Significance
Telomeres protect the ends of eukaryotic chromosomes. Telomeres shorten with age and serve as a biological clock that limits cell proliferation. Excessive telomere shortening accelerates aging, but telomere elongation may facilitate cancer. We found inherited mutations in the regulator of telomere elongation helicase 1 (RTEL1), which cause Hoyeraal–Hreidarsson syndrome, a fatal disease characterized by accelerated telomere shortening, immunodeficiency, and developmental defects. Introducing a normal RTEL1 gene into affected cells prevented telomere shortening and extended their lifespan in culture. The telomere defects, genomic instability, and growth arrest observed in RTEL1-deficient cells help in our understanding the central roles of telomeres in aging and cancer.
Keywords: dyskeratosis congenita, genomic instability, aging, telomeropathies
Abstract
Telomeres repress the DNA damage response at the natural chromosome ends to prevent cell-cycle arrest and maintain genome stability. Telomeres are elongated by telomerase in a tightly regulated manner to ensure a sufficient number of cell divisions throughout life, yet prevent unlimited cell division and cancer development. Hoyeraal–Hreidarsson syndrome (HHS) is characterized by accelerated telomere shortening and a broad range of pathologies, including bone marrow failure, immunodeficiency, and developmental defects. HHS-causing mutations have previously been found in telomerase and the shelterin component telomeric repeat binding factor 1 (TRF1)-interacting nuclear factor 2 (TIN2). We identified by whole-genome exome sequencing compound heterozygous mutations in four siblings affected with HHS, in the gene encoding the regulator of telomere elongation helicase 1 (RTEL1). Rtel1 was identified in mouse by its genetic association with telomere length. However, its mechanism of action and whether it regulates telomere length in human remained unknown. Lymphoblastoid cell lines obtained from a patient and from the healthy parents carrying heterozygous RTEL1 mutations displayed telomere shortening, fragility and fusion, and growth defects in culture. Ectopic expression of WT RTEL1 suppressed the telomere shortening and growth defect, confirming the causal role of the RTEL1 mutations in HHS and demonstrating the essential function of human RTEL1 in telomere protection and elongation. Finally, we show that human RTEL1 interacts with the shelterin protein TRF1, providing a potential recruitment mechanism of RTEL1 to telomeres.
Human telomeres are composed of tandem TTAGGG DNA repeats, ending with an essential single-stranded 3′-overhang (reviewed in refs. 1 and 2). This overhang can be elongated by the enzyme telomerase to make up for losses caused by incomplete DNA replication and degradation. The expression of the telomerase reverse-transcriptase subunit (hTERT) is suppressed in most human somatic tissues; consequently, telomeres gradually shorten with each cell division. Critically short telomeres activate the DNA damage response (DDR) and cause cell-cycle arrest or apoptosis. Thus, telomere length and integrity control cellular lifespan and provide a tumor-suppressing mechanism (3). Shelterin, a complex of six core proteins, assembles at mammalian telomeres to suppress DDR and regulate telomere length (4, 5). Shelterin was suggested to facilitate the formation of a telomere (T)-loop, via invasion of double-stranded telomeric DNA by the 3′ overhang, where it is inaccessible to DDR factors and to telomerase.
Dyskeratosis congenita (DC) and its severe form Hoyeraal–Hreidarsson syndrome (HHS) are hereditary disorders associated with severely shortened telomeres and diverse clinical symptoms (6–8). The major cause of death in DC and HHS is bone marrow failure, but mortality from cancer and pulmonary fibrosis also occurs at frequencies above normal. Mutations in genes encoding the telomerase subunits hTR, hTERT, dyskerin, NOP10, NHP2, TCAB1 (WRAP53), and the telomere proteins TIN2 and CTC1, account for 60–70% of DC and HHS cases. Thus, accelerated telomere shortening and consequent impairment of cell proliferation is thought to be the molecular basis of the pathology. The genetic defects causing DC and HHS in 30–40% of patients are still unknown.
We have been studying a family in which four of five siblings were diagnosed with HHS; three of them passed away at ages of 3–7, and the fourth died of pulmonary fibrosis 5 y after successful bone marrow transplantation (9) (Fig. 1A). Telomeres in blood cells derived from the patients were severely shortened, and lymphoblastoid cell lines (LCLs) grown in culture showed progressive telomere shortening until reaching senescence, despite the presence of active telomerase. Primary fibroblasts had normal average telomere length but nevertheless displayed telomere dysfunction-induced foci and grew substantially slower than normal fibroblasts (9). Ectopic expression of hTERT, a standard procedure for fibroblast immortalization, failed to stabilize telomere length and prevent senescence of the HHS fibroblasts. These observations indicate that telomeres in these fibroblasts, as in affected LCLs, cannot be extended by telomerase. In addition, fibroblast telomeres elicit DDR despite their normal average length.
Fig. 1.
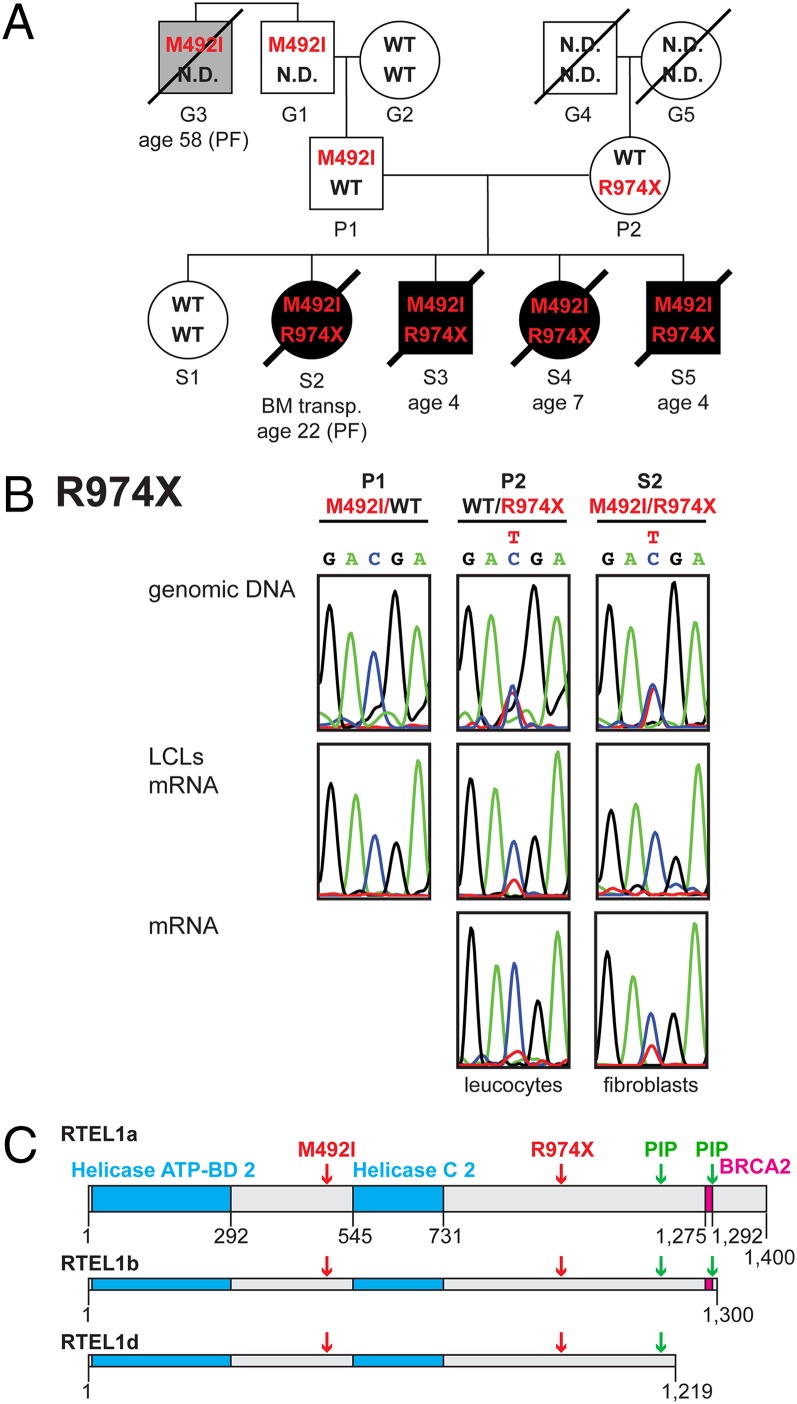
Compound heterozygous RTEL1 mutations were associated with HHS. (A) Genealogical tree of the family. Open circles and squares represent unaffected females and males, respectively. Black circles and squares represent affected females and males. A gray square indicates a family member who died from pulmonary fibrosis. Tilted lines indicate mortality, and the ages of mortality are indicated underneath. Patient S2 underwent bone marrow transplantation (BM transp.) but passed away 5 y later from pulmonary fibrosis. (B) PCR amplification and sequencing of exon 30 from genomic DNA validated the presence of the heterozygous R974X mutation in S2 and P2, but not P1. The results for the rest of the family members appear in Fig. S1. RT-PCR of the same exon from total RNA revealed lower level of the nonsense-carrying transcript. (C) Schematic illustration drawn to scale of the three splice variants of RTEL1 used in this study and listed in AceView as RTEL1a, -b, and -d (31). Indicated are the helicase type 2 ATP binding and C-terminus domains (cyan), a BRCA2 repeat (magenta) identified by searching PFAM (18), PIP boxes [green; identified by searching for the consensus (17)], and the mutations associated with HHS (red).
