Abstract
Nanoparticles have opened new exciting avenues for both diagnostic and therapeutic applications in human disease, and targeted nanoparticles are increasingly used as specific drug delivery vehicles. The precise quantification of nanoparticle internalization is of importance to measure the impact of physical and chemical properties on the uptake of nanoparticles into target cells or into cells responsible for rapid clearance. Internalization of nanoparticles has been measured by various techniques, but comparability of data between different labs is impeded by lack of a generally accepted standardized assay. Furthermore, the distinction between associated and internalized particles has been a challenge for many years, although this distinction is critical for most research questions. Previously used methods to verify intracellular location are typically not quantitative and do not lend themselves to high throughput analysis. Here we developed a mathematical model which integrates the data from high throughput flow cytometry measurements with data from quantitative confocal microscopy. The generic method described here will be a useful tool in biomedical nanotechnology studies. The method was then applied to measure the impact of surface coatings of vesosomes on their internalization by cells of the reticuloendothelial system (RES). RES cells are responsible for rapid clearance of nanoparticles, and the resulting fast blood clearance is one of the major challenges in biomedical applications of nanoparticles. Coating of vesosomes with long chain polyethylene glycol showed a trend for lower internalization by RES cells.
Keywords: nanoparticle internalization, drug delivery, reticuloendothelial system, flow cytometry, confocal laser scanning microscopy, vesosomes, polyethylene glycol
A number of techniques have been established to measure association of micro- and nanoparticles to cells. These include high throughput techniques such as Fluorescence Activated Cell Sorting/scanning (FACS), inductively coupled plasma (ICP) spectroscopies such as ICP mass spectrometry, and high sensitivity methods for certain nanoparticles such as absorption measurements of lysed cells.1 However, none of these techniques are able to differentiate internalized from externally adhered particles. Attempts to solve this problem have included the destruction of particles followed by removal through washes,2 the measurement of fluorescence in the presence and absence of fluorescence quenchers,3,4 detection of surface bound particles with secondary, non-cell penetrating reagents5 or use of fluorophores that require binding to nucleic acids.6 However, these methods are specific for certain particles and/or certain fluorophores, making comparisons of data between different types of nanoparticles difficult. Another frequently used technique is the parallel measurement at different temperatures, namely 4°C and 37°C and subtraction of the 4°C values from those obtained at 37°C.7 This technique is problematic for nanoscale particles which have been observed to enter the cell at low temperatures, potentially through an energy independent mechanism.8 Confocal laser scanning microscopy (CLSM) has been widely used to verify internalization, but is generally not considered a quantitative technique. A recently introduced technique, imaging flow cytometry (IFC) delivers both FACS data and microscopical images of cells.9,10 While it is very convenient to obtain visual data of the same cells, which are analyzed in FACS, it comes at the cost of lower sample throughput compared to conventional FACS and lower resolution compared to CLSM. IFC has been used to quantify internalization, but requires to permeabilize cells and co-stain internal cell compartments, thereby only measuring particles within the stained cell compartments. It is not a confocal technique, and other planes in the microscopic image contribute to the fluorescence; therefore typically only the upper quartile of pixels (based on intensity) is used for quantification. An elegant, flow cytometry-based approach to distinguish between adsorbed and internalized particles by labeling them with a pH-sensitive dye has been introduced by Semmling et al.11 In that study, particles fluoresced red in the slightly alkaline extracellular milieu and green in the acidic intracellular milieu, enabling a semi-quantitative evaluation. Conventional FACS and CLSM are often combined to obtain quantitative data on particle association with cells, and qualitative data on internalization, but the data from the two techniques has always been presented as independent sets. We describe here an approach to measure internalization quantitatively by CSLM and integrate this data with high throughput FACS data for a precise measurement of nanoparticle uptake. In addition, a correction factor is introduced, accounting for a potential change of fluorescence intensity upon internalization. Different mechanisms contribute to such changes: When fluorophores are internalized via endocytosis or phagocytosis, they travel down the endocytotic pathways in organelles with decreasing pH, ranging from 6.3 to 6.5 in early endosomes to below 4 in lysosomes.12 Commonly used fluorophores such as fluorescein and its derivatives, as well as seminaphtorhodafluors (SNARF) are very sensitive to pH changes.13,14 On the other hand, an increase of fluorescence upon entering the cytosol has been described for widely used fluorophores, such as fluorescein, Texas Red, Cy5 and Atto647.13 Furthermore, non-specific protein binding may alter the fluorescence intensity.15 Therefore, fluorescence values have to be corrected for accurate quantification. This applies both to microscopy-and FACS-derived data, a fact that to our knowledge has not been considered so far.
We first derived and validated the method described here, using polystyrene (PS) particles. We then applied it to an important biomedical question, i.e. the impact of different surface coatings of vesosomes on their uptake by cells of the reticuloendothelial system (RES). Vesosomes are lipid-based multicompartment drug delivery vehicles with structural similarities to eukaryotic cells. They are created by closure of two-dimensional lipid bilayer sheets and used to encapsulate smaller nanoparticles, containing reporter molecules or drugs.16 This results in more efficient containment of the encapsulated drugs.17 One major challenge for the biomedical application of nanoparticles, in general, is their rapid removal from the bloodstream through cells of the RES, particularly macrophages. Polyethylene glycol (PEG) is frequently used as a sterical stabilizer and has been reported to increase the circulation time of lipid bilayer vesicles after systemic injection into laboratory animals.18 While this increase of circulation time has been well documented, the exact mechanism of action is not entirely understood. A number of effects have been suggested as a cause: 1. Prevention of protein adsorption with the consequence of reduced opsonization; 2. promotion of the adsorption of specific proteins which might mask the particle (dysopsonization effect); 3. prevention of aggregation; 4. sterical inhibition of binding to RES cells; 5. stabilization by inhibition of lipid degradation (reviewed in 18). As a consequence of some of these factors, it has been suggested that PEG inhibits the recognition by cells of the RES, although this hypothesis is controversial.18,19 It is further known that results vary with the concentration and chain length of the incorporated PEG.19,20 We therefore investigated the impact of coating vesosomes with different concentrations and species of PEG on internalization by macrophages.
RESULTS AND DISCUSSION
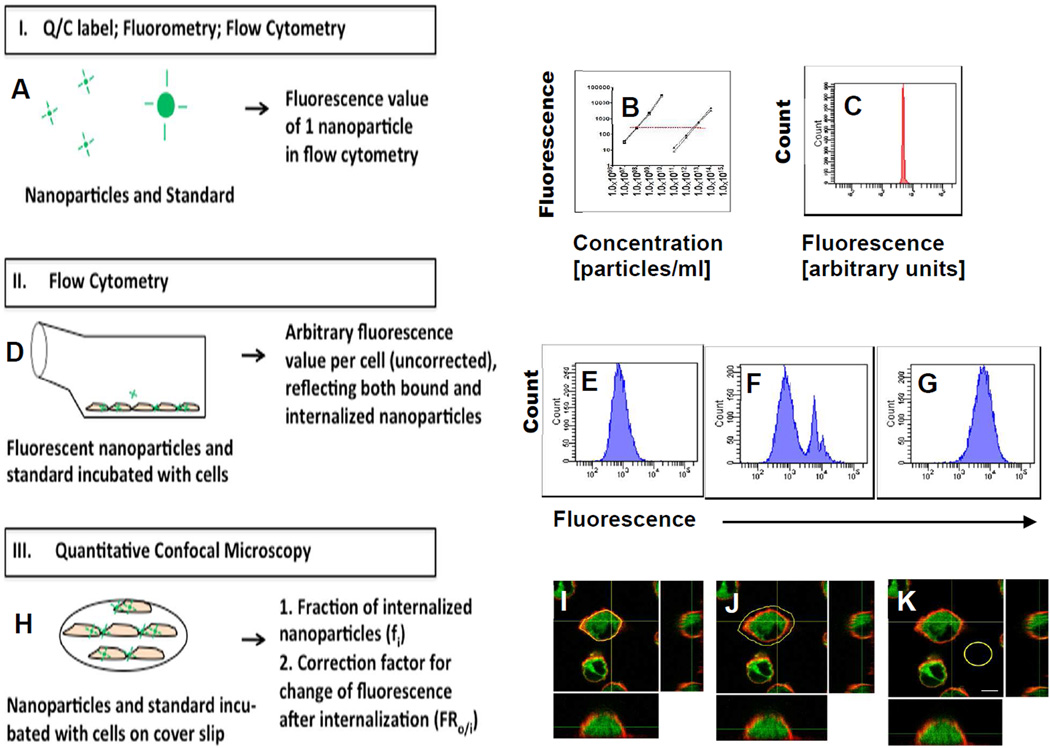
A method for quantification of internalized particles per cell was developed. An overview on the different steps of the method is illustrated in Figure 1. Briefly, fluorescence of one nanoparticle is determined by fluorimetry and FACS using a microparticle standard (Figure 1-I); fluorescence of cells incubated with nanoparticles is determined by FACS (Figure 1-II); and the internalized fraction as well as the amount of fluorescence intensity change after internalization is measured by CLSM (Figure 1-III). Equation 1.1 is then used to calculate the number of nanoparticles internalized per cell.
