Significance
Stapled α−helical peptides have emerged as a promising new modality for a wide range of therapeutic targets. Here, we describe the development of a stapled α−helical peptide lead molecule for the treatment of cancers that possess the intact p53 tumor suppressor protein but are resistant to drug therapy because of the overexpression of inhibitory proteins MDM2 and MDMX. The molecule ATSP-7041 is a highly potent dual inhibitor of both MDM2 and MDMX that is shown to effectively reactivate the p53 tumor suppressor pathway in a mechanism-dependent manner in p53-positive cancers in vitro and in vivo.
Abstract
Stapled α−helical peptides have emerged as a promising new modality for a wide range of therapeutic targets. Here, we report a potent and selective dual inhibitor of MDM2 and MDMX, ATSP-7041, which effectively activates the p53 pathway in tumors in vitro and in vivo. Specifically, ATSP-7041 binds both MDM2 and MDMX with nanomolar affinities, shows submicromolar cellular activities in cancer cell lines in the presence of serum, and demonstrates highly specific, on-target mechanism of action. A high resolution (1.7-Å) X-ray crystal structure reveals its molecular interactions with the target protein MDMX, including multiple contacts with key amino acids as well as a role for the hydrocarbon staple itself in target engagement. Most importantly, ATSP-7041 demonstrates robust p53-dependent tumor growth suppression in MDM2/MDMX-overexpressing xenograft cancer models, with a high correlation to on-target pharmacodynamic activity, and possesses favorable pharmacokinetic and tissue distribution properties. Overall, ATSP-7041 demonstrates in vitro and in vivo proof-of-concept that stapled peptides can be developed as therapeutically relevant inhibitors of protein–protein interaction and may offer a viable modality for cancer therapy.
The human transcription factor protein p53 induces cell-cycle arrest and apoptosis in response to DNA damage and cellular stress and thereby plays a critical role in protecting cells from malignant transformation (1, 2). Inactivation of this guardian of the genome either by deletion or mutation or through overexpression of inhibitory proteins is the most common defect in human cancers (1, 2). Cancers that overexpress the inhibitory proteins MDM2 and MDMX also possess wild-type p53 (p53WT), and thus pharmacological disruption of the interactions between p53 and MDM2 and MDMX offers the opportunity to restore p53-dependent cell-cycle arrest and apoptosis in this important class of tumors (3–6).
MDM2 negatively regulates p53 function through multiple mechanisms, including direct binding that masks the p53 transactivation domain, impairing nuclear import of the p53 protein, and ubiquitination and proteasomal degradation of the p53 protein (6, 7). Consequently, aberrant MDM2 overexpression and gene amplification contribute to accelerated cancer development and growth (1, 8). The other negative regulator, MDMX, possesses a similar p53-binding activity and also effectively inhibits p53 transcriptional activity. Amplification of MDMX is seen in many tumors, including melanoma, breast, head and neck, hepatocellular, and retinoblastoma, and, interestingly, amplification of MDMX appears to correlate with both p53WT status and an absence of MDM2 amplification (6, 9, 10). MDMX does not have the intrinsic E3 ubiquitin ligase activity of MDM2 and cannot affect p53 stability, but MDM2/MDMX heterodimers can increase ubiquitin ligase activity relative to the MDM2 monomer. Given these functional differences, MDM2 and MDMX are each unable to compensate for the loss of the other, and they regulate nonoverlapping functions of p53 (4, 6).
The first potent and selective small-molecule inhibitors of the p53–MDM2 interaction, the Nutlins, provided proof of concept that restoration of p53 activity is feasible and may have application in cancer therapy (11, 12). Although three different classes of small-molecule MDM2 antagonists are currently under clinical investigation, one potential limitation of these molecules is that they are all practically inactive against MDMX. Although the relative contributions of MDM2 and MDMX to regulation of p53 are not completely understood, several lines of evidence suggest that selective MDM2 antagonists will not be optimally effective in tumors that express high levels of MDMX (1, 6, 10, 13). Despite the structural similarity between MDM2 and MDMX, there is sufficient diversity in the p53-binding regions of these proteins to make the development of small-molecule dual antagonists challenging. We recently reported a potent and selective small molecule, RO-5963, that effectively inhibits p53 binding to both MDM2 and MDMX via a protein dimerization mechanism of action (14). However, the poor pharmacological characteristics of this molecule render it unsuitable for further development. Thus, despite intensive efforts, a therapeutically viable dual MDM2/MDMX inhibitor remains elusive.
One solution to this challenge may lie in converting the p53 α-helix from the native p53–MDM2/MDMX complex into a suitably stable, potent, and specific therapeutic agent (3, 15, 16). Stapled peptides were first described as a novel approach to creating macrocyclic α-helical peptides through the addition of an all-hydrocarbon cross-link between two α−methyl-substituted amino acids having terminal olefin side-chains to enable ring-closing metathesis using Grubbs ruthenium catalyst (17, 18). Historically, it is important to recognize the precedence macrocyclization of the O-allyl-modified peptides by the Grubbs research group (19, 20) as well as intrinsic helical-inducing properties of linear peptides incorporating α−methyl-substituted amino acids by the Toniolo research group (21, 22). Relative to a previously published stapled peptide, SAH-p53-8 (3, 15), the limited biological potency determined in our cellular assays using more stringent physiologic conditions (i.e., in the presence of 10% serum) inspired us to tackle this highly important cancer target and advance a more rigorously tested preclinical lead compound relative to both in vitro and in vivo efficacy. Here we describe the discovery of ATSP-7041 as the first highly potent and specific stapled peptide dual inhibitor of MDM2/MDMX that possesses robust drug-like properties and on-mechanism in vitro and in vivo activity. We provide the first high resolution (1.7Å) crystallographic structure of a stapled peptide bound to MDMX and further evaluate the biophysical, biochemical, cell penetrating and on-target cellular properties of ATSP-7041. Our results demonstrate that antagonizing both negative p53 regulators effectively suppresses the growth of human tumor xenografts overexpressing MDM2/MDMX and suggest that stapled peptides may offer a new modality for p53-activating cancer therapy.
Materials and Methods
ATSP-7041 and Related Stapled Peptide Synthesis.
Reagents used were as follows. Fmoc-protected α-amino acids (other than the olefinic amino acids Fmoc-S5-OH and Fmoc-R8-OH) were purchased from Advanced Chemtech or Novabiochem. The 2-(6-chloro-1-H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium hexafluorophosphate (HCTU), 6-Chloro-1-Hydroxy-1H-Benzotriazole (6-Cl-HOBt), (7-Azabenzotriazol-1-yloxy)tripyrrolidino-phosphonium hexafluorophosphate (PyAOP), 2-(7-Aza-1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HATU), and 1-Hydroxy-7-azabenzotriazole (HOAt) were purchased from Advanced Chemtech or Novabiochem. Rink Amide linker modified AmphiSpheres40 resin was from Agilent Technologies. Dimethylformamide (DMF), N-methyl-2-pyrrolidinone (NMP), N,N-diisopropylethylamine (DIEA), acetic anhydride, trifluoroacetic acid (TFA), dichloromethane (DCM), anisole, triisopropylsilane (TIS), 5-carboxyfluorescein (5-FAM), and piperidine were purchased from EMD Milipore Corporation and Sigma-Aldrich and used as supplied. The (1,3-bis-(2,4,6-trimethylphenyl)-2-imidazolidinylidene)dichloro(o-isopropoxyphenylmethylene)ruthenium (Hoveyda-Grubbs catalyst second-generation catalyst) was obtained from Materia Inc. The synthetic methodology used for the olefinic amino acids (R8 and S5) has been described elsewhere (17, 23).
The precursor linear peptides were synthesized by following the standard Fmoc peptide synthesis protocol with Rink amide linker modified AmphiSpheres 40 resin (substitution = 0.35–0.45 mmol/g) on a Tetras multichannel peptide synthesizer (Thuramed) or Focus XC (AAPPTec). For each regular coupling reaction, 5 equivalents (Eq) of Fmoc-amino acid, 5 Eq of HCTU, 5 Eq of 6-Cl-HOBt, and 10 Eq of N,N- diisopropylethylamine (DIEA) were used. For the coupling of the olefin building blocks, 2 Eq of amino acid, 2 Eq of HATU, 2 Eq of HOAt, and 4 Eq of DIEA were used. The coupling reaction was allowed to proceed for 180 min in N-Methylpyrrolidone (NMP). The Fmoc deprotection was accomplished by treating the peptide-bound resin with 20% (vol/vol) piperidine/DMF (for 10 min followed by treatment for 15 min). After the amino acids were assembled, the N-terminal amine was acetylated using a solution of acetic anhydride and DIEA (5/1) in DMF. For the stapled peptides ATSP-7041 1 and 2, ring-closing metathesis was performed on the peptide while still on the solid support in a disposable fritted reaction vessel. The peptide resin was cyclized in the presence of Hoveyda-Grubbs catalyst second-generation catalyst (1,3-bis-(2,4,6-trimethylphenyl)-2-imidazolidinylidene)dichloro(o-isopropoxyphenylmethylene)ruthenium) in 1,2-dichloroethane for 8 h at 50 °C. Completeness of the ring-closing metathesis (RCM) reaction was verified by liquid chromatography-mass spectrometry (LC-MS). The resin-bound peptide was washed six times with DMF and six times with DCM and dried under vacuum. In the case of fluorescein-labeled peptides, Fmoc-beta-Alanine was added as a spacer to the N terminus of the peptide followed by RCM reaction with (1,3-bis-(2,4,6-trimethylphenyl)-2-imidazolidinylidene)dichloro(o-isopropoxyphenylmethylene)ruthenium (Hoveyda-Grubbs catalyst second generation catalyst) as described above. Then Fmoc group was removed with 20% piperidine in DMF, and 5-Carboxyfluorescein was coupled with PyAOP/HOAt/DIEA in NMP (1:1:2 molar ratio, twofold excess).
