Abstract
An immature state of cellular differentiation—characterized by stem cell–like tendencies and impaired differentiation—is a hallmark of cancer. Using glioblastoma multiforme (GBM) as a model system, we sought to determine whether molecular determinants that drive cells toward terminal differentiation are also genetically targeted in carcinogenesis and whether neutralizing such genes also plays an active role to reinforce the impaired differentiation state and promote malignancy. To that end, we screened 71 genes with known roles in promoting nervous system development that also sustain copy number loss in GBM through antineoplastic assay and identified A2BP1 (ataxin 2 binding protein 1, Rbfox1), an RNA-binding and splicing regulator that is deleted in 10% of GBM cases. Integrated in silico analysis of GBM profiles to elucidate the A2BP1 pathway and its role in glioma identified myelin transcription factor 1-like (Myt1L) as a direct transcriptional regulator of A2BP1. Reintroduction of A2BP1 or Myt1L in GBM cell lines and glioma stem cells profoundly inhibited tumorigenesis in multiple assays, and conversely, shRNA-mediated knockdown of A2BP1 or Myt1L in premalignant neural stem cells compromised neuronal lineage differentiation and promoted orthotopic tumor formation. On the mechanistic level, with the top-represented downstream target TPM1 as an illustrative example, we demonstrated that, among its multiple functions, A2BP1 serves to regulate TPM1’s alternative splicing to promote cytoskeletal organization and terminal differentiation and suppress malignancy. Thus, in addition to the activation of self-renewal pathways, the neutralization of genetic programs that drive cells toward terminal differentiation may also promote immature and highly plastic developmental states that contribute to the aggressive malignant properties of GBM.
Keywords: oncogenomics, cancer stem cells
In cancer, the bases of classical biological hallmarks such as aberrant cell cycle control or apoptosis resistance are brought about and reinforced by multiple genetic events governing such processes (1). These cooperative genetic events in the same cancer cell typically involve both the activation of positive drivers and inactivation of suppressive factors needed to commandeer the redundant genetic controls underlying all biological processes present in normal cells. For example, deregulated cell cycle control in cancer often involves the cooperative impact of activation of D-type cyclins and loss of G1 cyclin–dependent kinase inhibitors in the same cancer cell (2).
A hallmark of cancer is the acquisition of a cellular differentiation state typified by maintenance of stem cell–like properties that maintain robust renewal activity and capacity to generate differentiated progeny, albeit with profound impairment in terminal differentiation. Substantial evidence has established that this highly de-differentiated and plastic state reflects acquisition of genetic events that actively promote stemness, such as activation of WNT, Shh, and Notch signaling pathways (3). Glioblastoma multiforme (GBM), the most common and lethal primary brain tumor in adults (4, 5), possesses both glioma stem cells (GSCs) (6–12) and more differentiated tumor cells, which manifest as significant morphological heterogeneity characterized by immature glial and neuronal features (4, 13–15). Current evidence has established that the GSC state is driven in part by WNT and Myc (16–18), which promote stemness yet allow for partial cellular differentiation capacity, a process thought to fuel cellular heterogeneity in GBM (13, 15). In this study, we sought to answer whether loss of terminal differentiation capacity simply reflects acquisition of enhanced self-renewal capability of GSCs or whether the genes that drive cells toward terminal differentiation are also actively neutralized through mutations/genomic alterations, and if so, whether such genetic events are essential in gliomagenesis.
Results
The differentiation hierarchy of human GBM is readily evidenced by stem, progenitor, and cell differentiation marker profiles of a panel of human GBM tissue microarray (TMA) samples (n = 71), revealing moderate to high levels of the neural stem cell marker Nestin, as well as more committed progenitor markers for astrocytes (GFAP), oligodendrocytes (Olig2), and neurons (Tuj1 and Dcx) (Fig. 1A). In sharp contrast, fewer GBM cells express terminal differentiated cell markers for oligodendrocytes (MBP) and neurons (NeuN) (Fig. 1A). In addition, GBM cells that express terminal differentiated astrocyte marker S-100β are also significantly fewer than the cells that express progenitor astrocyte maker GFAP (Fig. 1A). Together, this profile of differentiation markers reveals marked cellular heterogeneity dominated by an immature state of cell differentiation. Notably, the level of heterogeneity positively correlates with grades of glioma, which is reflected by an increased cell population of neuronal lineage in high-grade glioma (SI Appendix, Fig. S1). The cells of neuronal lineage in GBM are indeed tumor cells instead of normal cells trapped among tumor cells, because they express proliferating GBM cell marker Ki67 and a high level of EGFR (SI Appendix, Fig. S2).
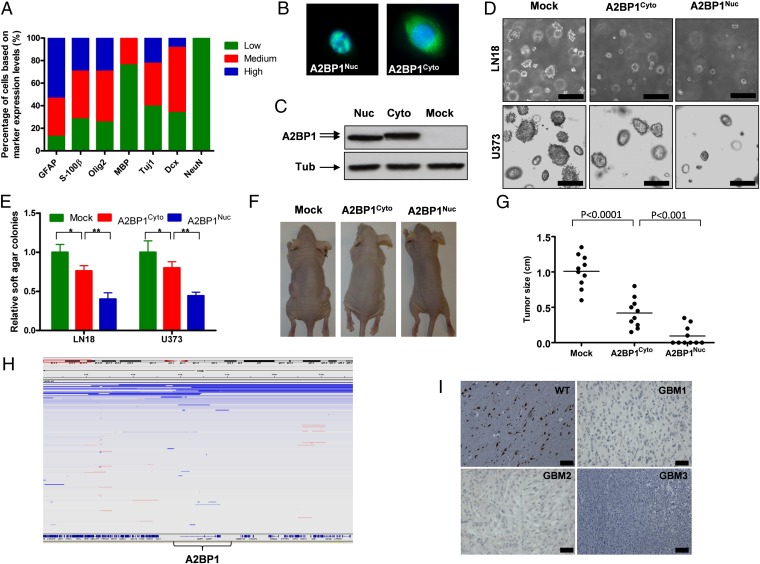
Fig. 1.
