Abstract
We have recently identified a novel population of activated low-density granulocytes (LDGs) in peripheral blood mononuclear cells of HIV seropositive patients. LDGs have a similar morphology to normal density granulocytes (NDGs), but are phenotypically different. Here we measured the expression levels of different phenotypic markers of granulocytes in the blood of HIV seropositive patients at different stages of HIV infection to determine whether the phenotype of NDGs and LDGs are affected by disease severity. Our results reveal that the phenotype of NDGs, but not that of LDGs, varies according to the severity of the disease.
Introduction
Neutrophils play a central role in the elimination of pathogens by using several strategies such as the production of reactive oxygen species, the release of antimicrobial peptides and neutrophil extracellular traps (NETs) (summarised in [1]). In HIV infections, both the number and the functions of neutrophils are impaired (summarised in [2]). Functional abnormalities of neutrophils include impaired phagocytosis and production of toxic oxygen species [2]. Furthermore, it has been recently shown that whereas NETs formation can capture and eliminate HIV, HIV can counteract this by inducing the production of IL-10 by dendritic cells and therefore inhibiting NETs formation [3].
Human neutrophils constitutively express arginase [4], an enzyme that catalyses the conversion of L-arginine into ornithine and urea [5], [6]. Recently, the metabolism of L-arginine by arginase has emerged as a crucial mechanism for the regulation of immune responses: increased catabolism of L-arginine by arginase results in the depletion of L-arginine from the microenvironment; since L-arginine is essential for efficient T cell activation, this decrease in L-arginine results in impaired T cell responses [7], [8], [9], [10]. Increased arginase activity has been described in malaria [11], tuberculosis [12], leishmaniasis [13], [14], [15] and HIV [16], [17].
We have recently shown that PBMCs from HIV seropositive patients with low CD4+ T cell counts expressed significantly more arginase activity as compared to patients with high CD4+ T cell counts or uninfected controls [16]. Higher arginase expression in PBMCs from HIV seropositive patients was associated with decreased levels of CD3ζ expression, a marker of T cell dysregulation [16]. The phenotype of arginase-expressing cells was identified as low-density granulocytes (LDGs) as these cells co-purify with PBMCs following density gradient centrifugation. This difference in density distinguishes this population from the remaining granulocytes that co-purify with the erythocyte fraction following density gradient centrifugation and thus have been named normal-density granulocytes (NDGs). LDGs purified from HIV+ patients display a similar morphology as NDGs, but have major phenotypic differences suggesting that LDGs were activated neutrophils that had degranulated and released arginase [17]. In the present study, we aim to determine whether the phenotype of NDGs and LDGs differs in different stages of HIV infection.
Materials and Methods
Subjects and samples
Twenty-one HIV seropositive (HIV+) treatment-naïve individuals were recruited from St Mary's Hospital (Table 1). Plasma HIV-1 viral RNA was quantified by real-time PCR (Bayer Quantiplex assay (bDNA) PCR test; lower detection level of 50 copies/mL). The standard T lymphocyte markers CD3, CD4, CD8 were determined by flow cytometry. The study was approved by the National Research Ethics Service (05/Q0410/93) and all individuals gave written, informed consent before participation.
Table 1. Clinical data.
| Patients | Age | Sex (M/F) | CD4 count | Viral load |
| 1 | 46 | M | 840 | 3.42 |
| 2 | 38 | M | 780 | 4.96 |
| 3 | 33 | M | 680 | 4.35 |
| 4 | 41 | M | 620 | 3.25 |
| 5 | 27 | M | 580 | 2.39 |
| 6 | 60 | M | 540 | 3.16 |
| 7 | 42 | M | 540 | 4.54 |
| 8 | 34 | M | 540 | 4.77 |
| 9 | 23 | M | 520 | 3.99 |
| 10 | 30 | F | 440 | 4.24 |
| 11 | 46 | M | 390 | 5.27 |
| 12 | 48 | M | 330 | 4.79 |
| 13 | 51 | M | 300 | 5.57 |
| 14 | 40 | M | 170 | 4.91 |
| 15 | 34 | M | 160 | 4.6 |
| 16 | 49 | M | 160 | 4.4 |
| 17 | 46 | M | 140 | 5.28 |
| 18 | 38 | M | 128 | 5.69 |
| 19 | 27 | F | 25 | 5.57 |
| 20 | 44 | M | 20 | 5.7 |
| 21 | 38 | F | 4 | 4.43 |
Age, sex, CD4+ T cell counts and viral load were recorded for HIV+ patients (n = 21).