The alkene metathesized resin-bound peptides were deprotected and cleaved from the solid support by treatment with TFA/H2O/TIS/anisole (85/5/5/5 vol/vol) for 2.5 h at room temperature. After filtration of the resin, the TFA solution was evaporated, and peptide was precipitated by addition of cold hexane/diethyl ether mixture (1:1, vol/vol) and centrifuged to yield the desired product as a solid. Purification of cross-linked compounds was achieved by mass triggered high performance liquid chromatography (HPLC; Waters) on a reversed-phase C8 column (Phenomenex; Luna C8) to yield the pure compounds. Chemical composition of the pure products was confirmed by LC-MS mass spectrometry (SQ Detector interfaced with Waters Acquity UPLC system). The analytical LC-MS chromatogram and mass spectral data of ATSP-7041 and its fluorescently-labeled analog (FAM-ATSP-7041), as well as the mass spectral data for each of the stapled peptides tested in this study, provided confirmation of purity and structural integrity (SI Text, Fig S1, and Tables S1 and S2).
Biophysical Properties: α−Helicity and X-ray Crystal Structure Determination.
The solution conformational properties of ATSP-7041 and the linear peptide ATSP-3748 were evaluated by using circular dichroism (CD) spectroscopy obtained on a spectropolarimeter (e.g., Jasco J-710) using standard measurement parameters (e.g., temperature, 20 °C; wavelength, 190–260 nm; step resolution, 0.5 nm; speed, 20 nm/s; accumulations, 10; response, 1 s; bandwidth, 1 nm; path length, 0.1 cm). The α-helical content of each peptide was calculated by dividing the mean residue ellipticity (e.g., [Φ]222obs) by the reported value for a model helical decapeptide (24).
Cloning, Expression, and Purification of Humanized Zebrafish MDMX.
The DNA sequence encoding zebrafish MDMX amino acid residues 15–106 with an N-terminal hexa-His tag and an enterokinase recognition sequence was amplified by PCR using an MGC cDNA clone as the template (MGC113995; Mammalian Gene Collection). This DNA fragment was coned into NcoI/NdeI predigested pET15b vector (Novagen). The L46V/V95L double mutant was generated by DNA site-directed mutagenesis using the QuikChange Kit (Stratagene). All DNA sequences were confirmed by sequencing. Plasmid DNA was transformed into BL-21 (DE3) cells for protein expression.
The Escherichia coli BL-21 (DE3) cells described above were grown at 37 °C and induced with 0.2 mM isopropyl β-D-1-thiogalactopyranoside at an OD600 of 0.7. Cells were then allowed to grow for 18 h at 23 °C. Protein was purified from harvested cells using Ni-NT Agarose followed by Superdex 75 buffered with 50 mM NaPO4, pH 8.0, 150 mM NaCl, and 2 mM TCEP and concentrated to 16 mg/mL.
For crystallization, the buffer of the protein stock solution was exchanged to 20 mM Tris, pH 8.0, 50 mM NaCl, 2 mM DTT. In preparation for cocrystallization with ATSP-7041, a stoichiometric amount of the compound from a 100 mM stock solution in DMSO was combined with the zebrafish MDMX protein solution and allowed to sit overnight at 4 °C before setting up the crystallization experiments. Crystals were obtained with the Jena Biosciences HTS I screen solution D8 [15% (wt/vol) polyethylene glycol 4000, 0.1 M sodium citrate, pH 5.6, 0.2 M ammonium sulfate] as the reservoir using the sitting drop vapor diffusion method. Drops were prepared by combining equal volumes (2 µL each) of the protein/peptide mixture and the reservoir solution and placed over the reservoir solution (500 µL) to allow vapor equilibration. Setups were stored at 5 °C, and crystals grew within 4–5 d.
Data Collection, Structure Solution, and Refinement.
Crystals grew as thin plates and could be cryoprotected by first passing them through a solution containing 20% (vol/vol) glycerol and 80% reservoir solution and then plunging them into liquid nitrogen. Data were collected at the Swiss Light Source beamline ×10SA equipped with a Pilatus 6M array detector. Data were processed with XDS (25) and were then scaled and reduced with the Scala and Truncate programs from the CCP4 Suite (26). The structure was solved by molecular replacement using the program Molrep (27) and the zebrafish structure with PDB ID code 2z5t (28) as the search model. All refinement was carried out with CNX (29). Details of the data collection and structure refinement are available (SI Text, Table S3).
Topochemical Property Analysis from Molecular Modeling Studies.
The 3D models of SAH-p53-8, ATSP-3900, and ATSP-7041 were derived from the X-ray crystal structures of MDMX complexed with each of these stapled peptides. The comparative topochemical properties of canonical α−helices of SAH-p53-8, ATSP-3900, and ATSP-7041 are shown relative to their solvent accessible surfaces as computationally derived using Discovery Studio (Accelerys Inc.). A more detailed analysis of the comparative conformational properties of ATSP-7041 and related peptides complexed with MDM2 or MDMX is provided (SI Text, Figs. S2 and S3) to further understand the impact of the staple moiety on the target protein binding.
Fluorescence Polarization Binding and Biacore Assays.
To determine dissociation constants for peptide–protein interaction, a fluorescence polarization assay (FPA) was used in which 10 nM of the fluorescently labeled analog of the linear peptide ATSP-3848 (specifically, FAM-β-Ala-Leu-Thr-Phe-Glu-His-Tyr-Trp-Ala-Gln-Leu-Thr-Ser-NH2 [FAM-ATSP-3848]) was incubated with 10 µM to 5 pM MDM2(1-138) and MDMX(1-134). Fluorescence polarization was measured after 3 h incubation on an Infinite F500 micro plate reader (Tecan). For competition assays, MDM2 or MDMX protein concentration to be used was calculated using the EC80 from the direct binding assay. The protein was then combined with a serial dilution of test stapled peptide (threefold dilution starting at 5 μM), followed by the addition of FAM-ATSP-3848 (10 nM). Fluorescence polarization was measured after 3 h incubation using an Infinite F500 micro plate reader (Tecan). IC50 values of the FAM-ATSP-3848 displacement were calculated by nonlinear regression analysis using Prism software (GraphPad Software).
The Ki values of competitive inhibitors were calculated using the method described in ref. 30 and described below:
where Ro = receptor concentration, Lo = total tracer concentration, and Lb = bound tracer concentration. The measured IC50 values and the Kd calculated from the direct binding to the protein are used for the above calculation. This equation is used instead of the Cheng-Prusoff as >10% of the tracer is bound to the receptor in the assay.
Biacore studies were performed as previously described (11). In short, competition assays were performed on a Biacore S51 (Biacore Inc.). A Series S Sensor chip CM5 was derivatized for immobilization of a Penta His antibody (Qiagen) for capture of the His-tagged p53. The level of capture was ∼200 response units (1 response unit corresponds to 1 pg of protein per mm2). The concentration of MDM2 or MMDX proteins was kept constant at 300 nM. The assays were run at 25 °C in running buffer [10 mM Hepes, 0.15 M NaCl, 2% (vol/vol) DMSO].
Imaging Fluorescently Labeled ATSP-7041 in HCT-116 Cells.
Human colorectal carcinoma cell line HCT-116 cells were seeded at 60,000 cells per well in four-well chambered cover-glass (Lab-Tek II) in normal medium [DMEM with 10% (vol/vol) FBS] and allowed to grow overnight. Cells were then incubated with 20 μM FAM-labeled ATSP-7041, 20 μM FAM-labeled-mt-ATSP-7041, or DMSO control for 4.5 h in Opti-MEM with 10% FBS. Cells were washed with Dulbecco's phosphate buffered saline and incubated with WGA-alexa 647 conjugate at 10 μg/mL for an additional 30 min to stain the cell membrane. Cells were washed, fixed with 4% (wt/vol) paraformaldehyde, and imaged using an LSM 510 Zeiss Axiovert 200M (v4.0) confocal microscope using the 63× oil lens. Images were analyzed using an LSM image browser.
Immunoprecipitation and Western Blot Analysis.
Immunoprecipitations were performed as previously described (14). Briefly, exponentially growing MCF-7 cells were treated with 10 μM ATSP-7041 for 4 h. For immunoprecipitation, mouse monoclonal anti-p53 (Santa Cruz) and rabbit polyclonal anti-MDMX (Bethyl Laboratories) were used. Immune complexes were then collected on protein G Sepharose beads at 4 °C overnight, and beads were washed five times with cold lysis buffer. Precipitated proteins were subjected to Western blotting with rabbit anti-p53 (Santa Cruz), mouse anti-MDMX (Sigma), mouse anti-MDM2 (Santa Cruz), and mouse anti-Actin (Sigma) antibodies. For Western blotting, cells were grown in triplicate in six-well plates (105 cells per well), lysed in 0.1–0.2 mL of radioimmunoprecipitation assay (RIPA) buffer, and analyzed as described previously (14).
Cell Lines and Cell Culture.
Human cancer cell lines (SJSA-1, RKO, HCT-116, MDA-MB-435, MCF-7, and SW-480) were purchased from American Type Culture Collection. The culture media and supplements were obtained from Life Technologies. All cell lines were maintained in indicated media supplemented with 10% FBS, 2 mM l-glutamine, 1 mM sodium pyruvate, and penicillin-streptomycin (100 IU/mL). The cultures were incubated at 37 °C in a humidified atmosphere of 5% CO2.