A2BP1 is a tumor suppressor that is deleted and down-regulated in GBM. (A) Protein expression profiles of various stem cell and lineage markers in human GBM. (B) Localizations of A2BP1 nuclear and cytoplasmic isoforms shown with immunofluorescence (IF). (C) Expression levels of ectopic A2BP1 nuclear and cytoplasmic isoforms shown with immunoblotting. (D) Representative images of anchorage independent soft agar growth of LN18 and U373 cells expressing nuclear and cytoplasmic isoforms of A2BP1. (E) Statistical analysis of D. *P < 0.001, **P < 0.0001. (F) Representative images of s.c. tumor formation of U373 cells expressing nuclear and cytoplasmic isoforms of A2BP1. (G) Statistical analysis of F. (H) IGV representation of deletion of A2BP1 locus in GBM. (I) Representative IHC images of A2BP1 in GBM samples and WT brain tissues. (Scale bars, 100 μm.) Error bars indicate SD.
In an effort to identify genetic events that may drive deactivation of terminal differentiation capacity, we surveyed The Cancer Genome Atlas (TCGA) GBM dataset for genes showing both recurrent copy number loss (GISTIC) (19, 20) and possessing known roles in promoting nervous system development (ingenuity pathway analysis and PubMed). The ORFs of 71 genes that scored in both categories (SI Appendix, Table S1) were transduced individually into the GBM cell line LN319 and audited for anchorage-independent activity in the soft agar assay, which reflects both cell renewal and malignant potential. One of the 71 genes, A2BP1, caused marked reduction in soft agar colony formation of LN319 cells (SI Appendix, Fig. S3). A2BP1 is an alternative splicing factor that functions in neural development, and its genomic alterations have been implicated with several neurodevelopmental and neuropsychiatric disorders including autism spectrum disorder (ASD) (21). A2BP1 encodes both cytoplasmic and nuclear isoforms (Fig. 1 B and C), and the antioncogenic activity of both A2BP1 isoforms was also documented with soft agar colony formation and s.c. tumor growth in an additional GBM cell line: U373 (Fig. 1 D–G). Notably, the nuclear isoform showed more potent soft agar and s.c. growth inhibition than the cytoplasmic isoform (Fig. 1 D–G).
The copy number profiling of 430 TCGA GBM tumor samples (TCGA, Firehose 5/25/2011) shows that A2BP1 is deleted in 10% of GBM tumor samples (Fig. 1H). Furthermore, immunohistochemistry (IHC) analysis showed that the A2BP1 protein is absent or down-regulated in >90% of GBM tumor TMA samples (Fig. 1I). Besides GBM, A2BP1 is also deleted in other nervous system tumors, including ∼26% of neuroblastomas and 18% of medullablastomas (22) (TCGA). Additionally, deletion of A2BP1 in 48% of colon cancer samples and 18% of sarcoma (22) samples suggests a major tumor suppressor function across multiple cancer types (TCGA).
A2BP1 is expressed exclusively in differentiated neurons as documented by costaining of A2BP1 and the mature neuronal cell marker NeuN, as well as pan-neuronal cell marker Tuj1 (labels both progenitor and mature neurons) and by lack of costaining of A2BP1 and stem cell markers Msi-1 and BLBP, astrocyte markers GFAP and ALDH1L1, oligodendrocyte markers Olig2, NG2, and A2B5, and neuronal progenitor cell marker Dcx (SI Appendix, Fig. S4). Costaining with α-BrdU and α-A2BP1 antibodies of actively developing brains (2-wk-old postnatal mice injected with BrdU) revealed no overlap, indicating restriction of A2BP1 expression in postmitotic terminally differentiated neurons (Fig. 2A). We then sought to test whether A2BP1 controls the terminal differentiation of NSCs by taking advantage of premalignant neural/progenitor stem cells (PM-NSCs) derived from p53L/L Pten+/L GFAP-Cre mice, which maintain full neuronal lineage differentiation capability (18). On placement in ENStem-A neuronal differentiation medium, A2BP1 knockdown by independent shRNAs (shA2BP1s) (SI Appendix, Fig. S5 A and B) led to marked reduction in the neuronal terminal differentiation marker NeuN and increase of BrdU incorporation and Dcx+ neuroblast cell population (Fig. 2 B and C; SI Appendix, Figs. S5 C–E and S7 A and B), yet exerted no impact on the pan-neuronal cell marker Tuj1 and MAP2 staining relative to nontargeting shRNA (shNT) control (SI Appendix, Figs. S5 F and G and S7 A and B). Under stem cell medium (SCM) conditions, shA2BP1 does not influence expression of the NSC marker Nestin (SI Appendix, Figs. S5 F and G and S7 A and B) or neurosphere formation (SI Appendix, Figs. S6 and S7C) relative to shNT control, further supporting that the A2BP1 shRNAs only operate in a more differentiated cell context. Notably, shA2BP1 also had no impact on the capacity for astrocytic differentiation of PM-NSCs cultured in astrocytic differentiation medium (1% FBS) (SI Appendix, Figs. S5 F and G and S7 A and B). Similarly, using the P19 embryonal carcinoma model system, A2BP1 knockdown in prodifferentiation conditions (retinoic acid) results in decreased NeuN-positive cell numbers and exerts no differences in Tuj1- and GFAP-positive cell numbers relative to shNT control (SI Appendix, Fig. S8 A and B). Thus, we conclude that A2BP1 plays a lineage-restricted, differentiation-specific role in promoting the terminal differentiation of neuronal lineage (Fig. 2 B and C).
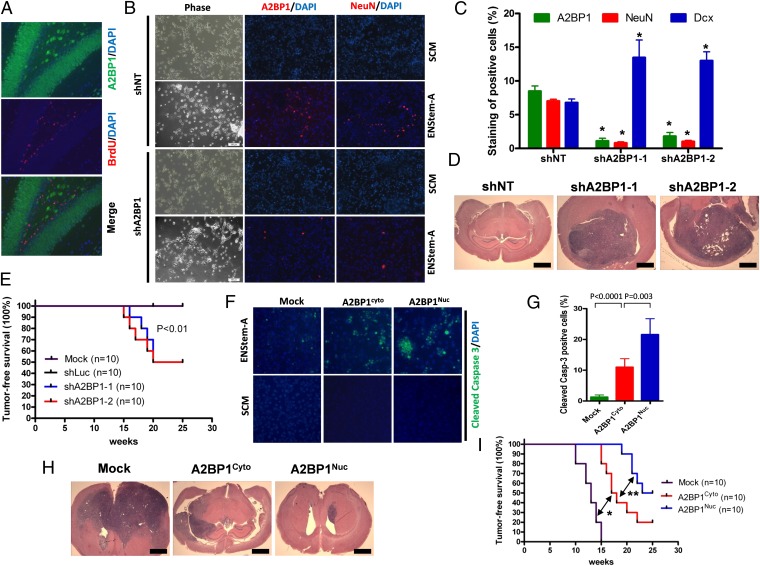
Fig. 2.