Figure 1. Determination of number of internalized particles per cell by combined fluorimetry, flow cytometry and quantitative confocal microscopy.
A-C: The mean fluorescence of one nanoparticle in the FACS assay was determined by fluorimetry and FACS, see text for details. A: Overview. B: Fluorimetry of nanoparticles in comparison to standard microparticles; datapoints show averages and standard deviations of triplicates; squares: 1 µm microbeads, two experiments; triangles: 50 nm nanoparticles, two experiments. Panel C: Flow cytometry of standard microbeads. D-G: To obtain the average fluorescence emitted by all nanoparticles associated with a cell, nanoparticles were incubated with J774 macrophages and then subjected to flow cytometry. The mean fluorescence of these cells and of control cells (no particles added) was determined. D: Overview. E: negative control cells; F: Cells after incubation with microbead standard (1-micron sized green fluorescent polystyrene particles); G: Cells after incubation with nanoparticles, here: green fluorescent polystyrene beads (50 nm). Note that a correction factor, FRo/i, for fluorophore change inside the cell may be required, which is determined by confocal microscopy (see text for details). H-K: The internalized fraction fi was measured by confocal microscopy. J774 macrophages were incubated with polystyrene nanoparticles (50 nm) and the membrane was counterstained with wheat germ agglutinin conjugated to the fluorophore Alexa 594; fluorescence area density inside the cell (I) and inside+outside the cell (J) was measured (see text for details); background subtractions for inside the cell were derived from a separate slide with negative control cells (no particles) and for the rim outside the cell from another area on the slide (K). Scale bar is 5 µm.
| (1.1) |
where
| n[Pin / cell] | is the average number of internalized particles per cell |
| fi | is the internalized fraction of particles associated with the cell, which is calculated as indicated in equation 1.4 |
| MFcell−MFneg_cell | is the mean fluorescence of cells incubated with particles minus the mean fluorescence of negative control cells (no particles), as derived by flow cytometry |
| FRo/i | is the ratio of fluorescence intensity of the fluorophore outside the cell over that inside the cell. This is measured by confocal microscopy. |
| MFP | is the mean fluorescence of one particle according to equation 1.2 |
| (1.2) |
where
| MFMP[FACS] | is the mean fluorescence of one microparticle as determined by FACS |
| cf | is the calibration factor according to equation 1.3. Note that micron-sized particles can be directly measured by FACS, in which case the fluorimetry is unnecessary and cf becomes 1. |
| (1.3) |
where
| MFMP[FLUO] | is the mean fluorescence of microparticles at defined concentrations A, B, C, measured by fluorimetry at the same excitation and emission conditions as the FACS for MFMP[FACS] |
| MFNP[FLUO] | is the mean fluorescence of nanoparticles at defined concentrations A, B, C, measured by fluorimetry at the same excitation and emission conditions as the FACS for MFMP[FACS] |
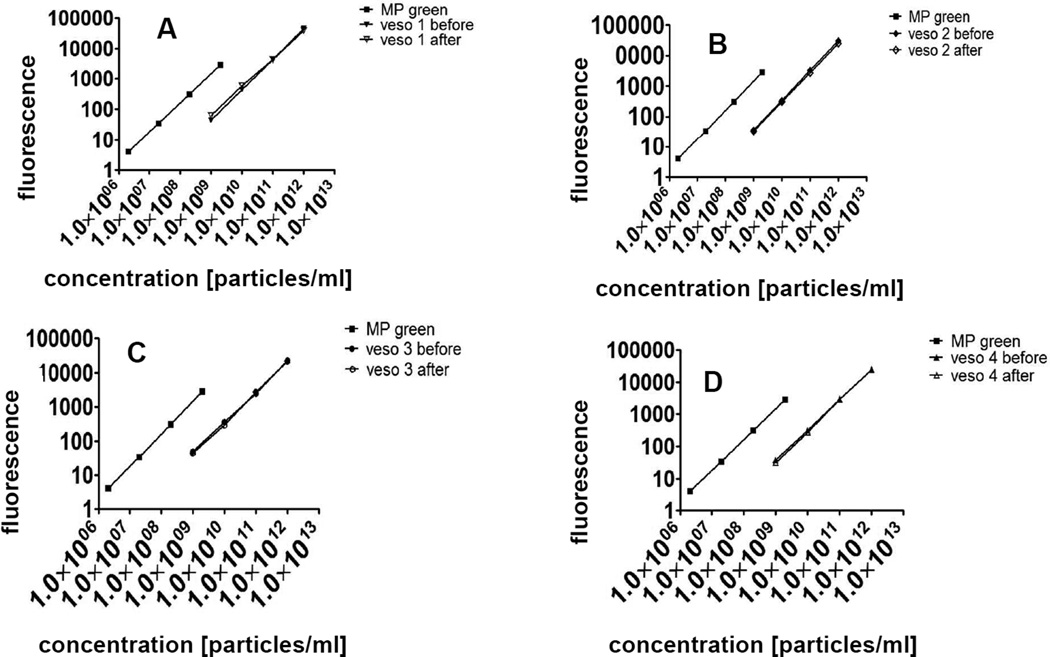
The method is now described in more detail: The first step was to determine the fluorescence value that one nanoparticle would exhibit in the flow cytometry experiment (Figure 1A). Due to fluidic, optical and sensor limitations, nanoparticles below a certain size cannot be measured as single entities by FACS. Therefore, a defined concentration of fluorescent nanoparticles was measured in a fluorimeter alongside a fluorescent microparticle standard under the same excitation and emission filter conditions as in the following FACS analysis. This yielded the calibration factor cf, by which the single microparticle fluorescence is brighter than the single nanoparticle fluorescence (Figure 1B, equation 1.3). For these experiments, a fluorimeter is required which is capable to operate with the same excitation wavelength and emission filter window as subsequently used in the FACS experiment. It is very important that there is no free fluorophore in the particle suspensions, both for this experiment as well as for the CLSM experiment. To confirm this, we performed a dialysis step of the particles and measured fluorescence before and after dialysis (Figure 2).
Figure 2. Dialysis test for free fluorophore.
Microparticles of 1 µm diameter (left lines in each panel, MP green) and vesosomes of 400 nm diameter before and after dialysis (right lines, veso 1–4 before, veso 1–4 after) at known concentrations were serially diluted and measured in triplicate on a fluorimeter at 488 nm excitation and 530/30 emission. A-D: Four different vesosome preparations. Graphs show average of triplicate samples with standard deviation error bars.
Figure 2 shows that the fluorescence curves of nanoparticles before and after dialysis were virtually identical, which ruled out free fluorophore in the suspension.
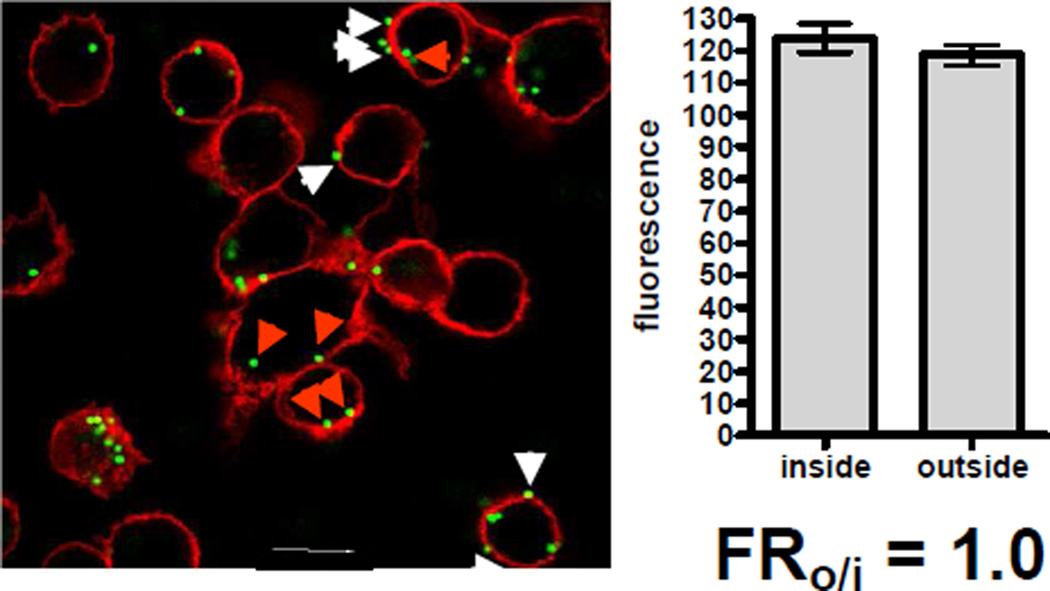
Next, we carried out a FACS analysis of cells incubated with fluorescent nanoparticles. The microparticle standard was also measured by FACS (Figure 1C), and used to calculate the fluorescence of one nanoparticle in FACS according to equation 1.2. In experiments with microparticles the fluorimetry step is unnecessary and cf becomes 1. For fluorophores that do not change fluorescence intensity after internalization, the data obtained gives the average number of nanoparticles associated with each cell (Figure 1D-G). Aggregation of particles may alter the amount of internalized particles, and different physiological buffers/additives can have distinct effects on the aggregation status. As further discussed below, qualitative data on the aggregation status of the particles can also be obtained in the FACS experiment. If a fluorophore is used that changes fluorescence after internalization, a correction factor, the fluorescence intensity ratio of the fluorophore outside the cell over inside the cell (FRo/i) is needed. FRo/i was determined by CLSM, as described in the Methods section, using a microparticle labeled with the same fluorophore as used for the nanoparticles (Figure 3). The determination of FRo/i should be done at the same time point as used in the internalization study. An early time point (90 min after addition of particles) was chosen, based on the assumption that endocytosed particles would still be located in early endosomes, and not distributed over various compartments in the cell. Ideally, a fluorophore is chosen, which is not very sensitive to the environment, such that FRo/i becomes 1, which greatly simplifies the calculation and reduces the uncertainty of the measurement (see below). Table 1 shows FRo/i values for the different fluorophores used in this study. These values should be determined specifically for each experiment, since they may be dependent on the particular particle preparation and the cell line used. The values given here pertain to J774 macrophages. The impact of the FRo/i factor on the integrity of adhesion/internalization data is demonstrated in Supplemental Figure 1.
