Significance
To be effective, gene therapy vehicles must be able to deliver their therapeutic payloads to widely dispersed tumor lesions after systemic administration. We describe novel nanoparticles that provided a >16,000-fold increase in the ratio of tumor to nontumor cell delivery over conventional formulations. However, only a small fraction of neoplastic cells expressed the transgene, and the extent of delivery varied with the tumor cell line, tumor site, and host mouse strain used. Although our nanoparticles represent a technical advance, they also illustrate the challenges that remain before nonviral gene therapy can be applied to cancer patients.
Abstract
Nanoparticle gene therapy holds great promise for the treatment of malignant disease in light of the large number of potent, tumor-specific therapeutic payloads potentially available for delivery. To be effective, gene therapy vehicles must be able to deliver their therapeutic payloads to metastatic lesions after systemic administration. Here we describe nanoparticles comprised of a core of high molecular weight linear polyethylenimine (LPEI) complexed with DNA and surrounded by a shell of polyethyleneglycol-modified (PEGylated) low molecular weight LPEI. Compared with a state-of-the-art commercially available in vivo gene delivery formulation, i.v. delivery of the core/PEGylated shell (CPS) nanoparticles provided more than a 16,000-fold increase in the ratio of tumor to nontumor transfection. The vast majority of examined liver and lung metastases derived from a colorectal cancer cell line showed transgene expression after i.v. CPS injection in an animal model of metastasis. Histological examination of tissues from transfected mice revealed that the CPS nanoparticles selectively transfected neoplastic cells rather than stromal cells within primary and metastatic tumors. However, only a small fraction of neoplastic cells (<1%) expressed the transgene, and the extent of delivery varied with the tumor cell line, tumor site, and host mouse strain used. Our results demonstrate that these CPS nanoparticles offer substantial advantages over previously described formulations for in vivo nanoparticle gene therapeutics. At the same time, they illustrate that major increases in the effectiveness of such approaches are needed for utility in patients with metastatic cancer.
Research on cancer genomes combined with functional and biochemical studies has led to the identification of signal transduction pathways whose component genes are altered in human tumors (1). Extraordinary efforts have been devoted to developing therapeutics that selectively target these altered pathways. These efforts have been remarkably successful, resulting in a new generation of agents that can shrink tumors without causing unacceptable side effects (reviewed in refs. 2–4). The effects of these targeted agents are generally short-lived, however, as a result of the inevitable outgrowth of cells with mutations that confer resistance (5–7). This phenomenon has spurred efforts to identify complementary approaches that are less likely to be subverted by mutations present in the tumor cells before therapy.
One of the most promising approaches to achieving this objective involves the exploitation of the abnormal vasculature in tumors. To maintain their high relative growth rate, tumors must recruit blood vessels, both afferent and efferent, to supply oxygen and nutrients. But given the abnormal organization of cancer cells with respect to the underlying stroma, the recruited vasculature is quite unlike that present in normal tissues (8). This tumor vasculature is characterized by chaotic organization, leakiness, and poor lymphatic drainage; thus, agents that target the abnormal vasculature should be particularly powerful weapons against cancers.
Nanoparticles represent one class of such weapons. When administered systemically, nanoparticles with unique size and charge characteristics may selectively accumulate in solid tumors, because they can more easily extravasate through the leaky tumor endothelium and less easily vacate through the tumor’s poorly developed lymphatic system (9, 10). This accumulation is known as the enhanced permeability and retention (EPR) effect (11).
Several types of nanoparticles have been developed to exploit the EPR effect in experimental model systems, some of which have demonstrated clinical value (12, 13). Nanoparticles consisting of nucleic acids complexed with cationic polymers also have been extensively evaluated for therapeutic purposes (reviewed in ref. 14). DNA-containing nanoparticles offer a wealth of therapeutic possibilities because they can encode highly toxic genes whose tumor-specific expression can be tailored via engineering of the nanoparticle itself or the molecular payload.
Although this potential has long been recognized, and much work has been done to optimize nanoparticle gene delivery formulations in vitro, relatively few studies have attempted to systemically deliver genes to experimental tumors in vivo (see refs. 15–17 for examples of in vivo work). In vivo studies have often used s.c. rather than internal tumors, global measures of transfection that can be achieved with whole-body imaging or whole-tissue lysates, and intratumoral rather than systemic delivery. However, for clinical relevance, it is critical not only to test systemic delivery of nanoparticle gene therapy formulations to internal tumors, but also to evaluate the nature and number of cells within tumors that express transfected reporters. Ultimately, to be clinically viable, gene delivery vehicles must be able to potently and selectively transfect metastatic tumor cells. Given that primary tumors often can be surgically excised, the treatment of metastatic disease is the most important clinical setting for nanoparticle therapeutics.
In the present study, we attempted to improve the efficiency and specificity of nanoparticle gene therapeutics, focusing on global and local measures of potency and specificity in internal tumors and their metastases. We also identified some bottlenecks limiting translation of these treatments to humans.
Results
Establishing a Benchmark in Internal Tumors.
Evaluation of in vivo-jetPEI in primary intrasplenic tumors.
Of the various transfection agents commercially available, only a few are able to safely and efficiently transfect cells in vivo. For systemic in vivo delivery, transfection agents based on polyethylenimine (PEI) have shown the most promise (18, 19). One of the most effective formulations, in vivo-jetPEI (Polyplus Transfection), has been evaluated for gene delivery in a number of cancer models in vivo (20–26), although rarely via systemic injection (27–29), as well as in several clinical trials for heart disease (30), HIV (31), and cancer (32). Because s.c. tumor models might not faithfully reflect the architecture of internal tumors, we elected to use a model established by intrasplenic injection of the HCT116 line of human colorectal cancer cells in NOD-SCID-IL-2Rγ–deficient mice (NOG mice) (33). In addition to primary tumors that formed in the spleen in this model, multiple metastases developed in the lungs and liver within 4 wk of tumor cell injection.
