Significance
Long-range electron transfer (ET) is vital in energy transduction pathways. Within metalloprotein ET active sites, the role of the axial ligand in the mononuclear, blue copper (BC), also called type 1 Cu, sites is well defined, whereas its role in the binuclear mixed-valent CuA sites is less clear. This study defines the axial interaction in the mixed-valent binuclear CuA active site and its role in ET. The axial S(Met) ligand is essential in tuning down the reduction potential while not increasing the inner-sphere reorganization energy, a similar role to that found for the S(Met) ligand in BC. Furthermore, much like BC, the S(Met) bond in CuA is weak and therefore under entatic control by the surrounding protein matrix.
Keywords: spectroscopy, reduction potential, energy transduction pathway
Abstract
Within Cu-containing electron transfer active sites, the role of the axial ligand in type 1 sites is well defined, yet its role in the binuclear mixed-valent CuA sites is less clear. Recently, the mutation of the axial Met to Leu in a CuA site engineered into azurin (CuA Az) was found to have a limited effect on E0 relative to this mutation in blue copper (BC). Detailed low-temperature absorption and magnetic circular dichroism, resonance Raman, and electron paramagnetic resonance studies on CuA Az (WT) and its M123X (X = Q, L, H) axial ligand variants indicated stronger axial ligation in M123L/H. Spectroscopically validated density functional theory calculations show that the smaller ΔE0 is attributed to H2O coordination to the Cu center in the M123L mutant in CuA but not in the equivalent BC variant. The comparable stabilization energy of the oxidized over the reduced state in CuA and BC (CuA ∼ 180 mV; BC ∼ 250 mV) indicates that the S(Met) influences E0 similarly in both. Electron delocalization over two Cu centers in CuA was found to minimize the Jahn–Teller distortion induced by the axial Met ligand and lower the inner-sphere reorganization energy. The Cu–S(Met) bond in oxidized CuA is weak (5.2 kcal/mol) but energetically similar to that of BC, which demonstrates that the protein matrix also serves an entatic role in keeping the Met bound to the active site to tune down E0 while maintaining a low reorganization energy required for rapid electron transfer under physiological conditions.
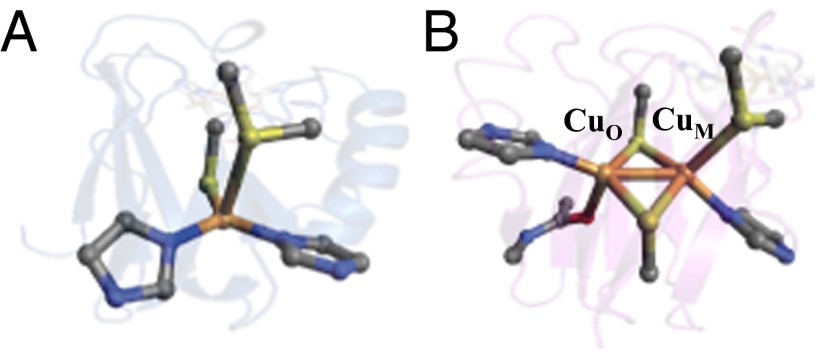
Long-range electron transfer (ET) is vital to a wide range of biological processes, including two key energy transduction pathways essential for life: H2O oxidation in photosynthesis and O2 reduction in respiration (1, 2). Nature has adapted a conserved cupredoxin fold motif (i.e., the Greek-key β barrel) to construct two evolutionarily linked, but structurally distinct Cu-containing ET proteins (3–5). These are the mononuclear type 1 (T1) or blue copper (BC) and binuclear purple CuA proteins. The first coordination sphere of the classic BC sites [e.g., plastocyanin (Pc) and azurin (Az)] consists of a trigonally distorted tetrahedral environment where Cu resides in an equatorial plane formed by one S(Cys) and two N(His) ligands and has an axial S(Met) ligand (Fig. 1A) (6, 7). The binuclear purple CuA site consists of two bridging S(Cys) ligands and two equatorial N(His) ligands as well as an axial polypeptide backbone carbonyl oxygen [O(Gln) on CuO] and an axial thioether sulfur [S(Met) on CuM] (Fig. 1B) (8–11). Both sites carry out rapid, efficient long-range ET with rates on the order of 103–105 s−1 (12, 13).
Fig. 1.
The active sites of two previously published Cu ET proteins: (A) the monomeric T1 Cu Az from Pseudomonas aeruginosa (PDB ID code 4AZU) and (B) the binuclear purple CuA from T. thermophilus (PDB ID code 2CUA).
Although BC proteins use a Cu+/Cu2+ redox couple, the binuclear CuA sites use a (Cu1+–Cu1+)/(Cu1.5+–Cu1.5+) redox cycle. The oxidized form of CuA is mixed-valent (MV), with a highly covalent Cu2S2 core that gives rise to its unique spectroscopic features. The unpaired electron is fully delocalized over the two Cu centers and exhibits a characteristic seven-line 63,65Cu hyperfine splitting pattern in electron paramagnetic resonance (EPR) spectroscopy (14, 15). Maintaining valence delocalization even in the presence of a low symmetry protein environment has been attributed to the large electronic coupling (HAB) resulting from a direct Cu–Cu σ bond and efficient superexchange facilitated by substantial Cu2–S(Cys)2 covalency. This strong electronic coupling between the two Cu’s leads to a Ψ → Ψ* (Cu–Cu σ → σ*) transition at ∼13,500 cm−1 (16). Excitation into this transition using resonance Raman (RR) yields a large excited state distortion in the totally symmetric Cu2S2 core “accordion” mode (ν1), a characteristic of Robin & Day class III MV delocalization (17–19). The two bridging S(Cys) ligands give rise to four in-plane S(p)-derived molecular orbitals (MOs) for S(Cys) → Ψ* charge transfer (CT) transitions. These have been assigned to absorption bands in the region of 20,000 cm−1. Laser excitation into these CT transitions gives rise to RR enhancement of three additional Cu2S2 core vibrations (SI Appendix, Fig. S1A). The functional advantage of a valence delocalization in terms of rapid, long-range ET at low driving forces (∼45 mV) has been ascribed to lowering the reorganization energy (λ) by distributing structural rearrangements associated with redox over two Cu centers (20).
