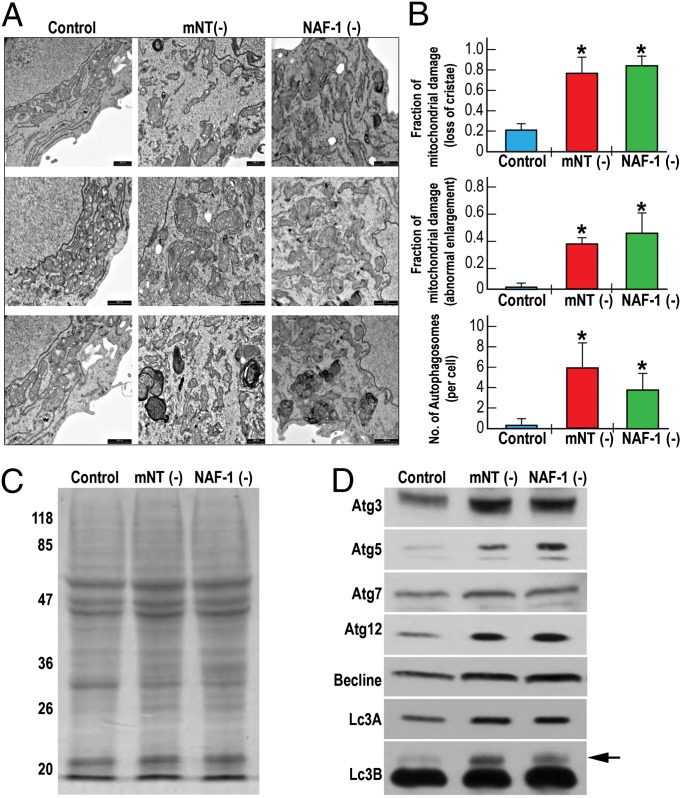
Accumulation of ROS in mitochondria is a known trigger of autophagy resulting in the removal of damaged mitochondria (26). To examine the degree of mitochondrial damage and the activation of autophagy in breast cancer cells with suppressed mNT or NAF-1 expression we conducted a TEM study of these cells. Human epithelial breast cancer cells with suppressed levels of mNT or NAF-1 accumulated damaged mitochondria with an elongated shape, many of which contained no crista (Fig. 3B). In addition, these cells contained high levels of autophagosomes (Fig. 3B), but did not display apoptotic bodies.
Fig. 3.
Activation of autophagy in human epithelial breast cancer cells with suppressed expression of mNT or NAF-1. (A) Three representative transmission electron microscopy (TEM) images of mitochondria from control cancer cells (Left) and cancer cells with suppressed expression of mNT (mNT−; Center) or NAF-1 (NAF-1−; Right). (B) Quantitative analysis of mitochondrial damage in the form of loss of crista (Top), abnormal elongation (Middle), and the accumulation of autophagosomes (Bottom) in TEM images from control and mNT− or NAF-1− cells. (C and D) Protein gel analysis (C) and protein blot analysis (D) showing accumulation of autophagy marker proteins in breast cancer cells with suppressed expression of mNT (mNT−) or NAF-1 (NAF-1−). Autophagy is shown to be activated in human epithelial breast cancer cells with suppressed expression of mNT or NAF-1. The arrow indicates the position of Lc3B. *P < 0.05.
To further confirm the activation of autophagy in mNT− or NAF-1− MCF-7 cells (Fig. 3 A and B), we performed protein blot analysis of control MCF-7 and mNT− or NAF-1− MCF-7 cells, following the level of known molecular markers of autophagy. Protein gels of total protein (Fig. 3C) and protein blots (Fig. 3D) of control MCF-7 and mNT− or NAF-1− cells indicate that compared with control MCF-7 cells, the level of many known protein markers for autophagy, including Atg3, Atg5, Atg12, and Lc3B, accumulate in mNT− and NAF-1− MCF-7 cells (representative blots are shown from two independent experiments; the arrow in the Lc3B panel indicates the autophagy-associated form).