Twenty ml of peripheral blood was collected in EDTA tubes and PBMCs were isolated by density gradient centrifugation on Histopaque®-1077 (Sigma). Neutrophils were isolated from the erythrocyte fraction by dextran sulphate sedimentation [17]. All experiments were performed on fresh cell, immediately after processing.
Flow cytometry
The following antibodies were used: CD14FITC, CD15PE (BD Pharmingen), Arginase1Alexa Fluor®647 (Hycult Biotechnology), CD11bPerCP-eFluro710, CD16eFluro450, CD33PE-Cy7 (eBioscience), CD13APC-Cy7 (Biolegend), CD66bFITC and CD63FITC (Beckman Coulter). 1×106 PBMCs were incubated with FcR blocking reagent (BD Pharmingen) and the antibodies against extracellular markers were added directly to cells. Cells were washed after 20 min, fixed, permeabilised and anti-arginase IAlexa FluorR 647 was added to the cells for 20 min as described in [18]. Analysis was performed on an FACS Canto II (BD Bioscience) and results were analyzed using FlowJo v8.7 (Tree Star, Ashland, OR).
Statistical analyses
Data were evaluated for statistical differences using a two-tailed Mann-Whitney test and a Spearman's rank test when appropriate (GraphPad Prism 5); differences were considered statistically significant at p<0.05. Results are expressed as median± SEM.
Results
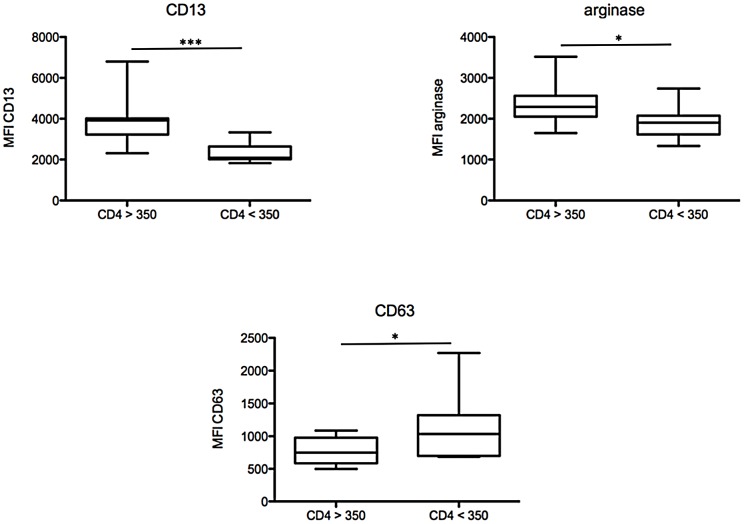
We first analysed the phenotype of NDGs to determine whether it changes with increased disease severity, as measured by CD4+ T cell counts and viral load. We measured the expression levels of CD11b, CD13, CD15, CD16, CD33, CD63, CD66b and arginase on NDGs from treatment naïve HIV+ patients with high (≥350 cells/µL, CD4high) and low (<350 cells/µL, CD4low) CD4+ T cell counts. The division of patients based on a CD4+ T cell count <350 cells/µL was chosen because 1) once the CD4+ T cell count falls below 350 cells/µL differences in clinical outcome increasingly appear [19] and 2) the initiation of antiretroviral therapy is recommended once CD4+ T cell count falls to <350 cells/µL [20]. Our results show that the MFIs of CD13 and arginase are statistically significantly lower (p = 0.0008 and p = 0.0048, respectively) and that of CD63 significantly higher (p = 0.0346) in the blood of CD4low HIV+ patients (Figure 1, Table 2) as compared to CD4high HIV+ patients. No significant difference was observed in the expression levels of CD11b (p = 0.9159), CD15 (p = 0.3072), CD16 (p = 0.5495), CD33 (p = 0.3787) and CD66b (p = 0.6985) between CD4high and CD4low HIV+ patients (Table 2). Of note, the expression levels of CD11b, CD13, CD15, CD16, CD33, CD63, CD66b and arginase 1 were homogenous (Figure S1).
Figure 1. Phenotypic analysis of NDGs.
