Abstract
XIST, a long non-coding RNA, plays an important role in triggering X chromosome inactivation in eutherians, and is used extensively for qualifying stem cells and cloned embryos. However, a porcine XIST has not yet been thoroughly identified despite its biological importance in a wide variety of research fields. Here, we present a full-length porcine XIST sequence assembled using known sequences (GenBank), RNA-Seq data (NCBI SRA), and PCR/sequencing. The proposed porcine XIST gene model encodes a 25,215-bp transcript consisting of 7 exons, including two conserved and two porcine-specific repeat regions. Transcription covering the entire XIST region was observed specifically in female cells, but not in male cells. We also identified eight transcription starting sites (TSSs) and evaluated CpG methylation patterns in the upstream (+2.0 kb) and downstream (−2.0 kb) regions. Sixty-seven CG di-nucleotides identified in the target region were considered to be candidate CpG sites, and were enriched in the following two regions: −284 to +53 bp (13 sites) and +285 to +1,727 bp (54 sites) from the selected TSS. Male 5` region of XIST (64.5 sites, 96.26%) had a higher level of CpG methylation than female DNA (33.4 sites, 49.85%). Taken together, our results revealed that the porcine XIST gene is expressed exclusively in female cells, which is influenced by the lower level of CpG methylation in the putative promoter region compared with male cells. The porcine XIST presented in this study represents a useful tool for related research areas such as porcine embryology and stem cell biology.
Introduction
X-chromosome inactivation (XCI) occurs early in embryo development, and involves silencing one of the X chromosomes in mammalian female cells to compensate gene dosage [1]. Different mammalian species use various strategies to induce XCI during early embryo development [2]. For example, it has been suggested that eutherians except rodents, like humans, employ random inactivation of the X chromosome [2], [3], whereas marsupials and rodents undergo imprinted inactivation of the paternal X chromosome during early embryo development [4], [5]. The X-chromosome inactivation-specific transcript, XIST/Xist, which is located at the X-inactivation center (Xic), has been suggested to trigger XCI in eutherian mammals by cis-coating one of the X chromosomes in female mammals [6], [7].
A gene model of XIST/Xist, which produces a long, non-coding RNA (lncRNA) [8], [9], showed that XIST/Xist consists of large first and last exons with several small exons between these large exons in both humans and mice. Identification of the XIST gene was a turning point in research on XCI in various eutherians. For example, the identification of XIST allowed for the evaluation of XCI in human, murine, and bovine species during early embryo development [3], [10], [11]. Recently, the importance of XIST RNA for early embryo development and embryonic stem cell research has been highlighted. For example, the identification of abnormal expression of Xist and X-chromosome-linked genes in a cloned mouse was reported [12], and RNA interference (RNAi)-mediated downregulation of Xist during embryo development in cloned mice was shown to dramatically elevate birth rates [13]. These reports suggest an important role for Xist in early embryo development. It has also been shown that Xist expression is associated with the pluripotency markers Oct4, Sox2, and Nanog, and that lower expression of Xist is maintained in undifferentiated mouse embryonic stem cells [14]. XIST research has also focused on the status of pluripotency, specifically naïve and primed status, because these two different stem cell types exhibit different patterns of X chromosome inactivation [15]. Therefore, XIST has been used as a marker for evaluating the status of pluripotent stem cells [15]. Although the importance of XIST has been highlighted in various research fields, only partial XIST sequences have been reported in pig [16]. And also, putative gene model resulted from aligning between pig genome and bovine XIST was suggested recently, only small fraction compared to the suggested draft gene model was validated just using RT-PCR analysis [17]. A recent paper that defined various genes and RNAs in the pig also identified partial sequences of porcine XIST, namely, 84 bp of exon 1 and 126 bp of exon 4 [18]. Although these findings suggest some information about porcine XIST RNA, but a large proportion of XIST RNA sequence was still unknown. While the identified porcine XIST sequences could be used for research related to early embryo development and stem cells, identification of a full XIST sequence using gene prediction methods may be difficult [18] because of the low sequence conservation of XIST genes among species [19], [20].
The identification of XIST/Xist has also led to verification of its promoter region [21], [22] and analysis of the CpG dinucleotide methylation patterns in this region [23]. The XIST/Xist promoter region has been suggested to regulate XIST expression by recruiting transcription factors [24], and the methylation pattern of the promoter region of Xist is highly related to its expression [23]. Analysis of methylation patterns has led to the suggestion that expression of Xist in the mouse is controlled by imprinting during preimplantation embryo development [23], [25]. In addition to the identification of CpG sites as differentially methylated regions (DMRs) and factors in CpG sites in the Xist promoter region have been used to determine the normality of early embryo development [26]. Specifically, abnormal methylation patterns in the Xist promoter have been reported in cloned mouse embryos [26]. Nevertheless, little is currently known about the CpG sites of porcine XIST.
In this study, we aimed to identify the candidate porcine XIST ncRNA encoding regions using BLAST homology searches. We verified our findings using reverse transcriptase (RT)-PCR, enzyme mapping, sequencing, and RNA-Seq alignment on the whole region. Based on the identified XIST sequence, we analyzed the methylation patterns of 67 CpG sites in the region ±2 kb from the transcription start site (TSS) to confirm the DMRs of porcine XIST. Our results showed that the porcine XIST RNA is nearly 24 kb in length, and contains previously reported partial sequences of porcine XIST. The 67 CpG sites that we analyzed exhibited sex-dependent methylation patterns similar to those found in other species.