We searched for the disease-causing mutations by whole-exome capture and deep sequencing and identified compound heterozygous mutations in the gene encoding regulator of telomere elongation helicase 1 (RTEL1). RTEL1 is an essential DNA helicase that belongs to a small family of iron-sulfur–containing DNA helicases, together with XPD, FANCJ, and DDX11/ChlR1. Mutations in the latter three cause the genome instability diseases Xeroderma pigmentosum, Fanconi anemia, and Warsaw breakage syndrome, respectively (10, 11). Rtel1 was originally identified as a dominant regulator of telomere length in mice (12). Mouse RTEL1 was suggested to resolve G-quadruplexes and T-loops during replication (12–15). However, the role of human RTEL1 in telomere biology remains unknown. The identification of deleterious mutations in RTEL1 in association with a telomere-dysfunction disease reported here helps to elucidate the telomeric role of human RTEL1.
Results
Compound Heterozygous Mutations in RTEL1.
We performed whole-exome capture and deep sequencing of genomic DNA samples from two of the patients, as described in Materials and Methods. A total of 113,917 single nucleotide variants (SNVs) and 7,266 small insertions or deletions (INDELs) were found, which deviated from the reference genome. After filtering out reported SNVs and INDELs, 1,022 novel SNVs and 498 novel INDELs remained that were common to both patients. We focused on a subset of 141 variants, which were potentially damaging to the encoded protein: stop gain, stop loss, frame-shifting INDELs, nonframe-shifting INDELs, change in splice site, or nonsynonymous SNVs predicted to be damaging to the protein by the Sorting Intolerant From Tolerant algorithm [SIFT value < 0.05 (16)]. In addition, we found 55 variants in noncoding RNAs (ncRNAs). Assuming recessive (homozygous or compound heterozygous) inheritance of the disease, we narrowed the list down to 33 protein-encoding and 18 ncRNA genes. None of the affected genes has been implicated previously in telomere function except for RTEL1 (12). RTEL1 harbored two novel heterozygous SNVs: a stop gain in exon 30, predicted to cause early termination of protein synthesis at amino acid 974 (NM_016434:c.C2920T:p.R974X), and a nonsynonymous SNV in exon 17, predicted to change the methionine at position 492 to isoleucine (NM_016434:c.G1476T:p.M492I). We examined the presence of the two RTEL1 SNVs in the other family members by PCR and conventional sequencing (Fig. 1 and Fig. S1). Parent P2 and the four affected siblings were heterozygous for R974X, and parent P1 and the four affected siblings were heterozygous for M492I. The healthy sibling S1 was homozygous WT for the two SNVs. These results were consistent with compound heterozygous mutations that cause a disease in a recessive manner: a maternal nonsense mutation, R974X, and a paternal missense mutation, M492I.
The R974X mutation resulted in translation termination downstream of the helicase domains, leaving out two proliferating cell nuclear antigen-interacting polypeptide (PIP) boxes (17) and a BRCA2 repeat identified by searching Pfam (18) (Fig. 1C). We examined the relative expression level of the R974X allele at the mRNA level by RT-PCR and sequencing. The chromatogram peaks corresponding to the mutation (T residue) were significantly lower than those of the WT (C residue) in RNA samples from patient S2 (LCL and skin fibroblasts) and parent P2 (LCL and leukocytes) (Fig. 1B). This result suggested that the R974X transcript was degraded by nonsense-mediated decay (NMD). Western analysis of cell extracts prepared from P1, P2, S1, and S2 with RTEL1-specific antibodies revealed three bands that may correspond to the three splice variants or to differentially modified RTEL1 proteins (Fig. 2C). All three forms of RTEL1 were reduced in the P2 and S2 LCLs (carrying the R974X allele) and no additional smaller protein was detected, consistent with the degradation of this transcript by NMD (Fig. 1B).
Fig. 2.
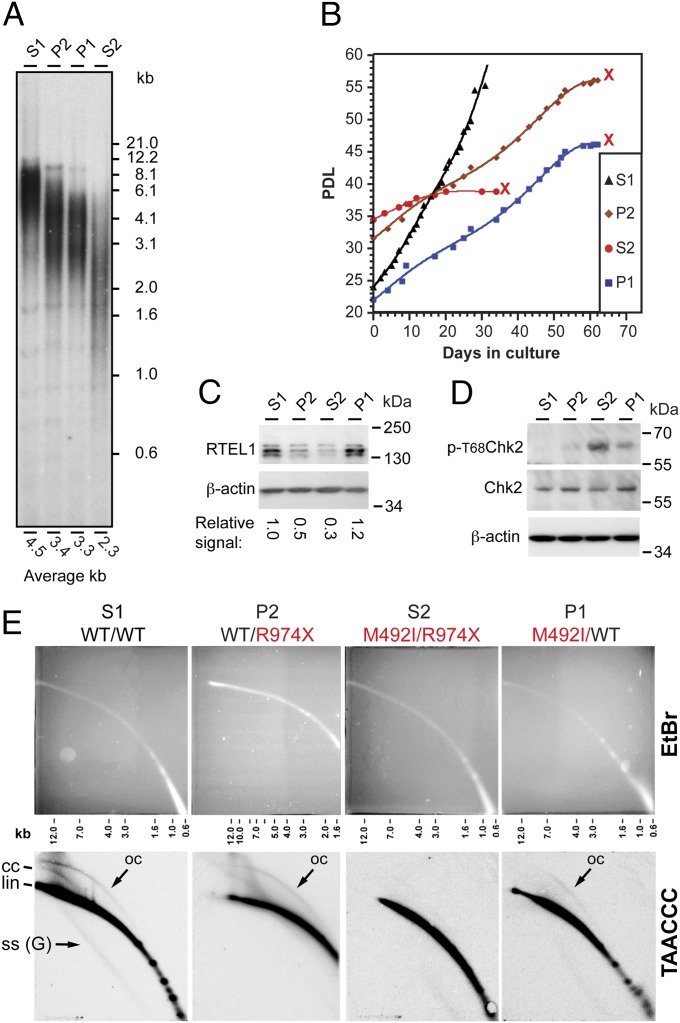
LCLs carrying the heterozygous RTEL1 mutations showed telomere shortening and senescence but no increase in T-circle formation. (A) Southern analysis shows the distribution of telomere restriction fragments in LCLs derived from the parents P1 and P2, the healthy sibling S1, and the affected sibling S2. Genomic DNA samples were prepared from LCLs at PDL ∼35, digested with AluI+MboI, blotted onto a membrane, and hybridized with a telomeric oligonucleotide C-rich probe. The average telomere length for each sample was calculated using MATELO (45) and indicated below the lane. (B) Growth curves showing the population doublings of the LCLs over time. All LCLs carrying RTEL1 mutations reached a stage of growth arrest (indicated by red “X”). (C) Western blot analysis with RTEL1 and β-actin (control) antibodies. The numbers below the lanes indicate the signal intensity of the bands corresponding to RTEL1 relative to β-actin, normalized to the RTEL1 in S1. (D) Western blot analysis with phospho-T68-CHK2, CHK2, and β-actin antibodies. (E) Genomic DNA samples prepared from the indicated LCLs were digested with AluI+MboI and analyzed by neutral–neutral 2D gel electrophoresis, separating first on the basis of size and then on the basis of conformation. Shown are gels stained with EtBr and blots hybridized with a C-rich telomeric probe. Indicated are linear (lin), closed (cc), and open (oc) T-circles, and G-rich single-stranded [SS (G)] forms of telomeric DNA.