NDGs were isolated from the blood of HIV+ patients with CD4+ T cell counts >350 (n = 11) or <350 cells/µL (n = 10) as described in materials and methods and the expression levels of phenotypic markers were determined by flow cytometry. Isotype controls: <1%. Statistical significance was determined by a two-tailed Mann-Whitney test. Box = interquartile range and median; whiskers = range.
Table 2. Phenotype of NDGs.
| CD4high | CD4low | p value | |
| (median ± SEM) | (median ± SEM) | ||
| CD11b | 2684±767 | 3418±610 | 0.9159 |
| CD13 | 3915±345 | 2091±152 | 0.0008 |
| CD15 | 2270±693 | 3732±812 | 0.3072 |
| CD16 | 10259±934 | 8731±743 | 0.5495 |
| CD33 | 2114±196 | 1644±241 | 0.3787 |
| CD63 | 751±57 | 1035±152 | 0.0346 |
| CD66b | 6326±771 | 6854±604 | 0.6985 |
| arginase | 2294±148 | 1904±124 | 0.0448 |
NDGs were isolated from the blood of HIV+ patients with CD4+ T cell counts >350 cells/µL (n = 11) or <350 cells/µL (n = 10) as described in materials and methods and expression levels of phenotypic markers were determined by flow cytometry.
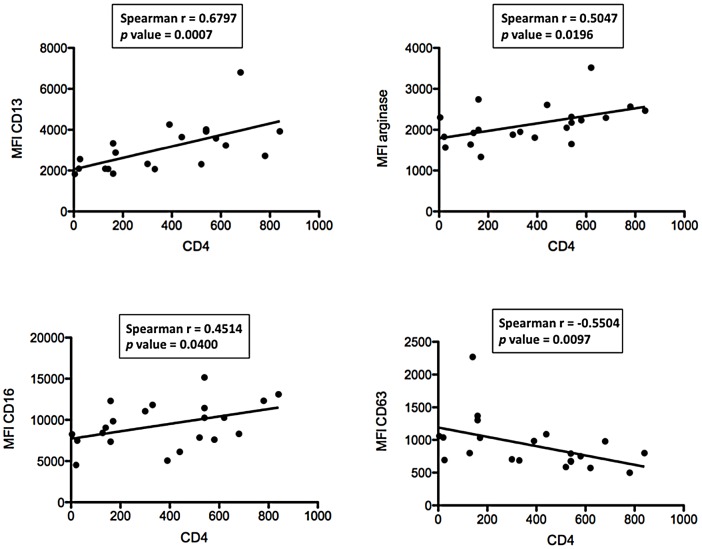
To characterise further the association between these markers and disease severity, we plotted their MFI values against CD4+ T cell counts. As shown in Figure 2, there are statistically significant positive correlations between CD4+ T cell counts and the MFIs of CD13 (p = 0.0007), arginase (p = 0.0196) and CD16 (p = 0.0400); and a significant negative correlation between CD4+ T cell counts and the MFI of CD63 (p = 0.0097) (Figure 2, Table 3). No significant correlation was observed between CD4+ T cell counts and CD11b (p = 0.5122), CD15 (p = 0.0580), CD33 (p = 0.0825), and CD66b (p = 0.6064) (Table 3).
Figure 2. Correlation between CD4+ T cells and phenotypic markers.
NDGs were isolated from the blood of HIV+ patients (n = 21) as described in materials and methods and the expression levels of phenotypic markers were determined by flow cytometry. Correlation between CD4+ T cell counts and phenotypic markers was determined by a Spearman's rank test.
Table 3. NDGs: Correlation between CD4+ T cell counts and MFIs.
| Spearman r | p value | |
| CD11b | −0.1522 | 0.5122 |
| CD13 | 0.6797 | 0.0007 |
| CD15 | −0.3707 | 0.0980 |
| CD16 | 0.4514 | 0.0400 |
| CD33 | 0.3876 | 0.0825 |
| CD63 | −0.5504 | 0.0097 |
| CD66b | −0.1213 | 0.6034 |
| arginase | 0.5047 | 0.0196 |
NDGs were isolated from the blood of HIV+ patients with CD4+ T cell counts >350 cells/µL (n = 11) or <350 cells/µL (n = 10) as described in materials and methods and the correlations between CD4+ T cell counts and phenotypic markers were determined by a Spearman's rank test.