Suppressed Growth of Tumors with Altered Expression of NEET Proteins.
Impaired mitochondrial function and activation of autophagy in human breast cancer cells with suppressed expression of NEET proteins (Figs. 1D, 2, and 3) resulted in suppressed proliferation of cells growing in cell culture medium (Figs. 1C and 4A and Figs. S2 and S3). Although these conditions could be similar to some of the conditions seen in early phases of cancer development, tumor formation and the various processes involved in tumor establishment differ greatly from processes seen in cells growing in culture.
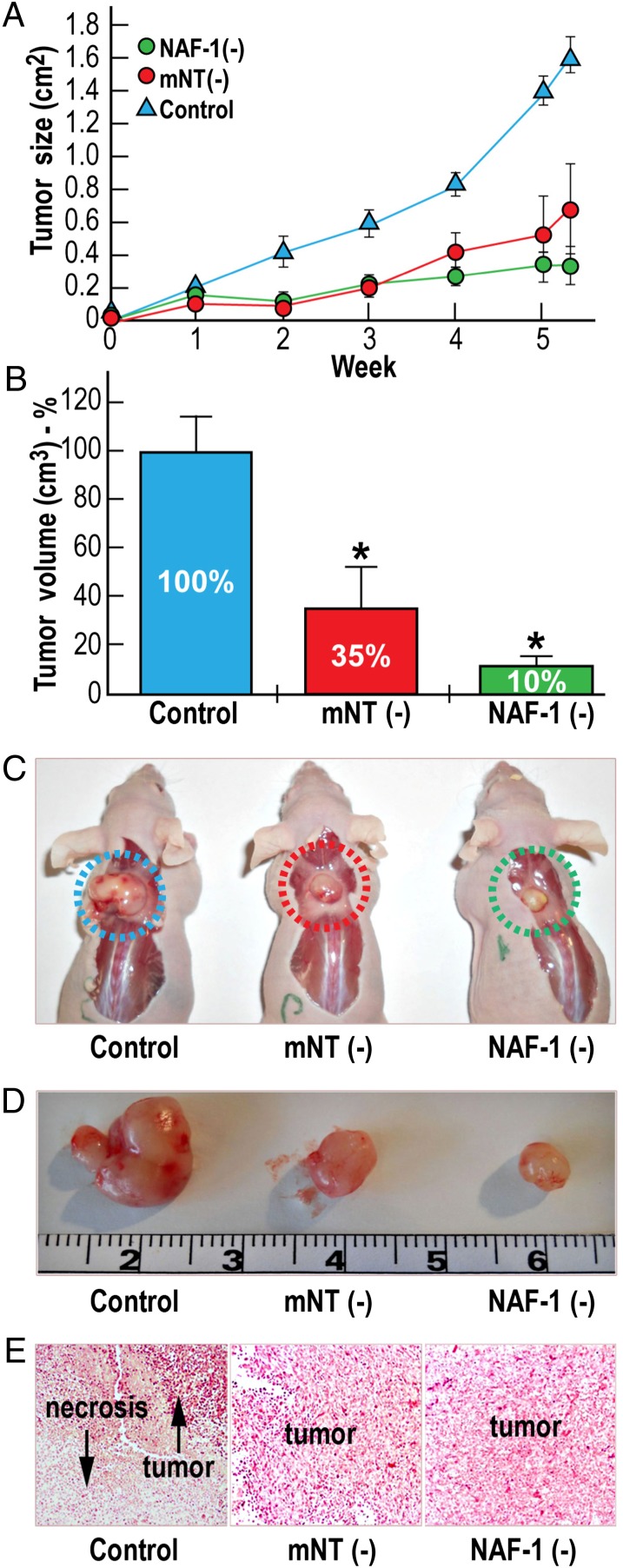
Fig. 4.