The M492I SNV is located between the helicase ATP binding domain and the helicase C-terminal domain 2 (Fig. 1C), and it is predicted to be damaging to the protein with a SIFT value of 0.02. Protein sequence alignment by ClustalX (19) revealed that methionine 492 is conserved in 32 vertebrate species examined, with only two exceptions: leucine in Felis catus (cat) and lysine in Mus spretus (Fig. S2A). RTEL1 orthologs from nonvertebrate eukaryotes mostly have leucine in this position (Fig. S2B). Leucine is predicted to be tolerated at this position (SIFT value = 1), but lysine, a charged residue (unlike methionine and leucine), is predicted to be damaging (SIFT value = 0.05). Interestingly, M. spretus has much shorter telomeres than Mus musculus (20). This difference had been exploited previously to search for loci associated with telomere length by crossing the two species, leading to the initial discovery of Rtel1 as a dominant regulator of telomere length (12, 21). The finding of a mutation associated with HHS in a position where M. spretus Rtel1 deviates from the conserved methionine suggests that in both cases the amino acid change contributes to telomere shortening.
Cells Harboring Heterozygous RTEL1 Mutations Show Telomere Defects.
The heterozygous parents, although healthy, had relatively short telomeres in leukocytes, with broader distribution of lengths compared with the paternal grandmother G2 who does not carry the RTEL1 mutation (9). The shorter telomeres in the younger parents suggest compromised telomere length maintenance as leukocyte telomeres normally shorten with age, and thus telomeres of children are expected to be longer than those of their parents. Another telomere defect found in leukocytes from both patients and heterozygous parents was a shorter than normal telomeric overhang (Fig. S3). These telomere phenotypes suggested that the cells of the heterozygous carriers of either RTEL1 mutation had a telomere defect, although it was not severe enough to cause a disease. The telomeres of paternal grandfather G1 were shorter than those of G2, suggesting that the genetic defect was transmitted from G1 to P1 and to the affected siblings (9). Sequencing confirmed that G1 and G3 carried the M492I mutation, whereas G2 was WT at this position. We have previously found normal telomere length in P1 spermatocytes, excluding the possibility that paternal inheritance of a dominant mutation combined with short telomeres in sperm caused the disease by way of anticipation (9). Altogether, the identified mutations and the telomere phenotypes are consistent with recessive compound heterozygous inheritance of HHS, with partial dominance of the single heterozygous mutations at the cellular phenotype level.
We studied the telomere phenotype of cell cultures derived from a patient and the heterozygous parents to gain insight in the molecular mechanism of RTEL1 function. Although normal LCLs express telomerase, maintain stable telomere length, and readily immortalize (22), LCLs derived from patient S2, although also expressing active telomerase, had very short telomeres and senesced at population doubling level (PDL) 40–50, as counted from their establishment (9) (Fig. 2 A and B). Interestingly, telomeres in LCLs derived from the parents, each carrying a single heterozygous RTEL1 mutation, were also shorter than those of the noncarrier S1 at a PDL of about 35 (Fig. 2A). The P2 LCL carrying the nonsense mutation (R974X) reached a temporary crisis at PDL 55–60 (with only 40% live cells remaining) (Fig. 2B). P1 LCL, carrying the missense mutation (M492I), reproducibly senesced at PDL 45–60 and failed to recover (Fig. 2B). Western blot analysis with specific antibodies against Thr68-phosphorylated CHK2 revealed the phosphorylation of CHK2, a substrate of the ATM kinase that is activated upon DNA damage and telomere uncapping (23), in LCLs from S2, P1, and to some extent in P2, but not S1 (Fig. 2D).
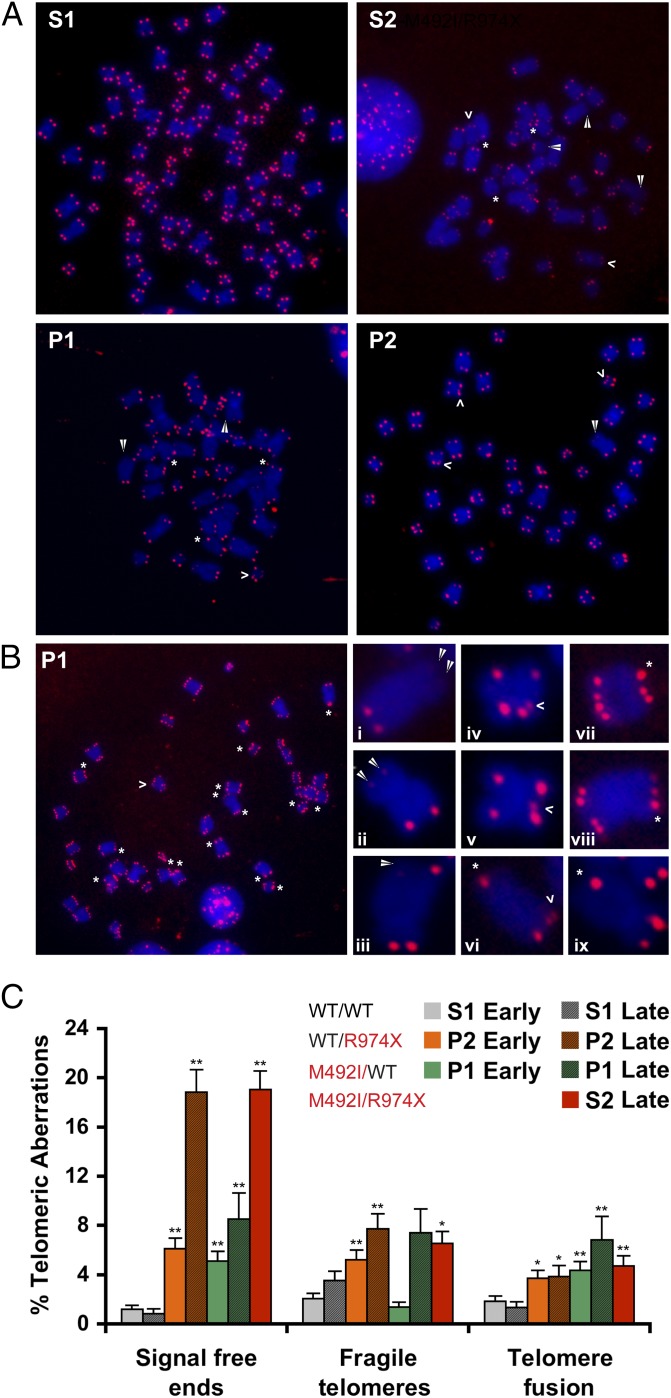
Next, we examined individual telomeres by FISH performed on metaphase chromosomes of LCLs (Fig. 3). We observed increased frequency of telomere defects in the cells of patient S2, compared with the healthy sibling S1. The most frequent defect was signal-free end (in 19% of the counted S2 chromosomes, compared with ∼1% of S1), but fragile telomeres and telomere fusions were also significantly elevated (Fig. 3C). The heterozygous P1 and P2 cells showed increased frequencies of these three types of defects even in early cultures (PDL ∼20; except for fragile telomeres that showed no increase in P1). In late P1 and P2 cultures (PDL ∼40) these events were more frequent and comparable (in most cases) to S2 (Fig. 3C). Interestingly, we observed three P1 cells (of about 80 P1 cells examined) with diplochromosomes (Fig. 3B). We did not see such cells in any of the other control or RTEL1-deficient cells. Persistent telomere damage, which activates DNA damage signaling, was shown previously to enable bypass of mitosis and endoreduplication in dividing cells with short telomeres, contributing to cancer development (24–26).
Fig. 3.
Metaphase chromosomes of RTEL1-deficient cells revealed telomere defects. (A) Metaphase chromosomes hybridized with a telomeric peptide nucleic acid probe reveal increased frequencies of signal-free ends (white arrowhead), fragile telomeres (open arrowhead), and telomere fusions (asterisk) in the RTEL1-deficient lymphoblastoid cells, compared with WT (S1). (A and B) Images were taken with a 100× objective. (B, Left) A P1 cell with diplochromosomes indicating endoreduplication. (B, Right) Enlargements of chromosomes with signal-free ends (i, ii, iii), fragile telomeres (iv, v, vi), and telomere fusion (vii, viii, ix). (C) Chart illustrating the frequency of telomere aberrations in early (PDL ∼20) and late (PDL ∼40) cultures of P1, P2 and S1, and PDL ∼35 of S2. Asterisks indicate significant difference by t test (*P < 0.05, and **P < 0.01). Early P1 and P2 cultures are compared with early S1, and late P1, P2, and S2, are compared with late S1. Total metaphase chromosomes counted are: 815, 787, 1,028, 176, 467, 658, and 596 for early P1, P2, S1, and S2, and late P1, P2, and S1, respectively. Statistical analysis was performed using two-tailed Student’s t test.