To monitor the development of the primary and metastatic lesions, we used HCT116-luc2, a line of HCT116 cells that constitutively express firefly luciferase (FLuc). We then evaluated the ability of nanoparticles based on in vivo-jetPEI to systemically deliver Renilla luciferase (RLuc)-encoding plasmids to intrasplenic tumors. We focused first on primary tumors to facilitate high-throughput formulation screening coupled with high confidence in tumor specificity. (Well-circumscribed primary tumors could be easily dissected away from normal spleens.) After i.v. injection of in vivo-jetPEI complexed with the RLuc plasmid, RLuc expression was observed in the primary tumor in the spleen at 10-fold higher levels than the background signals in intrasplenic tumors in mice not injected with RLuc plasmid (Fig. S1A). However, substantial RLuc activity was also evident in the spleen, liver, and lungs of nontumor-bearing injected mice, with particularly high activity in normal lungs (Fig. S1B).
PEGylation of in vivo-jetPEI.
High transfection of normal tissues after i.v. injection of in vivo-jetPEI is unacceptable if toxic genes are used for cancer gene therapy. Delivery of nanoparticles to normal tissues can be abrogated through surface modification of such vehicles with polyethylene glycol (PEG) to increase particle circulating half-life; to decrease uptake by phagocytes such as those present in the lungs, spleen, and liver (34); and, in the case of positively charged nanoparticles, to prevent aggregation in blood, which can lead to nonspecific accumulation in first-pass organs, such as the lungs (35). In an attempt to minimize the transfection of normal tissues by in vivo-jetPEI, we conjugated 5-kDa PEG (PEG5) to nitrogen groups present in the polymer after it was complexed with DNA. We found that PEGylation at a 10:1 weight ratio of PEG5 to in vivo-jetPEI did not impact tumor transfection, but did substantially reduce lung transfection (Fig. S1 C and D).
Improvements Leading to Core/PEGylated Shell Formulation.
LPEI88 as an equivalent to in vivo-jetPEI.
Although in vivo-jetPEI is thought to be a linear PEI (LPEI) derivative of ∼22 kDa, its precise composition is proprietary. Thus, to facilitate further optimization, we synthesized 88-kDa LPEI (LPEI88) as an in vivo-jetPEI substitute amenable to in-house characterization and modification. Sources and synthetic methods are specified in SI Materials and Methods. We found LPEI88 to be similar to in vivo-jetPEI in terms of molecular weight, toxicity, and transfection efficiency (Table S1 and Fig. S2).
Optimizing LPEI molecular weight.
We next sought to improve the results obtained with in vivo-jetPEI and LPEI88, aiming to reduce toxicity, preserve low levels of off-target lung transfection, and increase transfection of tumors. We first focused on toxicity by comparing maximum tolerated doses (MTDs) after i.v. injection of LPEIs of varying molecular weights, ranging from 2.5 kDa to 220 kDa, with 2.5-kDa LPEI (LPEI2.5) being the least toxic LPEI (Table S1). However, when LPEIs were complexed with DNA to form nanoparticles, the LPEI dose at the nanoparticle MTD varied substantially based on nanoparticle formulation (Table S2). For example, although the MTD of LPEI88 unbound to DNA was 2.4 mg/kg, its tolerable dose was as high as 9.9 mg/kg when administered as a nanoparticle formed at a nitrogen:phosphate (N:P) ratio of 2.5. Tolerable DNA doses also ranged widely based on N:P ratio, from as high as 30 mg/kg for N:P 2.5 to only 2.5 mg/kg for N:P 7.2.
Nanoparticle cores and shells.
The relatively low toxicity (i.e., high MTD) achieved with smaller N:P ratios can be explained by the salt-bridge interactions between PEI nitrogens and DNA phosphates that drive nanoparticle formation and the toxicity of free PEI (Table S1). At physiological pH, approximately one of every two to three nitrogen atoms in a single PEI chain is positively charged, and two to three nitrogen atoms are required to saturate each phosphate in the DNA (18). Thus, only at N:P ratios >2 are positively charged nanoparticles containing excess PEI formed.
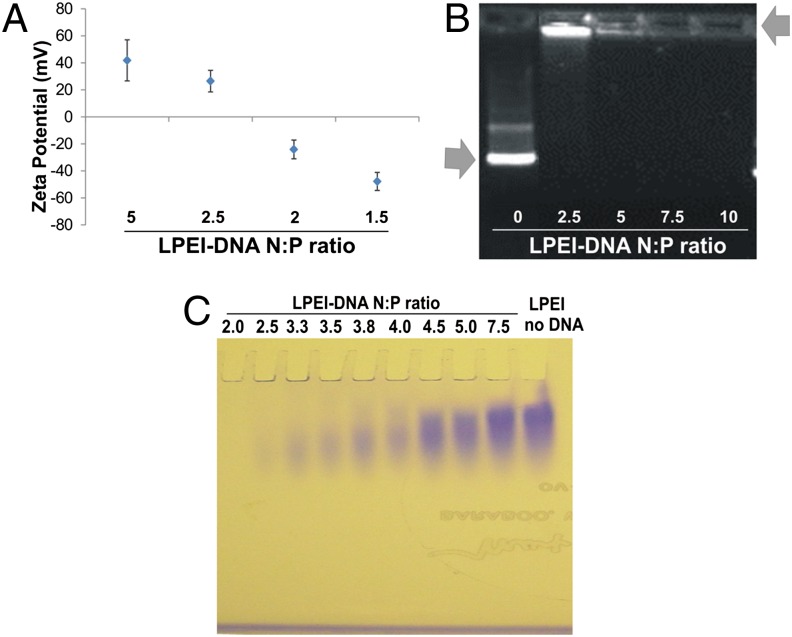
We experimentally confirmed this expectation by measuring the differences in zeta potential of nanoparticles assembled at various N:P ratios using a ZetaSizer, and found that the zeta potential reversed polarity from negative to positive at N:P ratios between 2 and 2.5 (Fig. 1A). DNA was also precluded from entering agarose gels when complexed with LPEI within this N:P range (Fig. 1B). Thus, an N:P ratio of 2–2.5 represents a critical “core” threshold at which PEI has saturated DNA to form a stable nanoparticle. At higher N:P ratios, PEI can be considered an excess “shell,” reversibly interacting with the core (Fig. 1A and Table S3).
