Abstract
Customized DNA-binding domains made using Transcription Activator-Like Effector (TALE) repeats are rapidly growing in importance as widely applicable research tools. TALE nucleases (TALENs), composed of an engineered array of TALE repeats fused to the FokI nuclease domain, have been used successfully for directed genome editing in multiple different organisms and cell types. TALE transcription factors (TALE-TFs), consisting of engineered TALE repeat arrays linked to a transcriptional regulatory domain, have been used to up- or down-regulate expression of endogenous genes in human cells and plants. Here we describe a detailed protocol for practicing the recently described Fast Ligation-based Automatable Solid-phase High-throughput (FLASH) assembly method. FLASH enables automated high-throughput construction of engineered TALE repeats using an automated liquid handling robot or manually using a multi-channel pipet. With the automated version of FLASH, a single researcher can construct up to 96 DNA fragments encoding various length TALE repeat arrays in one day and then clone these to construct sequence-verified TALEN or TALE-TF expression plasmids in one week or less. Plas-mids required to practice FLASH are available by request from the Joung Lab (http://www.jounglab.org/). We also describe here improvements to the Zinc Finger and TALE Targeter (ZiFiT Targeter) webserver (http://ZiFiTBeta.partners.org) that facilitate the design and construction of FLASH TALE repeat arrays in high-throughput.
Keywords: FLASH, TALEN, TALENs, engineered TAL effector nucleases, engineered TALE nucleases, REAL, REAL-Fast, protein engineering, DNA-binding domains, TALE-activators, TALE-TFs, TALE transcription factors
Introduction
Transcription activator-like effectors (TALEs) are DNA-binding proteins encoded by Xanthomonas bacteria that hijack host plant transcription machinery by mimicking eukaryotic transcription factors (Bogdanove and Voytas, 2011; Scholze and Boch, 2011). TALEs bind to DNA using small highly conserved repeat domains that are typically 33 to 35 amino acids in length. Each TALE repeat specifies one nucleotide of DNA with binding specificity dictated by the identities of residues at positions 12 and 13 of the repeat. Repeats have been described that recognize each of the four possible DNA nucleotides (Boch et al., 2009; Moscou and Bogdanove, 2009). Engineered TALE repeat arrays can be designed to recognize target sequences of interest by varying the number and composition of repeats. In the last few years, multiple groups have described the fusion of designer TALEs to various functional domains including nucleases (Briggs et al., 2012; Cade et al., 2012; Cermak et al., 2011; Christian et al., 2010; Hockemeyer et al., 2011; Huang et al., 2011; Li et al., 2011a; Li et al., 2011b; Liu et al., 2012; Mahfouz et al., 2011; Miller et al., 2011; Moore et al., 2012; Mussolino et al., 2011; Sander et al., 2011; Sun et al., 2012; Tesson et al., 2011; Wang et al., 2012; Watanabe et al., 2012; Wood et al., 2011), transcriptional activation domains (Blount et al., 2012; Bultmann et al., 2012; Cermak et al., 2011; Cong et al., 2012; Garg et al., 2012; Geissler et al., 2011; Miller et al., 2011; Morbitzer et al., 2011; Morbitzer et al., 2010; Tremblay et al., 2012; Wang et al., 2012; Weber et al., 2011; Zhang et al., 2011), and transcriptional repressor domains (Cong et al., 2012; Mahfouz et al., 2012).
TALE nucleases (TALENs), fusions of an engineered TALE repeat array to the non-specific FokI endonuclease domain, can be used to introduce precise double stranded breaks (DSBs) within the genomes of living cells or organisms. These targeted DSBs are repaired by either non-homologous end joining, resulting in the creation of insertion or deletion mutations that begin at the site of the break, or by homology-directed repair with a homologous “donor template”, which can be used to introduce single nucleotide alterations or larger insertions. To date, multiple groups have successfully used TALENs to modify the genomes of various organisms or cell types including yeast (Li et al., 2011b), fruit fly (Liu et al., 2012), zebrafish (Cade et al., 2012; Dahlem et al., 2012; Huang et al., 2011; Moore et al., 2012; Sander et al., 2011), nematode (Wood et al., 2011), cricket (Watanabe et al., 2012), rat (Tesson et al., 2011), plant (Cermak et al., 2011; Li et al., 2012), and various human cells (Hockemeyer et al., 2011; Miller et al., 2011; Mussolino et al., 2011; Reyon et al., 2012b).