NAF-1 or mNT are required to support tumor growth. Breast cancer cells (MDA-231) with or without suppressed expression of mNT (mNT−) or NAF-1 (NAF-1−) were injected s.c. into the back of female CD1 nude mice, and tumor growth was monitored over time. (A) Slower tumor growth of breast cancer cells (MDA-231) with suppressed expression of mNT (mNT−) or NAF-1 (NAF-1−) compared with control MDA-231 cells. (B) Reduction of mean tumor volume expressed as percentage of control MDA-231 in mNT− or NAF-1− MDA-231 cells. (C and D) Images of representative tumors of each group obtained at the end of the experiment. (E) Histological analyses of tumors using H&E staining showing large necrotic areas in tumors formed from the control MDA-231 cells compared with tumors formed from mNT− or NAF-1− MDA-231 cells. *P < 0.05.
To examine the effect of mNT or NAF-1 deficiency on tumor growth, we s.c. injected control MDA-231 cells and MDA-231 cells with suppressed expression of mNT or NAF-1 in nude mice and followed the rate of tumor growth. Compared with the mice injected with control MDA-231 cells, tumor size (Fig. 4A) and growth (Fig. 4 B–D) were significantly reduced in the mice injected with MDA-231 cells with suppressed mNT or NAF-1 expression. Along with their smaller size (Fig. 4 A–D), tumors from mNT− and NAF-1− MDA-231 cells were more homogeneous in their structure and did not contain a necrotic center (Fig. 4E). These results demonstrate that mNT and NAF-1 are important not only for the proliferation of cancer cells, but also for the development and growth of tumors.
Discussion
Our findings demonstrate that mNT and NAF-1 are required for human epithelial breast cancer cell proliferation and tumor growth. Interestingly, mNT and NAF-1 do not complement each other in the support of breast cancer cell proliferation or tumor growth (Figs. 1 and 4), and likely function via noncomplementary cellular routes. Deficiency in either mNT or NAF-1 disrupts mitochondrial function and enhances the accumulation of iron and ROS in mitochondria (Figs. 2 and 3), demonstrating that both proteins are required for maintenance of mitochondrial integrity, as well as iron and ROS homeostasis in cells. Moreover, deficiency in NAF-1 or mNT activates autophagy (Fig. 3), demonstrating that both proteins trigger a common downstream pathway in cancer cells. Thus, we propose that mNT and NAF-1 have similar, albeit noncomplementary, functions in breast cancer cells to support mitochondrial metabolism and maintain mitochondrial iron and ROS homeostasis. Although artificial overexpression of mNT has been suggested to trigger autophagy resistance in human breast cancer cells (23), the naturally occurring high levels of mNT protein found in the MCF-7 human breast cancer cell line (Fig. 1A) could not mitigate the activation of autophagy by NAF-1 deficiency (Fig. 3). On the other hand, NAF-1 has been proposed to interact with BCL-2 and Beclin-1 and suppress autophagy (12, 13). Nevertheless, the naturally occurring high levels of NAF-1 found in the different human breast cancer cell lines (Fig. 1A) could not mitigate the activation of autophagy by mNT deficiency (Fig. 3). These results could suggest that, at least in epithelial breast cancer cells, the function of mNT and NAF-1 in maintaining a balanced iron and ROS homeostasis outweighs their proposed function in autophagy or cell death regulation.
Our results demonstrate that mitochondrial function is essential for cancer cell proliferation and tumor growth. Only when mitochondrial function was suppressed in breast cancer cells as a result of mNT or NAF-1 deficiency did cancer cells turn to glycolysis for energy production (Fig. 1 D and E). The dependency of cancer cells on mitochondria, as well as on mNT or NAF-1 function, suggest that these proteins could be used as important targets for anticancer therapy. In this context, it is important to note that long-term treatment of diabetic patients with pioglitazone, a drug that binds NEET proteins and stabilizes their iron-sulfur clusters, reduces the occurrence of breast, colorectal, and liver cancers compared with placebo, with some conflicting reports on its effect on bladder cancer (27–29). Pioglitazone also proved effective in reducing the proliferation of human breast cancer cells grown in tissue culture (Fig. S6). These findings are also consistent with several clinical studies showing that the weak mitochondrial poison metformin decreases the onset of many different types of human cancers in diabetic patients (23, 30). Thus, mitochondria, NAF-1, and mNT should be considered as potential targets for anticancer therapy.