Annexin V Assay.
Cells were seeded in 24-well tissue culture plates (5 × 104 cells per well) 24 h before drug treatment and incubated with the drug for an additional 48 h. No treatment controls were established in parallel for each cell line. Culture medium that may contain detached cells was collected, and attached cells were trypsinized. Cells were combined with corresponding medium and collected by centrifugation at 240 × g for 10 min at 4 °C. Annexin V-positive cells were quantified using Guava Nexin kit and the Guava personal cell Analyzer (Guava Technologies) as recommended by the manufacturer.
Cell-Cycle Analysis.
For cell-cycle analysis, SJSA-1 and MCF-7 cells (1× 106 cells per T75 flask) were incubated with 0.3, 3, or 10 μM ATSP-7041 or an equivalent amount of solvent for 24 h. BrdU (20 μM; Sigma) was added during the last 2 h before fixation, and cells were processed and analyzed as previously described (14). Briefly, cell-cycle distribution was analyzed after propidium iodide/fluorescein isothiocyanate–antibody to BrdU staining. Apoptotic cells were detected by dual color (fluorescein/propidium iodide) flow cytometry using the APO-BRDU kit from Phoenix Flow Systems as recommended by the manufacturer.
Cell-Viability Assay.
Cell viability was assessed by addition of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) following the manufacturer’s protocol (Promega). The absorbance was measured at 570 nm using the Infinite F500 microplate reader (Tecan), and a reference wavelength of 650 nm was used. All assays were performed in triplicate, and data are normalized to vehicle treated control and analyzed using Prism software (GraphPad Software).
Antitumor Efficacy in Human Xenograft Models.
All animals were allowed to acclimate and recover from any shipping-related stress for a minimum of 72 h before experimental use. Autoclaved water and irradiated food (5058-ms Pico Chow; Purina) were provided ad libitum, and the animals were maintained on a 12-h light and dark cycle. Cages, bedding, and water bottles were autoclaved before use and were changed weekly. The health of all animals was monitored daily by gross observation and analyses of blood samples of sentinel animals. All animal experiments were performed in accordance with protocols approved by the Roche Animal Care and Use Committee.
For SJSA-1, 5 × 106 cells in 0.2 mL of 1:1 Matrigel:PBS were injected s.c. into the right lateral flank of athymic nude mice. For the MCF-7 human breast cancer xenograft model, which is estrogen-responsive and requires hormone supplementation for tumor establishment and growth, female BALB/c nude (CAnN.Cg-Foxn1nu/Crl; Charles River Laboratories) mice were implanted s.c. with 90 d 0.72 mg sustained release 17β-estradiol pellets (Innovative Research) on the nape of the neck, at least one day before cell implantation. Mice were implanted s.c. in the right flank with 1 × 107 MCF-7 cells suspended in a 0.2-mL volume of a 1:1 mixture of Matrigel:PBS. In both tumor models, when mean tumor volume reached ∼100–200 mm3, mice were randomized into treatment groups (n = 10 per group) and received either vehicle (50 mg/mL MPEGDSPE in histidine-buffered saline) i.v. daily (qd), or ATSP-7041 qd or every other day (qod) at the doses indicated. In the MCF-7 study, RG7112 was administered orally (p.o.) qd at the doses indicated in 1% (wt/vol) Klucel LF/ 0.1% Tween 80. Tumor volume was monitored by caliper measurement, and mouse body weights were recorded. Tumor volume and percent Tumor Growth Inhibition (TGI) was calculated as %TGI = 100 − [100 × (T − T0)/(C − C0)], in which T and C represent mean tumor volume on a specific day during the experiment of a treated group and the vehicle control group, respectively, and T0 and C0 represent mean tumor volume on the first day of treatment of the same treated group and the vehicle control group, respectively. Efficacy data were graphically represented as the group mean tumor volume ± SEM. Statistical analysis was determined by two-way ANOVA and Bonferroni post test (GraphPad Prism version 5.03). Differences between groups were considered significant when the probability value was ≤0.05.
RNA Extraction and RT-PCR.
Total RNA was extracted from tissue culture cells or tumor xenografts using an RNeasy Mini Kit (Qiagen) after homogenization by QIAShredder (Qiagen) according to the manufacturer’s instructions. For xenografts, tumors were first homogenized in lysis buffer using a motorized tissue homogenizer before the QIAShredder step. Total RNA was reverse transcribed using MultiScribe Reverse Transcriptase (Life Technologies), and real-time PCR was performed as previously reported (12).
Results
ATSP-7041 Is a Dual Inhibitor of MDM2 and MDMX.
Optimization of p53 stapled peptides was undertaken to improve both their biological and biophysical properties, and this optimization leveraged the sequence enhancements of a recently described phage display peptide (pDI) Ac-Leu17-Thr-Phe-Glu-His-Tyr-Trp-Ala-Gln-Leu-Thr-Ser28-NH2 (Fig. 1A) (31, 32). The prototype molecule Ac-Leu17-Thr-Phe-cyclo(R8-His-Tyr-Trp-Ala-Gln-Leu-S5)-Ser28-NH2 (ATSP-3900) achieved striking improvement in binding potency to MDM2 (Ki = 8 nM) and MDMX (Ki = 12.7 nM) compared with SAH-p53-8 (3, 15). The core binding triad of Phe19, Trp23, and Leu26 (enumeration based on human p53) are established molecular recognition elements in the binding of both the SAH-p53-8 stapled peptide and the linear pDI peptide to the target proteins MDM2 and MDMX. However, unique contributions also exist, including (i) the staple moiety of SAH-p53-8, which binds to a hydrophobic cleft of MDM2, and (ii) the Tyr22 of pDI, which has been identified as a fourth key amino acid by several phage display studies (31, 33, 34) and which has been shown to exhibit multiple hydrophobic, π−cation, and electrostatic interactions as well as water-mediated H-bonding within its binding sites in both MDM2 and MDMX. Through iterative lead optimization, ATSP-3900 was further modified to enhance target binding, biophysical properties (e.g., solubility), and cell potency. Specifically, His21 substitution by Ala and Glu provided a significant effect on solubility (i.e., approximately fourfold decreased and fourfold increased, respectively). Incorporation of Glu21 and modification of Leu26 by Cba and C-terminal extension with Ala-Ala yielded Ac-Leu17-Thr-Phe-cyclo(R8-Glu-Tyr-Trp-Ala-Gln-Cba-S5)-Ser-Ala-Ala30-NH2 (ATSP-7041), which exhibited high affinity binding to both MDM2 (Ki = 0.9 nM) and MDMX (Ki = 7 nM) (Fig. 1A). Overall, the iterative modifications of ATSP-3900 to advance ATSP-7041 exemplify improved biological potency with respect to MDM2 and MDMX, and a more detailed understanding of the subtleties of the structure–property relationship will be described elsewhere. A negative control analog of ATSP-7041 was rationally designed by incorporating a single mutation of Phe19 to Ala19 (ATSP-7342), which resulted in significantly diminished binding to both MDM2 (Ki = 536 nM) and MDMX (Ki >1,000 nM). By comparison, Nutlin-3a exhibits preferential binding to MDM2 (Ki = 52 nM) versus MDMX (Ki >1,000 nM).
Fig. 1.
The chemical structure of ATSP-7041 is shown highlighting the R8 (blue) and S5 (red) amino acids in the stapled peptide following ring-closing metathesis (and loss of ethylene). Note that each R8 and S5 is α-methylated and that the all-hydrocarbon linker consists of 11 carbon atoms. (A) Sequences of a series of stapled peptides and their binding properties to MDM2 and MDMX. Lead optimization of the linear phage display peptide (pDI) resulted in the highly potent lead stapled peptide ATSP-7041. ATSP-7041 exhibits high-affinity binding to both MDM2 and MDMX versus the small-molecule Nutlin-3a. The Phe19 to Ala19 mutant negative control analog (ATSP-7342) is essentially devoid of binding to MDM2 and MDMX as expected. (B) Biacore studies confirmed the high affinity of ATSP-7041 to MDM2 and MDMX and also reveal its fast on-rate but rather slow off-rate for target binding. (C) ATSP-7041 exhibits high potency in SJSA-1 cell viability assay in the presence of increasing serum in contrast to that of stapled peptides SAH-p53-8 and the pDI analog ATSP-3900.
Binding studies of ATSP-7041 using Biacore provided further insights into the high affinity of ATSP-7041 to MDM2 (KD = 0.91 nM) and MDMX (KD = 2.31 nM) and also indicated a fast rate of association and a rather slow rate of dissociation (Fig. 1B). By comparison, the dissociation rate (dissociation t1/2) of small-molecule inhibitors of MDM2 is in the order of ∼6 min compared with 43 min for the stapled peptide ATSP-7041. The resulting improved target binding properties of ATSP-7041 translated to a 25-fold enhancement of potency in SJSA-1 cells in the presence of 10% FBS relative to the prototype stapled pDI peptide ATSP-3900 and to a >50-fold enhancement of cellular potency relative to SAH-p53-8 (Fig. 1C). The observed >10-fold increased cell potency of ATSP-7041 versus ATSP-3900 likely reflects overall improved cell penetration efficiency due, in part, to its greater amphipathic α−helical nature (see next section) and solubility properties as related to the C-terminal Ala29-Ala30 extension and Glu21 modifications, respectively. Furthermore, we investigated the plasma albumin binding properties of ATSP-7041 and ATSP-3900. It was determined that each ATSP-7041 and ATSP-3900 were highly complexed to human albumin (98.2% and 97.2% fraction bound, respectively) and that such findings were comparable for ATSP-7041 relative to monkey, dog, rat, and mouse albumins (SI Text, Table S4).