In summary, each of the single heterozygous mutations was associated with relatively short telomeres and telomeric overhang, and increased frequencies of telomere signal-free ends, fragility, and fusion in LCLs grown in culture. Although none of the heterozygous carriers was affected with HHS or DC, the paternal great uncle G3 (carrying the M492I mutation) died of idiopathic pulmonary fibrosis at the age of 58 (Fig. 1A). Given the low prevalence of pulmonary fibrosis in the population [0.01–0.06% (27)] and its high prevalence in DC patients [∼20% (8)], this case of pulmonary fibrosis suggests that M492I is a predisposition mutation for pulmonary fibrosis. The R974X transcript is degraded, presumably by the NMD pathway (Figs. 1B and 2C), and thus likely causes disease by way of haploinsufficiency.
RTEL1 Dysfunction Is Not Associated with Increased T-Circle Formation.
Mouse RTEL1 had been suggested to function in T-loop resolution; Rtel1 deletion in mouse embryonic fibroblasts (MEFs) increased the amount of products in a rolling circle polymerization assay, which were attributed to extrachromosomal T-circles generated by improper resolution of T-loops (15). However, such an increase was not observed in mRtel1-deficient mouse embryonic stem cells by 2D gel electrophoresis (14). To detect T-circles we used 2D gel electrophoresis. As shown in Fig. 2E, LCLs derived from the compound heterozygous patient (S2) or heterozygous parents (P1, P2) did not show an increase in T-circle formation. If anything, the signal decreased, compared with LCL from the healthy sibling (S1). Hybridization with a C-rich probe, but not with a G-rich probe, revealed a population of single-stranded G-rich telomeric sequences (labeled “ss-G” in Fig. 2E). These single-stranded telomeric sequences were observed in S1 cells but they were diminished in P1 and P2 cells and not detected in S2, consistent with the duplex-specific nuclease analysis (Fig. S3). Finally, other forms of telomeric DNA, which may represent complex replication or recombination intermediates, appeared as a heterogeneous shadow above the main arc of linear double-stranded telomeric DNA. Similar migrating structures have been observed by 2D gel analyses of human ALT cells (28). These forms were not detected in P1 and S2 cells (Fig. 2E). In summary, we observed in normal cells various conformations of telomeric DNA, including T-circles, single-stranded DNA, and replication or recombination intermediates. These forms appeared reduced in the RTEL1-deficient cells.
Ectopic Expression of WT RTEL1 Suppresses the Short Telomere Phenotype of RTEL1-Deficient Cells.
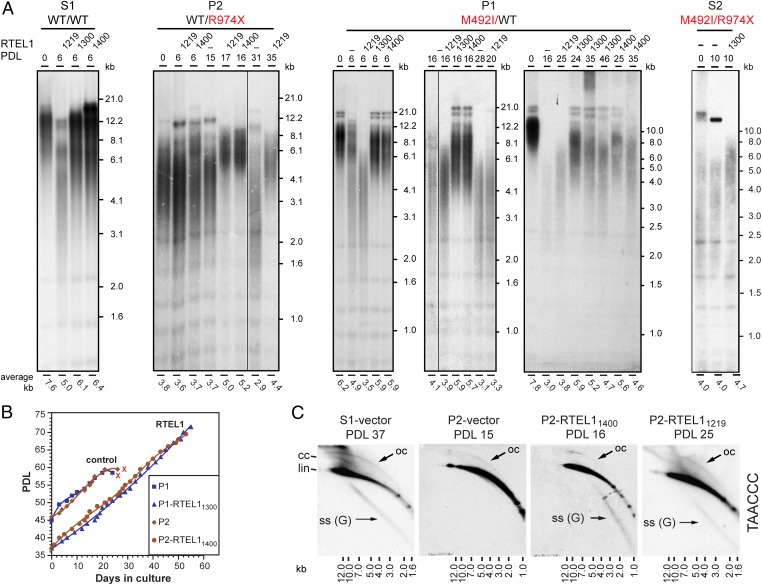
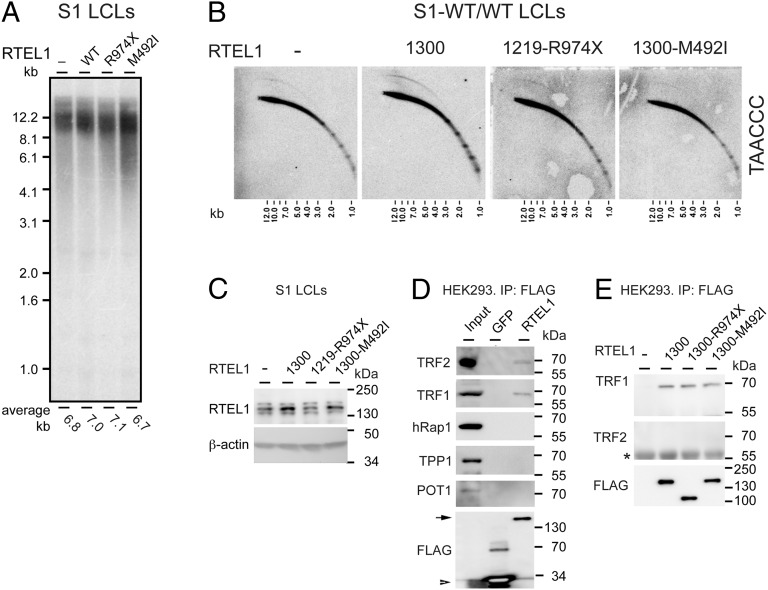
To validate the causal role of the RTEL1 mutations in HHS, we attempted to suppress the telomere defect by ectopic expression of WT RTEL1. The RTEL1 gene (originally termed novel helicase-like, NHL) resides in a four-gene cluster (29). It overlaps with M68/DcR3/TNFRSF6B, encoding a decoy receptor that belongs to the tumor necrosis factor receptor superfamily and suppresses cell death by competing with death receptors (30). Based on reported transcript sequences, the AceView program predicted at least 23 different splice variants in this complex locus (31). We cloned three splice variants (AceView variants aAug10, bAug10, and dAug10), encoding putative 1,400, 1,300, and 1,219 amino acid polypeptides, by RT-PCR of total RNA from normal human cells (Fig. 1C). We first attempted to express RTEL1 from lentiviral vectors using the strong promoters of cytomegalovirus (CMV) or human elongation factor-1 α. However, the transduction of both WT and RTEL1-deficient LCLs and primary fibroblasts with the viral vectors encoding RTEL1 (but not an empty vector) resulted in cell death, indicating that the high expression level of RTEL1 in these cells was toxic. Therefore, we replaced the CMV promoter with the weaker histone H4 promoter. We infected LCLs derived from the family members with the vectors encoding each of the three RTEL1 variants, or an empty vector. Transduction of healthy LCL (S1) with RTEL11219 caused significant telomere shortening, which is observed already at PDL 6 after transduction, and then telomeres reelongated to an intermediate length (Fig. 4A and Fig. S4). These observations suggest that telomere length regulation is sensitive to elevated levels of RTEL1, particularly the RTEL11219 variant, but the culture has an ability to adapt to this toxic effect or select for cells with lower expression level.
Fig. 4.
Ectopic expression of RTEL1 suppressed the telomere shortening phenotype of RTEL1-deficient cells. (A) LCLs derived from S1 (RTEL1WT/WT), P1 (RTEL1WT/M492I), P2 (RTEL1WT/R974X), and S2 (RTEL1M492I/R974X), were transduced with lentiviruses expressing one of the three splice variants or an empty vector (−), as indicated. Genomic DNA samples were prepared from the cultures at the indicated PDLs after transduction and puromycin selection, and analyzed by Southern blotting. PDL 0 indicates a sample taken at the time of transduction. S1 and P2 LCLs were transduced at late PDL (∼40), and P1 and S2 LCL at an early PDL (∼15 and 10, respectively). The average telomere length is indicated below the lanes. (B) Growth curves show the population doublings over time of selected LCLs. Although P1 and P2 cultures senesced at PDL ∼60 (indicated by red “X”), P1 expressing RTEL11300 and P2 expressing RTEL11400 continued to grow without reaching growth arrest as long as kept in culture. (C) Genomic DNA samples were prepared at the indicated PDL and analyzed by 2D gel electrophoresis. Shown are hybridizations with a C-rich telomeric probe. Indicated are linear (lin), closed (cc) and open (oc) T-circles, and G-rich single-stranded [SS (G)] forms of telomeric DNA.