Fig. 1.
Nanoparticle core and shell characterization. (A) Zeta potentials of LPEI88-DNA nanoparticles in double-distilled water (ddH2O) at various N:P ratios. Data represent means and standard deviation (SD) of more than two independent measurements. (B) DNA gel retardation assay. Nanoparticles were formed at various N:P ratios, and electrophoresed at 100 V for 40 min in a 0.5% agarose gel. Arrows point to free DNA and LPEI-complexed DNA. (C) PEI gel electrophoresis of LPEI88-DNA nanoparticles at various N:P ratios and free LPEI88. The LPEI that is not tightly bound to the DNA migrates into the gel and is stained, while the LPEI that is bound to the DNA remains in the well.
Further evidence supporting this conclusion was provided by PEI gel analysis. PEI can be evaluated via electrophoresis in polyacrylamide gels when run toward a negative electrode and stained with Coomassie blue. In such an assay, PEI strongly bound to DNA remained in the well (and was unstained), whereas loosely bound PEI did enter the gel and was stained with Coomassie blue (Fig. 1C). The extra shell, although facilitating transfection (Fig. 2A), is likely responsible for the extra toxicity observed with high N:P ratio nanoparticles (Table S2) (36).
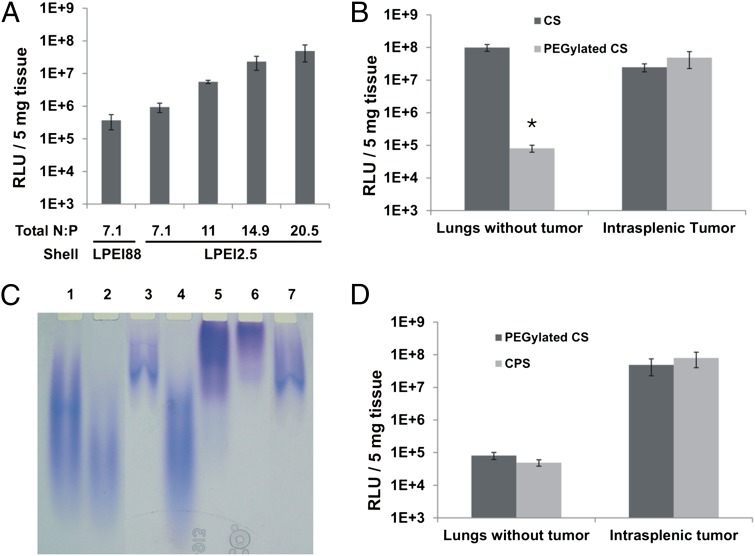
Fig. 2.
CPS formulation. (A) Relative Light Units (RLUs) generated by RLuc (RLuc RLUs) in intrasplenic tumors after transfection with nanoparticles containing LPEI88 as the shell or increasing amounts of LPEI2.5 as the shell. (B) RLuc RLUs in lungs or intrasplenic tumors after treatment with CS nanoparticles or PEGylated CS nanoparticles. (C) Gel electrophoretic analysis of PEGylation of nanoparticles generated with LPEI88 in both core and shell. Lane 1, free non-PEGylated LPEI; lane 2, LPEI purified from the shell of non-PEGylated nanoparticles; lane 3, LPEI purified from the core of non-PEGylated nanoparticles; note that the larger polymer molecules in the heterodisperse LPEI preferentially interacted with DNA and were therefore found in the core; lane 4, free LPEI incubated with non-reactive PEG as a control; lane 5, PEGylated free LPEI; lane 6, LPEI purified from the shell of PEGylated nanoparticles; lane 7, LPEI purified from the core of PEGylated nanoparticles (D) RLuc RLUs in lungs and intrasplenic tumors after transfection with PEGylated CS or CPS nanoparticles. Means and standard deviations of data collected from at least two mice per group are illustrated, * P <0.05.
Core/PEGylated shell formulation.
Given that the shell dose mediates both toxicity and transfection efficiency, we wondered whether replacing the LPEI88 in the shell with a less-toxic LPEI might reduce toxicity but preserve in vivo transfectability. Composite nanoparticles containing two types of cationic polymers described in the literature (37, 38), along with the data presented in Tables S1 and S2, inspired us to synthesize composite LPEI-based nanoparticles that take advantage of both the high transfectability of LPEI88 and the low toxicity of LPEI2.5. Thus, a nanoparticle core was formed by incubating DNA with LPEI88, and a shell around this core was formed by incubating this core with excess LPEI2.5. We found that a shell of LPEI2.5 was as effective as LPEI88 at conferring transfection efficiency (Fig. 2A). Furthermore, substitution of LPEI2.5 for LPEI88 in the shell allowed us to increase the dose, thereby increase transfection efficiency without increasing toxicity (Table S1 and Fig. 2A). The optimal nanoparticle, in terms of toxicity and transfection efficiency, contained a core of LPEI88 complexed with DNA at N:P ratio of 2.5 and a shell of LPEI2.5, yielding a total N:P ratio of 20.5 (Fig. 2A). We hereafter termed the nanoparticles with this specific formulation “CS” nanoparticles.