ATSP-7041 Binds to MDM2 and MDMX with the Same High-Affinity Conformation.
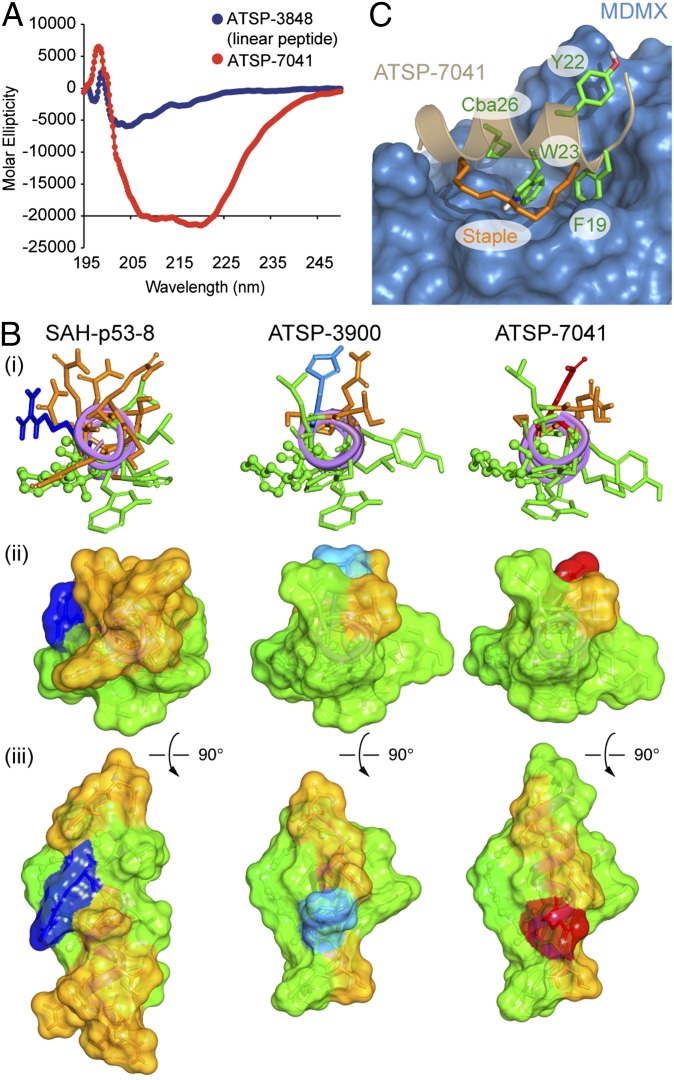
The biophysical properties of ATSP-7041 were also improved as confirmed by its solubility studies and analysis of its conformational and 3D structural properties. The solution conformation of ATSP-7041 was determined by CD studies to be highly α−helical (70% at pH 7.0; Fig. 2A) whereas the corresponding linear pDI peptide was weakly α−helical (11% at pH 7.0; Fig. 2A). Visualizing the comparative structural features of ATSP-7041, SAH-p53-8, and the prototype stapled pDI analog (ATSP-3900) as stick figure and space-filling models highlights the distinct topochemical relationships of these peptides (Fig. 2B). In contrast to SAH-p53-8, ATSP-3900 forms an idealized amphipathic helix with the hydrophobic amino acids Phe19, Tyr22, Trp23, and Leu26, forming a continuous hydrophobic patch that resembles the bound conformation of the p53WT peptide. Additionally, the deletion of the polar residues Gln14, Ser15, and Gln16 as well as the presence of Ala and Leu, respectively, at positions 17 and 24 further widen the hydrophobic patch and limit the polar face to only four polar amino acids (Thr18, His21, Gln25, and Ser28). However, studies with ATSP-3900 were somewhat limited by its solubility, as evidenced by peptide precipitation at 30 μM. Indeed, CD analysis of ATSP-3900 could not be performed at physiological pH without the addition of TFE. Thus, the analog ATSP-7041 was designed to retain the overall amphipathicity of the parent ATSP-3900 while possessing improved solubility to enable CD analysis to be readily performed in 50 mM phosphate, 100 mM KCl, pH 7.
Fig. 2.
Conformational and 3D-structural properties of ATSP-7041. (A) Comparative CD spectroscopy of pDI and stapled peptide ATSP-7041in pH 7 buffer illustrates their intrinsic α−helical properties. (B) The α−helical wheel projections (3D) of ATSP-7041 versus SAH-p53-8 and the stapled pDI analog ATSP-3900 show the extended hydrophobic surface contributing to the enhance amphipathicity of ATSP-7041. (i) The three stapled peptides are represented as fully α-helical peptides with the viewer facing the long axis of the helix by the N terminus. Side chains are represented in stick style whereas the olefin staple is in ball and stick style. Polar residues (Asn, Gln, Ser, and Thr) are colored orange; green is for hydrophobic, olefin staple, and aromatic residues (Ala, Val, Leu, Ile, Pro, Cba, Phe, Tyr, and Trp), blue for basic residues (Lys and Arg), red for acidic residues (Asp and Glu), and light blue for His. (ii) Solvent exposed surfaces representation of the three stapled peptides with the same view angle as in ref. 1 and the same coloring. (iii) Other view of the solvent exposed surfaces, after vertical rotation of 90°. (C) A high-resolution X-ray structure of ATSP-7041 bound to MDMX delineates its intermolecular contacts with the target protein. The side chain is colored in green: Phe19, Trp23, Cba26, and Tyr22 are the main interacting residues responsible for the high binding affinity whereas the staple olefin, also contributing to the binding affinity, is colored in orange (see Results for detailed discussion).
Specifically, incorporation of the Glu21 on the polar face and extending the helical structure of ATSP-3900 with two additional Ala residues at the C terminus resulted in ATSP-7041.
To determine the molecular basis for the unique high-affinity target-binding properties of ATSP-7041, we solved the crystal structure of ATSP-7041 bound to MDMX. Crystals of humanized zebrafish MDMX (15-106, L46V/V95L) bound to ATSP-7041 readily formed in solutions containing 15% (wt/vol) polyethylene glycol, enabling the collection of a high-quality dataset extending to 1.7 Å resolution. The resulting structure (Fig. 2C) shows the peptide bound into the p53 binding site of MDMX as expected, using the three key positions identified originally by Kussie et al. (35) and highlighted in Fig. 2 B and C. Interestingly, beyond the well-known triad of Phe19, Trp2, and Leu26 (mutated to Cba26), ATSP-7041 demonstrated additional interactions between Tyr22 and the staple moiety itself with the MDMX protein. The Tyr22 interacts with MDMX binding pocket through Van der Waals contact with Gln66, Arg67, Gln68, His69, Val89, and Lys90 as well as through water-mediated hydrogen bonds with Nε of the Lys90 side-chain and Nδ1 of the His68 side-chain. An extensive binding pocket for the staple exists on the MDMX protein, and a number of van der Waals contacts (to Lys47, Met50, His51, Gly54, Gln55, and Met58) were also observed. A distinguishing characteristic of this interaction between the staple moiety and the MDMX protein is a possible cation–π interaction between ND1 of His51 and the double bond in the staple. This type of interaction normally occurs over a distance of 4.5–5.1 Å (36), but, in this case, the nitrogen distances to the two carbon atoms are 3.8 and 4.0 Å. It is also noteworthy that close association of the staple moiety with the protein has also been observed in four other structures of proteins bound to stapled peptides, including SAH-p53-8 bound to MDM2 (37), MCL-1 SAHBD bound to MCL-1 (38), SP1 bound to the ERβ LBD, and SP2 bound to ERα (39), highlighting the potential of the hydrocarbon staple to further enhance binding affinity while preserving ligand/target selectivity.
ATSP-7041 Penetrates Cell Membranes and Exhibits a More Durable Effect on p53 Signaling than Small-Molecule MDM2-Selective Inhibitors.