Methods
Materials.
DFP (ferriprox; 1,2-dimethyl-3-hydroxypyridin-4-one) was obtained from Apo Pharma, and DHE was obtained from Sigma-Aldrich. The red fluorescent mitochondrial metal–sensor RPA was a kind gift from U. Rauen and R. Sustmann, University of Duisburg-Essen, Essen, Germany. TMRE was purchased from Molecular Probes. The Autophagy Antibody Kit was obtained from Cell Signaling Technology. MCF-7 and MDA-MB-231 epithelial breast cancer cells were obtained from American Type Culture Collection (CRL-10317) and from Professor Tamar Peretz-Yablonski, Sharett Institute of Oncology, Hadassah-Hebrew University Medical Center, Jerusalem, Israel, respectively.
Cell Culture and ShRNA Transfection.
MDA-MB-231 cells were grown in 5% CO2 RPMI medium 1640 supplemented with 10% FCS, l- glutamine, and antibiotics (Biological Industries). MCF7 cells were grown in complete DMEM (Life Technologies) supplemented with 10% FBS, 2 mM glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin, and 0.25 µg/mL amphotericin B. Cells were maintained in 100-mm Petri dishes (Sarstedt) in an incubator (HERAcell 150i; Thermo Scientific) controlled at 37 °C and 5% CO2. Cells were plated at 1 d before experimentation onto 12-well plates or onto microscopic slides glued to perforated 3-cm-diameter tissue culture plates. Four unique 29-mer pGFP-V-RS plasmid vectors were purchased from OriGene Technologies. The same plasmid with a scrambled sequence cassette was used as a negative control.
All shRNA transfections were performed with 1 μg DNA using Lipofectamine 2000 (Invitrogen) or Genejuice (EMD Millipore) as a transfection reagent according to the manufacturer’s instructions. Cells were treated with 1 μg/mL of puromycin for 48 h. Transfection efficiency was monitored by the expression of GFP at 24 h after transfection. Stable knockdown lines for mNT (CISD1) and NAF-1 (CISD2) were obtained by FACS after 3 mo of sorting based on GFP fluorescence. The stable lines isolated were characterized for the level of the NEET proteins in them by protein blots.
For autophagy protein blot analysis, control, mNT−, and NAF-1− cells were grown to full confluence. Medium was aspirated from cultures. Cells were washed with 1× PBS and immediately scraped off the plate into a microcentrifuge tube and processed. Protein gels were loaded based on equal protein levels and analyzed for autophagy markers according to the manufacturer’s instructions (Cell Signaling Technology). For growth curve analysis, 1 mL of cells were seeded in 24-well plates at 20,000 cells/well. Cells were counted using a Moxi Z cell counter and S cassettes (ORFLO Technologies) every day for 6–10 d.
OCR and ECAR were measured using a Seahorse XF24 analyzer with the XF Cell Mito Stress Test Kit and XF Glycolysis Stress Test Kit (Seahorse Bioscience), according to the manufacturer’s instructions. In brief, cells were grown to approximately 70–80% confluence in complete DMEM, trypsinized, and seeded in 100-μL volume at 80,000 cells per well in an XF24 cell culture microplate. Cells were then incubated at 37 °C with 5% CO2 for 3 h. The initial culture medium was exchanged, and the plates were incubated at 37 °C without added CO2 for 1 h before the assay. Plates were then transferred to the XF24 analyzer. The OCR was calculated after sequential additions of oligomycin A, FCCP, antimycin A, and rotenone using an XF Cell Mito Stress Test Kit. The ECAR was calculated after sequential additions of glucose, oligomycin, and 2-DG using an XF Glycolysis Stress Test Kit. All measurements were recorded at set interval time points. All compounds and materials were obtained from Seahorse Bioscience.