Next, we plotted expression levels of CD11b, CD13, CD15, CD16, CD33, CD63, CD66b and arginase against another marker of disease severity, viral load. A statistically significant negative correlation between VL and arginase (p = 0.0063, Figure 3) and a trend towards significance between VL and CD13 (p = 0.0553, Table 4) were observed, all other correlations were not statistically significant (Figure 3, Table 4).
Figure 3. Correlation between viral load and phenotypic markers.
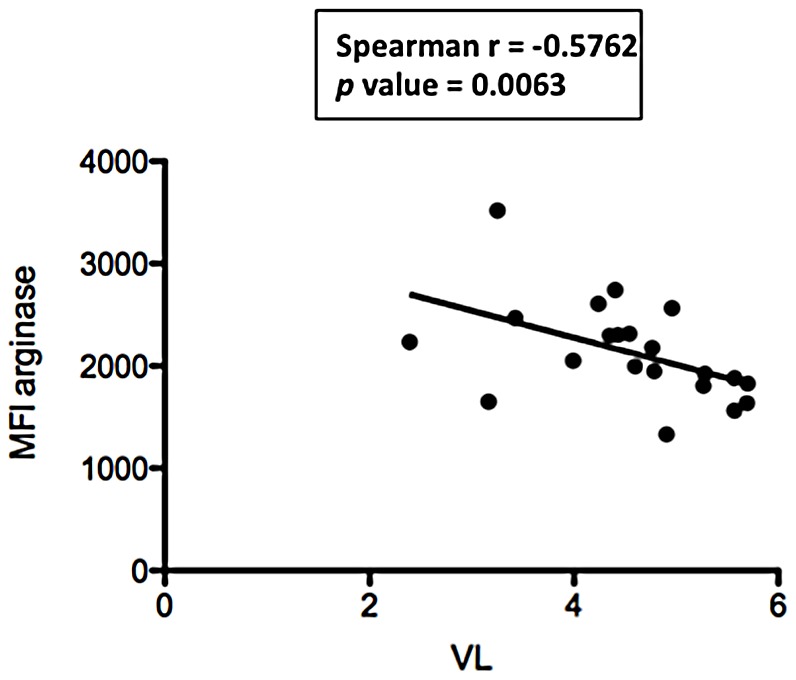
NDGs were isolated from the blood of HIV+ patients (n = 21) as described in materials and methods and the expression levels of phenotypic markers were determined by flow cytometry. Correlation between viral load and phenotypic markers was determined by a Spearman's rank test.
Table 4. NDGs: Correlation between viral loads and MFIs.
| Spearman r | p value | |
| CD11b | 0.1104 | 0.6337 |
| CD13 | −0.4242 | 0.0553 |
| CD15 | 0.1241 | 0.5921 |
| CD16 | −0.2014 | 0.3814 |
| CD33 | −0.1845 | 0.4234 |
| CD63 | 0.1358 | 0.5572 |
| CD66b | 0.1358 | 0.5574 |
| arginase | −0.5762 | 0.0063 |
NDGs were isolated from the blood of HIV+ patients with CD4+ T cell counts >350 cells/µL (n = 11) or <350 cells/µL (n = 10) as described in materials and methods and thes correlation between viral load and phenotypic markers were determined by a Spearman's rank test.
These results suggest that in HIV+ patients, the phenotype of NDGs varies according to the severity of the disease.
Our previous results have identified a novel population of low-density granulocytes (LDGs) in the PBMCs of HIV+ patients that are morphologically similar, but phenotypically different from NDGs. Here, we determined whether the MFIs value differ in this population of LDGs and whether there was a correlation between markers of disease severity and the expression levels of CD11b, CD13, CD15, CD16, CD33, CD63, CD66b and arginase: MFIs values were similar between LDGs from CD4low and CD4high HIV+ patients (Table 5) and none of the correlations were statistically significant (Tables 6 and 7).
Table 5. Phenotype of LDGs.
| CD4high | CD4low | p value | |
| (median ± SEM) | (median ± SEM) | ||
| CD11b | 4413±649 | 5148±890 | 0.2907 |
| CD13 | 2137±564 | 2368±366 | 0.9159 |
| CD15 | 10012±1277 | 8262±769 | 0.3072 |
| CD16 | 1528±1483 | 6932±1112 | 0.5035 |
| CD33 | 3575±285 | 2768±304 | 0.1300 |
| CD63 | 1474±178 | 1355±246 | 0.9717 |
| CD66b | 8899±2404 | 10198±1775 | 0.6472 |
| arginase | 1700±141 | 1533±123 | 0.2453 |
LDGs were isolated from the blood of HIV+ patients with CD4+ T cell counts >350 cells/µL (n = 11) or <350 cells/µL (n = 10) as described in materials and methods and the expression levels of phenotypic markers were determined by flow cytometry.