In P2 LCL, carrying the nonsense mutation R974X, ectopic expression of either RTEL11219 or RTEL11400 suppressed the telomere defect and enabled telomere elongation and continuous crisis-free growth (RTEL11300 was not examined) (Fig. 4 A and B, and Fig. S4). Telomere elongation was observed only at PDL 16 after transduction, suggesting that either telomere elongation is slow, or only a small population of cells was able to reverse the telomere defect, elongate the telomeres, and overgrow the rest of the culture. Interestingly, the elongated telomeres of P2 expressing RTEL11219 were comparable in length to the shortened telomeres of S1 expressing RTEL11219 (Fig. S4), supporting the dominant role of RTEL1 in setting telomere length. We examined the conformation of telomeric fragments by 2D gel electrophoresis and found that ectopic RTEL1 expression restored the appearance of G-rich single-stranded telomeric sequences (Fig. 4C). In the case of RTEL11219 expression, the signal presumably corresponding to complex replication or recombination intermediates also appeared, consistent with the resumption of normal growth (Fig. 4C). These results suggested that ectopic expression of RTEL1 restored sufficient levels of RTEL1 in P2 cells, consistent with the haploinsufficiency caused by the R974X mutation.
Suppression of the telomere defect in P1 LCL carrying the M492I mutation was more difficult. When starting with a culture of late PDL and short telomeres, none of the splice variants enabled telomere elongation or rescued the cells from senescence. Starting with a culture of early PDL and longer telomeres, the ectopic expression of RTEL11300 or RTEL11400, but not RTEL11219, stabilized telomere length, and extended the lifespan of the cells (Fig. 4 A and B). Examining these cultures up to PDL 25 and 35 after transduction revealed that telomere shortening was greatly slowed down but not completely prevented.
Initially we were unable to rescue patient S2 cells at a relatively late PDL (∼35), with severely shortened telomeres. However, lately we obtained an early PDL S2 LCL and show that ectopic expression of RTEL11300, resulted in telomere elongation at PDL10 after transduction (Fig. 4A). Taken together, these results confirmed the causal role of the RTEL1 mutations in the disease.
To gain further insight into the effects of the M492I and R974X mutations, we introduced the WT and mutant RTEL1 alleles in normal LCL (S1), primary foreskin fibroblasts (telomerase-negative), and the same fibroblast culture immortalized by hTERT. The ectopic expression of the RTEL1 alleles only caused minor changes in telomere length (Fig. 5A and Fig. S5A). The expression of WT and mutant RTEL1 in S1 LCL was examined by Western blotting (Fig. 5C). Although the middle band, presumably corresponding to RTEL11300, increased in signal in cells expressing WT and M492I RTEL1, relative to control, there was no obvious change in RTEL1 level in cells expressing the R974X mutant, consistent with the degradation of this transcript by NMD. Interestingly, telomere circles increased in both LCLs and hTERT-positive fibroblasts transduced with the WT RTEL11300-encoding lentivector, but not with the empty vector (Fig. 5B and Fig. S5B). These results suggest that functional RTEL1 contributes to T-circle formation, consistently with the apparently reduced T-circle formation in cells carrying RTEL1 mutations (Figs. 2E and 4C).
Fig. 5.
Ectopic RTEL1 induced T-circle formation and interacted with TRF1. LCLs derived from S1 were transduced with lentiviruses expressing WT or mutant (R974X or M492I) RTEL1, or an empty vector (−), as indicated. Genomic DNA samples were prepared from the cultures at day 13 after transduction and puromycin selection, and analyzed by Southern (A) and 2D gel electrophoresis (B). (C) Western blot analysis of the same LCLs as in A and B, using RTEL1 and β-actin antibodies. (D) 293 HEK cells expressing FLAG-GFP or FLAG-RTEL1 1300 were assayed by FLAG immunoprecipitation (IP) followed by Western blot with the indicated antibodies. Input shows nuclear extracts isolated from 293 HEK cells. Arrow indicates FLAG-RTEL11300, and arrowhead indicates FLAG-GFP. (E) 293 HEK cells were transfected with an empty vector (−), or vectors expressing WT or mutant FLAG-RTEL11300. Forty-eight hours posttransfection, cells were assayed by FLAG IP and Western blot with the indicated antibodies. For more stringent co-IP conditions in this co-IP experiment, two washes with 1× PBS were added after the regular washes in RIPA buffer. An asterisk indicates a nonspecific IgG band.
RTEL1 Interacts with the Shelterin Protein TRF1.
To examine how is RTEL1 recruited to telomeres, we tagged RTEL1 (WT and mutants) with an N-terminal FLAGx3 and overexpress it from a CMV promoter on a plasmid transfected into HEK 293 cells. We immunoprecipitated FLAG-tagged RTEL1 and analyzed the precipitate for the presence of the shelterin proteins TRF1, telomeric repeat binding factor 2 (TRF2), TPP1, POT1, and RAP1. Both TRF1 and TRF2 were found in association with RTEL1 and not with control GFP (Fig. 5D and Fig. S6A). However, increasing the wash stringency during immunoprecipitation led to the loss of TRF2 signal (Fig. 5E). In addition, in a reciprocal experiment using FLAG-tagged TRF1 and TRF2, only FLAG-TRF1 was found to immunoprecipitate RTEL1 (Fig. S6B). None of the mutations significantly affected the interaction of RTEL1 with TRF1 (Fig. 5E).
Discussion
DC and HHS are genetic diseases primarily caused by telomere dysfunction (reviewed in refs. 6–8). At first, disease-causing mutations were found only in telomerase subunits, suggesting that telomere shortening was the primary cause for the disease. More recently, mutations were found also in TINF2, encoding the shelterin protein TIN2 (32). These mutations were again suggested to cause the disease by compromising telomerase recruitment to telomere, leading to telomere shortening and the pathogenesis associated with DC and HHS (33). Lately, mutations in CTC1 and C16orf57 were found in DC patients, but the mechanism of pathogenesis is unclear (33–36). Disease-causing mutations have not been identified in about 30–40% of the DC and HHS patients (6, 8).
HHS in the investigated family is associated with excessive telomere shortening in blood cells, typical to DC and HHS. However, it also shows a unique feature of length-independent telomere defect in fibroblasts and inability of active telomerase to maintain stable telomeres in both fibroblasts and LCLs, pointing to a primary telomere defect that compromises both DDR suppression and telomerase recruitment or activation (9). We report that HHS in this family is caused by compound heterozygous mutations in RTEL1 (Fig. 1 and Fig. S1): a nonsense mutation, R974X, and a missense mutation, M492I, in an evolutionarily conserved residue (Fig. S2). Several observations suggest that each of the single heterozygous mutations, although not causing overt disease in the carriers, affected telomere maintenance: (i) telomeres in leukocytes of the parents were relatively short and exhibited a reduced single-stranded telomeric signal (9) (Fig. S3); (ii) pulmonary fibrosis, a rare disease with high frequency in DC and HHS patients, which caused the death of S2, also affected the paternal great uncle carrying the M492I mutation; (iii) LCLs derived from the parents, displayed short telomeres and increasing frequencies of signal-free ends, telomere fragility and fusions in culture (Figs. 2 and 3).
The R974X transcript is presumably degraded by the NMD pathway (Fig. 1B), and thus the heterozygous R974X mutation likely causes a telomere phenotype by haploinsufficiency. P1 cells carrying the M492I mutation displayed a more severe phenotype, manifested by the activation of the ATM pathway, endoreduplication, and the failure of P1 cells to immortalize (Figs. 2 and 3). Interestingly, methionine 492 is conserved across distant eukaryotes (Fig. S2). Only 1 of 32 vertebrate species, M. spretus, deviates from this conservation with a residue (lysine) that is predicted to damage the human protein if replacing M492. This finding is intriguing given the much shorter telomeres of M. spretus compared with M. musculus, and the identification of Rtel1 as responsible for this difference (12). It remains to be determined whether the deviation from the conserved methionine is indeed responsible for the shorter telomeres of M. spretus, and how does it tolerate such a change in a gene that is essential in human and M. musculus (12). Interestingly, endoreduplication, observed in P1 cells, was suggested previously as a mechanism for tetraploidization induced by telomere dysfunction in the early stage of tumorigenesis (25). Thus, endoreduplication provides a possible mechanistic explanation for the cancer predisposition observed in DC patients (8) and suggest that healthy heterozygous carriers of RTEL1 mutations may be at risk.