Microscopy.
Cells were cultured in glass-bottomed microscope dishes and analyzed using an epi-fluorescent microscope with a confocal (quality equivalent) opti-grid device (Nikon TE 2000 microscope equipped with a thermostatted stage and a Hamamatsu Orca-Era CCD camera) and driven by the Volocity 4 operating system (Improvision), which was used for both image data acquisition and analysis (31). The mitochondrial labile iron pool was measured by exposing cells for 15 min to 0.5 µM RPA, a mitochondrial-specific fluorescent probe that undergoes quenching on binding of iron. Owing to the irreversible iron-binding affinity of RPA, an iron chelator pretreatment can provide an indirect determination of mitochondrial labile iron level. Mitochondrial membrane potential (MMP) was measured microscopically with TMRE using Texas Red (excitation, 543 nm; emission, 633 nm) filter sets. Cell mitochondrial ROS production was determined by incubating cells at 37 °C with 10 μM DHE, which displays blue fluorescence in cell cytoplasm. The oxidized form (ethidium) of the probe was obtained by reaction with ROS, which turns the probe fluorescent red for microscopic analysis (excitation, 518 nm; emission, 605 nm). Pretreatment of cells with the chelator DFP (50–100 μM) was performed at indicated times.
Cell Viability and Metabolic Activity.
Cell viability and metabolic activity were determined as described previously (31). MCF-7 or MDA-231 cells in 12--well culture plates were supplemented with Alamar blue reagent (10% vol/vol), and fluorescence was measured on a plate reader after 1–4 h of incubation at 37 °C (excitation, 530–560 nm; emission, 590 nm).
Electron Microscopy and Morphometric Analysis of Mitochondrial Damage.
Cells were grown on eight-chamber slides to ∼70% confluence and fixed in 2.5% glutaraldeyde and 2% paraformaldehyde in 0.1 M cacodylate buffer (pH 7.4) for 2 h at room temperature. Cells were then rinsed four times for 10 min each in cacodylate buffer, postfixed, and stained with 1% osmium tetroxide and 1.5% potassium ferricyanide in 0.1 M cacodylate buffer for 1 h. Cells were then washed four times in cacodylate buffer, followed by dehydration in increasing concentrations of ethanol consisting of 30%, 50%, 70%, 80%, 90%, and 95% for 15 min at each step, followed by 100% anhydrous ethanol three times for 20 min each. After dehydration, the cells were infiltrated with increasing concentrations of Agar 100 resin in ethanol, consisting of 25%, 50%, 75%, and 100% resin, for 16 h at each step. The cells were then embedded in fresh resin and allowed to polymerize in an oven at 60 °C for 48 h. Embedded cells in blocks were sectioned with a diamond knife on an LKB 3 microtome, and ultrathin sections (80 nm) were collected onto 200-mesh, thin-bar copper grids. The sections on grids were sequentially stained with uranyl acetate and lead citrate for 10 min each and viewed with a Tecnai 12 100-kV transmission electron microscope (FEI) equipped with MegaView II CCD camera and Analysis version 3.0 software (SoftImaging Systems). Quantitative analysis of mitochondrial damage was performed by two investigators, who independently reviewed each enlarged electron microscopy image for the presence of structurally abnormal mitochondria. Structurally abnormal mitochondria were defined operationally as those with loss of cristae and integrity of mitochondrial membrane and the formation of autophagosomes. The number of damaged mitochondria per image was quantified and analyzed by ANOVA using Origin 8.1 (OriginLab Corp.)
Animal Studies.