Table 6. LDGs: Correlation between CD4+ T cell counts and MFIs.
| Spearman r | p value | |
| CD11b | −0.3507 | 0.1191 |
| CD13 | 0.1028 | 0.6576 |
| CD15 | 0.1502 | 0.5157 |
| CD16 | 0.0072 | 0.9754 |
| CD33 | 0.4039 | 0.0694 |
| CD63 | 0.1158 | 0.6173 |
| CD66b | −0.0134 | 0.9531 |
| arginase | 0.2738 | 0.7009 |
LDGs were isolated from the blood of HIV+ patients with CD4+ T cell counts >350 cells/µL (n = 11) or <350 cells/µL (n = 10) as described in materials and methods and the correlations between CD4+ T cell counts and phenotypic markers were determined by a Spearman's rank test.
Table 7. Correlation between viral loads and MFIs.
| Spearman r | p value | |
| CD11b | 0.2242 | 0.3286 |
| CD13 | 0.0071 | 0.9755 |
| CD15 | −0.2007 | 0.3830 |
| CD16 | −0.0084 | 0.9710 |
| CD33 | −0.3248 | 0.1509 |
| CD63 | −0.0364 | 0.8756 |
| CD66b | 0.0948 | 0.6826 |
| arginase | −0.4722 | 0.2235 |
LDGs were isolated from the blood of HIV+ patients with CD4+ T cell counts >350 cells/µL (n = 11) or <350 cells/µL (n = 10) as described in materials and methods and the correlations between viral load and phenotypic markers were determined by a Spearman's rank test.
These results suggest that the phenotype of NDGs, but not that of LDGs varies according to the severity of the disease.
Phenotype of NDGs and LDGs in CD4low HIV+ patients
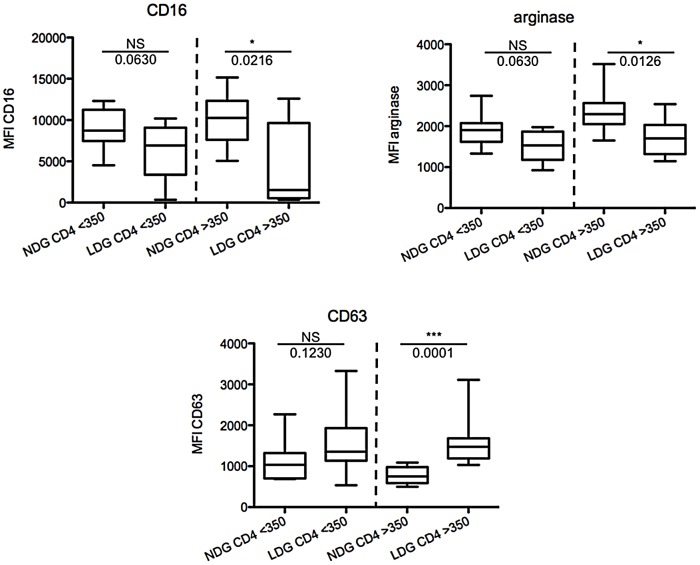
We have previously shown that LDGs are phenotypically different from NDGs, as they express increased levels of CD11b, CD15, CD33, CD66b, CD63 and decreased levels of CD16 and arginase 1 [17], suggesting that these cells are activated neutrophils that have degranulated and therefore change their density. Indeed, the results presented in Figure 2 suggest that NDGs get progressively more activated with increased disease severity, as measured by CD4+ T cell counts. In the next step, we assessed how the phenotype of NDGs differs from that of LDGs in CD4high and CD4low HIV+ patients. As shown in Figure 4 and Tables 8 and 9, the expression levels of CD16, arginase and CD63 are significantly different between LDGs and NDGs from CD4high HIV+ patients, but are similar in CD4low patients. Differences between CD11b, CD13 and CD66b MFIs remain non-significant and differences between CD15 and CD33 remain significant in CD4low and CD4high HIV+ groups.
Figure 4. Phenotypes of NDGs and LDGs in CD4low and CD4high HIV+ patients.