In nature, the S(Met) ligand of BC is sometimes found to be replaced by other protein residues. These can either coordinate to Cu [e.g., O(Gln) in stellacyanin (St)] or leave the axial position vacant (e.g., Leu in the fungal laccases) (21, 22). In BC, it was found that variation of the axial ligand from O(Gln) to S(Met) to nothing can tune E0 over a 300 mV range (23). In nitrite reductase (NiR), the Cu2+–S(Met) bond strength could be experimentally determined and was found to be weak (4.6 kcal/mol) as its loss is compensated by an increased S(Cys) donor interaction with Cu. The low strength of this bond suggested an important role of the protein in keeping the S(Met) ligand bound at physiological temperature. The contribution of the protein in stabilizing the active site structure has been referred to as an entatic/rack state in bioinorganic chemistry (24, 25). For BC sites, the protein matrix provides the negative free energy required to overcome the entropically favored S(Met) bond loss. This plays an important role in ET function as S(Met) binding stabilizes the oxidized more than the reduced state of the Cu site and lowers E0 by ∼200 mV.
In contrast to BC proteins, S(Met) is the only axial ligand found in naturally occurring CuA sites [cytochrome c oxidase (CcO), nitrous oxide reductase (N2OR), nitric oxide reductase (NOR), terminal oxidase in Sulfolobus acidocaldarius (SoxH)] (26). Interestingly, in contrast to BC, the Met to Leu mutation in the CuA Az only led to a 16 mV increase in E0 (compared with an 86 mV increase for this mutant in BC Az) (27). This apparent difference in the extent of the axial ligand contribution to E0 relative to previous studies on BC has led us to further explore its contribution to function in CuA and whether or not it is entatic as in BC. We use a combination of spectroscopic methods [low-temperature (LT) absorption and magnetic circular dichroism (MCD), RR, and EPR] coupled to density functional theory (DFT) calculations to investigate the geometric and electronic structures of CuA Az and a series of its axial ligand variants (M123X; X = Q, L, H). The influence of the axial ligand on the E0 and λ are evaluated and compared with these properties in the well-understood BC site. Furthermore, the proposed involvement of CuA in ET pathways (28) as well as the entatic/rack nature of the Cu–S(Met) bond in CuA are evaluated and discussed.
Results
Spectroscopic Features of Axial Perturbations.
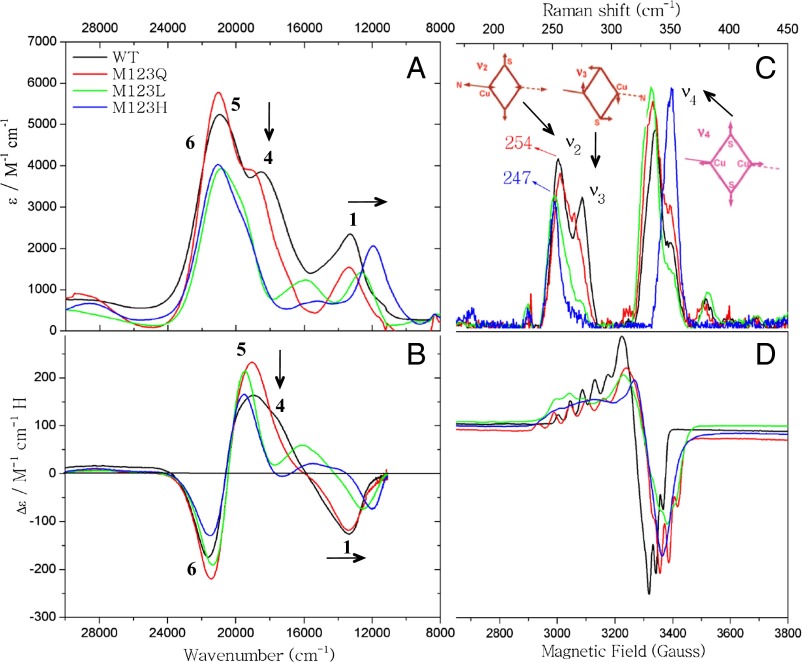
The LT absorption and MCD data for WT CuA Az and its axial ligand variants are given in Fig. 2 A and B [see SI Appendix, Fig. S2 and Table S1 for simultaneous Gaussian resolutions of the LT absorption, MCD, and circular dichroism (CD)]. The LT absorption and MCD data have been previously assigned, and we follow those assignments here (16, 18). From low to high energy, bands 1, 4, 5, and 6 have been assigned as a Ψ → Ψ* (Cu–Cu σ → σ*) transition and three S(Cys) → Cu ligand to metal charge transfer (LMCT) transitions, respectively. In going from WT (black) to M123Q (red), there are only minor changes in energies and intensities in the absorption and MCD spectra. However, for M123L (green) and M123H (blue), there is a redshift of band 1 (the Ψ → Ψ* transition) and a decrease in intensity of band 4 relative to bands 5 and 6 in absorption and MCD [a S(Cys) → Cu CT transition].
Fig. 2.
Spectroscopic characterization of WT CuA Az and the M123X (X = Q, L, H) variants: (A) absorption spectra (10 K), (B) MCD spectra (10 K), (C) RR spectra (77 K, λex = 476.5 nm), and (D) X-band EPR spectra (77 K).
The 77K RR data (λex = 476.5 nm) in the 175–450 cm−1 region for WT CuA Az and its axial ligand variants are given in Fig. 2C. These show RR enhancement of the mixed Cu–S/Cu–N stretching mode (ν2), the out-of-phase “twisting” Cu–S stretching mode (ν3), and the Cu2S2 core breathing mode (ν4) (29). The RR frequencies and intensities for M123Q (red) are very similar to WT CuA Az (black). However, both M123L and M123H show a decrease in the vibrational frequency of ν2 and a decrease in the intensity of ν3 relative to WT/M123Q.
The 77K X-band EPR data for CuA Az and its axial ligand variants are given in Fig. 2D. The spin Hamiltonian parameters are given in Table 1 (see SI Appendix, Fig. S3 and Table S2 for spectra and simulations). All show comparable hyperfine to both Cu’s, consistent with complete delocalization. However, although g|| for WT CuA Az and M123Q are similar and low (g|| = 2.177 and 2.174 for WT and M123Q, respectively), g|| for M123L and M123H increase to 2.215 and 2.255, respectively.
Table 1.