All surgical and experimental procedures and animal care were performed in accordance and compliance with the policies approved by the Hebrew University administrative panel on laboratory animal care (A. Hochberg laboratory). Animals were kept at the Hebrew University's animal facility, with water and food ad librum. Histopathological examinations of the different tumors were performed in consultation with a trained pathologist. Confluent MDA231 human breast carcinoma cells (control and shRNA stable lines of mNT− and NAF-1−) were trypsinized to a single-cell suspension and resuspended in PBS. Then 2 × 106 cells (in 200 μL volume) were s.c. injected into the backs of 6- to 8-wk-old female nonirradiated CD1 nude mice. At 7 d after cell inoculation, the developing tumors were measured in two dimensions. The mice were divided into different groups of the same size (n = 5) for each cell line injected. Tumor dimensions were measured every week, and tumor areas were calculated according to the formula width × length. The animals were killed at 6.5 wk after the injections, after which the tumors were excised and their ex vivo weight and volume measured. Samples of the tumors were fixed in 4% buffered formaldehyde and processed for histological examination for evidence of necrosis and tumor persistence.
Supplementary Material
Acknowledgments
We thank Dr. Birman T. for her assistance in the pathological work with nude mice. This work was supported by the Israel Science Foundation (ISF 863/09, to R.N.), the University of North Texas College of Arts and Sciences (to R.M.), Cell and Molecular Genetics Training Grant 2T32GM007240-29 (to A.R.C.), the National Institutes of Health (Grants GM54038 and GM101467, to P.A.J.), and The Cancer Prevention and Research Institute of Texas (to J.N.O.). J.N.O. is a Cancer Prevention Research Institute of Texas (CPRIT) Scholar.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313198110/-/DCSupplemental.
References
- 1.Tait SW, Green DR. Mitochondria and cell death: Outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol. 2010;11(9):621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y, Yang JM. Altered energy metabolism in cancer: A unique opportunity for therapeutic intervention. Cancer Biol Ther. 2013;14(2):81–89. doi: 10.4161/cbt.22958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12(10):685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491(7424):364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 5.Shoshan-Barmatz V, Mizrachi D. VDAC1: From structure to cancer therapy. Front Oncol. 2012;2:164. doi: 10.3389/fonc.2012.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fulda S, Galluzzi L, Kroemer G. Targeting mitochondria for cancer therapy. Nat Rev Drug Discov. 2010;9(6):447–464. doi: 10.1038/nrd3137. [DOI] [PubMed] [Google Scholar]
- 7.Torti SV, Torti FM. Iron and cancer: more ore to be mined. Nat Rev Cancer. 2013;13(5):342–355. doi: 10.1038/nrc3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiley SE, Murphy AN, Ross SA, van der Geer P, Dixon JE. MitoNEET is an iron-containing outer mitochondrial membrane protein that regulates oxidative capacity. Proc Natl Acad Sci USA. 2007;104(13):5318–5323. doi: 10.1073/pnas.0701078104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin J, Zhang L, Lai S, Ye K. Structure and molecular evolution of CDGSH iron-sulfur domains. PLoS ONE. 2011;6(9):e24790. doi: 10.1371/journal.pone.0024790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conlan AR, et al. Crystal structure of Miner1: The redox-active 2Fe-2S protein causative in Wolfram syndrome 2. J Mol Biol. 2009;392(1):143–153. doi: 10.1016/j.jmb.2009.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paddock ML, et al. MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone. Proc Natl Acad Sci USA. 2007;104(36):14342–14347. doi: 10.1073/pnas.0707189104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang NC, Nguyen M, Shore GC. BCL2-CISD2: An ER complex at the nexus of autophagy and calcium homeostasis? Autophagy. 2012;8(5):856–857. doi: 10.4161/auto.20054. [DOI] [PubMed] [Google Scholar]