TALEs can also be fused to activator or repressor domains to generate engineered TALE-transcription factors (TALE-TFs) capable of up- or down-regulating transcription of target endogenous genes. The ability of TALE-TFs to modulate expression provides a useful tool for studying the function of individual genes, for probing larger systems such as gene networks, and for creating synthetic circuits or networks. TALE repeat arrays have been fused to various transcriptional regulatory domains including the NF-KB p65, VP16, and VP64 domains for activation and the KRAB and SID domains for repression. TALE-TFs have been used successfully to alter the expression of target endogenous genes in plants (Cermak et al., 2011; Mahfouz et al., 2012), mouse cells (Bultmann et al., 2012), and human cells (Cong et al., 2012; Garg et al., 2012; Geissler et al., 2011; Miller et al., 2011; Tremblay et al., 2012; Wang et al., 2012; Zhang et al., 2011).
Here we describe a detailed protocol for making highly active TALENs and TALE-TFs using our recently described Fast Ligation-based Automatable High-throughput (FLASH) assembly method (Reyon et al., 2012b). FLASH enables high-throughput construction of TALE repeat arrays by assembling DNA fragments encoding these proteins on a solid-phase surface. Following assembly, DNA fragments are cleaved from the solid-phase support and cloned into expression vectors that contain additional TALE-derived sequences required for DNA-binding and a functional domain of choice. FLASH can be performed either manually, using a multichannel pipet, or in automated fashion using a liquid-handling robotic workstation. In a single day, one researcher can build DNA fragments for 24 or 96 TALE repeat arrays using manual or automated FLASH, respectively. These fragments can then be cloned into expression vectors and sequence-verified in one week or less.
To facilitate the use of FLASH for high-throughput assembly of TALE repeat arrays, we have added upgraded features to our web-based Zinc Finger and TALE (ZiFiT) Targeter software (http://ZiFitBeta.partners.org). These improvements enable users to find potential target sites within their sequences of interest in batch mode. After target sites are identified, users can export this data to a file together with details about the plasmids required for the construction of large numbers of TALEs using FLASH.
Strategic Planning
The use of FLASH to build TALE-based fusion proteins can be conceptually divided into three Basic Protocols: (1) identification of target sites and specific plasmids needed for construction of TALE repeat arrays that bind those sites using the web-based ZiFiT Targeter software (Sander et al., 2010); (2) construction of DNA fragments encoding TALE repeat arrays on a solid-phase support using serial digestions and ligation steps; and (3) cloning of the assembled DNA fragments encoding TALE repeat arrays into an expression vector of choice followed by sequence verification. Protocol (2) can be performed either with a liquid handling robot or manually with a multi-channel pipet.
Basic Protocol 1: Identification of target sites using ZiFiT Targeter Software
To facilitate the design of TALE repeat arrays in high-throughput, we have updated our web-based ZiFiT Targeter software (Sander et al., 2010) to enable users to submit queries in batch mode. Up to 96 query sequences in FASTA format can be uploaded into the program and then ZiFiT Targeter will identify and return a potential target site for each of the query sequences that users can download in CSV format. For all the target sites identified, ZiFiT Targeter also returns as part of the CSV output: (1) the names of specific plasmids from the archive of pre-assembled FLASH units required for assembly of TALE repeat arrays that bind the target sites and (2) the DNA sequence of the TALE repeat array constructs that will be assembled. As in previous versions of ZiFiT Targeter, users seeking to design TALENs can indicate the specific nucleotide targeted for sequence alteration by enclosing it in brackets. ZiFiT Targeter will attempt to design TALEN target sites such that the bracketed nucleotide falls within the spacer region where the FokI nuclease dimerizes and cleaves. If ZiFiT Targeter is unable to find an ideal TALEN target site, the brackets are ignored and the search is performed again. If ZiFiT Targeter is still unable to find a target site on the query sequence, design constraints based on our recent large-scale test of FLASH-assembled TALENs are relaxed and the search is performed again (these constraints limit spacer length to 16 to 18 base pairs and TALEN length to 14.5 to 16.5 TALE repeats). For TALE-TFs, by default, we identify target sites that are 20 base pairs in length because we have shown that TALE-TFs of length 18.5 function robustly in human cells (Maeder et al., manuscript in preparation). Users can alter or relax this constraint by entering any desired length in the textbox labeled “Length”.
Materials
ZiFiT web-based software (available at http://ZiFiTBeta.partners.org)
Repeat Masker web-based software (available at http://www.repeatmasker.org)
Genomic sequence of locus to be targeted.
Equipment
Computer with internet browser and internet connection
Protocol Steps
Visit the ZiFiT Targeter website at http://ZiFitBeta.partners.org.
Click on the ZiFiT option on the top menu. To design TALENs, click on the “Design TALE Nucleases (FLASH)” option under the “Nuclease Assembly” menu. Alternatively, click on the “Design TALE-TFs (FLASH)” under the “Monomer Array Assembly” to design TALE-TFs.