Experimental and calculated spin Hamiltonian parameters for WT CuA Az and the M123X (X = Q, L, H) variants
| Axial variants | Exp. | Calc. | ||||
| gz | AzCu1* | AzCu2 | gz | AzCu1 | AzCu2 | |
| (10−4⋅cm−1) | (10−4⋅cm−1) | |||||
| WT | 2.177 | 53 | 53 | 2.192 | 55 | 63 |
| M123Q | 2.174 | 61 | 57 | 2.190 | 60 | 68 |
| M123L | 2.215 | 35 | 35 | 2.209 | 48 | 59 |
| M123H | 2.255 | 58 | 42 | 2.214 | 52 | 55 |
Note that the experimental A-values are approximate and dependent on the fit protocol. Results of two fits are given in SI Appendix, Fig. S3.
From these data, we group WT/M123Q into one class and M123H/M123L into another. The following five spectroscopic trends are observed in going from WT/M123Q to M123L/H: (i) a decrease in the relative intensity of the S(Cys) → Cu LMCT transitions (band 4 relative to bands 5 and 6), (ii) a redshift in the Ψ → Ψ* transition (band 1), (iii) a decrease in intensity of ν3, (iv) a decrease in the frequency of ν2, and (v) an increase in g||. In D2h symmetry, the ground state of the CuA site is 2B3u (16). Four in-plane S(p) symmetry adapted linear combinations (SALCs) of the two bridged S(Cys) residues are predicted to give rise to two parity-allowed [S(px)g and S(py)g] and two parity-forbidden [S(px)u and S(py)u] S(Cys) → Ψ* LMCT transitions (SI Appendix, Fig. S1B). In addition, in D2h, the symmetry of ν3 is B1g and is therefore not enhanced via an A-term intensity mechanism and must gain intensity through mixing with other totally symmetric (A1g) modes. Thus, the decrease in intensity of the S(Cys) → Cu LMCT transition (band 4) in absorption and ν3 in RR indicate that the active sites of M123L/H have higher effective symmetry than WT/M123Q. This eliminates mixing between parity-allowed and -forbidden SALC MOs as well as the mixing between the B1g and energetically nearby A1g modes in D2h (SI Appendix, Fig. S1 A and B).
The Ψ → Ψ* band in CuA has been described as a σg to σu* transition. The energy separation between these MOs decreases upon weakening the Cu–Cu and Cu–N(His) bonds (i.e., a decrease in energy of the Ψ → Ψ* transition). In addition, the EPR g||-value has been correlated to the energy separation between the ground state, σu*, and the low-lying πu excited state (30). The g||-value is given by:
where ξ is the spin-orbit coupling parameter for Cu2+; α and β are the coefficients of Cu character in the σu* and πu orbitals, respectively; and ΔE is the energy separation between the πu excited and the σu* ground state. Therefore, the redshift of the Ψ → Ψ* transition and the larger g||-value in M123L/H both indicate that the Cu–Cu and Cu–N(His) bonds have weakened (the latter is also consistent with the lower frequency of the ν2 vibrational mode in RR) relative to WT and its M123Q variant. These spectroscopic differences reflect the fact that M123L/H have stronger axial ligand interactions than WT and M123Q, which become comparable in strength to the relatively strong carbonyl backbone ligand on CuO. This increases the effective symmetry of M123L/H closer to D2h, consistent with the decrease in intensity of the ν3 vibration mode and the S(Cys) → Cu LMCT transition. Because Leu is a noncoordinating ligand, this would suggest its replacement with H2O. Although His is a potentially good ligand, M123H is the only mutant that shows an additional pH dependence in EPR (pKa ∼6.5), indicating deprotonation of the axial His residue (SI Appendix, Fig. S4). We therefore assign the N(His) ligand as protonated and unbound at pH 5.5. Based on these spectroscopic trends for M123L and M123H (Fig. 2), H2O is assigned as the axial ligand in both, whereas O(Gln) is weakly coordinated in M123Q. These models are evaluated below. Note that there are some quantitative differences in going from M123L to M123H. These include a lower energy Ψ → Ψ* transition, a larger g||-value, and a higher ν4 vibrational frequency. These reflect a somewhat stronger axial H2O interaction in M123H, which is supported by DFT calculations presented below.
Spectroscopically Validated DFT Structures.
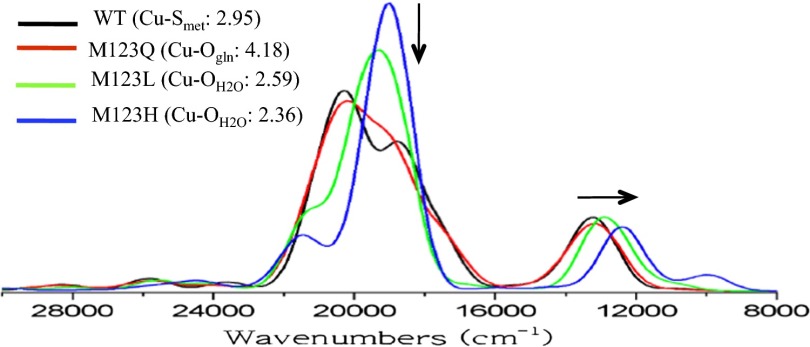
As a starting point for DFT calculations, a WT CuA Az model was constructed from the previously published 1.65 Å resolution X-ray structure [Protein Data Bank (PDB) ID code 1CC3] (SI Appendix, Fig. S5) (11). This model consists of the protein backbone loop connecting the two bridged S(Cys) residues as well as the equatorial His residues and both axial S(Met) and carbonyl backbone axial ligands to the Cu centers (93 total atoms). A partial geometry optimization was carried out with protein backbone and Cα constraints (see SI Appendix, Fig. S6 and Table S3 for structures and relevant optimized bond distances, respectively). This DFT optimized structure has a Cu–S(Met) distance of 2.95 Å, which agrees well with X-ray crystallography (2.98 Å) and previously reported DFT structures (18). The M123X (X = Q, L, H) variant structures for partial geometry optimization were constructed as indicated in Materials and Methods. For M123Q, O(Gln) is the axial ligand and remains at a long Cu–O distance [Cu–O(Gln), 4.18 Å].