Results
Identification of a Porcine XIST Gene Model and its RNA Sequence
To identify the porcine XIST gene and its RNA sequence, we aligned and compared the pig genomic DNA sequence (X-chromosome scaffold, GenBank accession No. NW_003612825.1) with mouse (NC_000086.7), human (NC_000023.10), and bovine XIST gene models (AC_000187.1) (Figure S1). The DNA sequence ranging from nucleotides 289,233 to 257,103 in the pig scaffold was well-aligned with the three gene models. Specifically, they matched the TSS and the last nucleotide of the human and bovine XIST genes, respectively (Figure 1A and Figure S1). Primer sets (Table S1) were designed to confirm whether the aligned pig sequence (candidate XIST) is transcribed in porcine embryonic fibroblast (PEF) cells (Figure 1A). RT-PCR using these primer sets confirmed that the candidate XIST gene was transcribed only in female PEFs, and the target regions were not detected in male PEFs (Figure S2). We also confirmed the amplicons by restriction enzyme mapping, which produced patterns consistent with our predictions (Table S2). All of the amplified regions were sequenced by Sanger sequencing, and most of the sequences were consistent with the porcine DNA sequence. However, one amplicon (E2EL) that partially covered the first and last exons of the candidate porcine XIST gene was shorter compared with the expected genomic DNA sequence (Figure S2), indicating the presence of introns and exons in the target DNA region. Two other amplicons (E1N1 and E1N2) exhibited longer bands than expected because of a gap (Ns) in the pig scaffold, which was filled with a 626-bp nucleotide sequence from the longer-than-expected amplicons (Figure S3). Finally, the sequenced amplicons were assembled into a porcine XIST gene model consisting of two long exons (exon 1 and exon 7) and 5 smaller exons (exon 2 - exon 6) between exons 1 and 7 (Figure 1B). The exons were located between introns with RNA-splicing recognition sequences (GT and AG, Table 1) at their 5′ and 3′ termini, respectively. Our results suggested that the porcine XIST gene shares an evolutionarily conserved region among mammals, indicating the reliability of our model.
Figure 1. Diagram of the candidate XIST expressing region in the pig.
(A) Designed XIST model and primer sets. Candidate XIST ncRNA expression regions on the pig X-chromosome scaffold (NW_003612825.1) determined by BLAST searches are represented by black boxes. Empty and filled arrowheads indicate the 289233rd and 257103rd nucleotides of the scaffold, which were aligned with the transcription start site (TSS) and the last sequence of human and bovine XIST, respectively. The designed primer sets are represented as black and gray lines: female-expressed primer sets are represented as black lines and regions that were not expressed in either sex (α and β) are represented as gray lines. Restriction enzymes used to digest each amplicon are shown. The diagram is scaled. (B) Diagram of the identified porcine XIST gene model. The porcine XIST expressing region was identified by sequencing. Filled rectangles indicate exons, and boxes with diagonal lines indicate undefined regions. Filled and empty arrowheads indicate candidate 5′ and 3′ end regions identified from alignment with human and bovine XIST, respectively. The diagram is scaled.
Table 1. Exons and introns information of Porcine XIST.
| Number | Sequence (5′ to 3′) | Length (bp) | |
| Exon 1 | *TATTTCTT | AGACTACT | 17,184 |
| Intron1 | ** GTAAGTAC | TTTTAAAG | 1,460 |
| Exon 2 | GGATGAAT | CTCCAAAG | 89 |
| Intron2 | GTGAATCT | TTCTCAAG | 2,271 |
| Exon 3 | GATATTCC | AGAAAAAG | 136 |
| Intron3 | GTAATTTA | TTCTCCAG | 2,136 |
| Exon 4 | ATCTTCCTC | CATCTGAG | 209 |
| Intron4 | GTGGGTAA | TCTTTTAG | 356 |
| Exon 5 | GAAAACAG | TACTCTAG | 329 |
| Intron5 | GTCAGTGG | TTCGGTAG | 390 |
| Exon 6 | CTCCTGAT | AGGATGAA | 129 |
| Intron6 | GTAAGTTG | TCTTCCAG | 989 |
| Exon 7 | TGATTGTC | AAAACTTA | 7,139 |
289,233rd nt of NW_003612825.1 scafford sequence was set to first sequence which were one of the identified TSS and matched to blast search in this study.
Bold sequence, GT/AG, indicate splicing recognition sequence.
Identification of the 5′ and 3′ end Regions of Porcine XIST
The transcript start sites (TSSs) of porcine XIST were identified by 5′-RACE PCR using the primer sets (Table S3). The 289,233rd nucleotide of the pig X chromosome scaffold (NW_003612825.1) was expected to be a porcine XIST TSS based on sequence alignment with the human and bovine XIST 5′ region (Figure S1), which has been suggested to be conserved among mammals [27]. We identified additional 7 TSSs in addition to the expected site (Figure S4). Two TSSs (−5 and −3) were found upstream of the expected TSS, while 5 TSSs (+16, +48, +72, +143, and +211) were located downstream of the expected TSS.
Transcription termination sites were also identified by 3′-RACE PCR. The 257,103rd nucleotide of the pig X chromosome scaffold (NW_003612825.1) was expected to be a porcine XIST termination site based on sequence alignment with the human and bovine XIST ncRNA sequence. We identified one transcription termination site (257,094th nucleotide) 10 bp downstream of the expected site. The 3′ region was confirmed by RACE analysis, which indicated the presence of an AATAAA poly A-tail signal sequence located 21 bp upstream of the termination site (Figure S4).