Based on the improved specificity afforded by PEG modification of in vivo-jetPEI (Fig. S1 C and D), we attempted to modify the CS nanoparticles with PEG5. We first PEGylated the fully assembled CS nanoparticle formulation and found that, like PEGylated in vivo-jetPEI, the PEGylated CS nanoparticle conferred stability in PBS and reduced lung transfection while preserving high tumor transfection (Table S3 and Fig. 2B). However, PEGylation performed on the fully formed nanoparticles at neutral pH required large amounts of reactive PEG and was difficult to control, characterize, and reproduce. Furthermore, based on retardation of PEGylated LPEI in PEI gel electrophoresis, we found that the majority of the conjugated PEG resided on the shell LPEI when the preformed nanoparticles were PEGylated (Fig. 2C). Thus, we PEGylated only the shell and removed the excess nonconjugated PEG before its addition to the core. The actual PEG5:LPEI2.5 ratios in the final preparations were determined by 1H-NMR. We found that nanoparticles containing a shell made from a PEG5-LPEI2.5 at a PEG:LPEI weight ratio of ∼1:1 were functionally equivalent to preformed nanoparticles (i.e., CS nanoparticles) PEGylated at a PEG5:LPEI2.5 weight ratio of 10:1 (Fig. 2D).
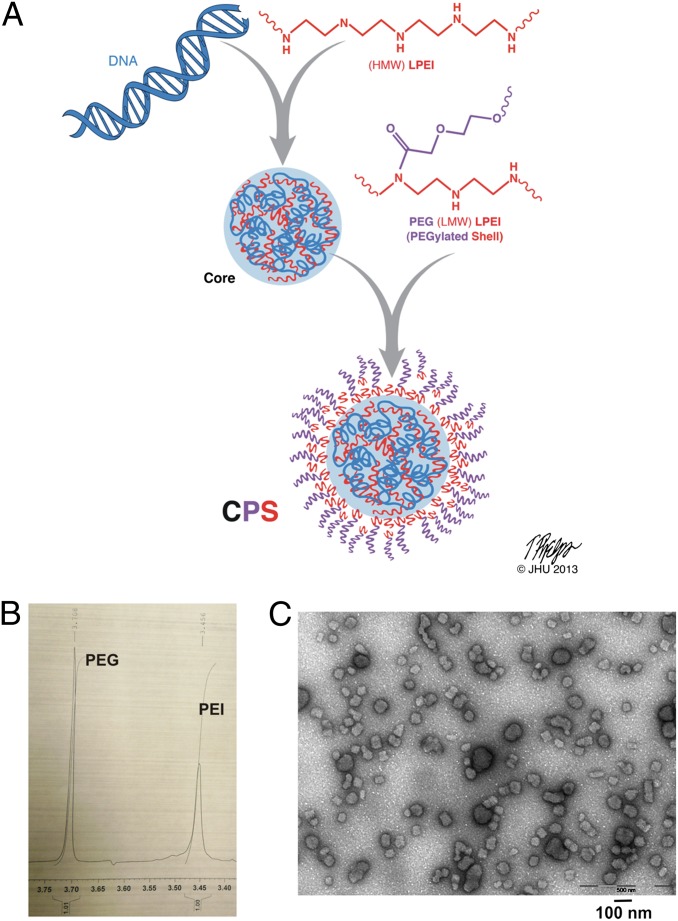
The optimized core/PEGylated-shell (CPS) nanoparticles (Fig. 3A) had a total N:P ratio of 20.5 (nitrogens in the core plus shell to phosphates in the core) and a PEG5:LPEI2.5 weight ratio of 1:1 as measured by 1H-NMR (Fig. 3B). We determined the composition of the CPS nanoparticles on the basis of input amounts of DNA, LPEI88, and PEG5-LPEI2.5, with particle numbers assessed by nanoparticle tracking analysis (NTA) on a NanoSight instrument (39, 40). We found that the average nanoparticle contained ∼60 DNA molecules bound to 600 LPEI88 molecules in the core plus 150,000 LPEI2.5 molecules and 76,0000 PEG5 molecules in the shell. The CPS nanoparticles averaged 70.8 nm in diameter, with a mean polydispersity index of 0.152 and a mean zeta potential of 35.2 ± 7.52 mV as measured by dynamic light scattering (DLS) (Table S3).
Fig. 3.
CPS nanoparticle characterization. (A) Proposed structure of a CPS nanoparticle. HMW, high molecular weight; LMW, low molecular weight. (B) 1H NMR of PEG5-LPEI2.5 as the shell in CPS. The chemical shifts corresponding to PEG and PEI are indicated. (C) Transmission electron microscopy of CPS.
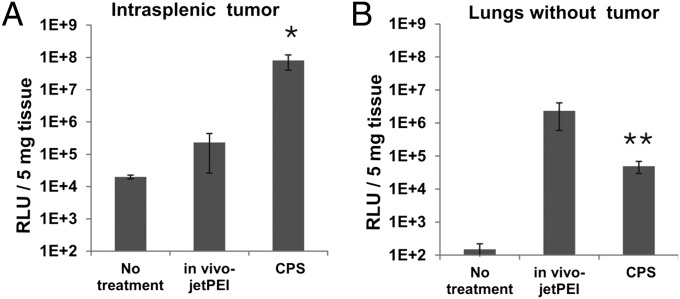
Unlike nanoparticles without a PEGylated shell, CPS nanoparticles are highly stable in physiological salt solutions. Electron microscopy studies revealed two populations of nanoparticles in CPS preparations, a dominant population of ∼40 nm and a minor population of ∼80 nm (Fig. 3C). The latter population could be removed by pelleting cores via centrifugation before CPS formulation. This procedure yielded more uniform 40-nm CPS nanoparticles (Fig. S3 A and B); however, the 40-nm CPS nanoparticles did not enhance transfection of intrasplenic tumors (Fig. S3C) or reduce transfection of normal tissues (Fig. S3D). A quantitative comparison of in vivo-jetPEI with the optimized CPS nanoparticle is shown in Fig. 4A; the improvement in transfection to the intrasplenic tumors was ∼300-fold. When coupled with the reduced transfection of normal lungs provided by the CPS nanoparticles (Fig. 4B), the specificity of DNA delivery was enhanced by ∼16,000-fold.