Loss of A2BP1 contributes to GBM tumorigenesis by causing compromised terminal differentiation. (A) Colocalization of A2BP1 and proliferation marker BrdU in mouse brain tissue shown with IF. (B) Representative IF staining of A2BP1 and NeuN in p53−/−Pten+/− premalignant neural stem cells (PM-NSCs) with A2BP1 knockdown or control shRNA cultured in stem cell medium (SCM) and neuronal differentiation medium EN-StemA. (C) Statistical analysis of staining of A2BP1, NeuN, and Dcx in p53−/−Pten+/− PM-NSCs with A2BP1 knockdown or control shRNA cultured in SCM and EN-StemA medium. *P < 0.0001 compared with shNT controls. (D) Representative H&E staining of intracranial tumor formation of p53−/−Pten+/− PM-NSCs with A2BP1 knockdown or control shRNA. (E) Kaplan–Meier survival curves (log-rank test) of mice intracranially injected with p53−/−Pten+/− PM-NSCs with A2BP1 knockdown or control shRNA. (F) Representative IF staining of cleaved caspase3 in p53−/−Pten−/− GSCs with A2BP1 overexpression or mock cultured in SCM and EN-StemA medium. (G) Statistical analysis of F. (H) Representative H&E staining of intracranial tumor formation of p53−/−Pten−/− GSCs with A2BP1 overexpression or mock. (I) Kaplan–Meier survival curves (log-rank test) of mice intracranially injected with p53−/−Pten−/− GSCs with A2BP1 overexpression or mock. (Scale bars, 2 mm.) *P < 0.0001, **P = 0.03. Error bars indicate SD.
The p53−/− Pten+/− PM-NSCs do not form tumors on orthotopic injection into the mouse brain (see below). To assess whether compromised neuronal terminal differentiation affected by A2BP1 extinction enhances gliomagenesis, p53−/− Pten+/− PM-NSCs transduced with shA2BP1 or shNT were implanted orthotopically and monitored for tumor formation. The shA2BP1 cohort generated brain tumors commencing at 15 wk, whereas PM-NSCs expressing shNT remained tumor free through 25 wk of observation (Fig. 2 D and E). Similarly, A2BP1 knockdown also significantly decreased the latency of intracranial tumor formation of P19 cells (SI Appendix, Fig. S8C). Histological analysis revealed that p53−/− Pten+/−-shA2BP1 orthotopic tumors express neuronal progenitor cell markers Tuj1 and Dcx, stem/progenitor cell marker Nestin, and astrocyte marker GFAP (SI Appendix, Fig. S9), suggesting that extinction of factors driving terminal differentiation can promote glioma tumors that exhibit a highly plastic and immature developmental state with varied lineage representation. To further verify the impact of A2BP1 on differentiation and tumorigenic potential, A2BP1 was overexpressed in p53−/− Pten−/− GSCs derived from gliomas arising in p53L/L Pten+/L GFAP-Cre mice (18). Although enforced expression of A2BP1 had no impact on GSCs maintained in stem cell medium, induction of neuronal differentiation in the setting of enforced A2BP1 expression resulted in increased cell death in neuronal lineage (Fig. 2 F and G; SI Appendix, Fig. S10 A–D). Supporting the function of A2BP1 in promoting terminal differentiation of neurons, ectopic expression of A2BP1 in NSCs increased NeuN/Synapsin double positive cell population in neuronal differentiation medium (SI Appendix, Fig. S10 E and F). Correspondingly, enforced A2BP1 expression also impaired tumor formation following orthotopic implantation of both mouse and human GSCs (Fig. 2 H and I; SI Appendix, Fig. S11). IF (immunofluorescence) costaining showed that a significant fraction (60%) of A2BP1-associated apoptosis occurred in the neuronal lineage, but only minimal apoptosis occurred in GSCs and the astrocytic lineage (SI Appendix, Fig. S12 A and B), whereas A2BP1 expression was induced equally in all cell populations (SI Appendix, Fig. S12C) of xenograft tumors derived from p53−/− Pten−/− GSCs.
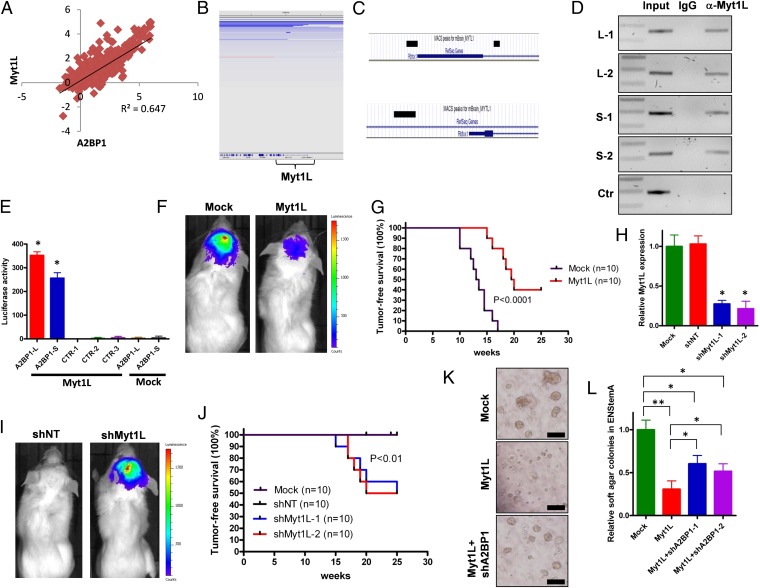
If neutralization of neuronal terminal differentiation is indeed required for gliomagenesis, we next asked whether other genetic components in the A2BP1 pathway are impacted in those GBMs with genomic retention of the A2BP1 locus. Because A2BP1 expression is suppressed in greater than 90% of the GBM samples yet deleted in only 10% of cases, we focused on the identification of potential transcriptional regulators of A2BP1 and assessed their genomic status by first correlating expression of A2BP1 with all other genes in the transcriptomic dataset of 537 TCGA tumor samples (TCGA, Firehose 5/25/2011). Myt1L was the top transcription factor whose expression levels positively correlate with A2BP1 expression levels in GBM (R2 = 0.647; Fig. 3A). Myt1L was found to be deleted in about 5% of GBM samples in a mutually exclusive manner with A2BP1 deletion (P < 0.01; Fig. 3B), and IHC analysis of GBM TMAs showed that the Myt1L protein is absent or down-regulated in >80% of samples (SI Appendix, Fig. S13A). Notably, Myt1L is one of the ingredients of the transcription factor mixture that can transdifferentiate fibroblasts into mature neurons (23–27). Using α-Myt1L CHIP-seq and confirmatory CHIP-PCR in mouse brain extracts, Myt1L was shown to bind to four distinct sites within two A2BP1 promoters (Fig. 3 C and D). Luciferase reporter assays showed that both A2BP1 promoter sequences, but not three control sequences, were strongly activated by enforced Myt1L expression (Fig. 3E).