Repeat Sequence Analysis in Porcine XIST RNA
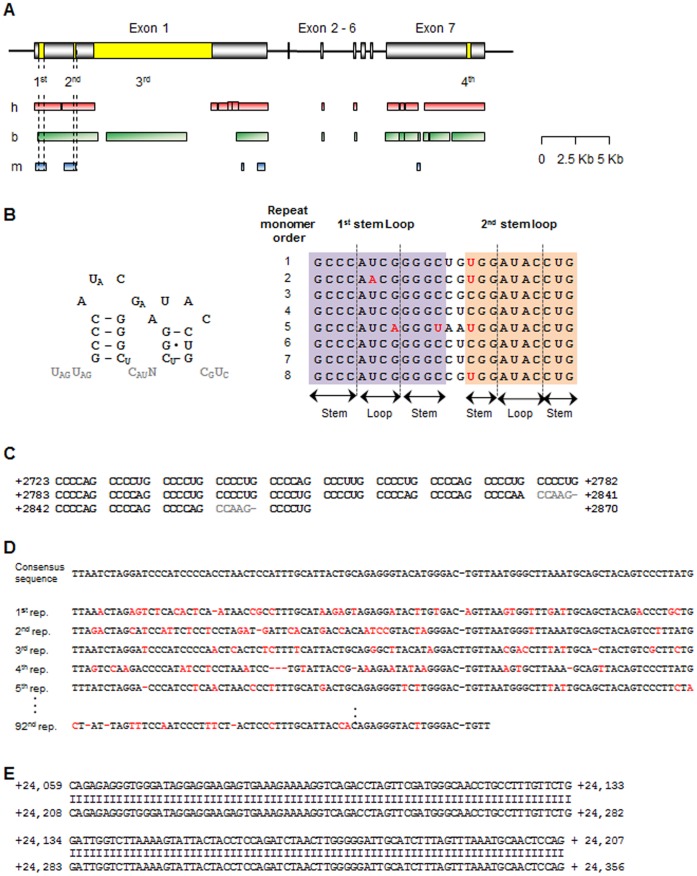
The repeated sequences in XIST/Xist RNA have been suggested to have a function in X chromosome silencing. Therefore, we examined whether these repeated sequences and conserved functional domains are present in the identified porcine XIST RNA sequence. Four repeat sequence regions were identified (Figure. 2A). Interestingly, the Xist A-repeats comprises copies of a 24-mer consensus sequence known to conserved between humans and mice [9] were detected in the first repeat region of porcine XIST. The consensus sequence of the first repeat region was predicted to have a secondary structure containing two stem-loops (Figure 2B), which is the same structure as that of mouse Xist [28]. The consensus 24-mer monomer was repeated 8 times in the +327 to +695 region (Figure 2B). The second repeat region contained a cytosine-rich 6-mer repeat that was located in the +2,723 to +2,870 region of the porcine XIST RNA as the cysteine-rich repeat-B region of mouse Xist [9]. In all, a total of 23 repeats were observed in this region (Figure 2C). We also searched for a consensus sequence of the mouse C-repeat sequence, but did not locate such a sequence in porcine XIST. Two novel repeat regions (the third and fourth regions in Figure 2A) were also identified. The third region comprised 91.7 copies of a 96-bp consensus sequence ranging from +4,302 to +13,102 in the porcine XIST gene (Figure 2D). The fourth region consisted of two copies of a 149-bp monomer ranging from +24,059 to +24,356 (Figure 2E). Interestingly, consensus sequences in the third region of porcine XIST were not present in either human or mouse repeat regions, while the fourth region was only partially aligned to a non-repeating region of human XIST. Taken together, the identified porcine XIST RNA contained not only conserved sequences, but also porcine-specific repeated sequences, suggesting conserved and hypothetically distinct functions of porcine XIST.
Figure 2. Analysis of repeat sequences in porcine XIST RNA.
(A) Distribution of repeated sequences in the porcine XIST gene model. Gray rectangles indicate exons, and the yellow boxes in the exons indicate repeat regions. The regions having homology with human, bovine, and mouse XIST/Xist are represented to red (h), green (b), and blue (m) boxes, respectively. The first and second repeat regions showed similarity with all accessed XIST/Xist sequence from other species (dashed lines). The diagram is scaled. (B) The first repeat region in porcine XIST. A monomer of the first repeat region in pigs (left panel) share a consensus sequence and a predicted secondary structure of the mouse [28]. Gray text indicates non-conserved sequences. Eight repeated monomers identified in the porcine XIST first repeat region (+327 to +695) were aligned with a consensus sequence (Right panel). Red character indicates different sequence compared to the consensus sequence in conserved stem-loop region. (C) The second repeat region (+2,723 to +2,870) is shown, with non-repeat sequences in gray. (D) The third repeat region (+4,302 to +13,102) fraction. The first five repeats and last repeat were aligned with the consensus sequence. Red characters mean the sequences which were not same to consensus sequence. (E) Alignment of the fourth repeat region (+24,059 to +24,356). Two copies of the monomer were perfectly matched.
Expression of XIST in Other Porcine Tissues
We assembled a full-length porcine XIST RNA sequence by PCR and sequencing and identified its 5′ and 3′ regions. Although assembly and identification of both ends of the RNA strand was performed using PEFs, it was not immediately clear whether XIST was also expressed in other porcine tissues in the same manner as our gene model. Thus, to confirm our gene model and verify the expression of XIST in other tissues, Illumina sequence reads from three different porcine tissues–liver, abdominal fat, and the longissimus dorsi muscle–of two female pigs were aligned with our porcine XIST gene model (Figure 3A). The Illumina sequence reads were well aligned onto the most of the region of the model, including the 5′ and 3′ regions (Figure 3 and S5), while none of the Illumina reads were aligned to the upstream/downstream regions of the assembled XIST RNA (Figure 3B). Thus, our results suggested that XIST is expressed in other porcine tissues as described above.
Figure 3. Alignment of Illumina reads to the porcine XIST gene model.
(A) Alignment of Illumina reads to the porcine XIST gene locus. Six Illumina reads from three tissues (liver, abdominal fat, and longissimus dorsi muscle) of two female pigs (F1 and F2) were aligned to the entire porcine XIST gene including introns and exons. Small peaks between exon 3 and 4 were identified to be non-specific alignment of reads. The diagram is scaled. The two dotted boxes indicate upstream/5′ and 3′/downstream regions of the XIST gene, respectively. (B) Alignment of Illumina reads to upstream/5′ and 3′/downstream regions. Arrows in the upstream and 5′ region indicate the transcription start sites (TSS) of the porcine XIST gene identified by 5′ RACE. The arrow at the 3′ and downstream region indicates the end of the porcine XIST RNA identified by 3′ RACE.
Analysis of Methyl CpG Sites in the Upstream Region of Porcine XIST
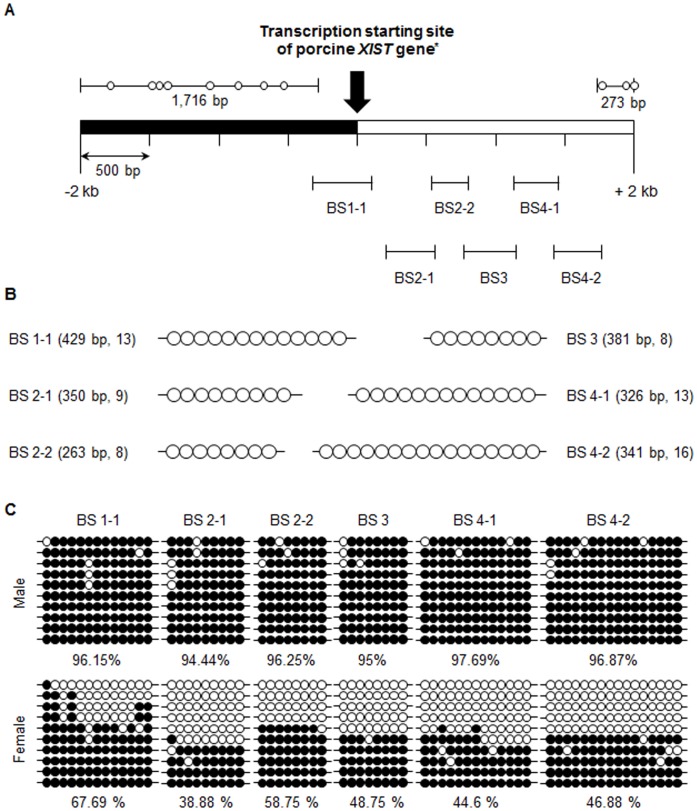
After identifying the TSSs of the porcine XIST gene, we next analyzed the methylation status of the CpG sites upstream of the XIST TSS. The search for CG dinucleotides was conducted using the ±2 kb region of the first porcine XIST sequence (289,233rd nucleotide of NW_003612825.1, Figure S4), which identified 78 CG dinucleotides, 67 of which were densely located between nucleotides −284 and +1,727 (Figure 4A). The methylation statuses of the 67 CpG sites in both male and female embryonic fibroblasts were analyzed using six different primer sets (Table S4, and Figure 4B). With the exception of five CpG sites, the analyzed regions were over 90% methylated in male PEFs, and the total methylation rate for male PEFs was 96.26% (Figure 4C). However, in female PEFs, although five CpG sites on the BS1-1 region were highly methylated (>70%), the remaining CpG sites were methylated at a rate of 40∼60%. Thus, the total methylation rate of the 67 CpG sites in female PEFs was 49.85% (Figure 4C). The results suggested the analyzed CpG sites were differentially methylated between the male and female.