Figure 3. Measurement of the fluorescence change of a fluorophore after internalization.
Polystyrene beads of 1 µm diameter, which were internally-dyed with Firefli™ green were incubated with macrophages. Macrophage cell membranes were counterstained with wheat germ agglutinin-Alexa-594 and cells were analyzed by confocal microscopy as described in Methods. The mean fluorescence of each particle was measured. The image shows examples for particles outside the cell (white arrows) and inside the cell (red arrows). The ratio of the average of mean fluorescence values outside the cells over inside the cells (FRo/i) was then calculated. A FRo/i of 1 represents no change in fluorescence intensity after internalization.
Table 1.
Change of fluorescence of different fluorophores after internalization into mammalian cells.
| Fluorophore | n[particles] | FRo/i | Fout Av (SD) |
Fin Av (SD) |
p |
|---|---|---|---|---|---|
| Firefli green (internally-dyed particles) | 17 | 1.0 | 119 (9) | 124(14) | 0.4 |
| Firefli red (internally-dyed particles) | 14 | 1.0 | 845 (107) | 823 (187) | 0.8 |
| DiO (surface label) | 25 | 0.4 | 138 (75) | 348 (177) | 0.001 |
| Intracellular GFP (internally expressed by bacteria) | 11 | 1.0 | 2025 (294) | 2049 (329) | 1.0 |
FRo/i: Fluorescence ratio of fluorophore outside over inside the cell. Fout: Fluorescence of particles outside the cell; Av (SD): average and standard deviation; Fin: Fluorescence of particles inside the cell; p: statistical significance of difference between Fout and Fin (Mann-Whitney test, 2-tailed).
To obtain quantitative data on the internalized fraction (fi), cells were seeded on glass coverslips and incubated with nanoparticles at the same particle-to-cell-ratios and under identical environmental conditions as used in the FACS experiment (Figure 1H-K). Cell membranes were counterstained with fluorescently labeled wheat germ agglutinin, and coverslips were mounted on microscopical slides. Control slides with unstained cells were also prepared. The area for internal and external fluorescence was chosen as follows: Images taken on the confocal laser scanning microscope were opened in the software Image J. The images were enlarged and the inside of the cell manually delineated by highlighting the membrane (see yellow line in Figure 1I). The outside of the cell was marked by highlighting the area around the cell, with enough distance to enclose any particles that may be bound to the cell membrane externally (see yellow line in Figure 1J). Background for inside the cell was measured similar to Figure 1I on a negative control slide, and background for the outside of the cell as in Figure 1K on the same slide. A sharp membrane stain is essential for proper identification of the regions of interest. The procedure described in the Methods section is a result of a number of optimization studies with regards to fixation and fluorophore selection. Wheat germ agglutinin stains the glycocalix, visualizing extensions on which externally adhered particles may reside. It is important to confirm that no bleedover between the channels occurs. Supplemental Figure 2 shows these and other negative controls. In addition, we confirmed that backgrounds on different slides were similar. Using fluorescent microbeads, we developed and validated the formula described below to determine fi. Briefly, weighted averages of mean fluorescence from interior cell areas were subtracted from those of whole cells after subtraction of respective background measurements. Typically, 20–50 cells were analyzed in this fashion and the raw measurements were exported into an excel file which contained a macro to calculate fi (Supplement 2). fi was computed according to equation 1.4 (Figure 4).
Figure 4. Calculation of fi (internalized fraction of associated particles).
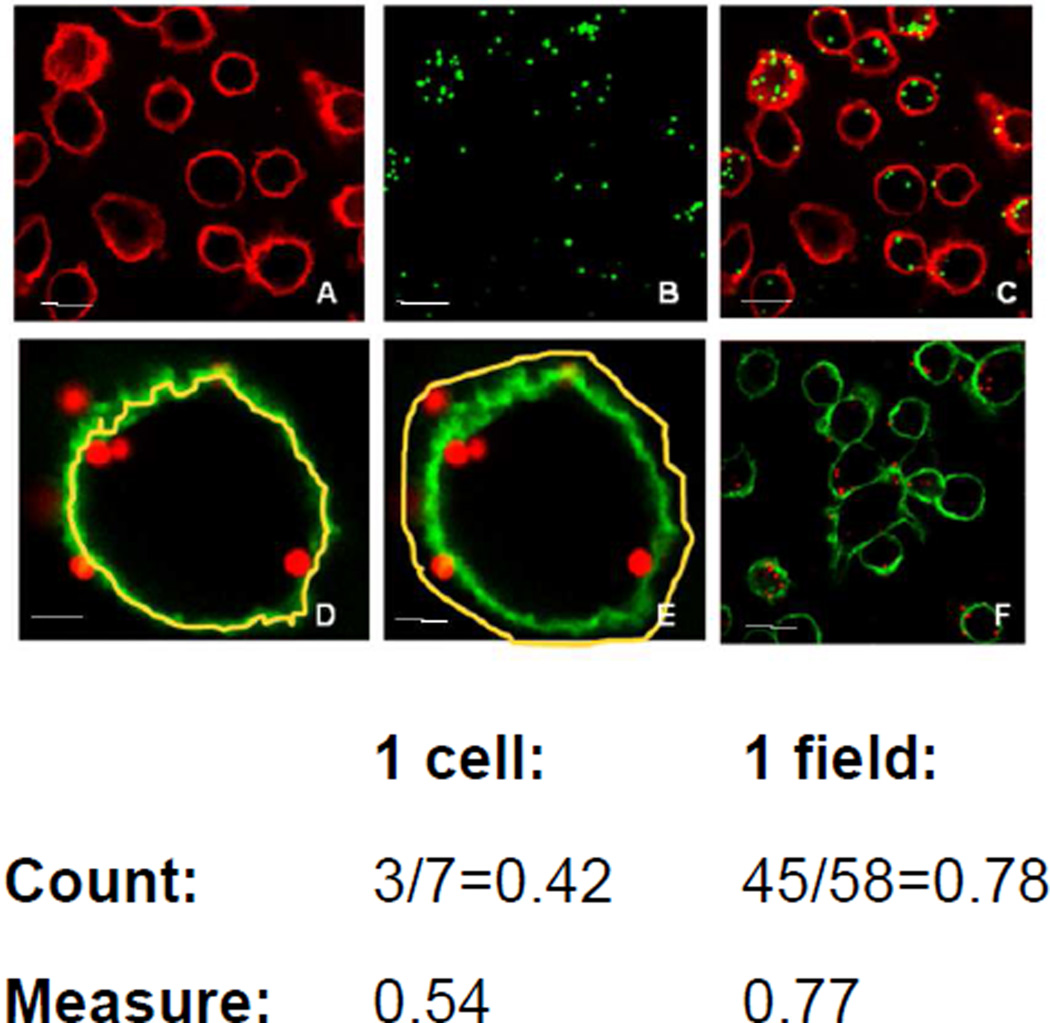
Validation of the method was carried out with fluorescent microbeads. Using 1-micron-sized particles we visually determined and counted the particles inside and outside of cells, and then compared to the data obtained by using the method described here. Figure 5 shows good correlation between the two methods, if one or more microscopic fields were counted.
The cellular uptake of internally-dyed fluorescent polystyrene particles of various sizes was then measured using this method (Table 2).
Table 2.
Number of internalized particles for polystyrene particles of different sizes.
| Particle Size (diameter in nm) |
Ratio Particles:Cells |
fi | MF_cells | MF_neg cells |
FRo/i | MF_Particle | n(Pin/cell) |
|---|---|---|---|---|---|---|---|
| 1000 | 102:1 | .86 (0.13) | 2564 (179)a | 222 (14) | 1.0 (0.1) | 3776 (189)a | .51 (0.09)b |
| 400 | 2 × 104:1 | .88 (0.02) | 202456 (14614) | 222 (14) | 1.0 (0.1) | 222 (20) | 771 (129)b |
| 250 | 8 × 104:1 | .86 (0.03) | 168329 (12502) | 222 (14) | 1.0 (0.1) | 77 (9) | 1806 (336)b |
Data from representative experiments with average and (standard deviation);fi: internalized fraction; MF_cells: mean fluorescence of sample cells (as determined by flow cytometry); MF_neg cells: mean fluorescence of negative control cells (as determined by flow cytometry); FRo/i: Fluorescence ratio of fluorophore outside over inside the cell; MF_Particle: mean fluorescence of one particle, as it would measure in flow cytometry; n(Pin/cell) : number of internalized particles per cell.
estimated;
standard deviations of n(Pin/cell) were calculated using the error propagation model (Supplement 3).