-
Paste up to 96 genomic DNA sequences in FASTA format into the text box labeled ‘Sequence’. Users scanning for TALEN targets can bracket a single nucleotide if they wish to identify TALEN target sites such that the spacer region encompasses that nucleotide. If a specific nucleotide is not selected ZiFiT will identify the first available target site as it scans the sequence from left to right. All characters other than [, ], A, C, G and T are ignored. When scanning for transcription factor targets, users can manually select the length of the target (the default is 20 bps).
Before using ZiFiT to identify TALEN target sites, users are strongly encouraged to ensure that there are no repetitive elements within the sequence of interest using the RepeatMasker webserver (http://www.repeatmasker.org).It is critical to ensure that the sequence entered is genomic DNA sequence. Use of non-genomic sequence such as cDNA sequence could result in the identification of targets from splice junctions not actually present in the genome.We strongly encourage users to sequence the region of genome to be targeted directly from the organism or cell type of interest to ensure that there are no unexpected polymorphisms present that might affect TALE binding. -
Click the “Submit” button to retrieve a tabular view of targets for the user-supplied FASTA sequences. For TALENs, the output table has 5 columns comprised of the following fields: 1) FASTA ID 2) left target site 3) spacer 4) right target site 5) ordered list of plasmid IDs required for TALEN assembly. For TALE-TFs, the output table has 3 columns with the following fields: 1) FASTA ID 2) target site 3) ordered list of plasmid IDs required for assembly of the TALE-TF.
By default, ZiFiT Targeter will scan for TALE-TFs sites of length 18 and TALEN sites consisting of a spacer 16–18 base pairs in length that lies between TALEN monomer half-sites 16–18 base pairs in length (i.e. targets for TALENs composed of 14.5–16.5 TALE repeats). TALEN target sites meeting these criteria are given priority because our recent large-scale test of FLASH-assembled TALENs demonstrated that following these criteria leads to nucleases with high activities and minimizes toxicities. Similarly, in a large scale analysis of TALE-TFs, we showed that TALE-TFs recognizing 20bp functioned robustly as activators. Click the “Save to CSV” button to export the data in the table and the corresponding DNA sequence of the assembled TALENs to a .csv text file.
Basic Protocol 2: FLASH assembly of DNA encoding TALE repeat arrays
As with our previously described REAL and REAL-Fast methods, FLASH assembly involves serial digestions and ligation steps (Figure 1). In this updated protocol, we now assemble DNA fragments in streptavidin-coated 96-well plates instead of on magnetic beads, resulting in improved yields for product. In addition, we have also added a “capping” step in which a hairpin oligo is added to block any ends that fail to undergo ligation in the previous ligation cycle.
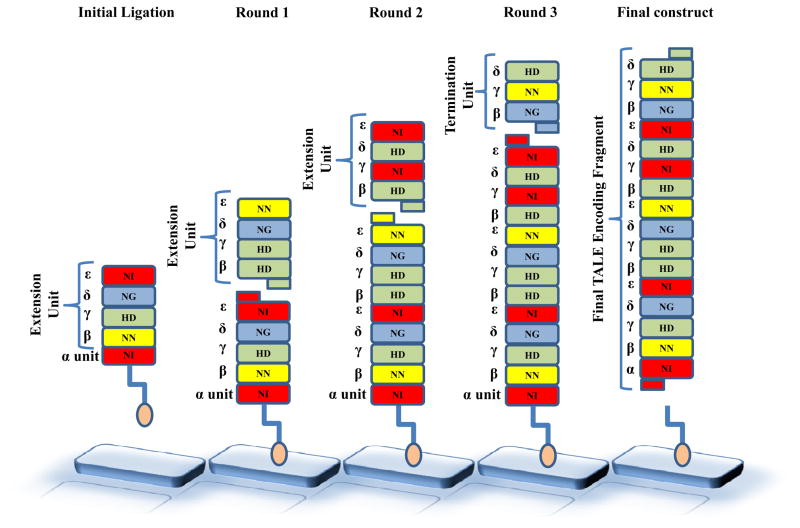
Figure 1. Schematic overview of FLASH assembly.
Assembly of a DNA fragment encoding a 16 TALE repeats is shown. A biotinylated repeat unit (α unit) is first ligated to a predigested extension unit (βγδε unit) in solution and then attached to a streptavidin coated plate. This fragment is uncapped using a restriction digest and the second extension unit (βγδε unit) is ligated. This process is repeated to extend the fragment to a length of 13 repeat arrays. Finally, a termination unit (βγδ unit) is attached and the full length fragment is cleaved away from the Biotin. This fragment is column purified and cloned into an expression vector of choice.