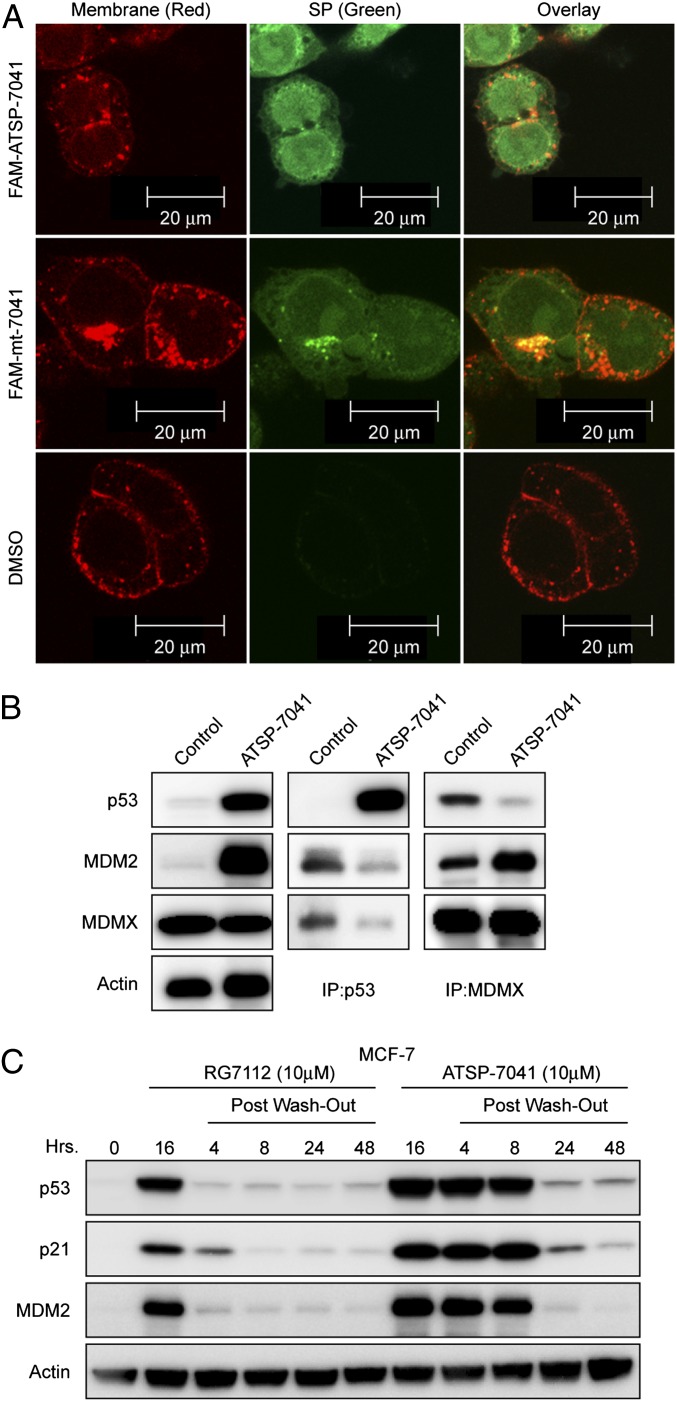
To investigate the cellular uptake of ATSP-7041, a fluorescent FAM moiety was appended via a β-alanine spacer to the N terminus of ATSP-7041. Importantly, FAM-ATSP-7041 was found to be equipotent to the parent in terms of MDM2 and MDMX binding and cellular potency, indicating that addition of the fluorophore does not significantly alter the biophysical/biological properties of ATSP-7041. HCT-116 cells treated with either 20 μM FAM-ATSP-7041 or FAM-mt-7041 (the FAM-labeled version of stapled peptide ATSP-7342, where a single point mutation drastically reduces binding to both MDM2 and MDMX) were stained with wheat germ agglutinin (WGA) membrane stain and imaged 4.5 h posttreatment (Fig. 3A). FAM-ATSP-7041 and FAM-mt-7041 showed a diffused intracellular localization, confirming efficient cellular penetration of both active and inactive control FAM-labeled stapled peptides.
Fig. 3.
ATSP-7041 penetrates cell membranes and disrupts p53-MDM2 and p53-MDMX binding. (A) A fluorescent FAM was linked via a β-alanine spacer to the N terminus of ATSP-7041. HCT116 cells were seeded at 60,000 cells per well and incubated with 20 μM stapled peptides or DMSO control for 4.5 h. Cells were imaged using an LSM 510 Zeiss Axiovert 200M (v4.0) confocal microscope using the 63× oil lens. The equipotent FAM-labeled ATSP-7041 showed a diffused intracellular localization, demonstrating efficient cellular penetration. Similarly the FAM-mt-7041 containing single F19 to A19 modification that exhibits no binding or cell activity also showed efficient cell penetration, confirming that its inactivity is due to lack of binding to the target proteins. (B) ATSP-7041 inhibits p53-MDM2 and p53-MDMX binding in cancer cells. MCF-7 cells were incubated with 10 μM ATSP-7041 for 4 h, and the levels of p53, MDM2, and MDMX were determined in protein complexes immunoprecipitated with anti-MDMX or anti-p53 antibodies by Western blotting. ATSP-7041 induces a dose-dependent decrease in the level of both MDM2 and MDMX associated with p53 in comparison with the vehicle control (DMSO). Similarly, IP of MDMX from ATSP-7041 dosed MCF-7 cells shows a significant dose-dependent decrease in the level of p53 in the complex in comparison with the vehicle control (DMSO). ATSP-7041 does not inhibit the complex between MDM2 and MDMX. (C) ATSP-7041induced a more durable effect on p53 signaling than small-molecule, MDM2-selective inhibitors. Induction of p53, p21, and MDM2 was unchanged up to 4 h after drug removal and continued to be elevated above the control up to 48 h in response to ATSP-7041treatment. In contrast, RG7112 lost most of its inhibitory effect at 4 h after media replacement and maintained p53 and target protein levels only slightly above basal levels.
To assess the ability of ATSP-7041 to inhibit p53 binding to MDM2 and MDMX, we incubated MCF-7 cells with the stapled peptide at 10 µM for 4 h and determined the levels of MDM2 and MDMX associated with p53 by immunoprecipitation (IP) from cell lysates followed by Western blotting. Our results revealed substantial reduction of both proteins bound to p53 in lysates from cells treated with ATSP-7041 compared with DMSO controls (Fig. 3B). These studies confirmed the ability of ATSP-7041 to penetrate living cancer cells in the presence of 10% FBS and inhibit the binding of its target proteins MDM2 and MDMX to p53.
Biacore experiments with ATSP-7041 indicated slow off-rate kinetics and suggested a prolonged inhibitory effect on MDM2 and MDMX binding (Fig. 1B). To assess whether these properties have an effect on the p53 activity in cells, we incubated exponentially growing MCF-7 cells with 10 µM ATSP-7041 for 16 h followed by washing and the addition of fresh drug-free media. As a comparator, the investigational small-molecule MDM2 inhibitor RG7112 was used (40). Samples were taken over the indicated time course, and the levels of p53, p21, and MDM2 were analyzed by Western blotting (Fig. 3C). Strikingly, the inhibitory effect of ATSP-7041 is strongly evident at 4 and 8 h after drug removal and persists above the control for up to 48 h. In contrast, RG7112 lost most of its inhibitory effect after 4 h. These data indicate that ATSP-7041 exerts a notably prolonged inhibitory effect compared with the small-molecule MDM2 antagonists.
ATSP-7041 Activates p53 Signaling in Cancer Cells.
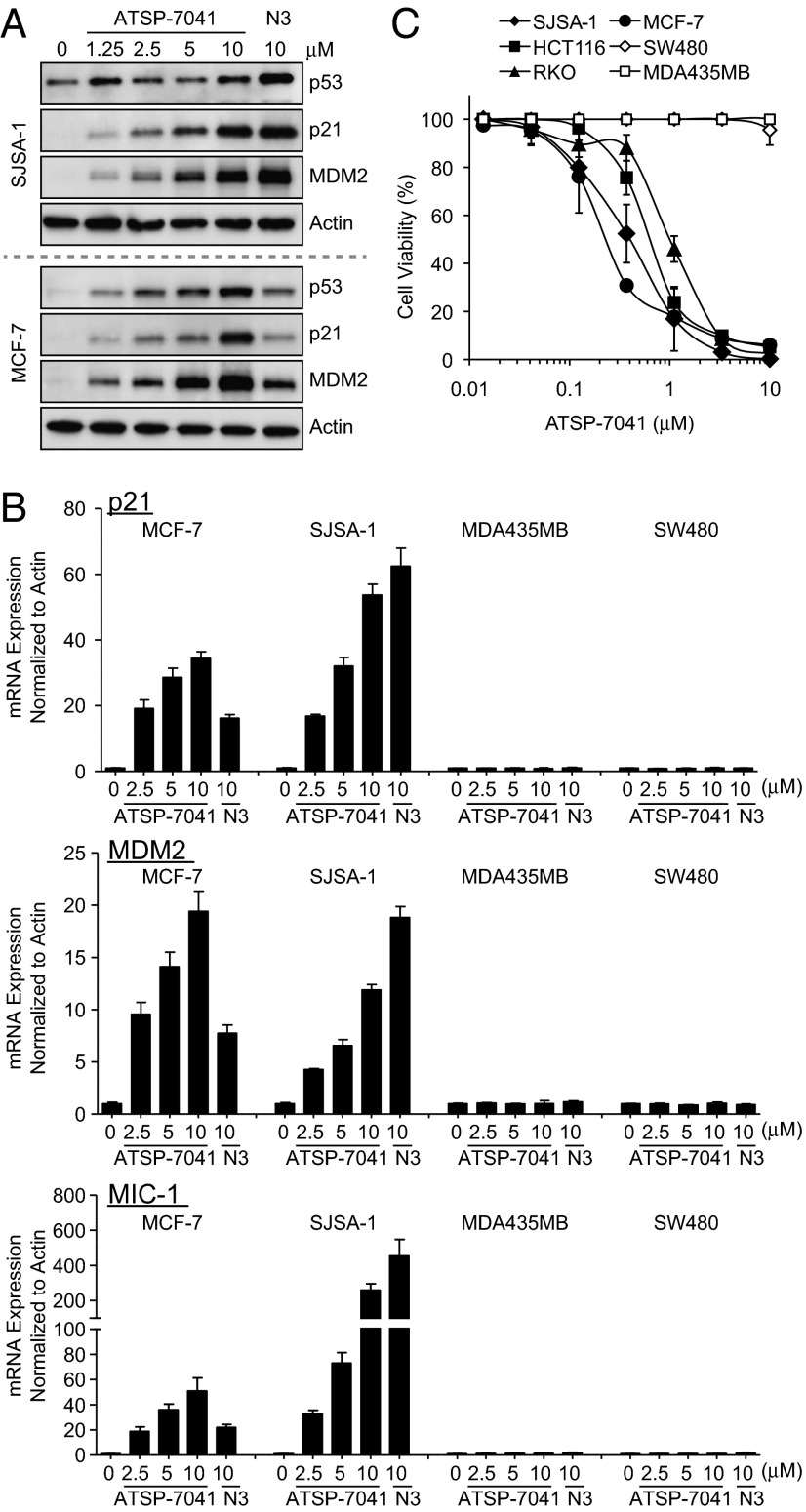
Inhibition of p53 binding to its negative regulators, MDM2 and MDMX, should stabilize p53 and activate the pathway. This pathway activation should occur only in cells that express the wild-type but not the mutant form of the p53 tumor suppressor. To assess the activity of ATSP-7041, we chose cancer cell lines representing two clinically relevant populations of tumors that overproduce MDM2 (SJSA-1 osteosarcoma) or MDMX (MCF-7 breast cancer). Treatment with ATSP-7041 led to a dose-dependent increase of p53 protein levels in both cell lines and concomitant elevation of the p53 transcriptional targets p21 and MDM2 (Fig. 4A). ATSP-7041–induced protein levels were comparable with that induced by the MDM2-specific inhibitor Nutlin-3a in SJSA-1 cells, but ATSP-7041–induced levels were significantly higher than Nutlin-3a in MCF-7 cells. This difference is expected because Nutlin does not affect p53-MDMX binding and cannot fully activate p53 in MCF-7 cells due to MDMX overexpression (24). Nutlin’s lack of MDMX activity does not manifest itself in SJSA-1 cells, which have amplification of the MDM2 gene and high MDM2 protein levels but very low levels of MDMX (24). The p53 activation effect of ATSP-7041 was demonstrated by the dose-dependent induction of mRNA of three p53 target genes, p21, MDM2, and MIC-1, and this induction was observed only in p53WT (SJSA-1 and MCF-7) but not p53 mutant (MDA-MB-435 and SW480) cancer cells (Fig. 4B). The activation of p53 correlated well with loss of cell viability as measured by the MTT assay (Fig. 4C). The effect was strictly dependent on p53 status, and a selectivity index of >30 was determined from the ratio of IC50 between p53 mutant (MDA-MB-435 and SW480) and p53WT (SJSA-1, MCF-7, RKO, HCT116) cancer cell lines (Fig. 4C).