Fig. 3.
Myt1L is a transcription factor that directly regulates A2BP1 expression under neural/glioma stem cell context. (A) Plotted correlation between A2BP1 expression levels and Myt1L expression levels in human GBM samples. (B) IGV representation of deletion of Myt1L locus in GBM. (C) Enrichment of Myt1L binding sequences (black bars) in two A2BP1 promoters identified with CHIP-Seq. (D) Verification of Myt1L binding sequences of A2BP1 promoters by CHIP-PCR with multiple sequences. L and S, A2BP1 promoters driving expressions of long and short transcripts, respectively. (E) Luciferase assay of A2BP1 promoters with overexpression of Myt1L or mock. L and S, A2BP1 promoters driving expression of long and short transcripts, respectively *P < 0.0001 compared with controls. (F) Representative images of intracranial tumor formation of p53−/−Pten−/− GSCs with Myt1L overexpression or mock. (G) Kaplan–Meier survival curves (log-rank test) of mice with intracranial injection of p53−/−Pten−/− GSCs with Myt1L overexpression or mock. (H) Knockdown of Myt1L in p53−/−Pten+/− PM-NSCs with shMyt1L or control shRNAs. *P < 0.0001 compared with mock or shNT controls. (I) Representative images of intracranial tumor formation of p53−/−Pten+/− PM-NSCs with shMyt1L or control shRNA. (J) Kaplan–Meier survival curves (log-rank test) of mice with intracranial injection of p53−/−Pten+/− PM-NSCs with shMyt1L or control shRNA. (K) Representative images of anchorage independent soft agar growth of p53−/−Pten+/− PM-NSCs with Myt1L overexpression with/without A2BP1 knockdown in EN-stemA medium. (L) Statistical analysis of K. (Scale bars, 50 μm.) *P < 0.001, **P < 0.0001. Error bars indicate SD.
To dissect the functions of Myt1L, we did Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis on 258 genes (SI Appendix, Table S2) with Myt1L binding sites within 10 kb from the transcription start site identified by CHIP-seq and found overrepresentation of genes regulating multiple biological functions including metabolism, cellular assembly and organization, and nervous system development (SI Appendix, Table S3), which partially overlap with the functions assigned to A2BP1 (SI Appendix, Table S5). Next, we sought to assess whether Myt1L exerts tumor suppressive and proneuronal differentiation activities similar to A2BP1. Enforced Myt1L expression in p53−/−Pten−/− GSCs increased cell death in the neuronal differentiation medium and exerted no impact on the survival of GSCs in astrocytic differentiation medium (SI Appendix, Fig. S13B) nor affected the self-renewal activity of GSCs maintained in stem cell medium (SI Appendix, Fig. S13 C and D). In addition, enforced Myt1L expression in GSCs inhibited orthotopic tumor formation (Fig. 3 F and G). Supporting the function of Myt1L in promoting neuronal terminal differentiation, enforced Myt1L expression in NSCs increased NeuN/Synapsin double positive cell population in neuronal differentiation medium (SI Appendix, Fig. S13 E and F). shRNA-mediated knockdown of Myt1L by two independent hairpins in p53−/−Pten+/− PM-NSCs reduced NeuN+ cell numbers and increased BrdU incorporation in neuronal differentiation medium (SI Appendix, Fig. S13 G and H) and induced orthotopic tumor formation at 15 wk, whereas shNT controls remained tumor free through 25 wk of observation (Fig. 3 H–J). Finally, to further substantiate the Myt1L–A2BP1 axis in gliomagenesis, we asked whether Myt1L inhibits tumorigenesis in part via A2BP1. A2BP1 knockdown significantly restored the soft agar formation reduced by ectopic Myt1L in neuronal differentiation medium (Fig. 3 K and L; SI Appendix, Fig. S14A), and ectopic A2BP1 significantly decreased soft agar and tumor formation induced by Myt1L knockdown (SI Appendix, Fig. S14B–E). Together, these epistasis studies support the view that A2BP1 is at least partly involved in Myt1L-dependent glioma suppression and control of neuronal differentiation but also point to nonoverlapping functions of A2BP1 and Myt1L in these processes.