For the M123L and M123H models, based on the above spectroscopic characterizations, H2O was placed near the CuM center to serve as the axial ligand. The optimized M123L/H structures have shorter Cu–Laxial distances than those in WT/M123Q [Cu–O(H2O) in M123L, 2.59 Å; Cu–O(H2O) in M123H, 2.36 Å; Cu–SMet, 2.95 Å; Cu–OGln, 4.18 Å]. These structures were used for time-dependent DFT (TD-DFT) calculations. The calculated absorption spectra are given in Fig. 3. These reasonably reproduce the experimentally observed trends in Fig. 2A. Specifically, in going from WT/M123Q to M123L/H, the calculations show a decrease in intensity of the S(Cys) → Cu LMCT transition (∼18,000 cm−1) and a redshift of the Ψ → Ψ* transition (∼12,000 cm−1) as indicated by the arrows in Fig. 3. In addition, the calculated EPR parameters follow the experimentally observed trend: M123L/H both have larger calculated g||-values than WT and M123Q (Table 1). This trend in the calculated g||-value correlates reasonably to the calculated energy separation between the σu* and πu states (from TD-DFT) and the calculated Cu character in these orbitals (using Eq. 1; SI Appendix, Table S5). Specifically, both M123L and M123H have smaller calculated σu*/πu energy separations and larger g||-values.
Fig. 3.
TD-DFT calculated absorption spectra of WT CuA Az and M123X (X = Q, L, H) models. The Gaussian-broadened spectra were simulated using the SWizard program with Gaussian bandshapes that have full-width at half maxima of 1,350 cm−1 (from fits to LT absorption spectra).
In summary, the DFT structures of WT CuA Az and its M123X variants reproduce the spectroscopic trends and support H2O binding as an axial ligand to the CuA core in the M123L/H variants and that S(Met) and O(Gln) are both relatively weakly interacting axial ligands in WT/M123Q, respectively. [Note we have also computationally evaluated possible Cu–O(H2O) vibration modes for M123L. The stretch mixed into several modes at ∼100 cm−1 and there is no significant calculated isotope shift for any of the resonance-enhanced ν2–ν4 vibrations (SI Appendix, Table S4)]. These structures are used below to evaluate the axial S(Met) ligand contributions to function in CuA relative to BC.
Analysis
Axial Ligand Influence on E0.
It has been previously reported that axial ligand variation in CuA azurin results in little change in E0 (27). This is in contrast to the much larger E0 changes for the analogous axial ligand mutations in BC. The smaller change in E0 for the CuA variants, and thus the potentially diminished influence of the S(Met) axial ligand, was attributed to the nature of the diamond core in the CuA center. To further understand the effects of axial ligand binding to CuA on modulating E0, we have calculated the ionization energies (IEs) of WT, Met to Leu, and H2O-bound DFT models of both BC (as a reference performed in the same manner) and CuA sites. Upon varying the axial ligand from S(Met) to Leu, the calculated IE increases by 130 and 100 mV for BC and CuA Az, respectively. For the BC model, this reflects the experimentally observed ΔE0 (exp, 86 mV; calc., 130 mV). It is important to note that previous spectroscopic characterization of the Leu mutation in BC indicated that H2O does not bind in the open axial ligand position (23, 31). In contrast, the experimental ΔE0 in CuA Az is 20 mV, which is much smaller than the 100 mV increase calculated with no axial ligand. However, the calculated IE of the l-H2O model only increases by 10 mV in CuA Az (20 mV for BC). This difference correlates well with the experimentally observed ΔE0 (exp, 20 mV; calc., 10 mV) and is consistent with the spectroscopic assignment that H2O coordinates to the CuA center in the M123L mutant. The calculated IEs for the series of WT, L, and l-H2O models in both CuA Az and BC indicate that the small change in the experimental E0 for M123L CuA relative to BC results from H2O binding to the Cu center in CuA, and that, in CuA, the axial ligand should influence E0 to an extent comparable to BC. Note that the changes in the calculated IE for the Met to Leu (without H2O bound) mutation are quite similar for BC and CuA (BC, 130 mV; CuA, 100 mV) even though the redox states of the H2O-bound Cu differ between the two active sites (Cu+/Cu2+ for BC; Cu+/Cu1.5+ for CuA). We therefore explore contributions to this calculated difference in E0 and evaluate the possibility of entatic control of the Cu–S(Met) bond by the protein environment in the CuA site relative to previous studies on BC.
Axial Met Bond Strength/Entatic State.
For BC, the thermodynamic contributions to the Cu2+–S(Met) bond have been determined experimentally (ΔH ∼ 4.6 kcal/mol). This indicated that the protein matrix and secondary environment in T1 Cu proteins can overcome the entropic gain of Cu–S(Met) bond rupture at physiological temperature. This is the entatic/rack state in T1 Cu proteins. Here, we use the experimental/computational results for parallel insight into CuA. The results of potential energy surface (PES) scans of Cu–S(Met) binding in CuA Az and BC (SI Appendix, Fig. S7) are compared in Table 2. The Cu2+–S(Met) binding energy in BC is calculated to be 7.5 kcal/mol and agrees well with the previously calculated (6.8 kcal/mol) and experimental (4.6 kcal/mol) values (24). The S(Met) binding energy for CuM1.5+ in CuA Az is calculated to be 5.2 kcal/mol, which is lower than BC, but not by half, which might be anticipated from the difference in oxidation state. The calculated Cu1+–S(Met) bond strengths in BC and CuM1+ are similar (1.8 and 1.1 kcal/mol, respectively).
Table 2.
Experimental and calculated Cu–S(Met) bond strengths in BC, delocalized, and localized CuA sites
| Model* | Blue copper | Deloclized CuA (CuO–CuM) | Localized CuA (ZnO–CuM) |
| Exp† | 4.6 | n.d. | n.d. |
| Reduced‡ | 1.8 | 1.1 | 4.6 |
| Oxidized§ | 7.5 | 5.2 | 16.8 |
| ΔΔE(ox-red) | 5.7 | 4.1 | 12.2 |
Constrained Met residue replaced by dimethyl thioether.
kcal/mol.
Energies are relative to a 10 Å CuM–S(thioether) distance. For Cu+ BC dCu–S(thioether)–red .is fixed at dCu–S(thioether)–ox.
Energies are obtained the same way as Cu+ BC.