As previously described, TALE repeat arrays constructed by FLASH consist of an iterative pattern of repeats that differ slightly in their amino acid sequences at positions other than the specificity-determining hypervariable residues. The first amino-terminal repeat in each array is encoded on a specific framework and we refer to these DNAs as “α units” (Figure 1). The α unit is then followed by repeats on the β, γ, δ, and ε frameworks, which we refer to collectively as an “extension unit” (Figure 1). Additional extension units (each comprising four repeats) are then added serially resulting in continuation of the pattern β, γ, δ, ε (Figure 1). (Note that the α and ε frameworks are nearly identical, differing only slightly in sequence at the 5′ end to enable restriction digest of that end of the α unit when releasing the final assembled fragment from the solid support). The final ligation is performed with a “termination unit”. If the final carboxy-terminal repeat in the array is a β or δ then the termination units will consist of a single β repeat or a triplet of βγδ repeats, respectively. If the final carboxy-terminal repeat in the array is a γ or ε then repeats on the slightly variant frameworks known as γ* or ε* must be used and therefore termination units will consist of double βγ* or δε* repeats; these variant framework repeats have slight differences in DNA sequence that provide a unique cloning site at the 3′ end of the assembled DNA fragment. (Note that use of the δε* termination unit requires ligation of a βγ extension unit in the prior ligation cycle to maintain the iterative pattern of frameworks within the array.) We have assembled an archive of 376 plasmids encoding α units, βγδε and βγ extension units, and β, βγδ, βγ*, and δε* termination units of all possible combinations of TALE repeats harboring NN, NI, HD, or NG at the hypervariable positions.
Because FLASH assembles DNA fragments on a solid-phase support, the iterative digestion and ligation process can be readily automated. Furthermore, DNA fragments encoding large numbers of TALE repeats can be made without the need to passage intermediate constructs through bacteria (as is the case for nearly all other methods published to date). FLASH can also be practiced in medium-throughput using a multichannel pipet. When assembly is completed, DNA fragments are released from the solid-phase surface by restriction digest, yielding a product that can be ligated into an expression vector of interest.
Materials
Digested α unit (See Support Protocol 2-A)
Digested FLASH extension units (encoding two or four repeats) and termination units (encoding one, two, or three TALE repeats) (See Support Protocol 2-B)
2X B&W buffer (see Solutions and Master Mixes below)
100X Bovine Serum Albumin (BSA) (10 mg/ml) (New England Biolabs [NEB] cat. no. B9001S)
-
Restriction enzymes from NEB
BbsI (cat. no. R0539L)
BsaI-HF (cat, no. R3535L)
-
10X buffers (included with enzymes from NEB)
NEBuffer 2
NEBuffer 4
T4 DNA Ligase (NEB cat. no. M0202L)
Quick Ligation Buffer (See Solutions and Master Mixes below)
MinElute® PCR Purification Kit (QIAGEN cat. no. 28006)
Oligo oDR49: 5′ CTTAGGACGCTGGCGCGGGTTTTCCCGCGCCAGCGTCC 3′
StrepMax Streptavidin-Coated Plates (Thermo Scientific cat. no. AB-1226)
Solutions and Master Mixes
2X B&W Buffer
10.0 mM Tris-HCl (pH 7.5)
1.0 mM EDTA
2.0 M NaCl
2X Quick Ligation Reaction Buffer
132 mM Tris-HCL
20 mM MgCl2
2 mM dithiothreitol (DTT)
2 mM ATP
15% Polyethylene glycol (PEG 6000)
pH 7.6 @25°C
STE Buffer
10 mM Tris pH 8.0
50 mM NaCl
1 mM EDTA
1X BSA (diluted in Nuclease-free water (Unit 4.1))
α unit master mix
-
Combine the following sterile solutions to prepare 31.5 μl of α unit master mix for use in a single reaction in step 2 of Basic Protocol 2:
Purified and digested α unit fragment (See Support Protocol 2-A) 2 μl 2X QLB buffer 27 μl T4 DNA Ligase (400 U/μl) 2.5 μl
BsaI-HF master mix (50 μl)
-
Combine the following sterile reagents to prepare 50 μl of BsaI-HF master mix for a single reaction in steps 6 and 12 of Basic Protocol 2:
NEBuffer 4 5 μl BsaI-HF (20 U/μl) 2 μl H2O 43 μl
BbsI master mix
-
Combine the following sterile reagents to prepare 50 μl of BbsI master mix for a single reaction in step 13 of Basic Protocol 2:
NEBuffer 2 5 μl BbsI (5 U/μl) 5 μl H2O 40 μl
Ligase master mix (27.5 μl)
-
Combine the following sterile reagents to prepare 27.5 μl of Ligase mater mix for a single reaction in step 8 of Basic Protocol 2:
2X QLB buffer 25 μl T4 DNA ligase (400 U/μl) 2.5 μl
Capping Oligo
Dissolve capping oligo oDR49 in STE Buffer to a concentration of 1000 ng/μl, heat to 94°C followed by rapid cooling down to 0°C. Dilute with Nuclease-free water (Unit 4.1) to a final concentration of 100 ng/μl
Equipment
Tabletop Centrifuge
For automated assembly: SciClone G3 (Caliper) or comparable liquid handling system
Multi-channel (12-channel) pipet (20–200 μl, 10–100 μl)
96-well PCR thermocycler
Orbital platform shaker with adjustable speed
Sterile bacterial culture tubes
1.5ml microcentrifuge tubes
Protocol Steps
-
Prepare the FLASH reagents identified by ZiFiT Targeter according to Support Protocol 2-A: Preparation of α unit for TALEN assembly and Support Protocol 2-B: Preparation of Extension and Termination Unit DNA for FLASH Assembly.