We expressed three splice variants of WT RTEL1 in LCLs derived from the family members. In P2 cells, carrying the nonsense mutation, both the short (RTEL11219) and the long (RTEL11400) variant enabled elongation of the short telomeres at late PDL (Fig. 4 and Fig. S4). RTEL11219 only has one PIP box; the longer variants contain two PIP boxes and a BRCA2 repeat (Fig. 1C). This finding suggests that for the telomere length maintenance function of RTEL1 two PIP boxes are not essential and one may be sufficient, even if not optimal. RTEL11219 caused telomere shortening in S1 (WT) cells, and did not rescue P1 cells (Fig. 4). RTEL11300 and RTEL11400 prevented telomere shortening in P1 cells when introduced at an early PDL, but failed to facilitate telomere elongation when introduced at a late PDL. Taken together, these results suggest that the defect in P1 cells is more severe and cannot be suppressed by the partially functional RTEL11219. Initially, we failed to rescue the patient S2 LCL when transduced at late PDL, close to senescence. However, we have recently obtained early passage S2 LCLs and were able to show that ectopic expression of RTEL11300 can elongate telomeres in these cells (Fig. 4A).
While this manuscript was under revision, three reports were published describing RTEL1 mutations in association with HHS (37–39). Two of these papers reported the R974X mutation described here, referred to as R998X in a 1,243-amino acid splice variant (NM_032957). This variant includes an alternative 24-amino acid exon not present in the three variants examined in our study (37, 39). AceView documented a cDNA clone encoding the 1,243-amino acid variant only in testis, whereas the three splice variants reported here were documented in a variety of tissues (31). Moreover, we did not detect the inclusion of this alternative exon in normal LCLs or fibroblasts by RT-PCR. Therefore, this splice variant is not likely to be relevant to the cell types examined in our research. Walne et al. (37) reported the same family described here but the healthy sibling, S1 in our work, is reported as a heterozygous carrier, whereas we found this sibling to be WT/WT for the RTEL1 mutations (Fig. S1).
Mouse Rtel1 had been suggested previously to resolve G-quadruplexes potentially forming by the G-rich strand of the telomere during DNA replication, which may cause replication fork collapse and telomere fragility (12, 13, 15). Indeed, we observed fragile telomeres in RTEL1-deficient cells derived from HHS patients or their parents, confirming the role of RTEL1 in preventing telomere fragility. However, RTEL1 is likely to have additional essential activities in telomere maintenance because we did not observe telomere fragility in early passage P1 cells, although they displayed telomere shortening, fusion, and endoreduplication. In addition, the chances for a breakage to occur in a telomere—as well as the amount of sequence loss in case of such an event—presumably correlates with telomere length. Therefore, as a telomere shortens one would expect that telomere fragility would be reduced to the point where telomerase is able to compensate for the loss and stabilize telomere length. However, we observed gradual telomere shortening that continued even after a portion of the telomeres in the population shortened below 1,000 bp (Fig. 2A), and eventually the cells senesced (Fig. 2B). Finally, ectopic expression of hTERT did not rescue either LCL or fibroblasts derived from S2 (9), indicating that loss of telomeric sequence by breakage is not the only defect associated with RTEL1 dysfunction. Taken together, our results point to a role of RTEL1 in facilitating telomere elongation by telomerase, as has been suggested for RTEL1 in mouse embryonic stem cells (14). Indeed, a major defect in telomere elongation is found in the vast majority of DC and HHS patients, carrying mutations in various telomerase subunits and accessory factors or in TINF2, suggesting a common etiology for the disease.
Mouse RTEL1 was suggested to function in the resolution of T-loops, based on the increase in T-circles observed upon Rtel1 deletion in MEFs (15). We failed to detect any increase in T-circle formation in the RTEL1-deficient human cells by 2D gel electrophoresis (Figs. 2E and 4C). Rather, we observed a decrease in T-circles in the RTEL1-deficient cells and an increase in T-circles in both telomerase-positive fibroblasts and LCLs upon ectopic expression of RTEL1 (Fig. 5B and Fig. S5B). The increased level of T-circles in RTEL1-deficient MEFs was observed by a rolling-circle amplification assay (15) and such an increase was not observed in RTEL1-deficient mouse embryonic stem cells by 2D gel electrophoresis (14). Thus, it is possible that RTEL1-deficiency manifests differently in different organisms and cell types, or that the different methods detect different forms of telomeric DNA. Walne et al. reported an increase in T-circles in genomic DNA from HHS patients carrying RTEL1 mutations, using the rolling-circle amplification assay (37). We did not see such an increase by 2D gel electrophoresis, suggesting that these two assays detect different species of telomeric sequences.
We observed by duplex-specific nuclease (Fig. S3) and 2D gels (Figs. 2E and 4C) a decrease in G-rich single-stranded telomeric sequences in cells carrying RTEL1 mutations. We also observed a decrease in other forms of telomeric DNA (Figs. 2E and 4C), which may include complex replication or recombination intermediates (28). Although we do not understand yet how these forms are generated, we noticed that they are generally associated with normal telomere length maintenance and cell growth; they are reduced in the RTEL1-deficient cells with short telomeres and reappeared in the rescued P2 cultures (Fig. 4C). If these structures are important for telomere function and if RTEL1 is involved in their generation, they may provide a clue to understanding the function of RTEL1 at telomeres. Alternatively, T-circles and other forms of telomeric DNA may be products of a telomere trimming mechanism preferentially targeting long telomeres (40), and their disappearance is not a direct consequence of RTEL1 dysfunction but of the short telomeres. Finally, we show by coimmunoprecipitation that human RTEL1 interacts with TRF1 (Fig. 5 D and E, and Fig. S6), providing a potential recruitment mechanism of RTEL1 to telomeres, as proposed previously (12, 13, 15).
In summary, the results reported here reveal several functions of RTEL1 that are compromised in the RTEL1-deficient cells: preventing telomere fragility, repressing DDR, and facilitating telomere elongation by telomerase. The use of the RTEL1-deficient cells and the functional complementation assay developed here will elucidate the function of RTEL1 in normal cells and disease.
Materials and Methods
This study was approved by the Helsinki Committee for Human Studies of Hadassah University Hospital. Informed written consent was obtained from the participants in this study (or their parents in cases of minors).
Agilent SureSelect Human All Exon and Sequencing.
Genomic DNA was subjected to the exome capture procedure using Agilent’s SureSelect Human All Exon Kit (G3362B) Protocol v1. Briefly, 3 μg of gDNA was sheared into the size range of 100–500 bp using the Covaris S-series System. Agilent’s Bioanalyzer DNA-1000 Assay (5067-1504) was used to assess the size range. The resulting fragments were prepared for paired-end sequencing by creating blunt ends, adding an A overhang, ligating the samples with Illumina’s paired-end adaptors, and PCR amplification of the ligated libraries. After PCR, the libraries were purified and 500 ng were hybridized to biotinylated RNA library “Baits” in an Eppendorf PCR machine at 65 °C for 24 h. The next day, the library-bait hybridizations were purified using streptavidin-coated magnetic beads (Dynabeads M-280 Streptavidin, Invitrogen, 112–05D), thus enriching for the exomic sequences contained in the libraries. The captured libraries were PCR amplified and purified, and quality and molarity determined by Agilent’s BioAnalyzer High Sensitivity DNA Assay (5067-4626). Each captured library was sequenced 100–115 bp paired-end on the Illumina GAIIx or HiSeq at a concentration of 5–6 pM.
Computational Analysis.
The sequencing output was analyzed using the CASAVA v1.7 pipeline (Illumina) and Mapping and Assembly with Quality (MAQ) 0.7.1. Because of CASAVA’s ELANDv2 aligning constraints, most of the samples had only 80 bp of the 100–115 bp (from each end) aligned to the University of California at Santa Cruz human genome build HG18 (National Center for Biotechnology Information build 36.1). This process allowed for more optimal phred-like quality output (≥30), compared with using the full sequenced length. The uniquely aligned sequence tags were used for SNV and INDEL calling via the CASAVA pipeline.
In addition, the raw ≥100-bp paired-end sequence tags were converted to Fastq format and aligned to HG18 using MAQ’s easyrun pipeline to call SNVs and INDELs. A 3′ adapter sequence was provided to allow MAQ to use reads <100 bp to help increase the coverage. The resulting SNVs and INDELs from each pipeline were filtered using ANNOVAR to help find the novel nonsynonymous SNVs that were not included in human dbSNP130 or the 1000 Genomes Project (41). Only SNVs and INDELs that were discovered by both aligners were used for further analysis. Each sample had ∼90% of the exonic bases sequenced at least ten times and had an average coverage of over 100×, which is ideal for confidently identifying functional mutations (42).
Construction of RTEL1 Containing Vectors.