Fig. 4.
Comparison of CPS nanoparticles with the benchmark. (A) RLuc RLUs in intrasplenic tumors after transfection with CPS or in vivo-jetPEI nanoparticles. (B) RLuc RLUs in normal lungs after transfection with CPS or in vivo-jetPEI nanoparticles. Data are means ± SD of values collected from at least two mice per group. *P < 0.05; **P < 0.01.
As part of the optimization process, we evaluated other components as shells. These included anionic and cationic lipids alone or lipids, peptides, and other functional groups preconjugated to LPEI (Fig. S4 A and B). We also combined CPS formulations with agents previously reported to enhance transfection or tumor delivery by other nonviral vehicles (Fig. S4 C and D). None of these LPEI substitutes or combination agents strikingly enhanced CPS-based gene transfection in vivo under the evaluated conditions.
Localizing and Counting Transfected Intrasplenic Tumor Cells.
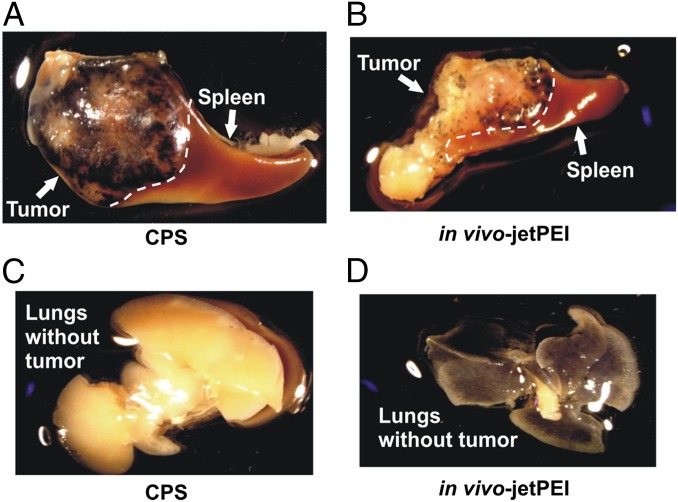
The dramatic improvement in potency and specificity of CPS nanoparticles was further confirmed via delivery of a β-galactosidase reporter plasmid. In CPS-treated NOG mice with intrasplenic HCT116 tumors, β-galactosidase activity was present in the primary tumors within the spleen, but not in the adjacent normal spleen (Fig. 5A). The level of β-galactosidase activity in the tumors was remarkably enhanced with the CPS nanoparticles compared with that produced by in vivo-jetPEI nanoparticles (compare Fig. 5A and Fig. 5B). Furthermore, β-galactosidase activity was absent from normal lungs after transfection of the CPS nanoparticles (Fig. 5C), whereas it was evident throughout the normal lungs with in vivo-jetPEI (Fig. 5D).
Fig. 5.
Gross visualization of in vivo transfection. Organs from animals transfected with nanoparticles bearing the β-galactosidase gene are shown. The dark-blue 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) product accumulates in transfected regions. (A) Intrasplenic tumors transfected with CPS and adjacent normal spleen. (B) Intrasplenic tumors transfected with in vivo-JetPEI and adjacent normal spleen. (C) Normal lungs after transfection with CPS. (D) Normal lungs after transfection with in vivo-JetPEI.
To confirm tumor specificity at the single cell level and determine the fraction of transfected cells, we generated intrasplenic tumors with HCT116 cells engineered to overexpress EGFP (SI Materials and Methods). After injection of a plasmid containing a red fluorescent protein (RFP) reporter plasmid incorporated into CPS nanoparticles, the tumors were harvested, and cells were dissociated and examined under a confocal microscope. On average, 99% of the individual cells emitting orange fluorescence from the transfection of RFP also emitted green fluorescence from the EGFP (Fig. S5A). This experiment demonstrated that tumor cells, and not stromal cells such as macrophages, had been successfully transfected in vivo. Quantification of cells dissociated from tumor-containing spleens showed that ∼8,400 of 1,000,000 tumor cells (0.84%) emitted orange fluorescence (Fig. S5B).
Transfection of Metastatic Tumors.