Support protocol 2-A typically generates enough α unit for 25 reactions. Support protocol 2-B typically generates enough extension unit or termination unit to use 8 times. Because the amount of each sample you use can vary greatly between runs, it is important to estimate the amount of each α-unit and extension and termination units that you will need before starting each TALEN assembly. -
Set up initial ligations (in 300 μl wells of a 96-well PCR plate) by mixing the following components and incubating for 10 minutes at room temperature:
α unit master mix 31.5 μl Digested extension unit 22.5 μl Cap the α units that did not ligate to the extension units by adding 200 ng of phosphorylated capping oligo and incubating for 5 minutes.
Bind the initial ligation products to wells in a streptavidin-coated 96-well plate by adding 54 μl 2X B&W buffer to the initial ligation reaction from the previous step and adding the mixture into a well on the plate. Incubate for 15 min at room temperature.
Aspirate the supernatant and prepare plate-bound ligation products for BsaI-HF digest by washing twice with 200 μl of 1X BSA.
Uncap the 3′ end of the plate-bound DNA fragments by adding 50 μl of BsaI-HF master mix and incubating at 50°C for 15 minutes.
Prepare the plate-bound fragment for ligation of the next extension unit by washing with 200 μl of 1X B&W buffer, followed by 2 washes with 200 μl of 1X BSA.
-
Ligate the next extension unit as follows:
Ligase master mix 27.5 μl Digested extension unit 22.5 μl (See Support Protocol 2-B) Incubate the preceding reaction at room temperature for 10 minutes. Cap the DNA fragments that did not ligate the previously added extension unit by adding 200ng of capping oligo and incubating for 5 min.
Prepare the construct for BsaI-HF digesting by aspirating the supernatant and washing with 200 μl of 1X B&W buffer, followed by 2 washes with 200 μl of 1X BSA.
Repeat steps 6 to 10 of Basic Protocol 2 to extend the TALE array construct.
Uncap the final extension unit by adding 50 μl of BsaI-HF master mix and incubating for 15 min at 50°C
Prepare the construct for ligation of termination unit by aspirating supernatant and washing with 200 μl of 1X B&W buffer, followed by 2 washes with 200 μl of 1X BSA.
-
Ligate the termination unit as follows:
Ligase master mix 27.5 μl Digested termination unit 22.5 μl (See Support Protocol 2-B) Incubate the preceding reaction at room temperature for 10 minutes. Prepare the construct for the final BsaI-HF digestion by aspirating supernatant and washing with 200 μl of 1X B&W buffer, followed by 2 washes with 200 μl of 1X BSA.
Perform the final BsaI-HF digestion to create compatible ends for cloning of the fragment into the expression vector by adding 50 μl of the BsaI-HF master mix and incubating at 50°C for 15 min.
Prepare constructs for final BbsI digest to release the fragments from the plate by aspirating the supernatant and washing with 200 μl of 1X B&W buffer followed by two washes with 200 μl of 1X BSA.
Release DNA fragments encoding the full-length TALE repeat array from the streptavidin coated well by adding 50 μl of BbsI master mix and incubating for 2 hours at 37°C.
Purify TALE constructs using MinElute® PCR Purification Kit according to manufacturer’s instructions and elute in 20 μl of 0.1X EB.