The cDNA encoding RTEL11219 (729-4606 of NM_016434) was amplified by RT-PCR using total RNA prepared from HeLa cells and cloned using the restriction endonucleases SpeI and SalI into a lentivirus vector (pLU-H4-TRE-puro) to generate pLU-H4-TRE-RTEL1v1-puro. The RTEL11300 ORF was cloned using EcoRI and HindIII into pCMV-Tag2B (Stratagene), and then an FseI-SalI fragment was subcloned into pLU-H4-TRE-RTEL1v1-puro to generate pLU-H4-TRE-RTEL1v2-puro. To generate a vector encoding RTEL11400 (pLU-H4-TRE-RTEL1v3-puro), an FseI-SalI fragment was amplified by RT-PCR from total RNA prepared from S1 LCLs and subcloned into pLU-H4-TET-RTEL1v1-puro. A vector expressing FLAG-RTEL11300 was generated by PCR amplification and cloning of RTEL11300 into EcoRI/NotI sites of pCMV-FLAG-puro vector (a gift of Ramin Shiekhattar, The Wistar Institute, Philadelphia, PA). All vectors were sequenced to verify the entire RTEL1 sequence.
Lentiviral Packaging and Transduction.
Lentiviral particles were produced by The Wistar Institute protein expression facility or in the laboratory, following ref. 43. One to two million lymphoblastoid cells were infected twice on consecutive days with 1 mL of the medium containing the lentiviral particles, by spin infection at 80 × g and 25–30 °C for 90 min. Next, 1 μg/mL puromycin was added 24 h after the second infection and medium was replaced every 2 d until selection was completed and the culture resumed growth (about a week). The integration of the plasmid and the ectopic expression of RTEL1 at the mRNA level were verified by PCR and RT-PCR amplification using an RTEL1-specific forward primer and a vector specific reverse primer.
Cell Culture.
EBV-infected LCLs were established in the Department of Human Genetics, Hadassah University Hospital, Ein Kerem, Jerusalem. LCLs were grown in RPMI Media 1640 supplemented with penicillin and streptomycin, 2 mM l-glutamine or GlutaMAX (Life Technologies), and 20% (vol/vol) FBS. For cultures growing poorly, the medium was further supplemented with 1 mM sodium pyruvate, 10 mM Hepes pH 7.2, and 2.25 g/L l-glucose (Sigma; G5500). Media and media supplements were purchased from Life Technologies or from Biological Industries. Primary fibroblasts or fibroblasts transduced with hTERT were cultured in DMEM media supplemented with penicillin and streptomycin, 2 mM l-glutamine or GlutaMAX, and 15% (vol/vol) FBS. HEK 293 cells were grown in the same medium but with 10% (vol/vol) FBS.
Genomic DNA and Total RNA Extraction.
Genomic DNA was prepared using a standard proteinase K phenol extraction or Wizard genomic DNA purification kit (Promega) and treated with RNase A. RNA was extracted from cell pellets using TRIzol reagent (Life Technologies) or EZ-RNA Total RNA isolation kit (Biological Industries), according to the manufacturers’ instructions.
PCR and RT-PCR.
cDNA synthesis was performed using Masterscript (5′ Prime) or SuperScript III (Life Technologies) reverse transcriptases and oligo dT or RTEL1-specific oligo. PCR for cloning purposes was done using Herculase (Agilent) or Q5 High Fidelity DNA polymerase (New England Biolabs). Sequencing was done at The Wistar Institute or the Center for Genomic Technologies, Hebrew University of Jerusalem.
Western Analysis.
Equal amounts of whole-cell extracts in 1× Laemmli buffer were electrophoresed on an 8–16% (wt/vol) Tris⋅Glycine gel (Life Technologies), electroblotted onto a nitrocellulose membrane, probed with the indicated antibodies, and visualized by ECL plus kit (GE Healthcare), according to the manufacturers’ instructions. Rabbit polyclonal antibodies to RTEL1 were raised against a recombinant C-terminus fragment of human RTEL1 and affinity-purified, phospho-Chk2 (Thr68) and Chk2 antibodies were from Cell Signaling, TPP1 antibody from Bethyl Laboratories, POT1 antibody from Santa Cruz, FLAG M2 antibody or agarose beads and monoclonal β-actin peroxidase conjugate were from Sigma-Aldrich. Rabbit antibodies to TRF1, TRF2, and hRap1 were generated against recombinant proteins and affinity-purified.
Immunoprecipitation.
About 1 × 107 293 HEK cells overexpressing FLAG-RTEL1 proteins or FLAG-GFP control (as indicated) were lysed in 1 mL of RIPA buffer [1% Nonidet P-40, 1% Deoxycholate, 0.1% SDS, 150 mM NaCl, 10 mM Tris⋅HCl, pH 7.5, 1 mM DTT, 1 mM PMSF, and 1× protease inhibitor mixtures (Sigma)] for 30 min at 4 °C. The lysates were cleared by centrifugation for 10 min at 20,000 × g, and the supernatants were precleared with protein G Sepharose beads for 1 h at 4 °C. The precleared lysates were immunoprecipitated with FLAG agarose beads (Sigma) overnight at 4 °C, washed four times with RIPA buffer for 10 min each, and subjected to Western blot analysis.
Southern Blot Analysis of Telomeric Restriction Fragments.
Genomic DNA (2–5 μg) was digested with AluI+MboI or AluI+HinfI restriction endonucleases, separated on a 0.7% agarose gel, denatured, and transferred to a Hybond N+ membrane (GE Healthcare). The blot was hybridized at 42 °C with a telomeric oligonucleotide probe, (TTAGGG)4 or (TAACCC)4 5′-end-labeled with T4 polynucleotide kinase (New England Biolabs) and 32P-γATP, and washed twice for 5 min with 0.2 M wash buffer [0.2 M Na2HPO4 pH 7.2, 1 mM EDTA, and 2% (wt/vol) SDS] at room temperature and once with 0.1 M wash buffer at 50 °C, following Church and Gilbert (44), and exposed to an X-ray film or visualized by Typhoon 9410 Imager (GE Healthcare). Average telomere length was calculated by the computer program MATELO (45).
Two-Dimensional Gel Electrophoresis.
Two-dimensional gel electrophoresis was modified from ref. 46. Equal amounts of AluI+MboI digested DNA (10–15 μg) was subjected to electrophoresis in a 0.4% agarose gel (first dimension) at room temperature and 30 V for 12–14 h, and then in a 1.2% (wt/vol) agarose gel (second dimension) containing 0.3 μg/mL ethidium bromide at 4 °C and 150 V for 6 h. The gel was processed as described above for the Southern analysis. In Fig. S5, 2 μg of ligated λ DNA HindIII fragments were electrophoresed together with the digested genomic DNA in 2D gels and hybridized with λ DNA probes generated by random prime labeling of λ DNA HindIII fragments with 32P-α-dCTP.
Metaphase Telomere FISH.
LCLs were subcultured into fresh medium and incubated at 37 °C for 24 h. Colcemid (0.1 μg/mL; Gibco) was added for 4 h to accumulate mitotic cells. Cells were collected by centrifugation at 112 × g for 10 min and suspended in 75 mM KCl hypotonic solution at 37 °C for 25 min before fixation in fresh 3:1 methanol/acetic acid three to four times. Fixed cells were dropped onto cold and wet glass microscope slides and allowed to dry slowly in a humid environment. Metaphase chromosome spreads were fixed in 4% (wt/vol) paraformaldehyde in 1× PBS for 3 min, treated with 1 mg/mL pepsin for 10 min at 37 °C, dehydrated in ethanol series [70%, 95%, 100% (vol/vol)], and air-dried. Slides were denatured for 5 min at 80 °C in hybridization mix [70% (vol/vol) formamide, 10 mM Tris⋅HCl (pH 7.2), and 0.5% blocking solution (Roche)] containing telomeric PNA-Tamra-(CCCTAA)3 probe. After denaturation, hybridization was continued for 2 h at room temperature in the dark. Slides were washed twice for 15 min with 70% (vol/vol) formamide, 10 mM Tris⋅HCl (pH 7.2), and 0.1% BSA, and then three times for 5 min each with 0.15 M NaCl, 0.1 M Tris⋅HCl (pH 7.2), and 0.08% Tween-20. Nuclei were counterstained with 0.1 μg/mL DAPI in 1× PBS and slides were mounted with VectorShield (Vector Laboratories). Images were taken with a 100× lens on a Nikon E600 Upright microscope (Nikon Instruments) using ImagePro Plus software (Media Cybernetics) for image processing. Statistical analysis was performed using two-tailed Student t Test.