Figure 4. Diagram of the CpG sites located in the ±2 kb region of the transcription start site of porcine XIST and the methylation status of XIST CpG sites in male and female porcine embryonic fibroblasts.
(A) One of the transcription start sites (TSS) identified by 5`-RACE-PCR was set as a +1 (asterisk, 289233rd nucleotide of NW_003612825.1). Black and empty bars indicate regions upstream and downstream of the TSS, respectively. Methylation of CpG sites in six regions (BS1-1, BS2-1, BS2-2, BS3, BS4-1, and BS4-2) was analyzed. The diagram is scaled. (B) Profiles of the six target regions. Each circle represents a single CG dinucleotide identified in the amplified region. The length of each amplicon was 250 bp –450 bp and contained between 8–16 CG dinucleotides. The diagram is not scaled. (C) The methylation status of 67 XIST CpG sites were analyzed in male and female embryonic fibroblasts. Each circle indicates a CpG dinucleotide. Filled and empty circles represent methylated and unmethylated CpG dinucleotides, respectively. The horizontal line represents one individual clone.
Discussion
To the best of our knowledge, this study is the first to report a gene model of porcine XIST that includes a ncRNA sequence and the CpG methylation status of its regulatory regions. The purpose of XCI is to balance the expression level of X-chromosome-linked genes between male and female eutherian mammals, occurs early during embryo development [1], [2], [29]. The initiation of XCI is induced by cis-binding of XIST/Xist onto the X chromosome [6], [7]. Interestingly, among the different strategies for XCI in eutherian mammals [2], [30], all appear to use XIST as a trigger for XCI. The importance of XIST/Xist has been a focus of not only XCI research, but also, more recently, stem cell research [14], [15] and embryo cloning research [12], [13]. Previous studies have indicated that XIST is associated with stem cell differentiation [14] and abnormalities of cloned embryos in the mouse [12]. Despite the importance of XIST and XCI in a number of research fields, only a partial sequence of porcine XIST has been previously reported [16]–[18]. Add to this, the porcine XIST expression and methylation status on its candidate CpG sites were analyzed in our previous report comparing porcine embryonic stem cells from different origins and induced pluripotent stem cells. However, the information about porcine XIST used for the study was restricted and kinds of putative ones, so identification of porcine XIST whole sequence and analysis of its regulating sites should be accessed to perform more accurate and detailed XIST analysis in pig pluripotent cells and embryos. This led us to generate a porcine XIST gene model, including its ncRNA sequence, and to examine the CpG methylation status of its regulatory regions.
Sequence alignment was used to identify candidate porcine XIST exons, which showed not only an evolutionarily conserved XIST gene model among mammals, but also species-specific sequence divergence. Several evolutionarily conserved regions, such as repeated sequences among mammalian XIST orthologs, have been previously reported [9], [27]. Indeed, the A-F regions are regarded as functional domains of Xist [9], [28], [31], some of which were present in our model of porcine XIST. On the other hand, divergence of XIST/Xist sequences among species due to rapid ncRNA evolution has been noted [19], [20]. Consistent with this observation, porcine XIST also exhibited sequence divergence in some regions compared with other mammalian XIST/Xist genes, and showed varying sequence similarity depending on the compared target species. However, certain regions, such as the 5′ region of exon 1, were highly conserved among species, as shown in Figure S1.
A previous study suggested that XIST/Xist originated from a protein-coding gene (Lnx3) and transposable elements [27], and specifically that the 5′ region of exon 1 and exons 5 through 7 of the consensus XIST sequence arose from a protein-coding gene. In this study, we showed that the same regions of human and bovine XIST were well-aligned to that of porcine XIST (Figure S1), suggesting that the highly conserved domains originating from the proposed protein-coding gene survived in pigs during evolution, and may have a role in porcine XIST function. In contrast, some regions originating from transposable elements are suggested to have integrated into XIST after mammalian taxa divergence, resulting in species-specific sequence variation among eutherian XIST/Xist [27]. Such sequence variation may account for the presence of porcine-specific regions that did not align to the XIST/Xist sequences of other species, and which may also contribute to species-specific modes of XIST function during XCI. Together, these evolutionarily properties of XIST/Xist support the reliability of the porcine XIST expression region.
Most of the RT-PCR target regions, except the α and β regions, were amplified exclusively in female porcine cells (Figure 1 and S2), which was highly consistent with previous observations in other mammals [2], [29], [32], [33]. In particular, primer pairs that amplified regions that were not selected as part of the first candidate model (E1N1 to E1H in Figure 1A) exhibited female-specific expression (Figure S2). These regions may have evolved in a porcine-specific manner as suggested previously [27], and may contribute to a novel role of XIST-associated XCI distinct from that in other species.
In the mammalian XIST/Xist gene model, the first and last exons are much longer than the central exons [8], [9]. As expected, the E2EL amplicon contained 5 small exons between the first and last exons, consistent with other species (Figure 1B). Furthermore, the identified exon 4 was conserved in pigs (Figure S1 and [34]). These results suggested that the porcine XIST ncRNA shares a common model with other mammalian XIST/Xist genes.