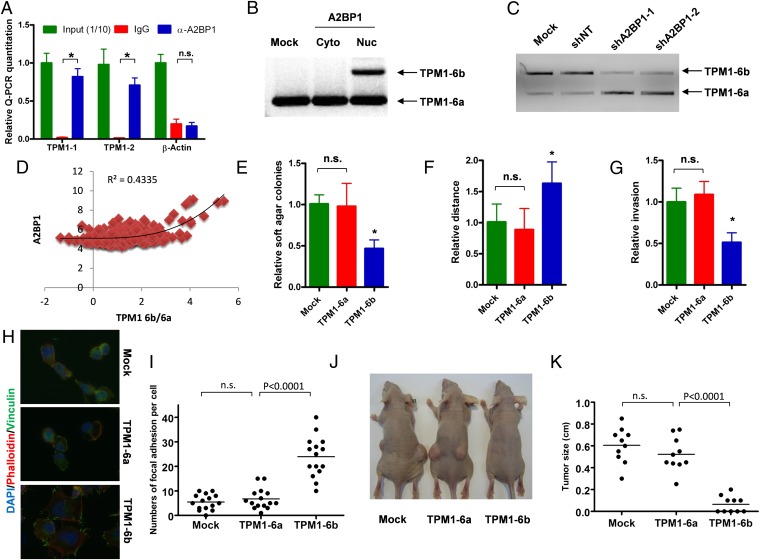
The potent antioncogenic activity of the A2BP1 nuclear isoform and the known role of this isoform in regulating alternative splicing (28) prompted us to comprehensively profile A2BP1 interactions with RNAs in an effort to fully understand the role of A2BP1 in neuronal differentiation and tumor suppression. To that end, A2BP1 protein–RNA complexes were immunoprecipitated from normal mouse brains with RNA-binding protein immunoprecipitation-microarray (RIP)-Chip protocol (29), and the associated RNAs were identified by exon array profiling (SI Appendix, Fig. S15A). The cutoff of fourfold enrichment by α-A2BP1 IP vs. control IgG IP identified A2BP1-associated mRNAs expressed by 2,464 genes. Examination of these genes for the well-characterized A2BP1’s binding motif (UGCAUG) (30, 31) within intronic 300 bp from the splicing site identified 7,061 putative targets of A2BP1 that are conserved between mouse and human (SI Appendix, Fig. S15B). Integration of A2BP1-associated RNAs identified by RIP-Chip and the genes with A2BP1 binding motifs identified in silico yielded a list of 906 genes potentially regulated by A2BP1 (SI Appendix, Fig. S15B and Table S4). KEGG pathway analysis identified 53 pathways significantly (P < 0.05) enriched in these A2BP1 targets (SI Appendix, Table S5). Sixty-three percent of these pathways belong to four major functions: cytoskeletion regulation (21%), neural development (11%), metabolism (21%), and signaling (15%). To check whether the antioncogenic activity of A2BP1 is governed in part through the alternative splicing of its targets identified in normal neural tissues, we focused on TPM1 as a working model because TPM1 was found to be the most enriched transcript (40- to 50-fold; Fig. 4A) in the α-A2BP1 immunoprecipates relative to IgG control.
Fig. 4.
A2BP1 regulates alternative splicing of exons 6a and 6b of TPM1. (A) RT-QPCR quantification of TPM1 mRNA associated with A2BP1 immunoprecipitated with anti-A2BP1 antibody in mouse brain. *P < 0.0001. (B) Alternative splicing of TPM1 exons 6a and 6b in LN18 cells with ectopic expression of A2BP1 nuclear or cytoplasmic isoforms or mock was shown with PCR. (C) Alternative splicing of TPM1 exons 6a and 6b in p53−/−Pten+/− PM-NSCs with A2BP1 knockdown and control shRNA in neuronal differentiation medium EN-StemA was shown with PCR. (D) Plotted correlation between A2BP1 expression levels and relative expression levels of TPM1 exon 6b vs. 6a in GBM samples. (E) Anchorage independent soft agar growth of LN18 cells with ectopic expression of TPM1-6a, TPM1-6b, or mock. *P < 0.01 compared with mock or TPM1-6a. (F) Wound healing assay of LN18 cells with ectopic expression of TPM1-6a, TPM1-6b, or mock. *P < 0.001 compared with mock or TPM1-6a. (G) Invasion chamber assay of cells with ectopic expression of TPM1-6a, TPM1-6b, or mock. *P < 0.001 compared with mock or TPM1-6a. (H) Representative images of focal adhesion sites and stress fiber of cells with ectopic expression of TPM1-6a, TPM1-6b, or mock were shown with IF. (I) Statistical analysis of H. (J) Representative images of s.c. tumor formation of cells with ectopic expression of TPM1-6a, TPM1-6b, or mock were shown with IF. (K) Statistical analysis of J. Error bars indicate SD.
TPM1 is an actin-binding protein (32), which belongs to the cytoskeleton regulation pathways. It has two UGCAUG binding motifs within the 200 bp flanking exons 6a and 6b, which are spliced mutually exclusively (33). EMSA confirmed that recombinant A2BP1 protein binds to in vitro transcribed TPM1 exon 6a and 6b fragments, and the stability of these nucleoprotein complexes is dependent on the presence of intact UGCAUG binding motifs (SI Appendix, Fig. S15C). To test whether A2BP1 regulates the splicing of TPM1 exon 6a and 6b, we overexpressed nuclear and cytoplasmic isoforms of A2BP1 in GBM cell line LN18. Although the cytoplasmic isoform did not alter TPM1 splicing, the nuclear isoform significantly increased the generation of exon 6b–containing transcripts (Fig. 4B; SI Appendix, Fig. S15D), which is consistent with the enrichment of exon 6b in PM-NSCs cultured in neuronal differentiation medium (SI Appendix, Fig. S15E). In addition, knockdown of A2BP1 also reduced exon 6b–containing transcripts in neuronal differentiated PM-NSCs (Fig. 4C). To check whether A2BP1 expression levels correlate with the relative expression of exon 6b vs. 6a, we took advantage of the exon array dataset of 194 TCGA GBM samples (20). Polynomial regression analysis (K = 3) showed a significant positive correlation between A2BP1 expression levels and the relative expression ratios of exon 6b/exon 6a (Fig. 4D; R2 = 0.4335), supporting that the mutually exclusive splicing of TPM1 exon 6a/6b regulated by A2BP1 also occurs in the human GBM context.
TPM1 is a well-established tumor suppressor, and the down-regulation of TPM1 and the resulting cytoskeletal reorganization are indispensable for Ras- and Src-mediated transformation (33–40), prompting us to assess whether switching TPM1 exon 6a to 6b will also have antioncologic effect. We ectopically expressed TPM1-6a (TM3) or TPM1-6b (TM2) in LN18 cells (SI Appendix, Fig. S15F), which were subsequently assessed in an array of cancer-relevant assays. TPM1-6b, but not TPM1-6a, resulted in significantly reduced soft agar colony formation and decreased invasive and migratory activity (Fig. 4 E–G). Consistent with the impact on cytoskeleton and cellular motility, analysis of focal adhesion marker Vinculin and stress fiber marker Phalloidin staining patterns showed that LN18 cells expressing TPM1-6b have significantly more focal adhesion sites and a higher order of stress fibers relative to cells expressing TPM1-6a or mock (Fig. 4 H and I). In line with A2BP1’s role in terminal differentiation, ectopic TPM1-6a but not 6b increased BrdU incorporation and decreased NeuN-positive cells in PM-NSCs (SI Appendix, Fig. S16 A and B). Finally, TMP1-6b–expressing cells but not TPM1-6a–expressing cells showed potent inhibition of s.c. tumor growth (Fig. 4 J and K). Supporting that A2BP1 inhibits gliomagenesis partly through regulating TPM1 alternative splicing, only ectopic TPM1-6b but not 6a significantly suppressed intracranial tumor formation induced by A2BP1 knockdown (SI Appendix, Fig. S16 C and D). Thus, although A2BP1 is likely to suppress gliomagenesis through regulation of many genes (SI Appendix, Tables S4 and S5), these studies identify one key mechanism underlying A2BP1’s tumor suppression activity via the control of TPM1 splicing isoforms in GBM cells.