- 13.Chang NC, Nguyen M, Germain M, Shore GC. Antagonism of Beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. EMBO J. 2010;29(3):606–618. doi: 10.1038/emboj.2009.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kusminski CM, et al. MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nat Med. 2012;18(10):1539–1549. doi: 10.1038/nm.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nechushtai R, et al. Characterization of Arabidopsis NEET reveals an ancient role for NEET proteins in iron metabolism. Plant Cell. 2012;24(5):2139–2154. doi: 10.1105/tpc.112.097634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu CY, et al. A persistent level of Cisd2 extends healthy lifespan and delays aging in mice. Hum Mol Genet. 2012;21(18):3956–3968. doi: 10.1093/hmg/dds210. [DOI] [PubMed] [Google Scholar]
- 17.Taminelli GL, et al. CISD1 codifies a mitochondrial protein upregulated by the CFTR channel. Biochem Biophys Res Commun. 2008;365(4):856–862. doi: 10.1016/j.bbrc.2007.11.076. [DOI] [PubMed] [Google Scholar]
- 18.Zuris JA, et al. Facile transfer of [2Fe-2S] clusters from the diabetes drug target mitoNEET to an apo-acceptor protein. Proc Natl Acad Sci USA. 2011;108(32):13047–13052. doi: 10.1073/pnas.1109986108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YF, Wu CY, Kirby R, Kao CH, Tsai TF. A role for the CISD2 gene in lifespan control and human disease. Ann N Y Acad Sci. 2010;1201:58–64. doi: 10.1111/j.1749-6632.2010.05619.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen YF, et al. Cisd2 deficiency drives premature aging and causes mitochondria-mediated defects in mice. Genes Dev. 2009;23(10):1183–1194. doi: 10.1101/gad.1779509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang NC, et al. Bcl-2-associated autophagy regulator Naf-1 required for maintenance of skeletal muscle. Hum Mol Genet. 2012;21(10):2277–2287. doi: 10.1093/hmg/dds048. [DOI] [PubMed] [Google Scholar]
- 22.Wiley SE, et al. Wolfram syndrome protein, Miner1, regulates sulphydryl redox status, the unfolded protein response, and Ca2+ homeostasis. EMBO Mol Med. 2013;5(6):904–918. doi: 10.1002/emmm.201201429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salem AF, Whitaker-Menezes D, Howell A, Sotgia F, Lisanti MP. Mitochondrial biogenesis in epithelial cancer cells promotes breast cancer tumor growth and confers autophagy resistance. Cell Cycle. 2012;11(22):4174–4180. doi: 10.4161/cc.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stelzer G, et al. In silico human genomics with GeneCards. Hum Genom. 2011;5(6):709–717. doi: 10.1186/1479-7364-5-6-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cortes DF, et al. Differential gene expression in normal and transformed human mammary epithelial cells in response to oxidative stress. Free Radic Biol Med. 2011;50(11):1565–1574. doi: 10.1016/j.freeradbiomed.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kongara S, Karantza V. The interplay between autophagy and ROS in tumorigenesis. Front Oncol. 2012;2:171. doi: 10.3389/fonc.2012.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dormandy J, Bhattacharya M, van Troostenburg de Bruyn AR. PROactive investigators Safety and tolerability of pioglitazone in high-risk patients with type 2 diabetes: An overview of data from PROactive. Drug Saf. 2009;32(3):187–202. doi: 10.2165/00002018-200932030-00002. [DOI] [PubMed] [Google Scholar]
- 28.Li MY, et al. Pioglitazone prevents smoking carcinogen-induced lung tumor development in mice. Curr Cancer Drug Targets. 2012;12(6):597–606. doi: 10.2174/156800912801784848. [DOI] [PubMed] [Google Scholar]
- 29.Bosetti C, et al. Cancer risk for patients using thiazolidinediones for type 2 diabetes: A meta-analysis. Oncologist. 2013;18(2):148–156. doi: 10.1634/theoncologist.2012-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anastasiou D. Metformin: A case of divide and conquer. Breast Cancer Res. 2013;15(2):306. doi: 10.1186/bcr3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sohn YS, Mitterstiller AM, Breuer W, Weiss G, Cabantchik ZI. Rescuing iron-overloaded macrophages by conservative relocation of the accumulated metal. Br J Pharmacol. 2011;164(2b):406–418. doi: 10.1111/j.1476-5381.2010.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.