The identified porcine XIST gene model covered a previously annotated partial porcine XIST sequence in the 3′ region of XIST (GenBank accession Nos. AJ429140 and EF619477.1). In addition, the recently annotated partial exon 1 (nannotator00001430) and exon 4 (nannotator00001519) sequences were also located in the expected porcine XIST exons (http://rth.dk/resources/rnannotator/susscr102/). The presence of a previously reported partial porcine XIST expression region on the defined XIST gene supports the reliability of our model of porcine XIST.
The repeat sequences in XIST/Xist have been suggested to have functional roles in XCI [8], [9]. Interestingly, some repeat regions were detected in the porcine XIST ncRNA sequence. The A repeat, which consists of 7.5 repeats of a 24-bp sequence, forms two stem-loop in mouse, and has been known to regulate processes essential for X chromosome silencing [28]. Interestingly, Wutz and colleagues showed that a mutated A region allele results in a failure to induce X chromosome silencing in a stem cell model without affecting Xist stability and localization [28]. The identified porcine XIST 5′ region included a total of 8 copies of the 24-bp sequence (Figure 2A and 2B), and this region was well-aligned to the 5′ region of mouse Xist (Figure 2A). This result suggested that, similar to the A-repeat in mice, the conserved first repeat region in porcine XIST may be involved in silencing X chromosomes, although this possibility needs to be further analyzed. Another repeat region, the C-repeat, which consists of 14 copies of an approximately 100-bp sequence in mice, has been suggested to be involved in Xist localization during XCI [31]. However, targeting of the C region with Locked nucleic acids (LNAs) does not affect the localization of human XIST [31], which has only one copy of the C-repeat [9], [34]. The porcine XIST sequence has a sequence similar to that of the human C-repeat sequence (49 bp downstream of the second repeat region), while only one copy was found in the pig. Thus, our results may suggest that porcine XIST has a novel mechanism of regulating XIST localization on the X chromosome compared with the mouse. Two other repeat regions of porcine XIST (regions 3 and 4 in Figure 2A) were identified as candidate functional domains. In particular, the third repeat region consisted of a highly repeated sequence (91.7 copies, Figure 2D) and was found to be well-aligned only with the bovine XIST sequence (Figure 2A). The D-repeat has been suggested to be the largest repeat, and is commonly found in exon 1 of XIST/Xist in many species [27], [34]. The D-repeat is hyper-variable with respect to both its sequence and length among species [34]. Thus, we hypothesized that the third repeat in porcine XIST may be either a D region or a porcine-specific repeat. While we identified four repeat regions in porcine XIST, their biological roles need to be clarified in future studies.
The alignment of Illumina reads (NCBI SRA accession numbers: SRX054582 - SRX054587) with our XIST gene model clearly demonstrated that the identified regions were expressed in other tissues according to the same model and that transcription start and termination sites at the 5′ and 3′ regions, respectively, were consistent among different porcine cell types (Figure 3 and Figure S5). Taken together, our results suggest that our model of the porcine XIST gene and the estimated 5′/3′ regions is reliable.
The promoter of XIST/Xist is important for regulating differential expression between female and male eutherians. Two representative XIST promoters, P1 and P2, have been reported. P1, a minimal promoter, is located close to the TSS [21], [22], while P2 is located nearly 1 kb from P1 [27], [35]. In our study, there was a region of high CG dinucleotide density from the −284th nucleotide to +53rd nucleotide and from the +285th nucleotide to +1727th nucleotide. The distribution patterns of CpG sites of porcine XIST were similar to P1 [21], [22] and P2 [27], [35] of other species. A previous study demonstrated that the Xist promoter is highly methylated on active X chromosomes in somatic cells, while inactive X chromosomes do not have a methylated promoter region [23]. Likewise, male porcine somatic cells carrying an active X chromosome were also highly methylated at XIST regulatory regions, while female porcine somatic cells carrying both active and inactive X chromosomes displayed a half methylation pattern (Figure 4C). Our results were consistent with those of previous reports, and support the hypothesis that the analyzed porcine CpG sites may be regulatory regions of XIST expression. Several transcription factors, such as CTCF, YY1, and TBP1, are known to regulate XIST/Xist expression at promoter regions [21], [22], [24], [36]. Interestingly, we confirmed a TBP1 consensus sequence in the candidate porcine P1 region (TTAAAG, −30 to −25), suggesting that TBP1 may be a regulator of porcine XIST expression. The presence of this TBP1 consensus sequence in 30bp upstream of one of the TSS, the 289,233rd nucleotide of the pig X chromosome scaffold (NW_003612825.1), suggest that the TSS which was applied to the first sequence of porcine XIST in this study is most reliable TSS of the porcine XIST RNA. Although we tried to find CTCF-binding sites in XIST gene and its upstream region, because of its diverse binding sites, it was hard to determine presence of suitable sites for CTCF binding. The YY1 of which consensus sequences, CCGCCATNTT, is also present in near the XIST TSS we identified (CCGCCATATT, −6 to +4), but it is unclear this region is actually recruited as an YY1 binding and functional site. So it should be analyzed these transcription factors are also functional in porcine XIST expression as in other species XIST/Xist expression. So the exact porcine XIST promoter regions also need to be further defined in order to clarify the relationship between CpG methylation and transcription factor binding on XIST expression using like chromatin immunopriciptation (Chip) - assay.
In this study, we identified the entire coding region of porcine XIST using sequence alignment, RT-PCR, Sanger sequencing, restriction enzyme mapping, and Illumina alignment. We proposed a porcine XIST ncRNA 25,215 bp in length and comprising seven exons (Table 1, GenBank accession No. KC753464). We also examined CpG methylation in the regulatory regions of porcine XIST. The porcine XIST gene model and CpG methylation profile reported here may be important for understanding the mechanism of XCI in pigs. Further studies will be needed to determine the implications of our model on the stem cell biology and embryology of pigs.
Materials and Methods
The pig experimental processes were performed strictly in accordance with the Guide for Care and Use of Research Animals in Teaching and Research. The experiment was approved by the Institutional Animal Care and Use Committee of Yonsei University, Wonju, Republic of Korea.
BLAST Search
The entire pig genomic sequence (Sus scrofa genome, version 10.2) was compared to the exons of mouse Xist and human and bovine XIST (Accession Nos: NC_000086.7, NC_000023.10, and AC_000187.1, respectively) using BLAST. Regions on the pig X chromosome that gave an alignment score of over 300 and that matched at least two Xist or XIST sequences from other species were considered to be candidate encoding regions of the porcine XIST ncRNA.