Since many different parameters are required to calculate the number of internalized particles per cell, which raises the concern of high standard deviations due to error propagation, we performed an error propagation analysis, using internalization data from green fluorescent 400 nm nanoparticles. An excel spreadsheet is provided with embedded formulas which can be used to calculate the final standard deviation evolving from uncertainties of all measured parameters (Supplement 3). This analysis shows that for the example used here the fractional uncertainty (percent standard deviation of average) is 17%SD. This appears acceptable for biological measurements. Using the spreadsheet and varying the uncertainties of the original parameters, it becomes obvious that variability in FRo/i has a big impact on the error of the final result, whereas fi has less impact. This further supports the statement made above, that choosing a fluorophore with minimal or no changes upon internalization into a cell would be highly preferred. It also highlights the necessity to experimentally confirm the value of FRo/i.
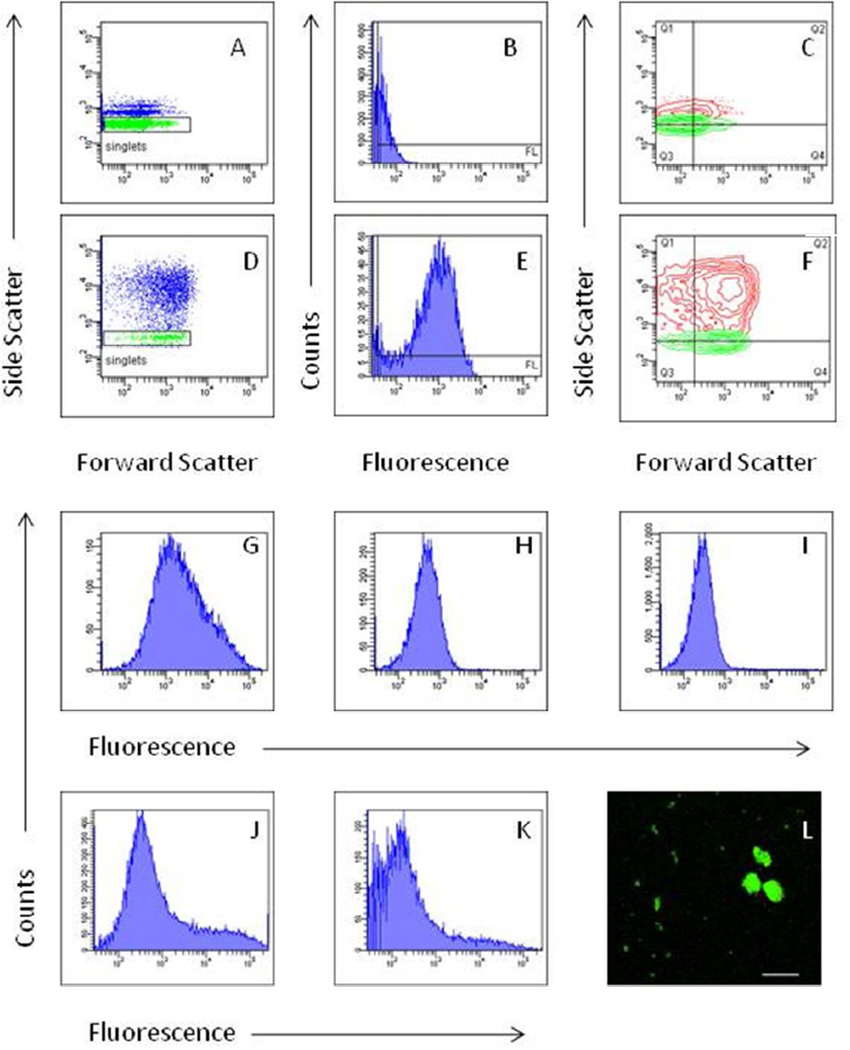
In addition to quantitative internalization data, the flow cytometry analysis can also yield qualitative information about aggregation status of particles and protein binding. This can conveniently be obtained in the same experiment using exactly identical experimental conditions as for the internalization measurement. When aggregation occurs, the forward and sideward scatter values increase, and the peak in the fluorescence histogram shows tailing or a complete shift to the right (Figure 6). Coincidence (measurement of more than one particle at a time) which also results in a one-sided broadening of the fluorescent peak can be distinguished from aggregation by analyzing different concentrations (Figure 6).
Figure 6. Aggregation of polystyrene (PS) particles and vesosomes in biological buffers.
Particles were diluted in physiological buffers as specified below and measured on a flow cytometer 24 h (PS) or 5 h (vesosomes) later. A-C: 400 nm PS particles in water; D-F: 400 nm PS particles in phosphate buffered saline with 2% w/v bovine serum albumin. Note the shift of the fluorescence peak to the right in E and the shift of the forward/sideward scatter plot to the right and up in D and F. G-K: Differentiation of coincidence and true aggregation. G-I: Non-PEGylated vesosomes labeled with 0.5 mol% dialkylcarbocyanine (DiO) and diluted at three different concentrations (neat, 1:10 and 1:100 from left to right). The shoulder of the peak in G disappears, when sample is diluted. Peak shoulder and shift to the right are due to coincidence (i.e. at high concentration, more than one particle is interrogated simultaneously by the laser). J, K: Non-PEGylated vesosomes with 5% 7-nitrobenzo-2-oxa-1,3-diazole (NBD) NBD at two different concentrations (neat and 1:10). The tail of the peak remains after dilution. O: Microscopic image of vesosomes modified with NBD verifies large aggregates as expected from the flow cytometry histogram. Size bar 100 µm.
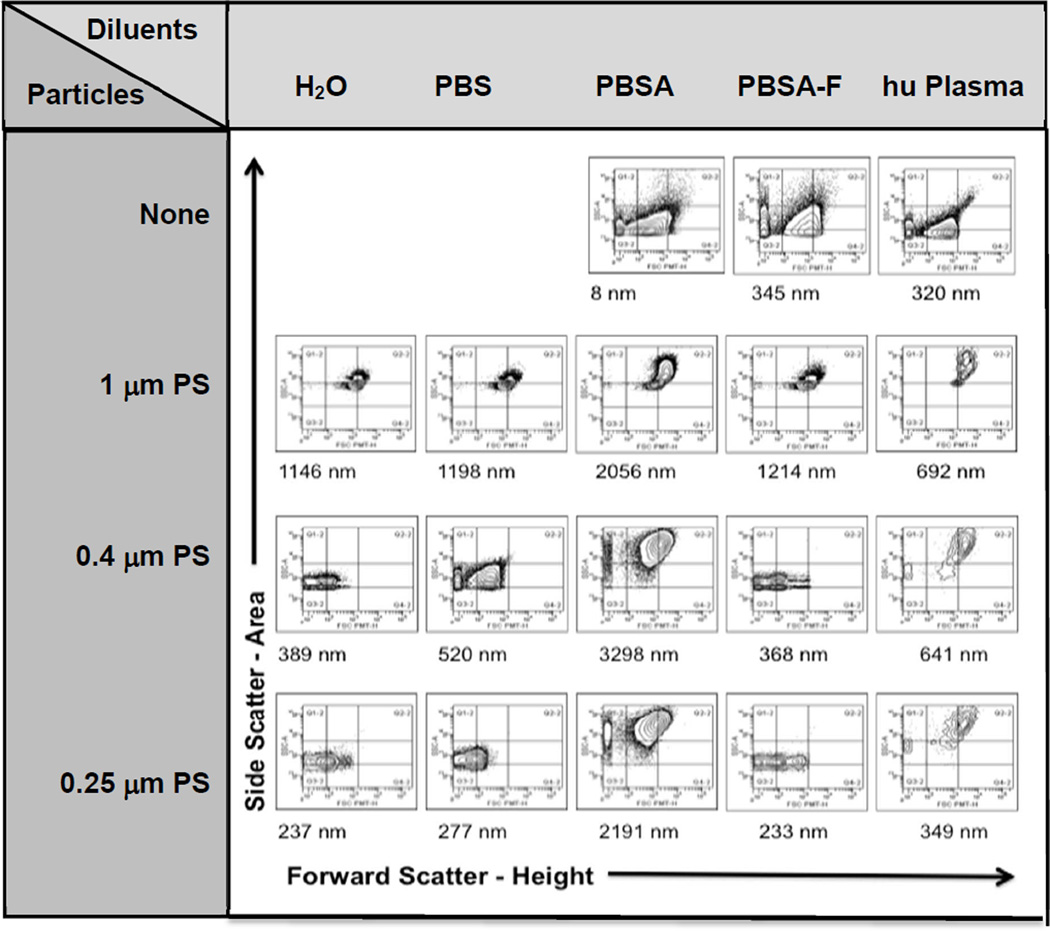
In complex solutions such as serum- or plasma-containing buffers, a high background exists for light scatter measurements, such as FACS and dynamic light scattering (DLS). Therefore it is an advantage that this non-fluorescent background can be eliminated through gating for fluorescence, to evaluate the scatter properties of the fluorescent population separately. Figure 7 illustrates an example: Polystyrene (PS) particles of various sizes undergo massive aggregation after dilution in phosphate buffered saline (PBS) containing 2% w/v bovine serum albumin (BSA), when measured 24 hours (but not 3 hours) later. However, when diluted in PBS containing 2% w/v BSA and 30% v/v fetal calf serum (FCS) this aggregation does not occur. It is well known that nanoparticles, when introduced into complex protein-containing solutions, adsorb various proteins to form a protein corona.21,22 The protein corona evolves over time,23–25 since highly abundant proteins appear to adsorb first, which are later replaced by less abundant proteins with higher affinity. The end result is a nanoparticle with a new, biological identity as opposed to its original, synthetic identity.26,27 Adsorption of proteins can promote colloidal stability28,29 or, conversely, can induce aggregation, as shown here and in other studies.30,31 The protein corona also has an impact on internalization rates. Conceptually, adsorbed proteins can either function as opsonins or as dysopsonins, thereby either promoting or inhibiting receptor-mediated endocytosis.27 Frequently, adsorbed proteins will inhibit unspecific binding and internalization. Our rationale to pre-incubate particles for 2h in PBS plus 2% BSA plus 30% FCS was to 1) prevent aggregation, and 2) inhibit unspecific binding and internalization, but not specific binding events. Indeed, in a previous study we could show that iron oxide nanoparticle binding and internalization is dependent on scavenger receptor A (SR-A), and that SR-A was still accessible and functional after dilution into this BSA- and FCS-containing buffer (Supplemental Figure 3). A recent review of 63 nanomaterials from 26 studies highlights that there is no one ‘universal” protein corona, but that the corona rather depends on the synthetic identity of the material, and that the ability of adsorbed proteins to stabilize colloids is nanomaterial-dependent.27 Our observations corroborate this finding. As an example, the aggregation seen with PS particles in PBS plus 2% BSA was not observed with vesosomes (Supplemental Figure 4). Furthermore, several studies report a stabilizing function of albumin for a variety of different nanomaterials.28,29 The PS particles also heavily aggregated after suspension in human plasma. This was already evident after 3 hours and remained until the 24 hour timepoint. Notably, this change was not reflected in the measurements of the effective diameter by DLS, since the scatter properties of the diluents dominated the averaged signal and therefore masked this change (Figure 7).