PBMCs and NDGs were isolated from the blood of HIV+ patients with CD4+ T cell counts >350 (n = 11) or <350 cells/µL (n = 10) as described in materials and methods and the expression levels of phenotypic markers were determined by flow cytometry. Isotype controls: <1%. Statistical significance was determined by a two-tailed Mann-Whitney test. Box = interquartile range and median; whiskers = range.
Table 8. LDGs and NDGs in HIV+ patients in CD4low HIV+ patients.
| LDGs | NDGs | p value | |
| (median ± SEM) | (median ± SEM) | ||
| CD11b | 5148±890 | 3418±610 | 0.2176 |
| CD13 | 2368±366 | 2091±152 | 0.9705 |
| CD15 | 8262±769 | 3732±812 | 0.0015 |
| CD16 | 6932±1112 | 8731±743 | 0.0630 |
| CD33 | 2768±304 | 1644±241 | 0.0288 |
| CD63 | 1355±246 | 1035±152 | 0.1230 |
| CD66b | 10198±1775 | 7671±604 | 0.0355 |
| arginase | 1533±123 | 1904±124 | 0.0630 |
LDGs and NDGs were isolated from the blood of HIV+ patients with CD4+ T cell counts <350 cells/µL (n = 10) as described in materials and methods. Expression levels of phenotypic markers were determined by flow cytometry.
Table 9. LDGs and NDGs in HIV+ patients in CD4high HIV+ patients.
| LDGs | NDGs | p value | |
| (median ± SEM) | (median ± SEM) | ||
| CD11b | 4413±694 | 2684±767 | 0.5994 |
| CD13 | 2137±564 | 3915±345 | 0.1007 |
| CD15 | 10012±1277 | 2270±693 | 0.0003 |
| CD16 | 1528±1483 | 10259±934 | 0.0216 |
| CD33 | 3575±285 | 2114±196 | 0.0016 |
| CD63 | 1474±178 | 751±57 | 0.0001 |
| CD66b | 8899±1775 | 7697±604 | 0.1679 |
| arginase | 1700±141 | 2294±150 | 0.0126 |
Ldgs And Ndgs Were Isolated From The Blood Of Hiv+ Patients With Cd4+ T Cell Counts >350 Cells/µL (N = 11) As Described In Materials And Methods. Expression Levels Of Phenotypic Markers Were Determined By Flow Cytometry.
Discussion
We have previously shown that arginase activity was significantly increased in the blood of CD4low HIV+ patients as compared to CD4high and healthy controls [16]. The phenotype of arginase-expressing cells in the PBMCs of HIV+ patients are a subset neutrophils, which were classified as low-density granulocytes (LDGs) [16], [17]. These cells have a similar morphology as normal density granulocytes (NDGs) [17]. However, LDGs differ from NDGs as i) they co-localise with the PBMCs and not the erythrocytic fractions, suggesting that their density is lower; ii) they express different levels of phenotypic markers of neutrophils. In addition, our results show that the frequency of LDGs is significantly higher in HIV+ patients with low CD4+ T cell counts and correlates with markers of disease severity in HIV+ patients [17]. We have already shown that the cells expressing arginase in PBMCs from HIV+ patients are LDGs, as they express CD15, but not CD14, and that the frequency of these cells increases with disease severity [16].
Our results show that there is no difference between the phenotype of LDGs from CD4low and CD4high HIV+ patients; however, there are clear phenotypic differences in the expression levels of CD13, CD63 and arginase in NDGs from HIV+ patients with low CD4+ T cell counts. Increased activation of neutrophils in HIV+ patients has already been described [21]. In this study, the expression levels of CD11b were shown to be increased on neutrophils from HIV+ patients as compared to HIV- controls. In our study we assessed whether the level of neutrophils' activation changes with disease severity, rather then comparing it to healthy controls. Our results are in agreement with the study by Elbim et al., as we did not find a change in CD11b expression on NDGs with lower CD4+ T cell counts.