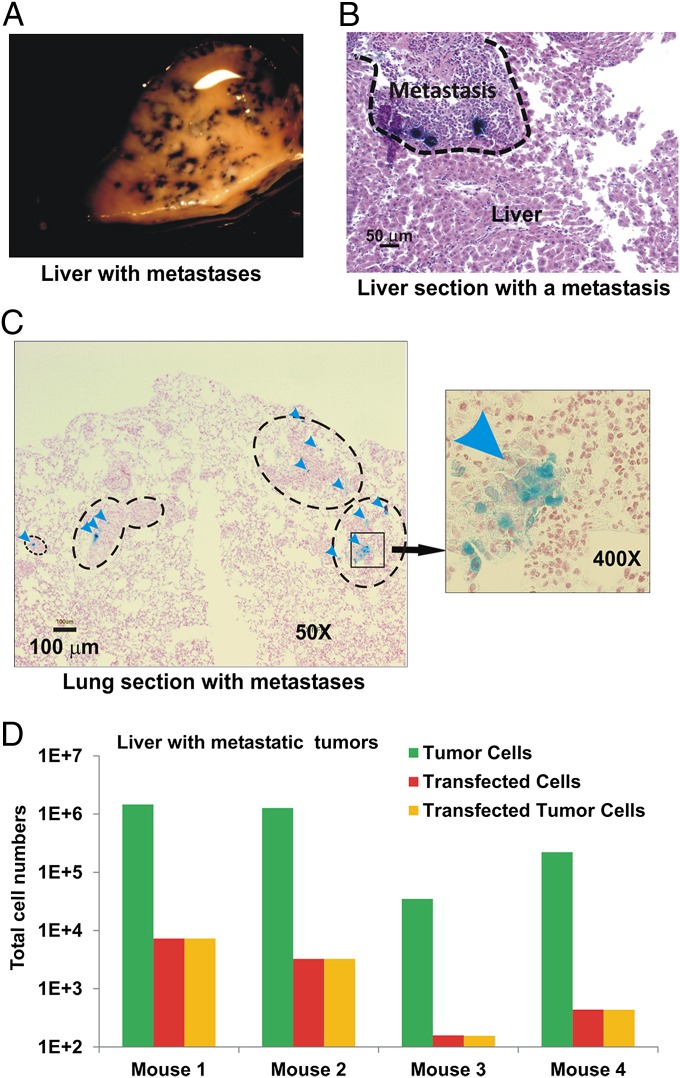
We used the same assays to evaluate the transfection of metastatic liver and lung lesions. After β-galactosidase gene delivery with CPS nanoparticles, every metastatic lesion in the liver was observed to express β-galactosidase under a dissecting microscope (Fig. 6A). After sectioning, β-galactosidase activity was observed in a small proportion of cells within liver and lung metastases (Fig. 6 B and C). The lung sections revealed that most metastatic lesions, no matter how small, contained at least some cells expressing β-galactosidase. Further, β-galactosidase activity in the liver and lung sections was confined to tumor cells in metastases. Finally, using the RFP reporter system, we found that on average, 0.35% of metastatic cells in the liver were transfected (Fig. 6D).
Fig. 6.
Transfection of metastases with CPS nanoparticles. NOG mice bearing HCT116-Luc2 (A–C) or HCT116-EGFP (D) tumors were transfected with CPS containing β-galactosidase (A–C) or RFP (D) plasmids. (A) Selected view of the transfected metastases in liver. The dark-blue X-gal product accumulates in transfected regions. (B) H&E-stained section of the transfected liver containing a cluster of the transfected metastases. Dark-blue X-gal product can be seen in the transfected tumor cells. (C) Nuclear fast red-stained section of lung containing metastases. Representative metastases and transfected cells (arrowheads) are indicated. (D) Numbers of tumor cells, transfected cells, and transfected tumor cells in the livers with metastases from four individual mice as determined by confocal scanning of dissociated cells.
Variation Among Tumor Models.
To test the generality of these results, we used several other experimental tumor models, including human tumors implanted in NOG mice and syngeneic mouse tumors in immunocompetent mice (Fig. S6). Intrasplenic tumors derived from HepG2 and Neuro2A cells in NOG mice were transfected ∼10-fold less efficiently than HCT116 cells, whereas other cancer cell lines were transfected much less effectively (Fig. S6A). Through the analysis of the same cell lines transfected with the same CPS nanoparticles in vitro, we determined that there was no tight correlation between in vitro and in vivo results (Fig. S6B); for example, HepG2 cells were transfected poorly in vitro but efficiently in vivo compared with HCT116 cells (Fig. S6 A and B).
The effectiveness of transfection also varied with the tumor site and mouse strain used. For example, primary s.c. tumors of HCT116 cells in NOG mice were transfected 20-fold less effectively than primary intrasplenic tumors in the same mouse or in other immunocompromised strains of mice (Fig. S6C). Moreover, tumor transfection efficiency was higher in immunodeficient mice compared with immunocompetent mice; for example, intrasplenic Neuro2A and CT26 tumors in severely immunocompromised NOG mice were transfected ∼700- and 300-fold more effeciently than the same tumors in their syngeneic, immunocompetent hosts (compare data in Fig. S6D with that in Fig. S6A).
Discussion
The CPS nanoparticle formulation described in this work has several notable properties. Chief among these are the low level of transfection of normal reticuloendothelial cells in the lung, liver, and spleen, coupled with a high level of transfection of both primary and metastatic tumors in mouse models. In the HCT116 cell model, the majority of metastatic lesions could be transfected, even metastases containing fewer than several hundred cells (Fig. 6). The efficiency of tumor transfection was much higher than achieved with the most commonly used commercial formulation, and the unwanted transfection of normal tissues was much lower. Furthermore, only the neoplastic cells, and not resident inflammatory or stromal cells within tumors, were transfected by the CPS nanoparticles. This is remarkable, considering that these nanoparticles do not contain any specific ligands that would facilitate their binding to tumor antigens.
The specificity of tumor transfection achieved with CPS nanoparticles apparently arises from altered tissue availability (e.g., EPR-like effects) (Fig. 2B and Fig. S1D). That specificity is innate to the CPS formulation and may provide a significant advantage over nanoparticle formulations dependent on tissue-specific promoters in their plasmid payloads. The latter formulations can be internalized by off-target cells and can cause toxicity if the promoters are even slightly leaky.