Supplementary Material
Acknowledgments
We thank the family affected by Hoyeraal–Hreidarsson syndrome for their generous assistance with samples and information, which made this research possible; Dirk Hockemeyer and Titia de Lange for help with antibodies, reagents, and advice; Aviva Yeheskel and Bella Meidan for establishing lymphoblast and fibroblast cell lines; Grace Heck and David Schultz from The Wistar Institute Protein Expression Facility for their advice and production of lentiviral vectors; Frederick Keeney from The Wistar Institute Microscopy Facility for his help with image processing; Fan Lai for FLAG-regulator of telomere elongation helicase 1 and FLAG-GFP transfections; Hagar Katzir and Sara Selig for help with estimating average telomere length by MATELO; and Ran Avrahami for assistance with statistical analysis and stimulating conversations. This work was supported by Grants 1355/08 and 1729/13 from the Israel Science Foundation (to Y.T.); Grant 2009204 from the United States-Israel Binational Science Foundation (to Y.T.); National Institutes of Health Grant R01CA140652 (to P.M.L.); Wistar Cancer Center Grant P30 CA10815 (to P.M.L.); American Heart Association Grant 11SDG5330017 (to Z.D.); and a Boehringer Ingelheim travel grant (to G.G.). Work in “Telomere and Cancer” laboratory is supported by the Institut Curie and La Ligue contre le Cancer.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300600110/-/DCSupplemental.
References
- 1.Jain D, Cooper JP. Telomeric strategies: Means to an end. Annu Rev Genet. 2010;44:243–269. doi: 10.1146/annurev-genet-102108-134841. [DOI] [PubMed] [Google Scholar]
- 2.O’Sullivan RJ, Karlseder J. Telomeres: Protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11(3):171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31(1):9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Lange T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19(18):2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 5.Liu D, O’Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem. 2004;279(49):51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 6.Nelson ND, Bertuch AA. Dyskeratosis congenita as a disorder of telomere maintenance. Mutat Res. 2012;730(1–2):43–51. doi: 10.1016/j.mrfmmm.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason PJ, Bessler M. The genetics of dyskeratosis congenita. Cancer Genet. 2011;204(12):635–645. doi: 10.1016/j.cancergen.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dokal I. Dyskeratosis congenita. Hematology (Am Soc Hematol Educ Program) 2011;2011:480–486. doi: 10.1182/asheducation-2011.1.480. [DOI] [PubMed] [Google Scholar]
- 9.Lamm N, et al. Diminished telomeric 3′ overhangs are associated with telomere dysfunction in Hoyeraal-Hreidarsson syndrome. PLoS ONE. 2009;4(5):e5666. doi: 10.1371/journal.pone.0005666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Lelij P, et al. Warsaw breakage syndrome, a cohesinopathy associated with mutations in the XPD helicase family member DDX11/ChlR1. Am J Hum Genet. 2010;86(2):262–266. doi: 10.1016/j.ajhg.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Y, Suhasini AN, Brosh RM., Jr Welcome the family of FANCJ-like helicases to the block of genome stability maintenance proteins. Cell Mol Life Sci. 2009;66(7):1209–1222. doi: 10.1007/s00018-008-8580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding H, et al. Regulation of murine telomere length by Rtel: An essential gene encoding a helicase-like protein. Cell. 2004;117(7):873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 13.Sfeir A, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138(1):90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uringa EJ, et al. RTEL1 contributes to DNA replication and repair and telomere maintenance. Mol Biol Cell. 2012;23(14):2782–2792. doi: 10.1091/mbc.E12-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149(4):795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 16.Sim NL, et al. SIFT Web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40(Web Server issue):W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warbrick E, Heatherington W, Lane DP, Glover DM. PCNA binding proteins in Drosophila melanogaster: The analysis of a conserved PCNA binding domain. Nucleic Acids Res. 1998;26(17):3925–3932. doi: 10.1093/nar/26.17.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chenna R, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31(13):3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starling JA, Maule J, Hastie ND, Allshire RC. Extensive telomere repeat arrays in mouse are hypervariable. Nucleic Acids Res. 1990;18(23):6881–6888. doi: 10.1093/nar/18.23.6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu L, et al. Telomere length regulation in mice is linked to a novel chromosome locus. Proc Natl Acad Sci USA. 1998;95(15):8648–8653. doi: 10.1073/pnas.95.15.8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kataoka H, et al. Immortalization of immunologically committed Epstein-Barr virus-transformed human B-lymphoblastoid cell lines accompanied by a strong telomerase activity. Differentiation. 1997;62(4):203–211. doi: 10.1046/j.1432-0436.1998.6240203.x. [DOI] [PubMed] [Google Scholar]
- 23.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448(7157):1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 24.Davoli T, de Lange T. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell. 2012;21(6):765–776. doi: 10.1016/j.ccr.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141(1):81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126(1):63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 27.Nalysnyk L, Cid-Ruzafa J, Rotella P, Esser D. Incidence and prevalence of idiopathic pulmonary fibrosis: Review of the literature. Eur Respir Rev. 2012;21(126):355–361. doi: 10.1183/09059180.00002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nabetani A, Ishikawa F. Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol Cell Biol. 2009;29(3):703–713. doi: 10.1128/MCB.00603-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai C, et al. Overexpression of M68/DcR3 in human gastrointestinal tract tumors independent of gene amplification and its location in a four-gene cluster. Proc Natl Acad Sci USA. 2000;97(3):1230–1235. doi: 10.1073/pnas.97.3.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitti RM, et al. Genomic amplification of a decoy receptor for Fas ligand in lung and colon cancer. Nature. 1998;396(6712):699–703. doi: 10.1038/25387. [DOI] [PubMed] [Google Scholar]
- 31.Thierry-Mieg D, Thierry-Mieg J. AceView: A comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(Suppl 1):S12.1–S12.14. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walne AJ, Vulliamy TJ, Beswick R, Kirwan M, Dokal I. TINF2 mutations result in very short telomeres: Analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood. 2008;112(9):3594–3600. doi: 10.1182/blood-2008-05-153445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang D, He Q, Kim H, Ma W, Songyang Z. TIN2 protein dyskeratosis congenita missense mutants are defective in association with telomerase. J Biol Chem. 2011;286(26):23022–23030. doi: 10.1074/jbc.M111.225870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasa GS, Ribes-Zamora A, Nelson ND, Bertuch AA. Three novel truncating TINF2 mutations causing severe dyskeratosis congenita in early childhood. Clin Genet. 2012;81(5):470–478. doi: 10.1111/j.1399-0004.2011.01658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I. Mutations in C16orf57 and normal-length telomeres unify a subset of patients with dyskeratosis congenita, poikiloderma with neutropenia and Rothmund-Thomson syndrome. Hum Mol Genet. 2010;19(22):4453–4461. doi: 10.1093/hmg/ddq371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walne AJ, et al. Mutations in the telomere capping complex in bone marrow failure and related syndromes. Haematologica. 2013;98(3):334–338. doi: 10.3324/haematol.2012.071068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walne AJ, Vulliamy T, Kirwan M, Plagnol V, Dokal I. Constitutional mutations in RTEL1 cause severe dyskeratosis congenita. Am J Hum Genet. 2013;92(3):448–453. doi: 10.1016/j.ajhg.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Guen T, et al. Human RTEL1 deficiency causes Hoyeraal-Hreidarsson syndrome with short telomeres and genome instability. Hum Mol Genet. 2013;22(16):3239–3249. doi: 10.1093/hmg/ddt178. [DOI] [PubMed] [Google Scholar]
- 39.Ballew BJ, et al. Germline mutations of regulator of telomere elongation helicase 1, RTEL1, in Dyskeratosis congenita. Hum Genet. 2013;132(4):473–480. doi: 10.1007/s00439-013-1265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pickett HA, Henson JD, Au AYM, Neumann AA, Reddel RR. Normal mammalian cells negatively regulate telomere length by telomere trimming. Hum Mol Genet. 2011;20(23):4684–4692. doi: 10.1093/hmg/ddr402. [DOI] [PubMed] [Google Scholar]
- 41.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng SB, et al. Exome sequencing identifies the cause of a Mendelian disorder. Nat Genet. 2010;42(1):30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1(1):241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 44.Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yehezkel S, Segev Y, Viegas-Péquignot E, Skorecki K, Selig S. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum Mol Genet. 2008;17(18):2776–2789. doi: 10.1093/hmg/ddn177. [DOI] [PubMed] [Google Scholar]
- 46.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop–sized deletions at human telomeres. Cell. 2004;119(3):355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.