Discussion
This work demonstrates that neutralization of the terminal differentiation program governed by the Myt1L–A2BP1 axis can contribute to gliomagenesis via cell autonomous mechanisms, although we cannot formally rule out additional non–cell autonomous effects of loss of Myt1L–A2BP1. These findings strongly support the concept that the promotion of an immature differentiation state in cancer is driven both by activation of pathways promoting cancer stemness and neutralization of those inducing terminal differentiation. That Myt1L–A2BP1 are major regulators in neoplastic differentiation processes in glioma is reinforced by the known potent transdifferentiation potential of Myt1L in neuron generation (23–27), the capacity of Myt1L to directly regulate A2BP1 transcription but not the A2BP1 paralogs Rbfox2 and Rbfox3 (SI Appendix, Table S2), the failure of Rbfox2/3 to rescue loss of the Myt1L–A2BP1 axis in tumorigenesis assays, and the tumorigenic impact of alternative splicing of a key A2BP1 target regulating cytoskeletal dynamics. Indeed, although our unbiased survey of A2BP1 targets identified its pleiotropic functionality in neural development, A2BP1’s regulation of cytoskeletal configuration during the neuronal terminal differentiation emerged as one of its prominent activities. In addition, low expression of A2BP1 is enriched in mesenchymal subtype, and the A2BP1 level is relatively higher in the proneural and neural subtypes (TCGA, P < 0.001). These findings are consistent with the importance of loss of cellular polarity and cell–cell adhesion, deregulated cytoskeletal dynamics, and enhanced cell motility in cancer, and represent a prominent phenotype typified by the epithelial–mesenchymal transition (EMT) phenomenon (41, 42). EMT-like cytoskeletal configuration changes have also been documented in diverse solid tumors including GBM (43, 44). Our study shows that loss of the Myt1L–A2BP1 axis can promote a gliomagenesis-prone cytoskeletal state through modulating alternative splicing of multiple cytoskeleton regulators including actin regulator TPM1.
GBM exhibits striking intratumoral heterogeneity, which may establish critical homotypic and heterotypic interactions across many cell types to form a cancerous community (13). Disease heterogeneity resides, not only in inter- and intratumoral genomic profiles (45–47), but also in the profound variability in cellular differentiation state of cancer cells. This genomic and biological variability has therapeutic implications as the state of differentiation is known to influence the function of specific genes and may therefore determine whether a drug target remains rate limiting for tumor maintenance. This principle is evident by the finding that enforced A2BP1 expression had no impact under stem cell conditions yet provoked apoptosis on differentiation (Fig. 2F; SI Appendix, Fig. S7).
Our work supports the view that the cellular heterogeneity and “multiforme” histological features defined by various differentiation states are contributed by the remaining intermediate differentiation capacities of GSCs and maintained by compromised terminal differentiation capability. Loss of terminal differentiation with a defective Myt1L–A2BP1 axis might explain the minimal impact of differentiation therapy (such as retinoic acid) in GBM, particularly in those cancers with genomic loss of these key prodifferentiation regulators (48, 49). Notably, other than genomic loss in 5% of GBM samples, mechanisms of Myt1L silencing in the majority of the GBM samples currently remain unclear. On the basis of our findings, we speculate that agents targeting both neutralization of stemness pathways (e.g., WNT) and activation of Myt1L–A2BP1-directed terminal differentiation may provide a more effective therapeutic approach in GBM. Besides neuronal lineage, compromised terminal differentiation of other lineages including astrocyte and oligodendrocyte is also observed in GBM (Fig. 1A; SI Appendix, Fig. S1). Similar approaches as described in this study can be used to identify key players driving terminal differentiation of the glial lineage in the context of gliomagenesis.
Materials and Methods
Mice.
p53L, PtenL, and hGFAP-Cre mice have been described previously (18), and were interbred and maintained on a FvB/C57Bl6 hybrid background in pathogen-free conditions at the Dana-Farber Cancer Institute and MD Anderson Cancer Center, monitored for signs of ill health every other day, and euthanized and necropsied when moribund. All manipulations were performed with Institutional Animal Care and Use Committee (IACUC) approval.
Histology and Antibodies.
Once killed, mice were perfused with 4% (wt/vol) paraformaldehyde (PFA), and brains were dissected, followed by overnight postfixation in 4% PFA at 4 °C. Serial sections were prepared at 5 μm for paraffin sections or 10 μm for cryostat sections, with every 10th slide stained by H&E (DF/HCC Research Pathology Cores). Primary antibodies used for IHC, IF, and immunoblotting are Gfap (Z0334; DAKO), Gfap (556330; BD Pharmingen), Nestin (MAB353; Chemicon; specifically for mouse), Nestin (MAB5326; Chemicon; specifically for human), Olig-2 (AB9610; Chemicon), Tuj-1 (MMS-435P; Covance), O4 (MAB1326; R&D), NeuN (MAB377; Millipore), Mbp (ab7349; Abcam), MAP2 (AB5622; Millipore), Phalloidin (Invitrogen), Vinculin (V4505 and V9131; Sigma), Caspase3 (Asp175) (9661; Cell Signaling), Dcx (ab18723; Abcam), BrdU (DAKO), S100 (RB044A0; LabSource), α-tubulin (T-9026; Sigma), V5 (Invitriogen), A2BP1 (N14) (Santa Cruz), A2BP1 (Genescript, against peptide sequence MAQPYASAQFAPPQN), A2BP1 (rat monoclonal antibodies; Dana Farber Monoclonal Antibody facility), Myt1L (AB093283; Abnova), and TPM1 (NB100-1908; Novus Biologicals).
Human GBM IHC Staining Grading.
Human GBM samples were stained with GFAP, S-100β, Olig2, MBP, Tuj1, Dcx, NeuN, and Nestin antibodies. Samples with >50%, 5–50%, and <5% positive cells were categorized as high, medium, and low, respectively.
Bioinformatic Approach to Get Candidate Genes Involved in Both Differentiation and Tumorigenesis.
The GISTIC algorithm was described previously (19). Briefly, deleted genes were defined by genes residing in regions that are deleted in >5% of GBM samples and contain <50 genes. Genes in nervous system development were selected by integration of ingenuity pathway analysis (IPA) and PubMed.