Support Protocol 2 - A: Preparation of α unit for use in FLASH assembly
With FLASH, an initial DNA fragment encoding an α unit ligated to a βγδε extension unit is anchored to a streptavidin-coated plate well and all subsequent digestion and ligation steps are performed on this solid-phase support. The DNA fragment encoding the α unit has a 5′ biotin, which binds to streptavidin with sub-picomolar affinity (Kuntz et al., 1999). This initial α unit is first ligated to a βγδε extension unit in solution. Once this ligated product is anchored, a series of iterative digestions and ligations can be safely performed to elongate the fragment that is eventually cleaved from the streptavidin surface using a BbsI digest (Basic Protocol 2: Step 14). The biotin is added onto the α unit DNA fragment when it is amplified by PCR (Support Protocol 2-A: Step 1). Following the purification (Support Protocol 2-A: Step 2), the α-unit is uncapped using a BsaI-HF digest (Support Protocol 2-A: Steps 3 and 4). To ensure that the digested a-unit is not in molar excess compared to the digested extension units in the initial ligation step (Basic Protocol 2: Step 2) we normalize the BsaI-HF digested α-unit to a concentration of 15 ng/μl. We note that the α unit is the only DNA fragment within the FLASH protocol that undergoes PCR amplification so users must be aware of the potential for the introduction PCR-induced errors and must pay special attention to the sequence of the α unit when verifying the final construct (Basic Protocol 3: Step 9).
MATERIALS
Plasmids encoding α units from the archive of plasmids required to practice FLASH (available from the Joung lab; see http://www.talengineering.org)
Biotinylated forward PCR primer oJS2581: 5′ Biotin–TCTAGAGAAGACAAGAACCTGACC – 3′
Reverse PCR primer oJS2582: 5′-GGATCCGGTCTCTTAAGGCCGTGG-3′
BamHI-HF (NEB cat. no. R3136L)
10X NEBuffer 4 (included with BamHI-HF from NEB)
QIAquick® PCR Purification Kit (QIAGEN cat. no. 28106)
Expand High-Fidelity PCR System kit (Roche cat. no. 11732641001)
Nuclease-free water (Unit 4.1)
Equipment
Thermocycler
1.5ml Microcentrifuge Tubes
Protocol Steps
-
Amplify DNA encoding an α unit using a biotinylated forward PCR primer and standard revers primer as follows:
dNTPs (10mM; 2.5mM each) 4 μl Expand Buffer 2 5 μl Plasmid encoding α-unit (1 ng/μl) 2 μl Expand Enzyme (3.5 U/μl) 1 μl Biotinylated forward primer (10 pmol/μl) 2 μl Reverse primer (10 pmol/μl) 2 μl Nuclease free H20 (Unit 4.1) 34 μl Cycling conditions:

Purify PCR reaction from the previous step using a QIAquick PCR Purification Kit according to the manufacturer’s instructions and elute in 40 μl 0.1 X EB buffer.
-
Digest the purified PCR product from the previous step with BsaI-HF enzymes by adding the following and incubating for 15 minutes at 50°C:
NEBuffer 4 5 μl BsaI-HF (20 U/μl) 5 μl α-unit PCR product 40 μl Purify digested DNA from the previous step using a QIAquick PCR Purification Kit according to the manufacturer’s instructions and elute in 40 μl 0.1X EB
Dilute digested α-unit in nuclease free water to a concentration of 15ng/μl.
Support Protocol 2 - B: Preparation of Extension and Termination Unit DNA for FLASH Assembly
The FLASH protocol involves the iterative addition of DNA fragments encoding TALE repeat domains, via a series of digestion and ligation steps (Basic Protocol 2: Steps 6 through 14), to construct a full length fragment encoding a TALE repeat array of desired specificity. To increase the speed and efficiency of this process, we use pre-assembled “extension units” (encoding two or four TALE repeats) and “termination” units (encoding one, two or three TALE repeats). DNA fragments encoding these extension or termination units are digested out from plasmid vectors. In total, four restriction enzymes are used to accomplish this. Two enzymes (BbsI and BamHI-HF) are used to release the fragment from the plasmid with appropriate sticky-end overhangs for FLASH assembly. The other two enzymes (XbaI and SalI-HF) are used to digest the remaining plasmid backbone so that it has overhangs that are incompatible with the overhangs on the fragment bearing the extension or termination unit. The generation of these incompatible overhangs eliminates the need to gel purify the fragment encoding the extension or termination unit away from the vector backbone. As summarized below, this quadruple digest is done in a two-step process. First, a BbsI digest is performed followed by a triple digest using XbaI, BamHI-HF, and SalI-HF. After these digests are completed, the fragments are cleaned up using a QIAquick PCR purification kit. Typically, this reaction as described provides enough product for use in eight FLASH assembly reactions.