To explore how electron delocalization in the MV binuclear CuA core influences the stabilization energy of Cu–S(Met) bond in the same ligand environment, the Cu ion in the CuO site was replaced by Zn2+ to localize the unpaired electron on the CuM center (i.e., a [Zn2+–Cu2+] core). The calculated difference in stabilization energy of CuM–S(Met) bond between the reduced [Zn2+–Cu1+] (4.6 kcal/mol) and oxidized [Zn2+–Cu2+] (16.8 kcal/mol) cores is 12.2 kcal/mol. Note that the additional increase in CuM–S(Met) stabilization energy above twice that of the delocalized CuA core (4.1 kcal/mol) reflects the additional positive charge of Zn2+ compared with Cu1+. [A parallel calculation with Ag+ gives 5.9 kcal/mol. However, the ground state wavefunction contains some delocalization, which may lower the calculated bond strength relative to twice that of the CuA core (8.2 kcal/mol) (SI Appendix, Fig. S8).]
From the above calculations, the Cu–S(Met) bond in CuA is weak, yet is energetically similar to that in BC. The small binding energy of the Cu–S(Met) bond in CuA implies that the protein matrix in CuA also serves an entatic role in keeping the Met bound to the active site. Furthermore, the comparable stabilization energy of the oxidized over the reduced state between CuA and BC (CuA ∼ 180 mV; BC ∼ 250 mV, from the ΔΔE’s in Table 2) indicates that the S(Met) likewise tunes down E0 in CuA. Thus, the S(Met) does have a significant influence on E0 even in the delocalized MV CuA site. The comparable function of the axial Met ligand on E0 can be attributed to the different ligand sets between the CuA [two bridged S(Cys) and one equatorial N(His) for each Cu] and BC [one equatorial S(Cys) and two equatorial N(His)] sites.
Reorganization Energy.
Maintaining a low reorganization energy (λ) is an important factor allowing the BC and CuA proteins to perform rapid long-range ET. The total reorganization energy (λT) of the ET process has inner-sphere and outer-sphere components (λi and λo, respectively). λT of engineered CuA Az has been determined to be roughly half that of BC Az (0.8 vs. 0.4 eV). This difference results in a threefold faster kET in CuA Az relative to BC (250 s−1 for BC and 650 s−1 for CuA) (32). It has been proposed that both λi and λo are lowered in CuA relative to BC due to electron delocalization in the MV Cu2S2 core and its larger charge radius relative to the mononuclear BC site (16, 33, 34). Here, we investigate the influence of the axial ligand and charge delocalization on the calculated λi relative to BC. The results of these calculations are summarized in Table 3. λi of the CuA Az and BC DFT models [both with an axial S(Met) ligand] are calculated to be 0.33 and 0.38 eV, respectively. These numbers are in agreement with previously reported values (18, 35–37). Upon removal of the axial S(Met) ligand, the calculated λi for both CuA Az and BC do not change (0.32 and 0.38 eV, respectively). This is consistent with previous considerations for axial ligand variants in stellacyanin (23).
Table 3.
Calculated λi with and without a thioether bound to Cu in BC, delocalized, and localized CuA sites
| Binding modes | Blue copper, eV | Deloclized CuA (CuO–CuM), eV | Localized CuA (ZnO–CuM), eV |
| On | 0.38* | 0.33 | 0.74 |
| Off† | 0.38 | 0.32 | 0.60 |
λi = (Eg = ox – Eg = red)reduced + (Eg = red – Eg = ox)oxidized, where “g=ox” and “oxidized” are the oxidation state of the geometry and wave function, respectively.
Cu–S(thioether) fixed at 10 Å.
We next explore the role of electron delocalization on λi by comparing the calculated values for the electron-delocalized [Cu1.5+–Cu1.5+] and electron-localized [Zn2+–Cu2+] cores. Without the axial S(Met) ligand, the calculated λi increases to 0.60 eV upon electron localization, which is almost twice that of the [Cu1.5+–Cu1.5+] model (0.32 eV) and consistent with the idea that delocalization reduces λi by roughly half. Upon binding of the S(Met) ligand to the localized Cu2+ center, the calculated λi increases to 0.74 eV. This 0.14 eV increase is in contrast to the negligible effect of the axial S(Met) on λi for the electron-delocalized [Cu1.5+–Cu1.5+] core. This difference in the calculated λi in the localized model is related to differences in structural distortions upon redox. S(Met) binding in the [Zn2+–Cu2+] core induces a significant Jahn–Teller distortion, which can be quantified by comparing the change in the angle between the S(Met)–Cu–S1(Cys) and N(His)–Cu–S2(Cys) planes upon redox (4° and 13° for delocalized [Cu1.5+–Cu1.5+] and localized [Zn2+–Cu2+] cores, respectively) (SI Appendix, Fig. S9) (38). Together, these results indicate that, as in BC, the S(Met) ligand contributes very little to λi upon redox for CuA Az. Also, as previously suggested, electron delocalization reduces λi by roughly half relative to a charge-localized model. However, it is additionally found here that it keeps the site from undergoing a Jahn–Teller distortion upon oxidation, which would increase λi.
ET Pathways.
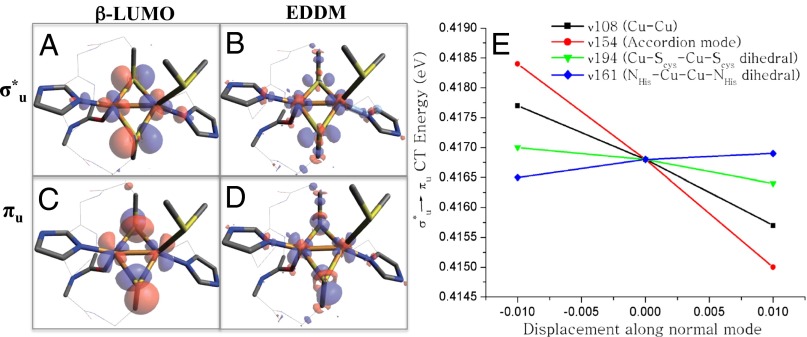
Ligand–metal covalency plays an important role in activating ET pathways and increasing kET through increased donor-acceptor coupling (HDA) (33, 39–41). The nature of the ET pathways coupling the CuA active site with its donor and acceptor has been the subject of much research. Recently, an alternative ET pathway for electron uptake from cytochrome c552 by the CuA site in Thermus thermophilus ba3 oxidase through the axial S(Met) ligand has been proposed (28). This involves a low-lying πu excited state, which has an estimated 10% axial S(Met) character in the highest occupied molecular orbital (HOMO). Here, we explore the possible S(Met) contribution to an ET pathway. The β-lowest unoccupied molecular orbitals (LUMOs) of the σu* ground state and the πu excited state are given in Fig. 4 A and B, respectively. (Note that the πu excited state was obtained by exchanging the electron occupation between σu* and πu orbitals in the σu* optimized structure followed by optimization of the SCF density.) For both σu* and πu wavefunctions, no S(Met) character is observed. We further explored the possibility of S(Met) contribution to redox for both σu* and πu wavefunctions using electron density difference maps (EDDMs) between the reduced and oxidized optimized total electron densities (Fig. 4 C and D). The EDDM contour plots for both σu* and πu states are qualitatively similar to the corresponding β-LUMOs. Importantly, as with the β-LUMOs, the EDDMs have no S(Met) character. We note that there is a predominantly S(Met)-based b1 orbital [HOMO-1, 42% S(p)(Met) character] that mixes into the HOMO [7% S(p)(Met) character] due to their close proximity in energy (SI Appendix, Fig. S10). This, however, involves two occupied levels and does not contribute to net bonding. Therefore, the previously reported 10% axial S(Met) character in the HOMO orbital appears to be a result of this occupied orbital mixing. We can therefore rule out the possibility that the axial S(Met) is a viable ET pathway in either the πu excited and σu* ground state.