Cell Culture.
The human glioma cell lines U373, LN18, and LN319 and the human embryonic kidney cell line HEK293T were purchased from American Type Culture Collection (ATCC) and maintained in DMEM containing 10% (vol/vol) FBS. The human GSC line TS603 was from Cameron Brennan of Memorial Sloan-Kettering Cancer Center (New York, NY).
Primary mouse PM-NSCs and GSCs were isolated from the brain subventricular zone (SVZ) of E13.5 mouse embryos and mouse brain tumors with the indicated genotypes as previously described, respectively (17, 18). PM-NSCs and GSCs were maintained in stem cell media (SCM) (05702; StemCell) supplemented with 20 ng/mL EGF (E4127; Sigma) and 10 ng/mL basic FGF (F0291; Sigma). Differentiation assays were carried out by plating the indicated cells in culture wells on coverslips precoated with 15 μg/mL poly-l-ornithine (P3655; Sigma) and 1 μg/mL fibronectin (F1141; Sigma); the cells were incubated in SCM supplemented with 1% FBS or EN-StemA medium for 7–10 d, and the differentiation capacities were examined under either a light or fluorescence microscope (Nikon).
P19 cells were differentiated into postmitotic neuronal cells as described previously (50). Briefly, P19 cells were grown in αMEM with 10% FBS. Differentiation was induced by transfer to medium containing αMEM, 5% FBS, and 0.5 μM all-trans retinoic acid (R-2625; Sigma Aldrich) in bacteriological Petri dishes at a density of 105/cm2 to promote cell aggregation. After 2 d, cell aggregates were transferred to fresh induction medium and cultured for another 2 d. The cell aggregates were then trypsinized and were plated at a density of 104/cm2 on tissue culture plates in αMEM plus 10% (vol/vol) FBS. The tissue culture plates were coated previously overnight with 0.1 mg/mL poly-l-lysine in 1.25% (wt/vol) boric acid/1.91% (wt/vol) sodium tetraborate solution (1:1; pH 8.2), and washed three times with sterilized water. Two days after plating, the medium was changed to Neurobasal-A medium (GIBCO) with 1× B27 supplement (GIBCO) and maintained through the experiment.
Subcutaneous and Orthotopic Cell Injections.
For s.c. injections, female nude mice (Charles River) aged 6–8 wk were anesthetized, and 10 million cells were injected into each flank of nude mice. Tumor sizes were obtained from average of the measurements at three dimensions. For intracranial injections, female SCID mice (Charles River) aged 6–8 wk were anesthetized and placed into stereotactic apparatus equipped with a z axis (Stoelting). A small hole was bored in the skull 0.5 mm anterior and 3.0 mm lateral to the bregma using a dental drill; 2 × 105 (Figs. 2 and 3; SI Appendix, Fig. S11) or 5 × 105 (SI Appendix, Figs. S9–11, S14, and S16) cells in Hanks buffered salt solution were injected into the right caudate nucleus 3 mm below the surface of the brain using a 10-μL Hamilton syringe with an unbeveled 30-gauge needle. The scalp was closed using a 9-mm Autoclip Applier. Animals were followed for the development of neurological deficits every other day. Animals transplanted with cells stably express luciferase activity were monitored with imaging system Lumina II (Xenogen) every other day. For doxycycline-induced expression, mice were injected and fed on regular water for tumor to grow, and mice were fed on doxycycline water for 3 d. All manipulations were performed with IACUC approval.
RNA-Binding Protein Immunoprecipitation–Microarray Profiling Analysis
Polysome lysis buffer was prepared as previously described (29). Briefly, 50 μL of 1 M Hepes, 500 μL of 1 M KCl, 25 μL of 1 M MgCl2, and 25 μL of Nonidet P-40 were added to 4.7 mL of RNase-DNase-free H2O. Fifty microliters of 1 M DTT, 12.5 μL of 100 U/mL RNase Out (Promega), and 200 μL of Protease inhibitor mixture (Roche Complete Mini) were added at the time of use. Two WT 8-wk-old mouse brains were homogenized in 10 mL of lysis buffer. Tissue lysate was precipitated, and A2BP1 antibody (Genscript)-coupled protein A/G beads (Santa Cruz) were added to the supernatant, which was subsequently incubated at 4 °C for 1 h. Beads were washed with lysis buffer three times, and RNAs were extracted with the RNeasy kit (Qiagen) and were subsequently profiled with mouse Exon Array (1.0 ST). RNA profiles were analyzed with dCHIP software.
Scratching Assay.
Cells were grown to subconfluence and then scratched with a 200-μL tip. Cells were subsequently cultured for 22 h. The distance between two borders of scratching was measured at 0 and 22 h. The distance of each scratch was measured six times at different sizes and averaged.
Expression and shRNA Plasmids.
ORFs of 71 candidate genes and the TPM1 gene were obtained from Open Biosystems and cloned into lentiviral plasmids pLenti6.3-V5/Dest, pLenti6-Ubc-V5/Dest, and pLenti4/TO-V5/Dest. Tet-O-FUW-Myt1L plasmid (24) was ordered from Addgene (Addgene plasmid 27152). PLKO-based lentiviral shRNA plasmids targeting A2BP1 and Myt1L were obtained from the DF/HCC DNA Resource Core. The targeting sequences are listed in SI Appendix, Table S6.
Chromatin immunoprecipitation analyses, the invasion assay, EMSA, and the Luciferase reporter assay are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank S. Jiang, R. Narurkar, and E. Fletcher-Sananikone for excellent mouse husbandry and care and all members of the DePinho and Chin laboratories for helpful discussion. We also thank H. Zhang and Y. Xiao for the bioinformatic analysis. J.H. is supported by National Institutes of Health (NIH) K99/R00 Pathway to Independence Award 5K99CA172700. J.H. and B.G. were supported by a Leukemia and Lymphoma Society fellowship. B.G. is supported by the Department of Defense Tuberous Sclerosis Complex Research Program (TSCRP) Career Transition Grant TS093049 and Prostate Cancer Research Program (PCRP) Exploration-Hypothesis Development Grant PC100356. H.Z. was supported by the Helen Hay Whitney Foundation. G.W. is supported by Juvenile Diabetes Research Foundation Grant 10-2012-240. L.C. and R.A.D. are supported by NIH Grants P01 5P01CA095616 and U01 5U01CA084313. R.A.D. is an Ellison Foundation for Medical Research Senior Scholar and an American Cancer Society Research Professor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See QnAs on page 14508.