Figure 7. Impact of common additives to physiological buffers on polystyrene particle aggregation.
Polystyrene (PS) particles of different diameters were diluted in water (H2O), phosphate buffered saline (PBS), PBS plus 2% w/v bovine serum albumin (PBSA), PBS plus 2% w/v bovine serum albumin plus 30% fetal calf serum (PBSA-F) or human plasma. Suspensions were analyzed 24 h after dilution by flow cytometry, and are shown as contour plots of forward scatter-height versus side scatter-area. Grids are provided to guide the eye. Row 1 (diluents alone) shows all events. For particle suspensions (rows 2–4), the fluorescent population was gated first, and this population is shown in the contour plots. Effective average diameters were determined by dynamic light scattering (numbers below the plots). A shift to the right and up in the contour plot is indicative of particle aggregation. PS particles of all three sizes showed aggregation 24h after dilution into PBSA and human plasma but not after dilution into PBSA-F.
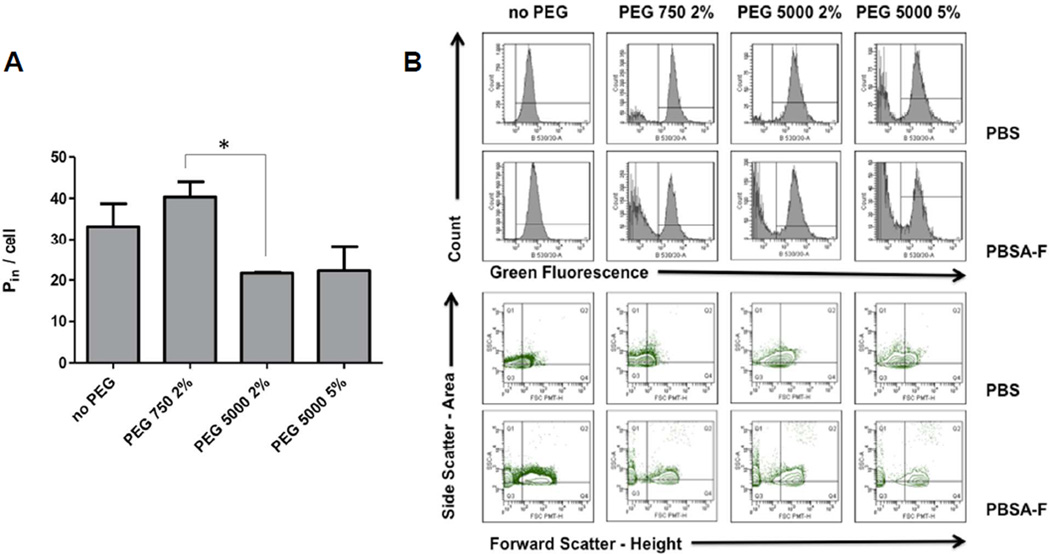
We then applied the method to a question of biomedical relevance, i.e. the impact of different surface coatings of vesosomes on the uptake by cells of the RES. Surface coatings have been reported to have a significant impact on protein adsorption, colloidal stability, cytotoxicity and cellular internalization.30–33 For example, citrate coated iron oxide nanoparticles destabilized in fetal calf serum-based buffers, and are rapidly absorbed and internalized, whereas poly(acrylic acid)-coated iron oxide nanoparticles were stable, and did not show any appreciable uptake into lymphoblastoid cells.30 Similarly, coating of iron oxide particles with polyvinyl alcohol, a mixture of polyvinyl alcohol and vinyl alcohol/vinylamine or with polyethylenimine had a significant impact on agglomeration in FCS-based media, and on cellular uptake, although in this case no clear correlation between aggregation and uptake was established.31 Surface coatings can also introduce positive or negative charges which have differential impact on stability and uptake, as in the example of polymer coated gold nanoparticles.34 In the current study, we coated vesosomes with PEG of different chain lengths at different concentrations, and determined the impact on the number of internalized particles per macrophage. We also investigated the aggregation status of the particles and indicators of protein binding. Fluorescent vesosomes were prepared with 2 mol% PEG750, 2 mol% PEG5000 and 5 mol% PEG5000. The uptake of particles by macrophages of the cell line J774 was analyzed using the method described above. While PEG750 did not inhibit macrophage uptake, PEG5000 at both concentrations reduced the uptake 30% (Figure 8A). Figure 8A shows the internalization data at a 8×104:1 particle-to-cellratio. The difference between PEG750 coated particles and PEG5000 2% -coated particles was statistically significant (p=0.036). Uptake at a particle-to-cell ratio of 2×104:1 displayed an identical pattern (not shown).
Figure 8. Cellular uptake of nanoparticles with different surface coatings.
J774 macrophages were incubated with vesosomes (400 nm) that had different surface coatings with polyethylene glycol (PEG, [mol%]). A. Uptake of nanoparticles by macrophages, given as average number of internalized particles per cell. One representative experiment at a particle-to-cell ratio of 8×104:1 is shown; averages and SEM. B. Particles without cells (same batch of vesosomes as in panel A) were diluted in various biological buffers, here: phosphate buffered saline (PBS) and PBS plus 2% w/v bovine serum albumin and 30% v/v fetal calf serum (PBSA-F, same buffer as used in internalization assay), and analyzed by flow cytometry 3h later for indications of aggregation and protein binding. Fluorescence peaks and contour plots of fluorescent population are shown for each particle suspension. Non-PEGylated vesosomes showed signs of aggregation in PBSA-F (shift to the right in fluorescence histogram and in contour plot), vesosomes coated with PEG 750 showed signs of protein binding (shift to the right in contour plot, but not in fluorescence histogram).
Aggregation status of the particles under identical conditions as those used in the internalization assay was investigated by FACS, DLS and fluorescence microscopy. Particles without cells (same batch of vesosomes as in Figure 8A) were diluted in various biological buffers, and analyzed by flow cytometry 3h later. The flow cytometry data supports the hypothesis that PEG750 coated vesosomes adsorb protein under assay conditions (i.e. dilution in PBS plus 2 % w/v BSA plus 30% v/v FCS, abbreviated as PBSA-F), while this effect was less pronounced for PEG5000 coated vesosomes. Figure 8B demonstrates that PEG750 coated vesosomes showed a significant increase in forward scatter without an increase of fluorescence, consistent with size increase due to protein binding. In contrast, vesosomes without PEG showed both an increase in forward scatter and an increase in mean fluorescence, consistent with aggregation. This interpretation was confirmed by fluorescence microscopy, which revealed significant aggregation for non-PEGylated particles but to a much lesser extent for PEGylated particles after suspension in PBSA-F. The change was not reflected in the measurements of the effective diameter by DLS.