Sample Preparation, RNA Extraction, and Reverse Transcription
The porcine embryonic fibroblasts (PEF, mixed breed) were obtained from the days post coitus (dpc) = 27 fetuses. Cultured male and female PEFs (passage = 5) were used in this study. PEF was cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; collected and processed in Canada; Hyclone, Logan, UT), 2 mM glutamax (Gibco Invitrogen, Carlsbad, CA), 0.1 mM ß-mercaptoethanol (Gibco Invitrogen, Carlsbad, CA), and 1× antibiotic/antimycotic (Gibco Invitrogen, Carlsbad, CA) as previously described methods [37]. Total RNA was extracted from cultured PEFs using TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Extracted RNA was diluted with sterile DEPC-treated water (pH = 8.0) and treated with recombinant DNase I (Takara Bio, Otsu, Shiga, Japan). DNase-treated RNA samples (2.5 µg) were converted to cDNA in 20 µl final volume using Superscriptase III (Invitrogen Carlsbad, CA, USA) following the manufacturer’s instrument.
PCR Amplification and RACE-PCR of 5′- and 3′- Region
One-micro litter of synthesized cDNAs was subjected to PCR using the primers listed in Table S1. PCR amplification was carried out using a 2× PCR master mix solution (iNtRON Bio Technology, Seongnam, Gyeonggi, Korea) containing 0.5 µM of each primer set in 10 µl reaction volume. Detailed amplification conditions were described in supplementary information. All 5′- and 3′-region amplifications were performed using RACE-PCR with the 5′-RACE Core Set and 3′-RACE Core Set (Takara Bio, Otsu, Shiga, Japan), respectively. The process for synthetizing cDNA was carried out in accordance with the manufacturer’s instructions and the amplification of synthesized cDNA samples were performed with modifications (see Material and method S1). Reverse-transcrition and amplification was performed using the primer pairs listed in Table S2.
Enzyme Cutting, Cloning, and Sequencing
Gel-extracted amplicons were purified using a commercial spin column (MEGAquick-spin™ Total Fragment DNA Purification Kit; iNtRON Bio Technology, Seongnam, Gyeonggi, Korea) for restriction enzyme mapping and cloning. Each purified PCR product was incubated with 2 U of the appropriate restriction enzyme listed in Table S3 at 37°C for 2 hours. Amplicons were then cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA) and transformed into E. coli (DH5α strain, Novagen, Madison, WI, USA). Plasmid samples were prepared using commercial spin columns (DNA-spin™ Plasmid DNA Purification Kit; iNtRON Bio Technology, Seongnam, Gyeonggi, Korea. Purified plasmid samples were sequenced with an ABI PRISM 3730 DNA Analyzer (Applied Biosystems, Foster, CA, USA) and compared with genomic scaffold sequences.
gDNA Extraction and Bisulfite Sequencing
Extraction of gDNA was performed using the G-spin Genomic DNA Extraction Kit for cell/tissue (iNtRON Bio Technology, Seongnam, Gyeonggi, Korea) according to the manufacturer’s instructions. Extracted gDNA was treated with bisulfite using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, USA) for methylation analysis of CpG dinucleotides according to the manufacturer’s protocol. Bisulfite treated gDNA was amplified with primer pairs listed on the Table S4 and a total of 67 CpG sites were analyzed following the condition described in supplementary information. Amplicons were cloned and sequenced as described above for enzyme cutting, cloning, and sequencing.
Illumina Sequences, Sequence Alignment, and Alignment Visualization
We used released Illumina sequences from one pair of female pig full-siblings (240 days old, breed: White Duroc X Erhualian F2) stored in the NCBI Sequence Read Archive (SRA) database (Accession Nos. SRX054582 - SRX054587). The reads contained paired-end 90-bp sequences (200-bp insert) generated using the Illumina HiSeq 2000 platform for three different tissues–liver, abdominal fat, and the longissimus dorsi muscle. Custom Perl scripts were used to enrich for higher quality (Q20) reads, which were subsequently aligned to XIST RNA sequences assembled manually using Bowtie2 [38]. Aligned reads were sorted using SAMtools [39] and visualized with Tablet [40].
Supporting Information
XIST / Xist homologue analysis of the pig genome by BLAST search. (A) Human XIST, (B) bovine XIST, and (C) Mouse Xist (GenBank accession Nos. NC_000023.10, AC_000187.1, and NC_000086.7 respectively) models are represented as black boxes (exons) and black lines (introns). Black dashed lines under each XIST/Xist gene indicate the pig X-chromosome scaffold (NW_003612825.1). Filled arrowheads indicate the transcription start sites (TSS) of each XIST/Xist gene. Empty arrowheads present on the defined regions of the counterparts of XIST/Xist gene indicate candidate porcine XIST TSSs based on BLAST alignment (289233rd nucleotide of the NW_003612825.1 scaffold). Arrows indicate the terminus of the XIST/Xist gene and the candidate last-sequence of the porcine XIST gene identified by aligning human/bovine XIST RNAs to the pig genome sequence. Colored lines are homologue regions in each XIST/Xist gene, and the colored boxes are the counterparts of the lines. Each color represents an alignment score. The diagram is scaled.
(TIF)
RT-PCR analysis of each candidate region of XIST PCR amplicon expression. (A) Amplified PCR products of designed primer pairs. Female PEF cDNA (1), male PEF cDNA (2), female PEF gDNA (3) and male PEF gDNA (4) were used for amplification using each designed primer pairs and distilled water (5) was used as a negative control. The asterisk, filled arrowhead, empty arrowhead, and arrow represent the 2000, 1500, 1000, and 750 bp DNA bands of the ladder, respectively. (B) Summary heat map for PCR detection of porcine XIST RNA and genomic DNA sequences. Filled gray boxes indicate the presence of PCR target regions. White blank boxes indicate the absence of PCR target regions. The E2EL region was detected only in cDNA templates from female cells.
(TIF)
Identified gap sequence in the pig X-chromosome scaffold, NW_003612825.1. A gap sequence region was amplified with primer pairs designed to span the gap region. An unknown region nearly 500 bp longer than the expected length (100 bp) was observed. The gray-boxed sequence (TTTAAA) indicates a DraI restriction site.