Fig. 4.
ATSP-7041 activates p53 signaling in cancer cells in p53WT cell lines. (A) ATSP-7041 stabilizes p53 and elevates protein levels of p53 targets p21 and MDM2. Log-phase SJSA-1 and MCF-7 cells were incubated with 1.25, 2.5, 5.0, or 10 μM ATSP-7041 or 10 μM Nutlin-3a for 24 h, and cell lysates were analyzed by Western blotting. (B) ATSP-7041 shows dose-dependent induction of p53 target genes in p53WT but not in mutant p53 cell lines. Exponentially growing p53WT (SJSA-1, MCF-7) and p53 mutant (SW480, MDA-MD-435) cancer cell lines were incubated with 2.5, 5, or 10 μM ATSP-7041 or 10 μM Nutlin-3a for 24 h, and mRNA levels of p53 targets, p21, MDM2, and MIC-1 were measured by quantitative PCR and expressed as fold increase. (C) Viability of four p53WT (SJSA-1, MCF-7, HCT-116, and RKO) and two mutant p53 (SW480 and MDA-MB-435) cancer cell lines was determined after incubation with ATSP-7041 and expressed as percentage of controls ± SD.
ATSP-7041 Reactivates the Main p53 Cellular Functions in Cancer Cells that Overexpress MDM2 or MDMX.
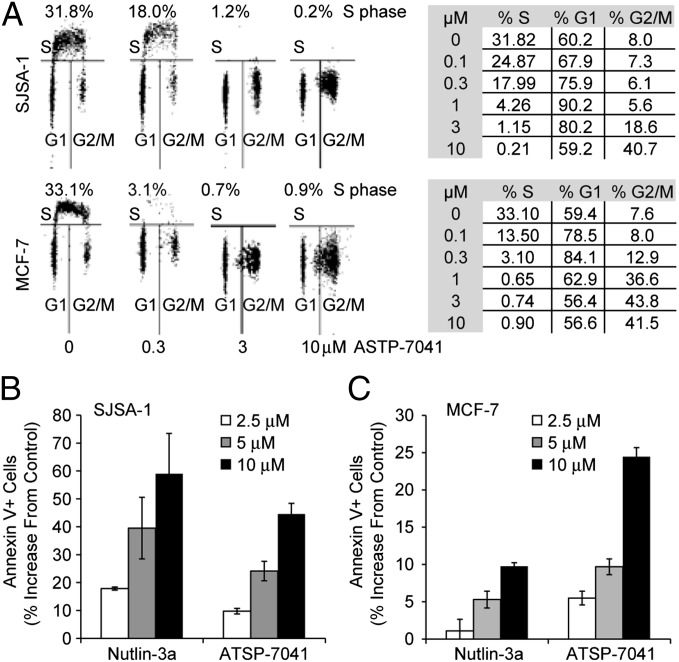
Among the many cellular functions assigned to p53, inductions of cell-cycle arrest and apoptosis are considered the most important for its tumor suppressor function. ATSP-7041 showed effective cell-cycle arrest in G1 and G2/M phase in both SJSA-1 and MCF-7 cells (Fig. 5A). This effect was dose-dependent and led to depletion of the S phase compartment. MCF-7 cells were more sensitive to ATSP-7041, consistent with the complete blockade of MCF-7 cell proliferation at concentrations between 0.3 µM and 1 µM. Next, the Annexin V assay was used to quantify the apoptotic effect of ATSP-7041 (Fig. 5 B and C). Dose-dependent induction of apoptosis was measured for both molecules, with slightly lower overall potency for ATSP-7041 in SJSA-1 cells. However, the apoptotic effect of ATSP-7041 was more pronounced in the MCF-7 cells compared with Nutlin-3a, reflecting the added MDMX inhibitory activity of the dual antagonist (Fig. 5C). MCF-7 cells are known to have caspase 3 deficiency and partially compromised apoptotic response, and the lower Annexin V levels are consistent with this understanding.
Fig. 5.
ATSP-7041 activates main p53 cellular functions in cancer cells. (A) ATSP-7041 arrests cell-cycle progression in cancer cells with p53WT. Exponentially growing SJSA-1 and MCF-7 cells were incubated with 0.3, 3, or 10 μM ATSP-7041for 24 h, and cell cycle distribution was determined by BrdU labeling and cell-cycle analysis. Numbers indicate the percentage of S-phase cells. (B) Apoptotic response to ATSP-7041 and Nutlin-3a in SJSA-1 MDM-2 sensitive cell line. SJSA-1 cells were exposed to ATSP-7041(2.5, 5, or 10 μM) and Nutlin-3a (2.5, 5, or 10 μM) for 48 h, and the percentage of apoptotic cells (±SD) was determined by the Annexin V assay. (C) Apoptotic response to ATSP-7041 and Nutlin-3a in the MCF-7 line. MCF-7 cells were exposed to ATSP-7041 (2.5, 5, or 10 μM) and Nutlin-3a (2.5, 5, or 10 μM) for 48 h, and apoptosis was determined by the Annexin V assay.
ATSP-7041 Suppresses Tumor Growth in Vivo of Multiple Human Xenograft Models and Increases Expression of p21 in Tumors.
ATSP-7041 was examined for efficacy in human cancer xenograft models harboring p53WT and elevated MDM2 or MDMX proteins, namely the SJSA-1 and MCF-7 human xenograft models. As shown in Fig. 6A, ATSP-7041 resulted in statistically significant tumor growth inhibition (TGI) against the MDM2-amplified osteosarcoma xenograft model SJSA-1; when dosed for 2 wk at either 15 mg/kg every day or 30 mg/kg every other day, ATSP-7041 induced a TGI of 61% for both dosing schedules. All dosing regimens were well tolerated. Having established that an intermittent dosing schedule was highly efficacious, we next investigated this dosing regimen in an MDMX overexpressing MCF-7 human breast cancer xenograft model. The treatment of ATSP-7041 for 23 d with 20 mg/kg or 30 mg/kg administered i.v. qod resulted in a TGI of 63% and 87%, respectively (Fig. 6B). In comparison, the selective MDM2 small-molecule inhibitor RG7112 (administered daily at 50 and 100 mg/kg p.o.) resulted in a TGI of 61% and 74%, respectively.
Fig. 6.
ATSP-7041 suppresses tumor growth in vivo of multiple human xenograft models and increases expression of p21. (A) ATSP-7041 induced statistically significant tumor growth inhibition for both qd and qod dosing schedules against the MDM2 amplified osteosarcoma xenograft model SJSA-1. Tumor volumes were calipered throughout the study, and data were plotted as mean ± SEM. *P < 0.05 vs. vehicle control. (B) Treatments resulted in dose-dependent and statistically significant tumor growth inhibition against the MDMX overexpressing MCF-7 human breast cancer xenograft model. Mice (n = 10 per group) bearing established s.c. xenografts received vehicle or 20 or 30 mg/kg ATSP-7041 solution i.v., qod, or 50 or 100 mg/kg of a p.o. suspension of RG7112 daily. *P < 0.05 vs. vehicle control. (C) ATSP-7041 induced greater p21 expression than the MDM2 selective small-molecule inhibitor RG7112. Tumors were removed from mice 4, 8, or 24 h after the last dose of either vehicle or ATSP-7041 at 20 or 30 mg/kg, and p21 mRNA levels were determined by quantitative RT-PCR. Induction of p21 was observed for tumors in response to ATSP-7041 treatment, with a robust 12- to 19-fold increase in p21 8 h post dose. In contrast, for RG7112, only modest induction of p21 was observed at all doses and time points.