To ensure that we use the same amount of digested elongation and termination units in each round we normalize the purified sequence-verified plasmids encoding the extension and termination units to 200 ng/μl prior to digestion.
MATERIALS
Plasmids encoding extension and termination units from the archive of 376 plasmids required to practice FLASH (available from the Joung lab; see http://www.TALengineering.org)
100X Bovine Serum Albumin (BSA) (10 mg/ml) (NEB cat. no. B9001S)
Restriction enzymes (NEB):
BamHI-HF (cat. no. R3136L)
XbaI (cat. no. R0145L)
BbsI (cat. no. R0539L)
SalI-HF (cat. no. R3138L).
10X restriction enzyme buffers included with enzymes from NEB
NEBuffer 2
NEBuffer 4
QIAquick® PCR Purification Kit (QIAGEN cat. no. 28106)
Equipment
Thermocycler
1.5 ml Microcentrifuge Tubes
Protocol Steps
-
Digest plasmids encoding FLASH extension or termination units with BbsI for two hours at 37°C as follows:
Plasmid (200 ng/μl) 50 μl NEBuffer 2 10 μl BbsI (5 U/μl) 10 μl Nuclease free H2O 30 μl -
Continue digesting the plasmid from the previous step with XbaI, BamHI-HF, SalI-HF by adding the following and incubating for an additional 15 minutes at 37°C:
NEBuffer 4 25 μl 100X BSA 2.5 μl XbaI (20 U/μl) 5 μl BamHI-HF (20 U/μl) 5 μl SalI-HF (20 U/μl) 5 μl Nuclease free H2O 107.5 μl These digestions release small pieces of DNA between both ends of the fragment encoding the TALE array and the plasmid backbone. Purification in the next step removes these two small pieces, preventing the fragment encoding the TALE array from being religated with the backbone and eliminating the need to gel purify the TALE encoding fragment away from the backbone. Purify the digestion from the previous step using a QIAquick PCR Purification Kit according to the manufacturer’s instructions and elute in 182 μl of 0.1X EB buffer.
-
Quantify the purified DNA from the previous step. Typical concentration obtained is ~40 ng/μl. Concentrations much lower than this can result in lower success rates of FLASH assembly performed with these units. Note that this purified preparation is hereafter referred to as an “extension unit” or as the “termination unit”.
The 5′ and 3′ overhangs of a digested termination fragment are different from the 5′ and 3′ overhangs of a digested extension fragment. The 3′ overhangs of extension units are incompatible with cloning into our TALEN or TALE-TF expression vectors. ZiFiT Targeter software automatically identifies which unit is the termination unit and returns the corresponding plasmid ID number to use.
Basic Protocol 3: Cloning and sequence verification of TALEN or TALE-TF expression vectors
FLASH produces DNA fragments encoding TALE repeat arrays that can be directly subcloned into expression vectors. Cloning into these vectors results in a fusion of the custom TALE with functional domains such as the FokI nuclease domain (to express TALENs) or a VP64 or NF-KB p65 transcriptional activation domain (to express TALE-TFs). Because each serial ligation step in the FLASH protocol is not 100% efficient, a population of different size fragments (e.g. - encoding N, N-4, N-8 repeats where N is the desired number of repeats) is produced in each assembly reaction. Therefore, after ligation of FLASH-assembled fragments into an expression vector, we use a colony PCR screening procedure to analyze approximately 6 clones and to identify those that contain the full length fragment (although in our experience, the majority of FLASH assembly reactions yield at least one clone of the correct length). For clones that contain the correct size fragment, we verify the sequence of the region encoding the TALE repeat arrays. A typical TALE repeat array assembled by FLASH will consist of 14.5 to 16.5 or more repeats and is therefore approximately 1500 to 1700 bps or greater in length. Due to the highly repetitive nature of the TALE repeats, accurately sequencing fragments of this length is not straightforward. We have developed a protocol that uses well-validated external primers as well as internal primers that exploit systematic DNA sequence variations within the TALE repeats we use in our architecture to enable routine sequencing of expression vectors. In addition, to help users analyze their raw sequence data, we have also updated ZiFiT Targeter to include new tools for sequence alignment and verification of TALE repeat-encoding DNA.