Our results suggest that during the course of HIV infection, there is no progressive activation of LDGs, as we found no correlation between CD4+ T cell counts or viral load and the expression levels of CD11b, CD13, CD15, CD16, CD33, CD63, CD66b and arginase 1. However, our results suggest that NDGs become progressively and systemically more activated and more degranulated, since decreased expression levels of CD13, CD16 and arginase and increasing expression levels of CD63 on NDGs correlate with decreasing CD4+ T cell counts. The degree of neutrophil activation is regulated by the intensity of the activating signal and occurs sequentially: 1) secretory granules; 2) gelatinous granules; 3) specific and 4) azurophilic granules. Arginase is found in gelatinous granules [22] and azurophilic granules [4] and upregulation of CD63 on neutrophils coincides with the release of azurophilic granule [23], [24]. Therefore, our result showing that CD63 is expressed at increased levels and arginase at lower levels in NDGs from patients with low CD4+ T cells suggest that NDGs get progressively more activated with increased disease severity. We hypothesise that as a result of degranulation, NDGs will change their density and become LDGs, and will be collected in the PBMC fraction following density gradient purification. Our results suggest that NDGs are activated and have already, at least partially, released azurophilic granules, as shown by increased CD63 and decreased arginase expression levels in CD4low HIV+ patients. Furthermore, our results show that the MFIs of CD16, CD63 and arginase are significantly different between LDGs and NDGs in CD4high HIV+ patients, but not any more in CD4low HIV patients; suggesting that there are less differences between the phenotype of LDGS and NDGs in CD4low as compared to CD4high HIV+ patients.
The origins of LDGs as well as the signals resulting in the degranulation of neutrophils remain unclear in HIV+ patients and we have not been able to activate NDGs to become LDGs. Stimulation of neutrophils with Phorbol 12-myristate 13-acetate (PMA) and/or N-formyl-methionyl-leucyl-phenylalanine (fLMP) has been described previously ([23]: whereas these stimuli result in activation and degranulation of neutrophils, the phenotype of these activated neutrophils differs from that of LDGs we described [17], as CD13 and CD16 are both downregulated on activated NDGs.
Further work into novel markers of immune suppression, such as activation of granulocytic cells, is warranted as this may result in improvement in the clinical management of patients with HIV infection through: 1) better evaluation of disease severity (including the stage and rate of progression of the disease) and 2) informing the timing and choice of treatment initiation so as to minimise morbidity associated with opportunistic infections, drug resistance and medication side effects.
Supporting Information
Phenotypic analysis of NDGs. NDGs were isolated from the blood of HIV+ patients with CD4+ T cell counts >350 (n = 11) or <350 cells/µL (n = 10) as described in materials and methods and the expression levels of phenotypic markers were determined by flow cytometry. Isotype controls: <1%. Statistical significance was determined by a two-tailed Mann-Whitney test. Box = interquartile range and median; whiskers = range.
(TIF)
Funding Statement
This work was supported by a grant from The Wellcome Trust (07664/Z/05/Z, PK) and TC is a recipient of a Imperial College London MB/PhD fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Drescher B, Bai F (2012) Neutrophil in viral infections, friend or foe? Virus Res [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kuritzkes DR (2000) Neutropenia, neutrophil dysfunction, and bacterial infection in patients with human immunodeficiency virus disease: the role of granulocyte colony-stimulating factor. Clin Infect Dis 30: 256–260. [DOI] [PubMed] [Google Scholar]
- 3. Saitoh T, Komano J, Saitoh Y, Misawa T, Takahama M, et al. (2012) Neutrophil extracellular traps mediate a host defense response to human immunodeficiency virus-1. Cell Host Microbe 12: 109–116. [DOI] [PubMed] [Google Scholar]
- 4. Munder M, Mollinedo F, Calafat J, Canchado J, Gil-Lamaignere C, et al. (2005) Arginase I is constitutively expressed in human granulocytes and participates in fungicidal activity. Blood 105: 2549–2556. [DOI] [PubMed] [Google Scholar]
- 5. Cederbaum SD, Yu H, Grody WW, Kern RM, Yoo P, et al. (2004) Arginases I and II: do their functions overlap? Mol Gen Metabol 81: S38–S44. [DOI] [PubMed] [Google Scholar]