See Commentary on page 14510.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308610110/-/DCSupplemental.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Wiedemeyer WR, et al. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proc Natl Acad Sci USA. 2010;107(25):11501–11506. doi: 10.1073/pnas.1001613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol. 2011;8(2):97–106. doi: 10.1038/nrclinonc.2010.196. [DOI] [PubMed] [Google Scholar]
- 4.Furnari FB, et al. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 5.Stupp R, et al. European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 6.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 7.Calabrese C, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11(1):69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 8.Hemmati HD, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100(25):15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 10.Rich JN, Eyler CE. Cancer stem cells in brain tumor biology. Cold Spring Harb Symp Quant Biol. 2008;73:411–420. doi: 10.1101/sqb.2008.73.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 12.Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4(5):440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonavia R, Inda MM, Cavenee WK, Furnari FB. Heterogeneity maintenance in glioblastoma: A social network. Cancer Res. 2011;71(12):4055–4060. doi: 10.1158/0008-5472.CAN-11-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inda MM, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24(16):1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholas MK, et al. Molecular heterogeneity in glioblastoma: Therapeutic opportunities and challenges. Semin Oncol. 2011;38(2):243–253. doi: 10.1053/j.seminoncol.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Nabeshima K, et al. Expression of c-Met correlates with grade of malignancy in human astrocytic tumours: An immunohistochemical study. Histopathology. 1997;31(5):436–443. doi: 10.1046/j.1365-2559.1997.3010889.x. [DOI] [PubMed] [Google Scholar]
- 17.Zheng H, et al. PLAGL2 regulates Wnt signaling to impede differentiation in neural stem cells and gliomas. Cancer Cell. 2010;17(5):497–509. doi: 10.1016/j.ccr.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng H, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455(7216):1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu J, et al. Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell. 2012;148(4):651–663. doi: 10.1016/j.cell.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fogel BL, et al. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum Mol Genet. 2012;21(19):4171–4186. doi: 10.1093/hmg/dds240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfisterer U, et al. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci USA. 2011;108(25):10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463(7284):1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambasudhan R, et al. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9(2):113–118. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoo AS, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476(7359):228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pang ZP, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476(7359):220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gehman LT, et al. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet. 2011;43(7):706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: The isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1(1):302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- 30.Jin Y, et al. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22(4):905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25(22):10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helfman DM, Flynn P, Khan P, Saeed A. Tropomyosin as a regulator of cancer cell transformation. Adv Exp Med Biol. 2008;644:124–131. doi: 10.1007/978-0-387-85766-4_10. [DOI] [PubMed] [Google Scholar]
- 33.Gimona M, Kazzaz JA, Helfman DM. Forced expression of tropomyosin 2 or 3 in v-Ki-ras-transformed fibroblasts results in distinct phenotypic effects. Proc Natl Acad Sci USA. 1996;93(18):9618–9623. doi: 10.1073/pnas.93.18.9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Safina AF, et al. Ras alters epithelial-mesenchymal transition in response to TGFbeta by reducing actin fibers and cell-matrix adhesion. Cell Cycle. 2009;8(2):284–298. doi: 10.4161/cc.8.2.7590. [DOI] [PubMed] [Google Scholar]
- 35.Shah V, Bharadwaj S, Kaibuchi K, Prasad GL. Cytoskeletal organization in tropomyosin-mediated reversion of ras-transformation: Evidence for Rho kinase pathway. Oncogene. 2001;20(17):2112–2121. doi: 10.1038/sj.onc.1204291. [DOI] [PubMed] [Google Scholar]
- 36.Shah V, Braverman R, Prasad GL. Suppression of neoplastic transformation and regulation of cytoskeleton by tropomyosins. Somat Cell Mol Genet. 1998;24(5):273–280. doi: 10.1023/b:scam.0000007130.08611.fc. [DOI] [PubMed] [Google Scholar]
- 37.Janssen RA, Veenstra KG, Jonasch P, Jonasch E, Mier JW. Ras- and Raf-induced down-modulation of non-muscle tropomyosin are MEK-independent. J Biol Chem. 1998;273(48):32182–32186. doi: 10.1074/jbc.273.48.32182. [DOI] [PubMed] [Google Scholar]
- 38.Prasad GL, Fuldner RA, Cooper HL. Expression of transduced tropomyosin 1 cDNA suppresses neoplastic growth of cells transformed by the ras oncogene. Proc Natl Acad Sci USA. 1993;90(15):7039–7043. doi: 10.1073/pnas.90.15.7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prasad GL, Masuelli L, Raj MH, Harindranath N. Suppression of src-induced transformed phenotype by expression of tropomyosin-1. Oncogene. 1999;18(11):2027–2031. doi: 10.1038/sj.onc.1202264. [DOI] [PubMed] [Google Scholar]
- 40.Takenaga K, Nakamura Y, Sakiyama S. Suppression of synthesis of tropomyosin isoform 2 in metastatic v-Ha-ras-transformed NIH3T3 cells. Biochem Biophys Res Commun. 1988;157(3):1111–1116. doi: 10.1016/s0006-291x(88)80988-4. [DOI] [PubMed] [Google Scholar]
- 41.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 43.Kaur H, et al. Cadherin-11, a marker of the mesenchymal phenotype, regulates glioblastoma cell migration and survival in vivo. Mol Cancer Res. 2012;10(3):293–304. doi: 10.1158/1541-7786.MCR-11-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mikheeva SA, et al. TWIST1 promotes invasion through mesenchymal change in human glioblastoma. Mol Cancer. 2010;9:194. doi: 10.1186/1476-4598-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467(7319):1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Levin VA, et al. Combination chemotherapy with 13-cis-retinoic acid and celecoxib in the treatment of glioblastoma multiforme. J Neurooncol. 2006;78(1):85–90. doi: 10.1007/s11060-005-9062-4. [DOI] [PubMed] [Google Scholar]
- 49.See SJ, Levin VA, Yung WK, Hess KR, Groves MD. 13-cis-retinoic acid in the treatment of recurrent glioblastoma multiforme. Neuro-oncol. 2004;6(3):253–258. doi: 10.1215/S1152851703000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee JA, Tang ZZ, Black DL. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev. 2009;23(19):2284–2293. doi: 10.1101/gad.1837009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.