(TIF)
Identification of transcription start sites and the termination site of porcine XIST . (A) Transcription start sites (TSSs) of the porcine XIST gene were identified by 5′ RACE-PCR. Eight putative TSSs were confirmed (red color characters), and the asterisk-marked sequence was determined to be the candidate TSS based on a BLAST homology search (289,233rd nucleotide of the pig X-chromosome scaffold sequence, NW_003612825.1). The TSS (289,223rd nucleotide of NW_003612825.1, asterisk-marked “T”) was set as +1. (B) The last sequence of porcine XIST was defined by 3′ RACE-PCR. The italic sequence (“T”, 257,103rd nucleotide of NW_003612825.1) is expected to be the last sequence based on BLAST alignment. The sequence marked with an asterisk (“A”, 257094th nucleotide of NW_003612825.1 and 32,817th nucleotide from TSS) indicates the last sequence identified by 3′ RACE-PCR. The gray-boxed region (AATAAA) represents one of the poly A-tail signal sequences.
(TIF)
Illumina sequencing coverage of porcine XIST mRNA expression. The entire region of XIST mRNA and upstream/downstream regions was used as a reference sequence for Illumina sequence alignment to show mRNA sequence distribution. The Illumina RNA-Seq was obtained from three tissues of different female individuals (F1 and F2). Sequencing depth was normalized using the total number of aligned reads. Several extreme peaks were observed because of sequence similarity among repeated sequences within the XIST gene. This peak could be lowered by increasing alignment stringency.
(TIF)
The List of primer pairs sequence for reverse transcription PCR.
(DOCX)
List of restriction enzymes used for each amplicon and the predicted sizes of the digested fragments.
(DOCX)
List of primer pair sequences for 5`- and 3`- RACE-PCR.
(DOCX)
List of primer pairs for bisulfite sequencing.
(DOCX)
PCR amplification condition.
(DOCX)
Funding Statement
This research was supported by the Next BioGreen 21 program (PJ009493), Rural Development Administration, and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2012R1A1A2006276), Republic of Korea. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lyon MF (1961) Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372–373. [DOI] [PubMed] [Google Scholar]
- 2. Okamoto I, Heard E (2009) Lessons from comparative analysis of X-chromosome inactivation in mammals. Chromosome Res 17: 659–669. [DOI] [PubMed] [Google Scholar]
- 3. Brown CJ, Robinson WP (1997) XIST expression and X-chromosome inactivation in human preimplantation embryos. Am J Hum Genet 61: 5–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sharman GB (1971) Late DNA replication in the paternally derived X chromosome of female kangaroos. Nature 230: 231–232. [DOI] [PubMed] [Google Scholar]
- 5. Huynh KD, Lee JT (2003) Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature 426: 857–862. [DOI] [PubMed] [Google Scholar]
- 6. Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N (1996) Requirement for Xist in X chromosome inactivation. Nature 379: 131–137. [DOI] [PubMed] [Google Scholar]
- 7. Wutz A, Jaenisch R (2000) A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell 5: 695–705. [DOI] [PubMed] [Google Scholar]
- 8. Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, et al. (1992) The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542. [DOI] [PubMed] [Google Scholar]
- 9. Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, et al. (1992) The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71: 515–526. [DOI] [PubMed] [Google Scholar]
- 10. De La Fuente R, Hahnel A, Basrur PK, King WA (1999) X inactive-specific transcript (Xist) expression and X chromosome inactivation in the preattachment bovine embryo. Biol Reprod 60: 769–775. [DOI] [PubMed] [Google Scholar]
- 11. Kay GF, Penny GD, Patel D, Ashworth A, Brockdorff N, et al. (1993) Expression of Xist during mouse development suggests a role in the initiation of X chromosome inactivation. Cell 72: 171–182. [DOI] [PubMed] [Google Scholar]
- 12. Inoue K, Kohda T, Sugimoto M, Sado T, Ogonuki N, et al. (2010) Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science 330: 496–499. [DOI] [PubMed] [Google Scholar]
- 13. Matoba S, Inoue K, Kohda T, Sugimoto M, Mizutani E, et al. (2011) RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos. Proc Natl Acad Sci U S A 108: 20621–20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nichols J, Smith A (2009) Naive and primed pluripotent states. Cell Stem Cell 4: 487–492. [DOI] [PubMed] [Google Scholar]
- 16. Cepica S, Masopust M, Knoll A, Bartenschlager H, Yerle M, et al. (2006) Linkage and RH mapping of 10 genes to a QTL region for fatness and muscling traits on pig chromosome X. Anim Genet. 37: 603–604. [DOI] [PubMed] [Google Scholar]
- 17. Bischoff SR, Tsai SQ, Hardison NE, Motsinger-Reif AA, Freking BA, et al. (2013) Differences in X-Chromosome Transcriptional Activity and Cholesterol Metabolism between Placentae from Swine Breeds from Asian and Western Origins. PLoS One 8: e55345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Groenen MA, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, et al. (2012) Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491: 393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hendrich BD, Brown CJ, Willard HF (1993) Evolutionary conservation of possible functional domains of the human and murine XIST genes. Hum Mol Genet 2: 663–672. [DOI] [PubMed] [Google Scholar]
- 20. Pang KC, Frith MC, Mattick JS (2006) Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet 22: 1–5. [DOI] [PubMed] [Google Scholar]
- 21. Pillet N, Bonny C, Schorderet DF (1995) Characterization of the promoter region of the mouse Xist gene. Proc Natl Acad Sci U S A 92: 12515–12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hendrich BD, Plenge RM, Willard HF (1997) Identification and characterization of the human XIST gene promoter: implications for models of X chromosome inactivation. Nucleic Acids Res 25: 2661–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Norris DP, Patel D, Kay GF, Penny GD, Brockdorff N, et al. (1994) Evidence that random and imprinted Xist expression is controlled by preemptive methylation. Cell 77: 41–51. [DOI] [PubMed] [Google Scholar]
- 24. Sheardown SA, Newall AE, Norris DP, Rastan S, Brockdorff N (1997) Regulatory elements in the minimal promoter region of the mouse Xist gene. Gene 203: 159–168. [DOI] [PubMed] [Google Scholar]