Although the CPS nanoparticles represent a technological advance, our findings also demonstrate that this nanoparticle gene transfection technology is far from being clinically applicable. The actual fraction of transfected cells in primary tumors or metastatic lesions was low, always <1% (Fig. 6D and Fig. S5B). Why only a small fraction of tumor cells are transfected in vivo is unknown, although it is possible that this number is an underestimate, given two factors. First, sensitivity limitations of the detection methods used might have prevented the recognition of tumor cells transfected at low levels. Second, it is possible that high local concentrations of CPS nanoparticles are toxic, and that some of the most highly transfected cells were destroyed. Regardless, the most conservative interpretation of our data are that even after extensive optimization, LPEI-based nanoparticles cannot transfect a major fraction of tumor cells in experimental models. This result is consistent with the general inability of nanoparticles of 40 nm to penetrate very deeply into the tumor parenchyma after extravasation from the leaky tumor endothelium (41). This low fractional transfection poses a major challenge for eradicating tumors via gene therapy. Gene cargos that impart extraordinary bystander effects represent a potential strategy for circumventing this challenge in the future.
Another vexing challenge is highlighted by the results obtained in different tumor models in vivo (Fig. S6). It is evident from this survey that tumors vary remarkably—by orders of magnitude—in the ability to be transfected in vivo. Their ability was not highly related to the cells’ ability to be transfected in vitro, underscoring the importance of optimization of formulations in animal models rather than simply in vitro. No known theory compellingly explains the differences that we observed among cell lines, among tumor sites within the same mouse, and among different mouse strains. If we assume that the tumor specificity of transfection with CPS nanoparticles was related entirely to EPR effects, then the differences in in vivo transfection must be attributed to differences in vascular systems that are both host- and tumor cell-dependent. One potential clue was provided by the observation that tumors in immunocompetent mice were more difficult to transfect compared with identical tumors in immunodeficient mice (Fig. S6). This finding suggests that the innate immune system plays a role in limiting gene transfection, which is unfortunate, given that humans with cancer are generally immunocompetent.
The variable transfection efficiency with different experimental tumor systems that we observed has important implications not only for gene therapy, but also for any cancer therapy that uses nanoparticles or targets the vasculature. An abnormal vasculature is always found in tumors and, coupled with advances in nanoparticle-based technologies, offers extraordinary therapeutic potential. However, this study emphasizes that much more basic research into the factors governing the interface between tumor cells and this vasculature is needed before this potential is likely to be realized in practice.
Materials and Methods
Materials and methods used in polymer synthesis and characterization, plasmid preparation, formulation and analysis of nanoparticles, and in vitro and in vivo transfections are detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Tim Phelps for the artwork; Hai-Quan Mao, Yong Ren, George Sgouros, and Hong Song for sharing their laboratory instruments; E. Latice Watson and Qiang Liu for technical support; and Yuan Qiao, Surojit Sur, and Ian Cheong for inspiring discussions. This project was supported by the Virginia and D. K. Ludwig Fund for Cancer Research and National Institutes of Health Grants CA062924 and CA043460.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313330110/-/DCSupplemental.
References
- 1.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papadopoulos N, Kinzler KW, Vogelstein B. The role of companion diagnostics in the development and use of mutation-targeted cancer therapies. Nat Biotechnol. 2006;24(8):985–995. doi: 10.1038/nbt1234. [DOI] [PubMed] [Google Scholar]
- 3.Irshad S, Ashworth A, Tutt A. Therapeutic potential of PARP inhibitors for metastatic breast cancer. Expert Rev Anticancer Ther. 2011;11(8):1243–1251. doi: 10.1586/era.11.52. [DOI] [PubMed] [Google Scholar]
- 4.Chapman PB, et al. BRIM-3 Study Group Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sequist LV, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3(75):75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson TR, et al. Widespread potential for growth-factor–driven resistance to anticancer kinase inhibitors. Nature. 2012;487(7408):505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diaz LA, Jr, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486(7404):537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain RK, Duda DG. Vascular and interstitial biology of tumors. In: Abeloff MD, Armitage JO, Niederhuber JE, Kastan MB, McKenna WG, editors. Abeloff’s Clinical Oncology. 4th Ed. Vol 1. Philadelphia: Churchill Livingstone Elsevier; 2008. pp. 105–124. [Google Scholar]
- 9.Yuan F, et al. Vascular permeability in a human tumor xenograft: Molecular size dependence and cutoff size. Cancer Res. 1995;55(17):3752–3756. [PubMed] [Google Scholar]
- 10.Dellian M, Yuan F, Trubetskoy VS, Torchilin VP, Jain RK. Vascular permeability in a human tumour xenograft: Molecular charge dependence. Br J Cancer. 2000;82(9):1513–1518. doi: 10.1054/bjoc.1999.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maeda H, Fang J, Inutsuka T, Kitamoto Y. Vascular permeability enhancement in solid tumor: Various factors, mechanisms involved and its implications. Int Immunopharmacol. 2003;3(3):319–328. doi: 10.1016/S1567-5769(02)00271-0. [DOI] [PubMed] [Google Scholar]
- 12.Green MR, et al. Abraxane, a novel Cremophor-free, albumin-bound particle form of paclitaxel for the treatment of advanced non–small-cell lung cancer. Ann Oncol. 2006;17(8):1263–1268. doi: 10.1093/annonc/mdl104. [DOI] [PubMed] [Google Scholar]