The method described here can be extended to other applications. One example is the study of phagocytosis of bacteria or other microorganisms by macrophages. Bacteria, with a size of 1–2 µm, can also be visually detected and counted by microscopy using overlay images. Determination of fi using the method described in this study, however, is not only more convenient but also more accurate, because signals can easily be masked when looking at overlap images. Drawing the region of interest in the channel for the membrane stain and then analyzing the particle number in the channel for particle stain eliminates this problem (Supplemental Figure 5). As an example, the internalized fraction of fluorescent bacteria expressing different peptides on their surface was measured (Supplemental Figure 6). The method can be applied to a wide variety of nanoparticles, as long as the particles can be efficiently labeled with a fluorophore (Supplemental Figure 7). The sensitivity depends on the brightness of the label and – if particles per cell are used as a unit – on the particle size. For example, one particle per cell of the internally-dyed PS microparticles (diameter 1 µm) is easily detected, both by flow cytometry and by microscopy. This translates to a calculated detection limit for flow cytometry of ≤23=8 particles per cell for 500nm PS microparticles (with the same label), and ≤203=8000 particles per cell for 50nm particles. Our observations are consistent with this calculation, since 50 nm PS particles were easily detected at 10,000 particles per cell. Jiang et al. were able to detect fluorescently labeled FePt particles (5nm diameter) by confocal microscopy at starting concentrations of as low as 1 nM.35
Further applications of the assay are the determination of nanoparticle internalization into other cell types, for example target cells such as cancer cells or endothelial cells (Supplemental Figure 8). Table 3 lists the advantages and limitations of the proposed methodology.
Table 3.
Advantages and Limitations of Proposed Technique
| Advantages: | Limitations: |
|---|---|
| Generally applicable to different materials, as long as they can be efficiently labeled with fluorophore | Requires fluorescent labeling: Fluorescence may change upon internalization and fluorophore may alter uptake |
| Combines high throughput data on adsorption+internalization with quantitative internalization data | Sensitivity for smaller particles is lower |
| Allows for comparison of data from different labs if a standard particle is defined that is run alongside in parallel | Requires combination of three techniques: flow cytometry, fluorometry and confocal microscopy |
| Qualitative observations of aggregation phenomena in same assay | |
| No assumptions made on intracellular distribution | |
| Visual confirmation by high resolution microscopy technique |
CONCLUSION
Here we report a generic technique for precise determination of nanoparticle uptake into cells, with a readout of average number of nanoparticles internalized per cell. For the first time, quantitative CLSM imaging data, which gives definite information on internalization, is mathematically integrated with high throughput FACS data, which yields information about cellular association. This mathematical correlation can, in theory, also be applied to a combination of label-free techniques. In addition, a correction factor, FRo/i, is introduced which has an impact on confocal microscopy as well as on FACS measurements. We view the technique and the mathematical model described here as a starting point which could be further refined, e.g. by using automated analysis of microscopic images,35,36 or by further quantifying the subcellular localization of nanoparticles as described by Schweiger et al.37 Furthermore, if the nanoparticle community could agree on one set of standard particles, then data derived in this fashion could be easily standardized as a percent uptake in comparison to the standard particle(s). This would make internalization data from different laboratories comparable.
METHODS
Particles
Vesosomes
Vesosomes were prepared via the interdigitated phase transition as described17,38 except that instead of encapsulating smaller vesicles, for this study we only encapsulated water, thus creating particles with the same structure as vesosomes but without specific encapsulated content. Briefly, the dry Dipalmitoylphosphatidylcholine (DPPC) lipid was hydrated by reverse osmosis treated water and vortexed at 55°C. DPPC unilamellar vesicles (50 nm in diameter) were prepared by sonication at room temperature using a 60 Sonic Dismembrator (Fisher Scientific, Atlanta, GA, USA) for 4 minutes at a power of 4 W. Secondly, interdigitated bilayer phase was induced by the addition of 3 M/L ethanol to a 50 mg/mL DPPC vesicle suspension. After incubating at 4°C for overnight, the interdigitated sheets were centrifuged and dispersed in reverse osmosis treated water 3 times to remove ethanol. Thirdly, the pellet of interdigitated DPPC sheets was mixed with water, and then heated at 50°C for 2 hours under vortex mixing, driving the sheets to close to form the interdigitation-fusion vesicles. Dried Dialkylcarbocyanine (DiO) or Distearoyl-phosphatidyl-ethanolamine-Polyethylenglycol (DSPE-PEG) was mixed with interdigitated DPPC sheets before heating, and the subsequent heating drives the incorporation of DiO or DSPE-PEG inside the lipid bilayer. The size of vesosomes was controlled by extruding vesosomes through a 400 nm polycarbonate membrane or, for FRo/i measurements, through a 1 micron polycarbonate membrane.
Polystyrene micro- and nanoparticles
Polystyrene micro- and nanoparticles at different sizes (from 50 nm to 1 micron) labeled with Firefli™ green or Firefli™ red were purchased from Duke Scientific Corporation (Fremont, CA, now part of Thermo Fisher Scientific, Waltham, MA).
Iron oxide nanoparticles
Green fluorescent iron oxide nanoparticles labeled with FITC were a kind gift of E. Ruoslahti, Burnham Institute for Medical Research at University of California Santa Barbara (UCSB).
Cell and bacterial culture
Murine macrophages (J774, EACC 85011428) were cultured in RPMI-1640 with 10% v/v fetal calf serum (FCS) at 37°C and 5% CO2 atmosphere. Fluorescent E. coli bacteria expressing green fluorescent protein (GFP) internally and recombinant peptides on the surface under an arabinose inducible promoter, were picked from single colonies and starter cultures were grown overnight in LB medium substituted with 100 µg/ml chloramphenicol and 1% w/v glucose (LB-CM-glc). Cultures were then diluted 1:250 in LB-CM-glc and grown until they had an optical density of A600=0.4, as determined by spectrophotometry. Bacterial suspensions were centrifuged and resuspended in LB-CM plus 0.02% w/v arabinose, and cultured for 2.5 h at room temperature. After expression, they were centrifuged and resuspended at the desired concentration for a 100:1 bacteria-to-cell ratio in RPMI-1640 (without glucose) with 100 µg/ml ampicillin, 10% v/v fetal calf serum and 0.02% w/v arabinose for confocal imaging studies.
Internalization assay
A. Flow cytometry
J774 cells were seeded in T25 tissue culture flasks at 4×106 cells per flask one day prior to the assay. Particles were diluted in phosphate buffered saline (PBS), pH=7.4 containing 2% w/v bovine serum albumin (BSA) and 30% v/v FCS 2 h prior to the assay to block unspecific binding and inhibit aggregation (see Figure 7). Cells were washed with PBS, particles were added at indicated particle:macrophage ratios and incubated at 37°C for 90 minutes. This time point was chosen from a series of pilot experiments, and should be determined for each cell line. Generally, a time point where significant internalization has taken place, but where most of the particles are still in the early endosomal compartment is desirable. After incubation, cells were put on ice to stop internalization, washed twice with ice-cold PBS (centrifuging at 4°C), detached with 0.08% w/v trypsin, washed twice again and analyzed on a flow cytometer, using the sample cooling function. Excitation was at 488 nm and emission filter was 530/30. Mean fluorescence of macrophages after incubation with nanoparticles and with standard polystyrene microparticles was measured, as well as the mean fluorescence of one standard microparticle.
B. Fluorimetry
Nanoparticles and standard polystyrene microparticles were diluted and measured in triplicates at six different concentrations on a Tecan fluorimeter under the same excitation and emission conditions as used for flow cytometry. The ratio of fluorescence of one microparticle over one nanoparticle (cf) was calculated and the fluorescence of 1 microparticle as measured by FACS was divided by cf, to obtain the theoretical fluorescence value of one single nanoparticle as it would occur in FACS.
C. Confocal Microscopy
J774 murine macrophages were seeded on round coverslips at 6×104 cells per slide one day prior to the assay. Particles were diluted prior to the assay as described for flow cytometry. Cells were washed with PBS, and particles in respective buffers were added at the same particle:macrophage ratios as used in the flow cytometry assay. After incubation at 37°C for 90 minutes, cover slips were washed with cold PBS containing 0.1% Tween 20, fixed with 2% paraformaldehyde, freshly diluted in PBS, for 1 h at 0°C, washed, and counterstained with wheat germ agglutinin coupled to Alexa 594 (Invitrogen-Molecular Probes, Carlsbad, CA) at a final concentration of 10 µg/ml in PBS + 1%w/v BSA + 10% v/v FCS. A sharp membrane stain is essential for exact determination of cell interior. Wheat germ agglutinin (WGA) stains the glycocalix, and a conjugate of WGA and Alexa Fluor 594 was our preferred label, after we tested several different membrane stains (Concanavalin A, anti-F4/80 antibody, anti-E-Cadherin antibody) or fluorescein diacetate for cell body staining. Images were acquired on an Olympus IX81 microscope using Fluoview 500 software. A blue argon laser (488 nm) and a green helium neon laser (543 nm) were used for excitation, emission filters were standard filters for FITC and Cy3.5. Images were analyzed using Image J software.