To demonstrate that tumor growth inhibition results from p53 activation by ATSP-7041, the expression of p21 was examined as a downstream pharmacodynamics marker of p53 activation in the MCF-7 xenograft study. Tumors were removed 4 h, 8 h, or 24 h after the last dose, and p21 mRNA levels were measured by quantitative RT-PCR. As shown in Fig. 6C, at 4 h post dosing a four- and fivefold increase in p21 expression was noted in the tumors treated with 20 mg/kg and 30 mg/kg of ATSP-7041, respectively. By 8 h, this increase in p21 elevated even further to a 12- and 19-fold in the 20- or 30-mg/kg ATSP-7041 treatment groups, respectively. Consistent with the negative feedback loop of the p53 pathway, this elevation in p21 was reduced yet remained above basal levels in the tumors of mice killed 24 h after dosing. Interestingly, for the reference compound RG7112, the maximum increase in p21 was only fivefold at all doses and time points examined.
ATSP-7041 Exhibits Favorable DMPK and Tissue Distribution Properties.
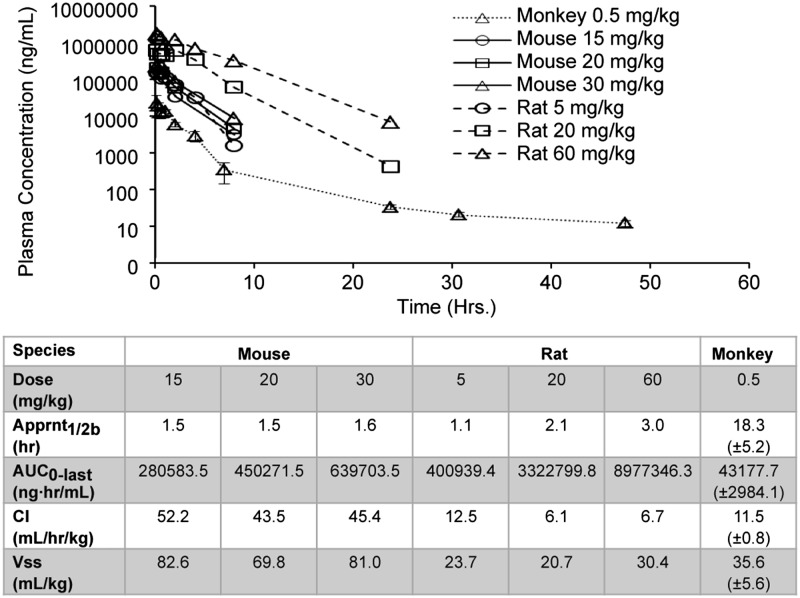
ATSP-7041 was readily formulated for i.v. administration and demonstrated well-behaved plasma pharmacokinetic profiles in mouse, rat, and cynomolgus monkey (Fig. 7). In mouse and rat, ATSP-7041 exposure (AUC0-last) increases in a dose-dependent manner. The elimination kinetics of ATSP-7041 demonstrates durable plasma half-lives of ∼1.5 h, 2.1 h, and 18.3 h in mouse, rat, and monkey, respectively. ATSP-7041 also has a low rate of clearance ranging from 43.5 mL⋅h−1⋅kg−1 to 52.2 mL⋅h−1⋅kg−1 in mouse, 6.1–12.5 mL⋅h−1⋅kg−1 in rat, and 11.5 mL⋅h−1⋅kg−1 in cynomolgus monkey. The steady-state volume of distribution (Vss) was found to be low, ranging from 69.8 mL/kg to 82.6 mL/kg in mouse, 20.7–30.4 mL/kg in rat, and 35.6 mL/kg in cynomolgus monkey, respectively. The low clearance and extended half-life, particularly in higher species, suggest that efficacious exposure levels are achievable at therapeutic doses in man.
Fig. 7.
ATSP-7041 exhibits favorable DMPK properties. Shown are mean plasma pharmacokinetic profiles of ATSP-7041 via i.v. administration in female nude mice, Sprague–Dawley rat, and cynomolgus monkeys. Female nude mice are represented by solid lines (sparse sampling with n of 2 at each time point): 15 mg/kg (circles), 20 mg/kg (squares), and 30 mg/kg (triangles). Sprague–Dawley rats are represented by dashed lines (serial sampling with n = 2 at each time point): 5 mg/kg (circles), 20 mg/kg (squares), and 60 mg/kg (triangles). Cynomolgus monkeys are represented by dotted lines (serial sampling with n = 3 at each time point): 0.5 mg/kg (triangles).
The tissue distribution, pharmacokinetics, and excretion of radiolabeled [3H]-ATSP-7041 were evaluated in rat after a single i.v. dose. Representative autoradioluminograms with radiolabeled [3H]-ATSP-7041 from this quantitative whole-body autoradiography (QWBA) study (Fig. 8) showed broad tissue distribution and clearance, with no accumulation in any subset of tissues. The calculated PK parameters for extravascular tissues (Tables S5 and S6) indicate that [3H]-ATSP-7041 distributes extensively throughout the body, with the exception of brain and CNS tissues, which was limited, as was erythrocyte penetrance. In addition, it was determined (SI Text, Table S6) that the majority of the administered radioactive dose was eliminated in the bile after 48 h, with less than 3% of the total dose recovered in urine. The extensive tissue coverage enables access of the drug to the tissues most critical for treating solid tumors and hematological malignancies, and the clearance mechanism suggests predictable elimination pathways and exposure in man.
Fig. 8.
ATSP-7041 exhibits favorable tissue distribution properties. Mean representative whole body autoradioluminograms showing tissue distribution of [3H]ATSP-7041–derived radioactivity at 0.5 (A) and 6 h (B) after an i.v. dose of [3H]ATSP-7041 at 5 mg/kg to male Long–Evans rats. Highly vascularized tissues, including essential target tissues for the treatment of solid and hematologic tumors, showed the greatest tissue-to-plasma AUC0-t ratios.
Discussion
A critical role of MDMX in cancer is becoming increasingly clear (3–6, 10, 41). MDMX has been shown to act as an oncogene when constitutively overexpressed, and MDMX overexpression correlates with both the presence of p53WT and relatively low levels of MDM2; therefore, in some tumors, MDMX appears to be the primary suppressor of the p53 pathway (3, 4, 42). Recently, Gembarska et al. (9) reported that ∼90% of primary melanoma tumors harbor p53WT and that MDMX is highly overexpressed in 70% of these p53-positive patient samples. Gembarska et al. (9) also demonstrated that knock-down of MDMX dramatically impaired the growth and survival of melanoma cell lines and that cells derived from primary melanoma tumors were particularly sensitive to a reduction in MDMX. Thus, in the case of melanoma, it appears that MDMX, and not MDM2, is a critical suppressor of p53 activity. Interestingly, Gembarska et al. (9) also reported that MDMX mRNA levels did not correlate with observed MDMX protein levels in the primary melanoma tumors. This apparent disconnect from MDMX message to protein is not well understood but suggests that the relevance of MDMX to cancer pathogenesis could be even broader than previously anticipated. In recent reviews by Wade and Wahl (6) and Macchiarulo et al. (10), the authors highlighted MDMX as a bona fide target in cancer therapy by summarizing reported amplification and/or overexpression of MDMX mRNA in human tumors. A diversity of solid tumor types, ranging from retinoblastoma to breast and lung, was cited as possessing amplified or overexpressed MDMX, with retinoblastoma, neuroendocrine tumor, head and neck squamous carcinoma, and hepatocellular exhibiting ∼50% or greater MDMX overexpression (6, 10). However, for melanoma, only a 14% incidence rate of overexpression of MDMX at the message level was noted, yet MDMX protein levels were discovered to be high and a critical driver of human melanoma. Taken together along with recent studies from Gembarska et al. (9), these findings suggest that advancing a dual inhibitor of MDM2/MDMX may provide significant opportunities for clinical translation in a broad spectrum of human cancers that retain p53WT.
Despite the growing interest in MDMX as a therapeutic target to treat p53-positive cancers, the only compounds to reach clinical development to date have been selective inhibitors of the p53–MDM2 protein–protein interaction, and it is likely that such agents will possess a limited therapeutic scope and also encounter chemoresistance as a result of MDMX overexpression in cancers. Such MDM2 selectivity has been found for small molecules despite the fact that p53 binds with high affinity to both MDM2 and MDMX via its transactivation domain α-helix and that only the interaction between p53 and MDM2 has thus far proven to be amenable to direct inhibition with drug-like low molecular weight organic compounds (13, 15). The solution to this challenge of developing a dual MDM2/MDMX inhibitor therefore lies in leveraging the p53 α-helix from the native p53–MDM2/X complexes and converting the physiologically unstable α-helical peptide ligand into a suitably potent and specific therapeutic agent.
Stapled peptides were first described (17) as a novel chemistry approach to create macrocyclic α−helical peptides via all-hydrocarbon cross-links using ruthenium-catalyzed olefin metathesis between two α−methyl–substituted, nonproteogenic amino acids bearing olefinic side chains. Numerous studies have shown that stapled peptides can modulate such intracellular protein–protein interactions, including BH3, Notch, Hif-1α, β−catenin, and Ras (43–46), to exemplify some key targets for cancer therapy. With respect to p53, Bernal et al. (3, 15) applied this strategy to generate a 16-residue stabilized α-helical peptide derived from p53 (SAH-p53-8) that exhibits a high affinity toward MDM2 and MDMX. Although SAH-p53-8 provided a proof-of-concept for a dual peptide inhibitor of MDM2 and MDMX, fundamental limitations of this progenitor molecule were evident, and further drug development was not supported.