- 13.Barenholz Y. Doxil®—the first FDA-approved nano-drug: Lessons learned. J Control Release. 2012;160(2):117–134. doi: 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 14.Davis ME. Non-viral gene delivery systems. Curr Opin Biotechnol. 2002;13(2):128–131. doi: 10.1016/s0958-1669(02)00294-x. [DOI] [PubMed] [Google Scholar]
- 15.Ogris M, Wagner E. Targeting tumors with non-viral gene delivery systems. Drug Discov Today. 2002;7(8):479–485. doi: 10.1016/s1359-6446(02)02243-2. [DOI] [PubMed] [Google Scholar]
- 16.Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nat Rev Drug Discov. 2005;4(7):581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 17.Park TG, Jeong JH, Kim SW. Current status of polymeric gene delivery systems. Adv Drug Deliv Rev. 2006;58(4):467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Boussif O, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc Natl Acad Sci USA. 1995;92(16):7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh YK, et al. Prolonged organ retention and safety of plasmid DNA administered in polyethylenimine complexes. Gene Ther. 2001;8(20):1587–1592. doi: 10.1038/sj.gt.3301516. [DOI] [PubMed] [Google Scholar]
- 20.Lavergne E, et al. Fractalkine mediates natural killer-dependent antitumor responses in vivo. Cancer Res. 2003;63(21):7468–7474. [PubMed] [Google Scholar]
- 21.Lavergne E, et al. Intratumoral CC chemokine ligand 5 overexpression delays tumor growth and increases tumor cell infiltration. J Immunol. 2004;173(6):3755–3762. doi: 10.4049/jimmunol.173.6.3755. [DOI] [PubMed] [Google Scholar]
- 22.Hua H, et al. Inhibition of tumorigenesis by intratumoral delivery of the circadian gene mPer2 in C57BL/6 mice. Cancer Gene Ther. 2007;14(9):815–818. doi: 10.1038/sj.cgt.7701061. [DOI] [PubMed] [Google Scholar]
- 23.Jeudy G, et al. Polyethylenimine-mediated in vivo gene transfer of a transmembrane superantigen fusion construct inhibits B16 murine melanoma growth. Cancer Gene Ther. 2008;15(11):742–749. doi: 10.1038/cgt.2008.42. [DOI] [PubMed] [Google Scholar]
- 24.Kang Y, et al. Tumor-directed gene therapy in mice using a composite nonviral gene delivery system consisting of the piggyBac transposon and polyethylenimine. BMC Cancer. 2009;9(1):126. doi: 10.1186/1471-2407-9-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amit D, Hochberg A. Development of targeted therapy for bladder cancer mediated by a double promoter plasmid expressing diphtheria toxin under the control of H19 and IGF2-P4 regulatory sequences. J Transl Med. 2010;8(1):134. doi: 10.1186/1479-5876-8-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hine CM, Seluanov A, Gorbunova V. Rad51 promoter-targeted gene therapy is effective for in vivo visualization and treatment of cancer. Mol Ther. 2012;20(2):347–355. doi: 10.1038/mt.2011.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caldas H, Jaynes FO, Boyer MW, Hammond S, Altura RA. Survivin and Granzyme B-induced apoptosis, a novel anticancer therapy. Mol Cancer Ther. 2006;5(3):693–703. doi: 10.1158/1535-7163.MCT-05-0423. [DOI] [PubMed] [Google Scholar]
- 28.Chumakova OV, et al. Composition of PLGA and PEI/DNA nanoparticles improves ultrasound-mediated gene delivery in solid tumors in vivo. Cancer Lett. 2008;261(2):215–225. doi: 10.1016/j.canlet.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 29.Bhang HE, Gabrielson KL, Laterra J, Fisher PB, Pomper MG. Tumor-specific imaging through progression elevated gene-3 promoter-driven gene expression. Nat Med. 2011;17(1):123–129. doi: 10.1038/nm.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taljaard M, et al. Rationale and design of Enhanced Angiogenic Cell Therapy in Acute Myocardial Infarction (ENACT-AMI): The first randomized placebo-controlled trial of enhanced progenitor cell therapy for acute myocardial infarction. Am Heart J. 2010;159(3):354–360. doi: 10.1016/j.ahj.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 31.Lisziewicz J, et al. Single DermaVir immunization: Dose-dependent expansion of precursor/memory T cells against all HIV antigens in HIV-1–infected individuals. PLoS ONE. 2012;7(5):e35416. doi: 10.1371/journal.pone.0035416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sidi AA, et al. Phase I/II marker lesion study of intravesical BC-819 DNA plasmid in H19 overexpressing superficial bladder cancer refractory to bacillus Calmette–Guerin. J Urol. 2008;180(6):2379–2383. doi: 10.1016/j.juro.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 33.Ericson K, et al. Genetic inactivation of AKT1, AKT2, and PDPK1 in human colorectal cancer cells clarifies their roles in tumor growth regulation. Proc Natl Acad Sci USA. 2010;107(6):2598–2603. doi: 10.1073/pnas.0914018107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris JM, Chess RB. Effect of pegylation on pharmaceuticals. Nat Rev Drug Discov. 2003;2(3):214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- 35.Goula D, et al. Polyethylenimine-based intravenous delivery of transgenes to mouse lung. Gene Ther. 1998;5(9):1291–1295. doi: 10.1038/sj.gt.3300717. [DOI] [PubMed] [Google Scholar]
- 36.Boeckle S, et al. Purification of polyethylenimine polyplexes highlights the role of free polycations in gene transfer. J Gene Med. 2004;6(10):1102–1111. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- 37.Zhou J, et al. Biodegradable poly(amine-co-ester) terpolymers for targeted gene delivery. Nat Mater. 2012;11(1):82–90. doi: 10.1038/nmat3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashley CE, et al. The targeted delivery of multicomponent cargos to cancer cells by nanoporous particle-supported lipid bilayers. Nat Mater. 2011;10(5):389–397. doi: 10.1038/nmat2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Filipe V, Hawe A, Jiskoot W. Critical evaluation of nanoparticle tracking analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm Res. 2010;27(5):796–810. doi: 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhise NS, Shmueli RB, Gonzalez J, Green JJ. A novel assay for quantifying the number of plasmids encapsulated by polymer nanoparticles. Small. 2012;8(3):367–373. doi: 10.1002/smll.201101718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perrault SD, Walkey C, Jennings T, Fischer HC, Chan WC. Mediating tumor targeting efficiency of nanoparticles through design. Nano Lett. 2009;9(5):1909–1915. doi: 10.1021/nl900031y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.