D. Determination of fi
Images of cells seeded on cover slips after incubation with particles and membrane counterstain, and of negative control cells (no particle addition) were taken with the Olympus confocal microscope (see C.) using a 60X oil immersion objective. The High-Low function in the initial set up was used to set the threshold as low as possible, and to avoid saturation. A too high threshold can skew the results, since weak signals would appear disproportionally low. All acquisition parameters (laser power, gain, offset, gamma, confocal aperture, laser intensity) were kept constant for samples and respective controls. Images were imported into Image J software. In Image J, the area, mean grey value and integrated density were recorded for regions of interest on sample slides (area inside cells, whole cells, cell free area) and on negative control slides (area inside cells, cell free area). Regions of interest were outlined as follows: In Image J, cells of interest were enlarged and then the inside of the cell was manually delineated by highlighting the membrane (see Figure 1I). The area, mean and integrated density were recorded. The outside of the cell was then marked by highlighting the area around the cell, with enough distance to enclose any particles that may be bound to the cell membrane externally (see Figure 1J), and measured. Background for inside the cell was measured similar to Figure 1I on a negative control slide, and background for the outside of the cell in a cell-free area on the same slide, as in Figure 1K. The respective background fluorescence on cell free areas of sample slides and control slides was compared as an internal control, and only control slides with the same or very close background values in these areas were used. Typically, 20–50 cells were analyzed. Raw data was pasted into the excel macro, which is provided in Supplement 2 for convenient calculation of fi, according to equation 1.4 (Figure 4). Supplement 2 also contains detailed instructions on sheet 2 and an example calculation on sheet 3.
E. Determination of FRo/i
Images of cells seeded on cover slips after incubation with particles and membrane counterstain were acquired using a 60X oil immersion objective. Saturation was avoided and all acquisition parameters were kept constant for samples and respective controls as described in D. Images were taken as xyz stacks at 0.5µm step size and imported into Image J software. In Image J mean grey values were recorded for regions of interest (particles inside and outside the cells, area inside the cell without particles, cell free area). For easiest operation, images were opened in Fluoview and Image J at the same time and a stacked overlay image was obtained in Fluoview to give an overview about all particles. Each particle was then analyzed in Fluoview for position: inside versus outside the cell. Then its mean fluorescence was measured in Image J in the z-section that provided the highest fluorescence. Generally, analyzing 20–30 particles yielded a normal distributed population.
F. Computation of n[Pin/cell]
After obtaining the FACS-derived mean fluorescence of sample and control cells, the mean fluorescence of one particle from FACS and fluorimetry, and FRo/i as well as fi from CSLM, the average number of internalized particles per cell, n[Pin/cell], was calculated according to equation 1.1.
Aggregation studies
Aggregation of nanoparticles in biological buffers was analyzed by flow cytometry, dynamic light scattering and fluorescence microscopy. For flow cytometry, particles were diluted in the indicated buffers, measured on a flow cytometer and analyzed as scatter plots. Initially, dot plots and contour plots of the parameters forward scatter area versus side scatter area were used (as in Figure 6). In agreement with the work of Tzur et al.,39 we found that forward scatter height gave a better discrimination of size, which was subsequently used. Dynamic light scattering measurements were performed on a Brookhaven Laser Light scattering instrument at 90°C angle, using BIC DLS software. For fluorescence microscopy analysis, particles were diluted in the indicated buffers and visually analyzed for aggregation at various time points after dilution.
Statistical Methods
Test for Gaussian distribution was carried out by Shapiro Wilk test (or KS test for fewer particles), using the graph pad prism software (version 5.0). The difference between mean fluorescence of a fluorophore inside the cell and outside the cell (FRo/i) and for internalized particle numbers of different vesosome preparations was tested for statistical significance using the unpaired, two-tailed t-test (with Welch’s correction for non-Gaussian sample populations) or with two-tailed Mann-Whitney test. An alpha-level of 0.05 was assumed for statistical significance.
Error propagation calculations
Fractional uncertainties were calculated according to the following standard formulas: For addition or subtraction of measured quantities x and y with standard deviations Δx and Δy, the standard deviation Δz of the calculated quantity z is:
For multiplication or division of measured quantities with standard deviations Δx and Δy, the standard deviation Δz of the calculated quantity z is:
An example calculation in excel format with embedded equations is provided in Supplement 3. The blank form on sheet 2 can be used to calculate the standard deviation Δz for internalized particles per cell after input of measurement data.
Supplementary Material
Figure 5. Validation of determination of internalized fraction via confocal microscopy.
Macrophages were incubated with green fluorescent polystyrene microbeads (1 µm diameter), and counterstained with wheat germ agglutinin coupled to Alexa-594. A-C show confocal images of the cells in the Cy3.5 channel (A), the FITC channel (B) and as overlay (C). D-E show images imported into Image J after selection of regions of interest within the yellow line for inside of the cell (D) and whole cell (E). Determination of the internalized fraction (fi) with manual counting was compared to using the quantitative fluorescence measurement method described here, either per single cell (D, E) or per microscopic viewing field (F). Even if only one microscopic view field is used, the counted fi is very close to the measured fi (see Supplement 2 for the raw data and fi-calculation for this example). The uncertainty for fi, if counting more than one field is typically between 1 and 15%SD (percent standard deviation of average). Scale bars: 10 µm for A-C and F; 2 µm for D and E.
Acknowledgment
We thank B. Matsumoto for help with confocal microscopy, and A. Cleland for guidance with error propagation calculations. Financial support was provided from NIH grant RO1EB012637. The confocal microscope was purchased from a NCRR Shared Instrument Grant # 1S10RR017753-01.
Footnotes
ASSOCIATED CONTENT
Supporting Information Available: Three supplemental files are available. Supplement 1 consists of eight additional Figure 1. Example calculation to demonstrate impact of correction factor FRo/i (fluorescence ratio outside the cell over inside the cell); 2. Negative controls for laser scanning microscopy experiments; 3. Cell binding and uptake in the presence of specific macrophage receptor inhibitors; 4. Lack of aggregation in PBS-BSA buffer for vesosomes; 5. Illustration of loss of data when using overlay images; 6. Application of this method to study internalized fraction of micron-sized recombinant bacteria; 7. General applicability of this method to a wide variety of nanoparticles. 8. Comparison of PS particle uptake by cancer cells and macrophages. Supplement 2 (excel file) contains a macro for facile determination of internalized fraction fi, after raw data from confocal imaging has been obtained. Raw data can be pasted into the reserved cells, and fi is calculated automatically. Instructions are provided on sheet 2 of the excel file, and an example in sheet 3. Supplement 3 contains a macro for calculation of standard deviation of n(Pin/cell), using an error propagation model. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Galimard A, Safi M, Ould-Moussa N, Montero D, Conjeaud H, Berret J-F. Thirty-Femtogram Detection of Iron in Mammalian Cells. Small. 2012;8:2036–2044. doi: 10.1002/smll.201102356. [DOI] [PubMed] [Google Scholar]
- 2.Lee RJ, Low PS. Delivery of Liposomes into Cultured KB Cells via Folate Receptor-mediated Endocytosis. J. Biol. Chem. 1994;269:3198–3204. [PubMed] [Google Scholar]
- 3.Sahlin S, Hed J, Runfquist I. Differentiation between Attached and Ingested Immune Complexes by a Fluorescence Quenching Cytofluorometric Assay. J. Immunol. Methods. 1983;60:115–124. doi: 10.1016/0022-1759(83)90340-x. [DOI] [PubMed] [Google Scholar]