Fig. 4.
The EDDMs and β-LUMOs of σ*u ground and πu excited states in the σ*u optimized geometry (A–D) and the calculated πu excited state slopes along normal modes ν108 (black), ν154 (red), ν194 (green), and ν161 (blue) (E).
Above we considered the electronic structure of the πu excited state in the optimized σu* ground state structure; the geometric changes related to the πu excited state are now evaluated. We had previously found that elongation of the Cu–Cu bond results in a structure with a πu ground state that is ∼300 cm−1 higher in energy than the corresponding σu* ground state structure (30). Recently, other structural coordinates have been emphasized (28). To further explore structural contributions that could lead to stabilization of either a σu* or πu state, a series of geometrically perturbed CuA structures have been taken as starting points for geometry optimizations (SI Appendix, Fig. S11). These distortions include: (i) the Cu–Cu distance (from 2.5 to 3.2 Å), (ii) the Cu–S(Cys)–Cu–S(Cys) dihedral angle (from 0 to 30°), (iii) the N(His)–Cu–Cu–N(His) dihedral angle (from –180 to –150°), (iv) distortion of the [Cu2S2] core along the accordion mode (ν1), (v) elongation of the Cu–N(His) bonds, and (vi) elongation of the Cu–S(Cys) bonds. These distortions sample a large fraction of the CuA active site PES. All different starting structures optimized back to either the previously reported σu* (Cu–Cu ∼2.5 Å) or πu (Cu–Cu ∼3.1 Å) geometries. In addition, possible distortions of the πu excited state relative to the σu* ground state geometry were probed by monitoring the TD-DFT calculated σu*/πu energy change upon +/− displacements along normal modes (i.e., possible excited state distorting forces). The modes evaluated were taken from the frequency analysis of the CuA Az geometry [e.g., the Cu–Cu stretch (ν108), the accordion distortion of the Cu2S2 core (ν154), the NHis–Cu–Cu–NHis dihedral mode (ν161), and the Cu–SCys–Cu–SCys dihedral mode (ν194)]. As shown in Fig. 4E, the slopes (and thus the degree of excited state distortion) of the two dihedral modes (ν161 and ν194) are relatively flat compared with the normal modes associated with Cu–Cu elongation (ν108 and ν154, both of which lower the energy of the πu state by Cu–Cu elongation). The ground state optimizations and the magnitudes of excited state distorting forces indicate that Cu–Cu elongation is the preferential mode of distortion in the low-lying πu excited state.
Discussion
From the above spectroscopic and computational results and analyses, the role of the axial S(Met) ligand in CuA is to tune down E0 without significantly affecting λi. The contribution to lowering E0 is especially important for CuA due to the narrow redox window (∼90 mV) between CuA and its redox partners (i.e., cyt c for electron uptake and heme a for electron delivery in CcO). These functions of the axial ligand are similar to those in BC. Furthermore, the lack of S(Met) character in the EDDMs and β-LUMOs of either the σu* ground state or the low-lying πu excited state indicates that the S(Met) ligand is not involved in an ET pathway. This is supported by kinetic results on CcO from Paracoccus denitrificans, which show that the ET rate from cyt c to CuA is unperturbed by the axial Met to Ile mutation (42). The Cu1.5+–S(Met) bond in CuA was calculated here to be weak and slightly weaker than the Cu2+–S(Met) bond in BC (BC, 7.5 kcal/mol; CuA, 5.2 kcal/mol). This finding indicates that, much like BC, the surrounding protein matrix of the CuA active site must impose an entatic/rack state to overcome the entropically favored Cu–S(Met) bond rupture at physiological temperature.
Although the role of the axial Met in CuA is quite similar to BC, the binuclear CuA core has an intrinsic advantage relative to the mononuclear BC in terms of lowering λi and λo. The presence of two highly covalent Cu–S(Cys) bonds in CuA is also important for activating multiple ET pathways. In particular, the electron entry pathway to the CuA site needs to be efficient, as this active site is relatively buried in the protein matrix. This is in contrast to BC, where the electron entry point is a surface-exposed His ligand with little covalent character in the redox active molecular orbital. These factors facilitate rapid and efficient long-range ET with a low driving force (∼45 mV) by the CuA active site.
In summary, a combination of LT absorption, MCD, RR, and EPR spectroscopies on WT CuA and its M123X (X = Q, L, H) axial ligand variants has demonstrated that Cu2S2 active cores in M123L/H are in a more symmetric environment. Spectroscopically validated DFT calculations indicate that the S(Met) ligand is essential in tuning down E0 but not increasing λi, a similar role to that found for the S(Met) ligand in BC. The smaller experimental ΔE0 for the Met to Leu mutation in CuA azurin relative to that in BC is not found to be a consequence of electron delocalization. Rather, this difference is attributed to the presence of a H2O ligand in the M123L mutant of CuA, which is not present in the analogous mutant of BC. Furthermore, much like BC, the S(Met) bond to the active site in CuA is weak and therefore under entatic control by the surrounding protein matrix. This study demonstrates that a detailed spectroscopic characterization of metalloprotein active sites and their perturbed forms is imperative to provide molecular level insight into understanding geometric and electronic structure contributions to function.