Here, we provide an example of a potent and specific dual inhibitor of MDM2 and MDMX, ATSP-7041, able to induce p53-dependent apoptosis and inhibit cell proliferation in multiple MDM2 and MDMX overexpressing tumors in vitro and in vivo. We demonstrate that ATSP-7041 not only preserves the biologically active α-helical conformation found in the p53 protein but also possesses favorable drug-like properties, including efficient cell penetration, specific high-affinity binding to both target proteins, and excellent stability in vivo. ATSP-7041 is chemically and biologically unique relative to previously described molecules (e.g., small-molecule MDM2-selective inhibitors such as Nutlin-3a or the original p53-derived stapled peptide SAH-p53-8). The high-affinity binding and slow dissociation rate of ATSP-7041 can be attributed to the multiplicity of intermolecular contacts that it makes with both MDM2 and MDMX, as evidenced by a high-resolution X-ray structure of ATSP-7041 in complex with MDMX. The biophysical properties of ATSP-7041 also contribute to its unique profile, and its potency in cellular assays and in vivo tumor models can be attributed to its optimized α−helical stability, resistance to proteolytic degradation, and cell penetration properties. In addition, the durable pharmacodynamic response seen in vitro and in vivo further confirms its mode of action and highlights the potential for intermittent administration in the clinic. Finally, the favorable PK properties and extensive tissue coverage enables access of the drug to the tissues most critical for treating solid tumors and hematological malignancies. Overall, ATSP-7041 highlights the capacity to transform prototype stapled peptides for intracellular protein–protein interaction targets into bona fide preclinical candidates for further drug development. In this case, ATSP-7041 provides a unique therapeutic lead molecule to be interrogated in a broad diversity of p53-positive tumors driven by MDM2 and MDMX.
Supplementary Material
Acknowledgments
We thank Dr. Loren Walensky for insightful discussion and input into the manuscript, and Santina Russo and Joachim Diez of Expose GmbH for synchrotron data collection.
Footnotes
Conflict of interest statement: All authors are employees of either Aileron Therapeutics, Inc. or Hoffmann-La Roche, Inc. and, in each case, are shareholders of the company for which they work.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303002110/-/DCSupplemental.
References
- 1.Cheok CF, Verma CS, Baselga J, Lane DP. Translating p53 into the clinic. Nat Rev Clin Oncol. 2011;8(1):25–37. doi: 10.1038/nrclinonc.2010.174. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Bernal F, et al. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell. 2010;18(5):411–422. doi: 10.1016/j.ccr.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marine JC. MDM2 and MDMX in cancer and development. Curr Top Dev Biol. 2011;94:45–75. doi: 10.1016/B978-0-12-380916-2.00003-6. [DOI] [PubMed] [Google Scholar]
- 5.Migliorini D, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol. 2002;22(15):5527–5538. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wade M, Wahl GM. Targeting Mdm2 and Mdmx in cancer therapy: better living through medicinal chemistry? Mol Cancer Res. 2009;7(1):1–11. doi: 10.1158/1541-7786.MCR-08-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 8.Onel K, Cordon-Cardo C. MDM2 and prognosis. Mol Cancer Res. 2004;2(1):1–8. [PubMed] [Google Scholar]
- 9.Gembarska A, et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nat Med. 2012;18:1239–1247. doi: 10.1038/nm.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Macchiarulo AGN, Carotti A, Moretti F, Pellicciari R. Expanding the horizon of chemotherapeutic targets: From MDM2 to MDMX (MDM4) Med Chem Commun. 2011;2:455–465. [Google Scholar]
- 11.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303(5659):844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 12.Tovar C, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: Implications for therapy. Proc Natl Acad Sci USA. 2006;103(6):1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fry DC, Graves B, Vassilev LT. Development of E3-substrate (MDM2-p53)-binding inhibitors: Structural aspects. Methods Enzymol. 2005;399:622–633. doi: 10.1016/S0076-6879(05)99041-1. [DOI] [PubMed] [Google Scholar]
- 14.Graves B, et al. Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization. Proc Natl Acad Sci USA. 2012;109(29):11788–11793. doi: 10.1073/pnas.1203789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J Am Chem Soc. 2007;129(9):2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown CJ, et al. 2013. Stapled peptides with improved potency and specificity that activate p53. ACS Chem Biol 8(3):506–512.
- 17.Schafmeister CJ, Po J, Verdine GL. An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides. J Am Chem Soc. 2000;122:5891–5892. [Google Scholar]
- 18.Kim YW, Grossmann TN, Verdine GL. Synthesis of all-hydrocarbon stapled α-helical peptides by ring-closing olefin metathesis. Nat Protoc. 2011;6(6):761–771. doi: 10.1038/nprot.2011.324. [DOI] [PubMed] [Google Scholar]
- 19.Blackwell HE, et al. Ring-closing metathesis of olefinic peptides: Design, synthesis, and structural characterization of macrocyclic helical peptides. J Org Chem. 2001;66(16):5291–5302. doi: 10.1021/jo015533k. [DOI] [PubMed] [Google Scholar]
- 20.Boal AK, et al. Facile and E-selective intramolecular ring-closing metathesis reactions in 3(10)-helical peptides: A 3D structural study. J Am Chem Soc. 2007;129(22):6986–6987. doi: 10.1021/ja071148m. [DOI] [PubMed] [Google Scholar]
- 21.Benedetti E, et al. Peptaibol antibiotics: A study on the helical structure of the 2-9 sequence of emerimicins III and IV. Proc Natl Acad Sci USA. 1982;79(24):7951–7954. doi: 10.1073/pnas.79.24.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crisma M, Formaggio F, Moretto A, Toniolo C. Peptide helices based on alpha-amino acids. Biopolymers. 2006;84(1):3–12. doi: 10.1002/bip.20357. [DOI] [PubMed] [Google Scholar]
- 23.Williams RM, Im M-N. Asymmetric synthesis of mono-substituted and alpha, alpha-disubstituted alpha-amino acids via diastereoselective glycine enolate alkylations. J Am Chem Soc. 1991;113:9276–9286. [Google Scholar]
- 24.Yang JT, Wu CS, Martinez HM. Calculation of protein conformation from circular dichroism. Methods Enzymol. 1986;130:208–269. doi: 10.1016/0076-6879(86)30013-2. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Popowicz GM, et al. Molecular basis for the inhibition of p53 by Mdmx. Cell Cycle. 2007;6(19):2386–2392. doi: 10.4161/cc.6.19.4740. [DOI] [PubMed] [Google Scholar]
- 29.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Kenakin T. Synoptic receptor function. Trends Pharmacol Sci. 1993;14(12):431–432. doi: 10.1016/0165-6147(93)90178-m. [DOI] [PubMed] [Google Scholar]
- 31.Hu B, Gilkes DM, Chen J. Efficient p53 activation and apoptosis by simultaneous disruption of binding to MDM2 and MDMX. Cancer Res. 2007;67(18):8810–8817. doi: 10.1158/0008-5472.CAN-07-1140. [DOI] [PubMed] [Google Scholar]
- 32.Phan J, et al. Structure-based design of high affinity peptides inhibiting the interaction of p53 with MDM2 and MDMX. J Biol Chem. 2010;285(3):2174–2183. doi: 10.1074/jbc.M109.073056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pazgier M, et al. Structural basis for high-affinity peptide inhibition of p53 interactions with MDM2 and MDMX. Proc Natl Acad Sci USA. 2009;106(12):4665–4670. doi: 10.1073/pnas.0900947106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Böttger V, et al. Comparative study of the p53-mdm2 and p53-MDMX interfaces. Oncogene. 1999;18(1):189–199. doi: 10.1038/sj.onc.1202281. [DOI] [PubMed] [Google Scholar]
- 35.Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274(5289):948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 36.Bissantz C, Kuhn B, Stahl M. A medicinal chemist’s guide to molecular interactions. J Med Chem. 2010;53(14):5061–5084. doi: 10.1021/jm100112j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baek S, et al. Structure of the stapled p53 peptide bound to Mdm2. J Am Chem Soc. 2012;134(1):103–106. doi: 10.1021/ja2090367. [DOI] [PubMed] [Google Scholar]
- 38.Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat Chem Biol. 2010;6(8):595–601. doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phillips C, et al. Design and structure of stapled peptides binding to estrogen receptors. J Am Chem Soc. 2011;133(25):9696–9699. doi: 10.1021/ja202946k. [DOI] [PubMed] [Google Scholar]
- 40.Ray-Coquard I, et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: An exploratory proof-of-mechanism study. Lancet Oncol. 2012;13(11):1133–1140. doi: 10.1016/S1470-2045(12)70474-6. [DOI] [PubMed] [Google Scholar]
- 41.Marine JC, Dyer MA, Jochemsen AG. MDMX: From bench to bedside. J Cell Sci. 2007;120(Pt 3):371–378. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- 42.Ramos YF, et al. Aberrant expression of HDMX proteins in tumor cells correlates with wild-type p53. Cancer Res. 2001;61(5):1839–1842. [PubMed] [Google Scholar]
- 43.Danial NN, et al. Dual role of proapoptotic BAD in insulin secretion and beta cell survival. Nat Med. 2008;14(2):144–153. doi: 10.1038/nm1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walensky LD, et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305(5689):1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patgiri A, Yadav KK, Arora PS, Bar-Sagi D. An orthosteric inhibitor of the Ras-Sos interaction. Nat Chem Biol. 2011;7(9):585–587. doi: 10.1038/nchembio.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moellering RE, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462(7270):182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.