- 6. Munder M (2009) Arginase: an emerging key player in the mammalian immune system. Br J Pharmacol 158: 638–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ochoa AC, Zea AH, Hernandez C, Rodriguez PC (2007) Arginase, prostaglandins, and myeloid-derived suppressor cells in renal cell carcinoma. Clin Cancer Res 13: 721s–726s. [DOI] [PubMed] [Google Scholar]
- 8. Popovic PJ, Zeh HJ 3rd, Ochoa JB (2007) Arginine and immunity. J Nutr 137: 1681S–1686S. [DOI] [PubMed] [Google Scholar]
- 9. Bronte V, Zanovello P (2005) Regulation of immune responses by L-arginine metabolism. Nat Rev Immunol 5: 641–654. [DOI] [PubMed] [Google Scholar]
- 10. Munder M, Choi BS, Rogers M, Kropf P (2009) L-arginine deprivation impairs Leishmania major-specific T cell responses. European Journal of Immunology 39: 2161–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weinberg JB, Lopansri BK, Mwaikambo E, Granger DL (2008) Arginine, nitric oxide, carbon monoxide, and endothelial function in severe malaria. Curr Opin Infect Dis 21: 468–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zea AH, Culotta KS, Ali J, Mason C, Park HJ, et al. (2006) Decreased Expression of CD3 zeta and Nuclear Transcription Factor kappa B in Patients with Pulmonary Tuberculosis: Potential Mechanisms and Reversibility with Treatment. J Infect Dis 194: 1385–1393. [DOI] [PubMed] [Google Scholar]
- 13. Abebe T, Hailu A, Woldeyes M, Mekonene W, Bilch K, et al. (2012) Local increase of arginase activity in lesions of patients with cutaneous leishmaniasis in Ethiopia. PLoS NTD 6: e1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takele Y, Abebe T, Weldegebreal T, Hailu A, Hailu W, et al. (2012) Arginase activity in the blood of patients with visceral leishmaniasis and HIV infection. PLoS NTD 7 (1) e1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abebe T, Takele T, Weldegebreal T, Cloke T, Closs E, et al. (2013) Arginase: a marker of disease status in patients with visceral leishmaniasis. PLoS NTD in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cloke T, Garvery L, Choi BS, Abebe T, Hailu A, et al. (2010) Increased arginase activity correlates with disease severity in HIV seropositive patients. Journal of Infectious Diseases 202: 374–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cloke T, Munder M, Taylor GP, Müller I, Kropf P (2012) Characterization of a novel population of low-density granulocytes associated with disease severity in HIV seropositive patients. PLOS ONE 7: e48939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kropf P, Baud D, Marshall SE, Munder M, Mosley A, et al. (2007) Arginase activity mediates reversible T cell hyporesponsiveness in human pregnancy. Eur J Immunol 37: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Phillips A, Pezzotti P (2004) Short-term risk of AIDS according to current CD4 cell count and viral load in antiretroviral drug-naive individuals and those treated in the monotherapy era. AIDS 18: 51–58. [DOI] [PubMed] [Google Scholar]
- 20. Gazzard BG (2008) British HIV Association Guidelines for the treatment of HIV-1-infected adults with antiretroviral therapy 2008. HIV Med 9: 563–608. [DOI] [PubMed] [Google Scholar]
- 21. Elbim C, Prevot MH, Bouscarat F, Franzini E, Chollet-Martin S, et al. (1994) Polymorphonuclear neutrophils from human immunodeficiency virus-infected patients show enhanced activation, diminished fMLP-induced L-selectin shedding, and an impaired oxidative burst after cytokine priming. Blood 84: 2759–2766. [PubMed] [Google Scholar]
- 22. Rotondo R, Bertolotto M, Barisione G, Astigiano S, Mandruzzato S, et al. (2011) Exocytosis of azurophil and arginase 1-containing granules by activated polymorphonuclear neutrophils is required to inhibit T lymphocyte proliferation. J Leukoc Biol 89: 721–727. [DOI] [PubMed] [Google Scholar]
- 23. Kuijpers TW, Tool AT, van der Schoot CE, Ginsel LA, Onderwater JJ, et al. (1991) Membrane surface antigen expression on neutrophils: a reappraisal of the use of surface markers for neutrophil activation. Blood 78: 1105–1111. [PubMed] [Google Scholar]
- 24. Martin-Martin B, Nabokina SM, Blasi J, Lazo PA, Mollinedo F (2000) Involvement of SNAP-23 and syntaxin 6 in human neutrophil exocytosis. Blood 96: 2574–2583. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phenotypic analysis of NDGs. NDGs were isolated from the blood of HIV+ patients with CD4+ T cell counts >350 (n = 11) or <350 cells/µL (n = 10) as described in materials and methods and the expression levels of phenotypic markers were determined by flow cytometry. Isotype controls: <1%. Statistical significance was determined by a two-tailed Mann-Whitney test. Box = interquartile range and median; whiskers = range.
(TIF)