Materials and Methods
Expression and purification of WT CuA Az and the variants studied here were performed using previously published protocols (27). UV-vis data were recorded in ammonium acetate buffer (pH 5.5) on a Cary 500 spectrophotometer. MCD was performed on Jasco J-730 and J-810 spectropolarimeters equipped with Oxford Instruments SM-4000 superconducting magnets. RR spectra were collected by detecting with an Andor Newton charge-coupled device detector cooled to –80 °C. Excitation was provided by a Coherent Innova Sabre 25/7 Ar+ CW ion laser (476.5 nm, ∼20 mW). EPR spectra were obtained by using a Bruker EMX spectrometer, ER 041 XG microwave bridge, and ER 4102ST cavity. DFT calculations were performed with Gaussian 03/09 and ORCA. For spectroscopic and computational details, see SI Appendix.
Supplementary Material
Acknowledgments
This work was funded by National Science Foundation Grants CHE-0948211 (to E.I.S.) and CHE-1058959 (to Y.L.) and National Institutes of Health Grant DK-31450 (to E.I.S.). M.-L.T. received support from the Postdoctoral Research Abroad Program sponsored by the National Science Council, Taiwan (Republic of China), and R.G.H. acknowledges a Gerhard Casper Stanford Graduate Fellowship and the Achievement Rewards for College Scientists Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314242110/-/DCSupplemental.
References
- 1.DeVault D, Parkes JH, Chance B. Electron tunnelling in cytochromes. Nature. 1967;215(5101):642–644. doi: 10.1038/215642a0. [DOI] [PubMed] [Google Scholar]
- 2.Ramirez BE, Malmström BG, Winkler JR, Gray HB. The currents of life: The terminal electron-transfer complex of respiration. Proc Natl Acad Sci USA. 1995;92(26):11949–11951. doi: 10.1073/pnas.92.26.11949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu Y. In: Electron Transfer: Cupredoxins Biocoordination Chemistry, Comprehensive Chemistry II: From Biology to Nanotechnology. McCleverty JA, Meyer TJ, Que LJ, Tolman WB, editors. Vol 8. Oxford: Elsevier; 2004. pp. 91–122. [Google Scholar]
- 4.Wilson TD, Savelieff MG, Nilges MJ, Marshall NM, Lu Y. Kinetics of copper incorporation into a biosynthetic purple Cu(A) azurin: Characterization of red, blue, and a new intermediate species. J Am Chem Soc. 2011;133(51):20778–20792. doi: 10.1021/ja205281t. [DOI] [PubMed] [Google Scholar]
- 5.Chacón KN, Blackburn NJ. Stable Cu(II) and Cu(I) mononuclear intermediates in the assembly of the CuA center of Thermus thermophilus cytochrome oxidase. J Am Chem Soc. 2012;134(39):16401–16412. doi: 10.1021/ja307276z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guss JM, Freeman HC. Structure of oxidized poplar plastocyanin at 1.6 A resolution. J Mol Biol. 1983;169(2):521–563. doi: 10.1016/s0022-2836(83)80064-3. [DOI] [PubMed] [Google Scholar]
- 7.Nar H, Messerschmidt A, Huber R, van de Kamp M, Canters GW. Crystal structure analysis of oxidized Pseudomonas aeruginosa azurin at pH 5.5 and pH 9.0. A pH-induced conformational transition involves a peptide bond flip. J Mol Biol. 1991;221(3):765–772. doi: 10.1016/0022-2836(91)80173-r. [DOI] [PubMed] [Google Scholar]
- 8.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376(6542):660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 9.Williams PA, et al. The CuA domain of Thermus thermophilus ba3-type cytochrome c oxidase at 1.6 A resolution. Nat Struct Biol. 1999;6(6):509–516. doi: 10.1038/9274. [DOI] [PubMed] [Google Scholar]
- 10.Brown K, et al. Revisiting the catalytic CuZ cluster of nitrous oxide (N2O) reductase. Evidence of a bridging inorganic sulfur. J Biol Chem. 2000;275(52):41133–41136. doi: 10.1074/jbc.M008617200. [DOI] [PubMed] [Google Scholar]
- 11.Robinson H, et al. Structural basis of electron transfer modulation in the purple CuA center. Biochemistry. 1999;38(18):5677–5683. doi: 10.1021/bi9901634. [DOI] [PubMed] [Google Scholar]
- 12.Winkler JR, Malmström BG, Gray HB. Rapid electron injection into multisite metalloproteins: Intramolecular electron transfer in cytochrome oxidase. Biophys Chem. 1995;54(3):199–209. doi: 10.1016/0301-4622(94)00156-e. [DOI] [PubMed] [Google Scholar]
- 13.Solomon EI, et al. Electronic structure of the oxidized and reduced blue copper sites: Contributions to the electron transfer pathway, reduction potential, and geometry. Inorg Chim Acta. 1996;243(1):67–78. [Google Scholar]
- 14.Beinert H, Griffiths DE, Wharton DC, Sands RH. Properties of the copper associated with cytochrome oxidase as studied by paramagnetic resonance spectroscopy. J Biol Chem. 1962;237(7):2337–2346. [PubMed] [Google Scholar]
- 15.Fee JA, et al. Multi-frequency EPR evidence for a binuclear CuA center in cytochrome c oxidase: Studies with a 63Cu- and 65Cu-enriched, soluble domain of the cytochrome ba3 subunit II from Thermus thermophilus. Biochem Biophys Res Commun. 1995;212(1):77–83. doi: 10.1006/bbrc.1995.1938. [DOI] [PubMed] [Google Scholar]
- 16.Gamelin DR, et al. Spectroscopy of mixed-valence CuA-type centers: Ligand-field control of ground-state properties related to electron transfer. J Am Chem Soc. 1998;120(21):5246–5263. [Google Scholar]
- 17.Wallace-Williams SE, et al. Far-red resonance raman study of copper A in subunit II of cytochrome c oxidase. J Am Chem Soc. 1996;118(16):3986–3987. [Google Scholar]
- 18.Xie X, et al. Perturbations to the geometric and electronic structure of the CuA site: Factors that influence delocalization and their contributions to electron transfer. J Am Chem Soc. 2008;130(15):5194–5205. doi: 10.1021/ja7102668. [DOI] [PubMed] [Google Scholar]