- 25. McDonald LE, Paterson CA, Kay GF (1998) Bisulfite genomic sequencing-derived methylation profile of the xist gene throughout early mouse development. Genomics 54: 379–386. [DOI] [PubMed] [Google Scholar]
- 26. Nolen LD, Gao S, Han Z, Mann MR, Gie Chung Y, et al. (2005) X chromosome reactivation and regulation in cloned embryos. Dev Biol 279: 525–540. [DOI] [PubMed] [Google Scholar]
- 27. Elisaphenko EA, Kolesnikov NN, Shevchenko AI, Rogozin IB, Nesterova TB, et al. (2008) A dual origin of the Xist gene from a protein-coding gene and a set of transposable elements. PLoS One 3: e2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wutz A, Rasmussen TP, Jaenisch R (2002) Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet 30: 167–174. [DOI] [PubMed] [Google Scholar]
- 29. Wutz A (2011) Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet 12: 542–553. [DOI] [PubMed] [Google Scholar]
- 30. Okamoto I, Patrat C, Thepot D, Peynot N, Fauque P, et al. (2011) Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 472: 370–374. [DOI] [PubMed] [Google Scholar]
- 31. Sarma K, Levasseur P, Aristarkhov A, Lee JT (2010) Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci U S A 107: 22196–22201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Augui S, Nora EP, Heard E (2011) Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet 12: 429–442. [DOI] [PubMed] [Google Scholar]
- 33. Chow J, Heard E (2009) X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol 21: 359–366. [DOI] [PubMed] [Google Scholar]
- 34. Yen ZC, Meyer IM, Karalic S, Brown CJ (2007) A cross-species comparison of X-chromosome inactivation in Eutheria. Genomics 90: 453–463. [DOI] [PubMed] [Google Scholar]
- 35. Johnston CM, Nesterova TB, Formstone EJ, Newall AE, Duthie SM, et al. (1998) Developmentally regulated Xist promoter switch mediates initiation of X inactivation. Cell 94: 809–817. [DOI] [PubMed] [Google Scholar]
- 36. Pugacheva EM, Tiwari VK, Abdullaev Z, Vostrov AA, Flanagan PT, et al. (2005) Familial cases of point mutations in the XIST promoter reveal a correlation between CTCF binding and pre-emptive choices of X chromosome inactivation. Hum Mol Genet 14: 953–965. [DOI] [PubMed] [Google Scholar]
- 37. Park J-K, Kim H-S, Uh K-J, Choi K-H, Kim H-M, et al. (2013) Primed Pluripotent Cell Lines Derived from Various Embryonic Origins and Somatic Cells in Pig. PLoS One 8: e52481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Milne I, Bayer M, Cardle L, Shaw P, Stephen G, et al. (2010) Tablet–next generation sequence assembly visualization. Bioinformatics 26: 401–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
XIST / Xist homologue analysis of the pig genome by BLAST search. (A) Human XIST, (B) bovine XIST, and (C) Mouse Xist (GenBank accession Nos. NC_000023.10, AC_000187.1, and NC_000086.7 respectively) models are represented as black boxes (exons) and black lines (introns). Black dashed lines under each XIST/Xist gene indicate the pig X-chromosome scaffold (NW_003612825.1). Filled arrowheads indicate the transcription start sites (TSS) of each XIST/Xist gene. Empty arrowheads present on the defined regions of the counterparts of XIST/Xist gene indicate candidate porcine XIST TSSs based on BLAST alignment (289233rd nucleotide of the NW_003612825.1 scaffold). Arrows indicate the terminus of the XIST/Xist gene and the candidate last-sequence of the porcine XIST gene identified by aligning human/bovine XIST RNAs to the pig genome sequence. Colored lines are homologue regions in each XIST/Xist gene, and the colored boxes are the counterparts of the lines. Each color represents an alignment score. The diagram is scaled.
(TIF)
RT-PCR analysis of each candidate region of XIST PCR amplicon expression. (A) Amplified PCR products of designed primer pairs. Female PEF cDNA (1), male PEF cDNA (2), female PEF gDNA (3) and male PEF gDNA (4) were used for amplification using each designed primer pairs and distilled water (5) was used as a negative control. The asterisk, filled arrowhead, empty arrowhead, and arrow represent the 2000, 1500, 1000, and 750 bp DNA bands of the ladder, respectively. (B) Summary heat map for PCR detection of porcine XIST RNA and genomic DNA sequences. Filled gray boxes indicate the presence of PCR target regions. White blank boxes indicate the absence of PCR target regions. The E2EL region was detected only in cDNA templates from female cells.
(TIF)
Identified gap sequence in the pig X-chromosome scaffold, NW_003612825.1. A gap sequence region was amplified with primer pairs designed to span the gap region. An unknown region nearly 500 bp longer than the expected length (100 bp) was observed. The gray-boxed sequence (TTTAAA) indicates a DraI restriction site.
(TIF)
Identification of transcription start sites and the termination site of porcine XIST . (A) Transcription start sites (TSSs) of the porcine XIST gene were identified by 5′ RACE-PCR. Eight putative TSSs were confirmed (red color characters), and the asterisk-marked sequence was determined to be the candidate TSS based on a BLAST homology search (289,233rd nucleotide of the pig X-chromosome scaffold sequence, NW_003612825.1). The TSS (289,223rd nucleotide of NW_003612825.1, asterisk-marked “T”) was set as +1. (B) The last sequence of porcine XIST was defined by 3′ RACE-PCR. The italic sequence (“T”, 257,103rd nucleotide of NW_003612825.1) is expected to be the last sequence based on BLAST alignment. The sequence marked with an asterisk (“A”, 257094th nucleotide of NW_003612825.1 and 32,817th nucleotide from TSS) indicates the last sequence identified by 3′ RACE-PCR. The gray-boxed region (AATAAA) represents one of the poly A-tail signal sequences.
(TIF)
Illumina sequencing coverage of porcine XIST mRNA expression. The entire region of XIST mRNA and upstream/downstream regions was used as a reference sequence for Illumina sequence alignment to show mRNA sequence distribution. The Illumina RNA-Seq was obtained from three tissues of different female individuals (F1 and F2). Sequencing depth was normalized using the total number of aligned reads. Several extreme peaks were observed because of sequence similarity among repeated sequences within the XIST gene. This peak could be lowered by increasing alignment stringency.
(TIF)
The List of primer pairs sequence for reverse transcription PCR.
(DOCX)
List of restriction enzymes used for each amplicon and the predicted sizes of the digested fragments.
(DOCX)
List of primer pair sequences for 5`- and 3`- RACE-PCR.
(DOCX)
List of primer pairs for bisulfite sequencing.
(DOCX)
PCR amplification condition.
(DOCX)