- 4.Nuutila J, Lilius E-M. Flow Cytometric Quantitative Determination of Ingestion by Phagocytes Needs the Distinguishing of Overlapping Populations of Binding and Ingesting Cells. Cytometry A. 2005;65A:93–102. doi: 10.1002/cyto.a.20139. [DOI] [PubMed] [Google Scholar]
- 5.Sveum RJ, Chused TM, Frank MM, Brown EJ. A Quantitative Fluorescent Method for Measurement of Bacterial Adherence and Phagocytosis. J. Immunol. Methods. 1986;90:257–264. doi: 10.1016/0022-1759(86)90083-9. [DOI] [PubMed] [Google Scholar]
- 6.Neumeyer A, Bukowski M, Veith M, Lehr C-M, Daum N. Propidium Iodide Labeling of Nanoparticles as a Novel Tool for the Quantification of Cellular Binding and Uptake. Nanomedicine. 2011;7:410–419. doi: 10.1016/j.nano.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Johnstone SA, Masin D, Mayer L, Bally MB. Surface-associated Serum Proteins Inhibit the Uptake of Phosphatidylserine and Poly(ethylene glycol) Liposomes by Mouse Macrophages. Biochim. Biophys. Acta. 2001;1513:25–37. doi: 10.1016/s0005-2736(01)00292-9. [DOI] [PubMed] [Google Scholar]
- 8.Kostarelos K, Lacerda L, Pastorin G, Wu W, Wieckowski S, Luangsivilay J, Godefroy S, Pantarotto D, Briand J-P, Muller S, et al. Cellular Uptake of Functionalized Carbon Nanotubes is Independent of Functional Group and Cell Type. Nat. Nanotechnol. 2007;2:108–113. doi: 10.1038/nnano.2006.209. [DOI] [PubMed] [Google Scholar]
- 9.Basiji DA, Ortyn WE, Liang L, Venkatachalam V, Morrissey P. Cellular Image Analysis and Imaging by Flow Cytometry. Clin. Lab. Med. 2007;27:653–670. doi: 10.1016/j.cll.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barteneva NS, Fasler-Kan E, Vorobjev IA. Imaging Flow Cytometry: Coping with Heterogeneity in Biological Systems. J. Histochem. Cytochem. 2012;60:723–733. doi: 10.1369/0022155412453052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Semmling M, Kreft O, Mu0000F1;oz Javier A, Sukhorukov GB, Käs J, Parak WJ. A Novel Flow-Cytometry-Based Assay for Cellular Uptake Studies of Polyelectrolyte Microcapsules. Small. 2008;4:1763–1768. doi: 10.1002/smll.200800596. [DOI] [PubMed] [Google Scholar]
- 12.Pillay CS, Elliott E, Dennison C. Endolysosomal Proteolysis and its Regulation. Biochem. J. 2002;363:417–429. doi: 10.1042/0264-6021:3630417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen AK, Cheng Z, Behlke MA, Tsourkas A. Assessing the Sensitivity of Commercially Available Fluorophores to the Intracellular Environment. Anal. Chem. 2008;80:7437–7444. doi: 10.1021/ac8011347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. [accessed 050713];Molecular Probes Handbook. 2013 Online link: www.invitrogen.com/site/us/en/home/References/Molecular-Probes-The-Handbook.html;
- 15.Graber ML, DiLillo DC, Friedman BL, Pastoriza-Munoz E. Characteristics of Fluoroprobes for Measuring Intracellular pH. Analytical Biochemistry. 1986;156:202–212. doi: 10.1016/0003-2697(86)90174-0. [DOI] [PubMed] [Google Scholar]
- 16.Walker SA, Kennedy MT, Zasadzinski JA. Encapsulation of Bilayer Vesicles by Self-assembly. Nature. 1997;387:61–64. doi: 10.1038/387061a0. [DOI] [PubMed] [Google Scholar]
- 17.Boyer C, Zasadzinski J. Multiple Lipid Compartments Slow Vesicle Contents Release in Lipases and Serum. ACS Nano. 2007;1:176–182. doi: 10.1021/nn7002025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabizon A, Papahadjopoulos D. Liposome Formulations with Prolonged Circulation Time in Blood and Enhanced Uptake by Tumors. Proc. Natl. Acad. Sci. U.S.A. 1988;85:6949–6953. doi: 10.1073/pnas.85.18.6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen T, Sapra P, Moase E. Use of the Post-insertion Method for the Formation of Ligand-coupled Liposomes. Cell. Mol. Biol. Lett. 2002;7:889–894. [PubMed] [Google Scholar]
- 20.Dos Santos N, Allen C, Doppen A-M, Anantha M, Cox K, Gallagher R, Karlsson G, Edwards K, Kenner G, Samuels L, et al. Influence of Poly(ethylene glycol) Grafting Density and Polymer Length on Liposomes: Relating Plasma Circulation Lifetimes to Protein Binding. Biochim. Biophys. Acta (BBA) 2007;1768:1367–1377. doi: 10.1016/j.bbamem.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 21.Cedervall T, Lynch I, Lindman S, Bergg rd T, Thulin E, Nilsson H, Dawson KA, Linse S. Understanding the Nanoparticle-protein Corona using Methods to Quantify Exchange Rates and Affinities of Proteins for Nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cedervall T, Lynch I, Foy M, Berggård T, Donnelly SC, Cagney G, Linse S, Dawson KA. Detailed Identification of Plasma Proteins Adsorbed on Copolymer Nanoparticles. Angew. Chem., Int. Ed. 2007;46:5754–5756. doi: 10.1002/anie.200700465. [DOI] [PubMed] [Google Scholar]
- 23.Vroman L. Effect of Absorbed Proteins on the Wettability of Hydrophilic and Hydrophobic Solids. Nature. 1962;196:476–477. doi: 10.1038/196476a0. [DOI] [PubMed] [Google Scholar]
- 24.Lundqvist M, Stigler J, Cedervall T, Berggard T, Flanagan MB, Lynch I, Elia G, Dawson K. The Evolution of the Protein Corona around Nanoparticles: A Test Study. ACS Nano. 2011;5:7503–7509. doi: 10.1021/nn202458g. [DOI] [PubMed] [Google Scholar]
- 25.Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V. Time Evolution of the Nanoparticle Protein Corona. ACS Nano. 2010;4:3623–3632. doi: 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]
- 26.Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA. What the Cell “Sees” in Bionanoscience. J. Am. Chem. Soc. 2010;132:5761–5768. doi: 10.1021/ja910675v. [DOI] [PubMed] [Google Scholar]
- 27.Walkey CD, Chan WCW. Understanding and Controlling the Interaction of Nanomaterials with Proteins in a Physiological Environment. Chem. Soc. Rev. 2012;41:2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 28.Gebauer JS, Malissek M, Simon S, Knauer SK, Maskos M, Stauber RH, Peukert W, Treuel L. Impact of the Nanoparticle-Protein Corona on Colloidal Stability and Protein Structure. Langmuir. 2012;28:9673–9679. doi: 10.1021/la301104a. [DOI] [PubMed] [Google Scholar]
- 29.Sund J, Alenius H, Vippola M, Savolainen K, Puustinen A. Proteomic Characterization of Engineered Nanomaterial - Protein Interactions in Relation to Surface Reactivity. ACS Nano. 2011;5:4300–4309. doi: 10.1021/nn101492k. [DOI] [PubMed] [Google Scholar]
- 30.Safi M, Courtois J, Seigneuret M, Conjeaud H, Berret JF. The Effects of Aggregation and Protein Corona on the Cellular Internalization of Iron Oxide Nanoparticles. Biomaterials. 2011;32:9353–9363. doi: 10.1016/j.biomaterials.2011.08.048. [DOI] [PubMed] [Google Scholar]
- 31.Petri-Fink A, Steitz B, Finka A, Salaklang J, Hofmann H. Effect of Cell Media on Polymer-coated Superparamagnetic Iron Oxide Nanoparticles (SPIONs): Colloidal Stability, Cytotoxicity, and Cellular Uptake Studies. Eur. J. Pharm. Biopharm. 2008;68:129–137. doi: 10.1016/j.ejpb.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 32.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle Size and Surface Properties Determine the Protein Corona with Possible Implications for Biological Impacts. Proc. Natl. Acad. Sci. U.S.A. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Safi M, Sarrouj H, Sandre O, N M, J-F B. Interactions between Sub-10-nm Iron and Cerium Oxide Nanoparticles and 3T3 Fibroblasts: the Role of the Coating and Aggregation State. Nanotechnology. 2010;21:145103. doi: 10.1088/0957-4484/21/14/145103. [DOI] [PubMed] [Google Scholar]
- 34.Huehn D, Kantner K, Geidel C, Brandholt S, De Cock I, Soenen SJH, Rivera Gil P, Montenegro J-M, Braeckmans K, Muellen K, et al. Polymer-Coated Nanoparticles Interacting with Proteins and Cells: Focusing on the Sign of the Net Charge. ACS Nano. 2013;7:3253–3263. doi: 10.1021/nn3059295. [DOI] [PubMed] [Google Scholar]
- 35.Jiang X, Weise S, Hafner M, Roecker C, Zhang F, Parak WJ, Nienhaus GU. Quantitative Analysis of the Protein Corona on FePt Nanoparticles Formed by Transferrin Binding. J. R. Soc. Interface. 2010;7(Suppl 1):S5–S13. doi: 10.1098/rsif.2009.0272.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kriete A, Papazoglou E, Edrissi B, Pais H, Pourrezaei K. Automated Quantification of Quantum-dot-labelled Epidermal Growth Factor Receptor Internalization via Multiscale Image Segmentation. J. Microsc. 2006;222:22–27. doi: 10.1111/j.1365-2818.2006.01564.x. [DOI] [PubMed] [Google Scholar]
- 37.Schweiger C, Hartmann R, Zhang F, Parak W, Kissel T, Rivera Gil P. Quantification of the Internalization Patterns of Superparamagnetic Iron Oxide Nanoparticles with Opposite Charge. J. Nanobiotechnology. 2012;10:28–38. doi: 10.1186/1477-3155-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kisak ET, Coldren B, Zasadzinski J. Nanocompartments Enclosing Vesicles, Colloids and Macromolecules via Interdigitated Lipid Bilayers. Langmuir. 2002;18:284–288. [Google Scholar]
- 39.Tzur A, Moore JK, Jorgensen P, Shapiro HM, Kirschner MW. Optimizing Optical Flow Cytometry for Cell Volume-Based Sorting and Analysis. PLoS One. 2011;6:e16053. doi: 10.1371/journal.pone.0016053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.