- 19.Robin MB, Day P. Mixed valence chemistry–A Survey and Classification. In: Emeleus HJ, Sharpe AG, editors. Advances in Inorganic Chemistry and Radiochemistry. Vol 10. New York and London: Academic Press; 1968. pp. 247–422. [Google Scholar]
- 20.Solomon EI, Xie X, Dey A. Mixed valent sites in biological electron transfer. Chem Soc Rev. 2008;37(4):623–638. doi: 10.1039/b714577m. [DOI] [PubMed] [Google Scholar]
- 21.Hart PJ, et al. A missing link in cupredoxins: Crystal structure of cucumber stellacyanin at 1.6 A resolution. Protein Sci. 1996;5(11):2175–2183. doi: 10.1002/pro.5560051104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Germann UA, Muller G, Hunziker PE, Lerch K. Characterization of two allelic forms of neurospora crussa laccase. J Biol Chem. 1988;263(2):885–896. [PubMed] [Google Scholar]
- 23.DeBeer George S, et al. Spectroscopic investigation of stellacyanin mutants: Axial ligand interactions at the blue copper site. J Am Chem Soc. 2003;125(37):11314–11328. doi: 10.1021/ja035802j. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh S, et al. Thermodynamic equilibrium between blue and green copper sites and the role of the protein in controlling function. Proc Natl Acad Sci USA. 2009;106(13):4969–4974. doi: 10.1073/pnas.0900995106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray HB, Malmström BG, Williams RJP. Copper coordination in blue proteins. J Biol Inorg Chem. 2000;5(5):551–559. doi: 10.1007/s007750000146. [DOI] [PubMed] [Google Scholar]
- 26.Wilson TD, Yu Y, Lu Y. Understanding copper-thiolate containing electron transfer centers by incorporation of unnatural amino acids and the CuA center into the type 1 copper protein azurin. Coord Chem Rev. 2013;257(1):260–276. [Google Scholar]
- 27.Hwang HJ, Berry SM, Nilges MJ, Lu Y. Axial methionine has much less influence on reduction potentials in a CuA center than in a blue copper center. J Am Chem Soc. 2005;127(20):7274–7275. doi: 10.1021/ja0501114. [DOI] [PubMed] [Google Scholar]
- 28.Abriata LA, et al. Alternative ground states enable pathway switching in biological electron transfer. Proc Natl Acad Sci USA. 2012;109(43):17348–17353. doi: 10.1073/pnas.1204251109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andrew CR, et al. Identification and description of copper-thiolate vibrations in the dinuclear CuA site of cytochrome c oxidase. J Am Chem Soc. 1996;118(43):10436–10445. [Google Scholar]
- 30.Gorelsky SI, Xie X, Chen Y, Fee JA, Solomon EI. The two-state issue in the mixed-valence binuclear CuA center in cytochrome C oxidase and N2O reductase. J Am Chem Soc. 2006;128(51):16452–16453. doi: 10.1021/ja067583i. [DOI] [PubMed] [Google Scholar]
- 31.Palmer AE, Randall DW, Xu F, Solomon EI. Spectroscopic studies and electronic structure description of the high potential type 1 copper site in fungal laccase: Insight into the effect of the axial ligand. J Am Chem Soc. 1999;121(30):7138–7149. [Google Scholar]
- 32.Farver O, Lu Y, Ang MC, Pecht I. Enhanced rate of intramolecular electron transfer in an engineered purple CuA azurin. Proc Natl Acad Sci USA. 1999;96(3):899–902. doi: 10.1073/pnas.96.3.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeBeer George S, et al. A quantitative description of the ground-state wave function of Cu(A) by X-ray absorption spectroscopy: Comparison to plastocyanin and relevance to electron transfer. J Am Chem Soc. 2001;123(24):5757–5767. doi: 10.1021/ja004109i. [DOI] [PubMed] [Google Scholar]
- 34.Hupp JT, Zhang XL. Solvational barriers to interfacial electron transfer: Minimization via valence delocalization. J Phys Chem. 1995;99(3):853–855. [Google Scholar]
- 35.Hadt RG, Xie X, Pauleta SR, Moura I, Solomon EI. Analysis of resonance Raman data on the blue copper site in pseudoazurin: Excited state π and σ charge transfer distortions and their relation to ground state reorganization energy. J Inorg Biochem. 2012;115:155–162. doi: 10.1016/j.jinorgbio.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Olsson MHM, Ryde U. Geometry, reduction potential, and reorganization energy of the binuclear Cu(A) site, studied by density functional theory. J Am Chem Soc. 2001;123(32):7866–7876. doi: 10.1021/ja010315u. [DOI] [PubMed] [Google Scholar]
- 37.Olsson MHM, Ryde U, Roos BO. Quantum chemical calculations of the reorganization energy of blue-copper proteins. Protein Sci. 1998;7(12):2659–2668. doi: 10.1002/pro.5560071220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LaCroix LB, et al. Electronic structure of the perturbed blue copper site in nitrite reductase: Spectroscopic properties, bonding, and implications for the entatic/rack state. J Am Chem Soc. 1996;118(33):7755–7768. [Google Scholar]
- 39.Beratan DN, Onuchic JN, Betts JN, Bowler BE, Gray HB. Electron-tunneling pathways in ruthenated proteins. J Am Chem Soc. 1990;112(22):7915–7921. [Google Scholar]
- 40.Gurbiel RJ, et al. Detection of two histidyl ligands to CuA of cytochrome oxidase by 35-GHz ENDOR: 14,15N and 63,65Cu ENDOR studies of the CuA site in bovine heart cytochrome aa3 and cytochromes caa3 and ba3 from Thermus thermophilus. J Am Chem Soc. 1993;115(23):10888–10894. [Google Scholar]
- 41.Lyons JA, et al. Structural insights into electron transfer in caa3-type cytochrome oxidase. Nature. 2012;487(7408):514–518. doi: 10.1038/nature11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zickermann V, et al. Perturbation of the CuA site in cytochrome-c oxidase of Paracoccus denitrificans by replacement of Met227 with isoleucine. Eur J Biochem. 1995;234(2):686–693. doi: 10.1111/j.1432-1033.1995.686_b.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.