Materials
Chemically competent bacterial strain XL-1 Blue (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F’ proAB lacIq lacZDM15 Tn10 (TetR)]; Stratagene cat no. 200249)
Carbenicillin (Sigma, cat. No C1389)
LB medium powder (Difco, cat No. 244620)
LB agar medium powder (Difco, cat No 244520)
Quick Ligation Buffer (See recipe)
Sequencing/PCR Primer oSQT1: 5′ AGTAACAGCGGTAGAGGCAG 3′
Sequencing Primer oSQT3: 5′ ATTGGGCTACGATGGACTCC 3′
Sequencing Primer JDS2980: 5′ TTAATTCAATATATTCATGAGGCAC 3′
Sequencing/PCR Primer oSQT38: TTCGGGAATACGGCGATTG
PCR Primer oSQT34: 5′ GACGGTGGCTGTCAAATACCAAGATATG 3′
PCR Primer oSQT35: 5′TCTCCTCCAGTTCACTTTTGACTAGTTGGG 3′
Agencourt® AMPure® XP beads (Agencourt/Beckman Genomics cat. no. A63881)
QIAprep® Spin Miniprep Kit (QIAGEN cat. no. 27106)
Agarose (Genesee cat. no. 20–102)
Phusion® High-Fidelity DNA polymerase (NEB cat. no. M0530L)
T4 DNA Ligase (NEB cat. no. M0202L)
BsmBI (cat. no. R0580L)
NEBuffer 3 (included with enzymes from NEB)
TALEN and/or TALE activator expression vectors available through Addgene (See Table 1)
Table 1.
TALEN and TALE-activator expression vectors
| 0.5 Domain in Vector | p65 Activation vector | VP64 Activation vector | Plasmid name |
|---|---|---|---|
| NI | pMLM2581 | pMLM3367 | pJDS70 |
| HD | pMLM2583 | pMLM3585 | pJDS71 |
| NN | pMLM2585 | pMLM3587 | pJDS74 |
| NG | pMLM2579 | pMLM3583 | pJDS78 |
Equipment
Thermocycler
SPRIplate® 96-Ring magnet
1.5ml microcentrifuge tubes
Gel electrophoresis apparatus
Protocol Steps
-
Digest the TALEN or TALE-activator expression vector harboring the desired carboxy-terminal 0.5 TALE repeat domain (Table 1) with BsmBI restriction enzyme as detailed below. Incubate the reaction at 55°C for 8 hours.
TALEN or TALE-activator Expression Vector 5 μg 10x Buffer (NEB Buffer #3) 5 μl BsmBI (10U/μl) 5 μl Add nuclease-free water to a total volume of 50 μl Isolate the vector backbone from the restriction digest in the previous step using AMPure XP beads for the TALEN backbones or by gel electrophoresis for the TALE-activator backbones. For AMPure purification, add 90 μl of AMPure XP beads to the 50 μl digest and purify according to manufacturer’s instructions. For gel purification, run digests on either an agarose or polyacrylamide gel and isolate 6091bp VP64 backbones or 6739bp p65 backbones.
Dilute the purified digested plasmid from the previous step in nuclease-free H2O to a final concentration of 5 ng/μl for cloning.
-
Ligate the purified DNA fragment isolated in Step 15 of Basic Protocol 2 into the purified TALEN expression vector isolated in the previous step as described below. Also perform a control ligation using vector backbone alone (i.e.--without fragment). Allow the ligation reactions to incubate for 15 minutes at room temperature.
Purified vector backbone (from previous step) 1 μl Purified fragment (Basic Protocol 2: Step 15) or water for control 3 μl Quick Ligase Buffer (NEB) 5 μl T4 DNA Ligase (400U/μl) 1 μl Transform 5 μl of the ligation from previous step into 45 μl chemically competent XL1-Blue cells. Mix ligations with competent cells and leave on ice for 5 minutes. Perform heat shock at 42°C for 1 minute then return transformations to ice for 1 minute. Add 450 μl LB and recover with agitation for 45 minutes at 37°C. Spin cells down, remove 470 μl of media, resuspend the cells in the remaining 30 μl of media, and plate the entire transformation on a 12 well LB agar plate supplemented with 100 μg/ml carbenicillin. Incubate overnight at 37°C for 12 – 16 hours.
-
Provided that the actual ligations yield more colonies than the vector only control, pick six single colonies and resuspend each in 50 μl of ddH20. Use 20 μl of the 50 μl suspension to inoculate 1 ml of LB supplemented with carbenicillin 100 mg/ml and grow overnight with agitation at 37°C. To check which clones have the desired length TALE repeat array-encoding fragments, use 5 μl of the 50 μl cell suspension to set up colony PCR reactions as follows:
5X Phusion HF Buffer 4 μl 10mM dNTPs 0.4 μl oSQT 34 for TALENs or oSQT1 for TALE-activators (5 uM) 1 μl oSQT 35 for TALENs or oSQT38 for TALE-activators(5 uM) 1 μl Suspension of bacterial cells 5 μl Phusion 0.2 μl Nuclease free H20 8.4 μl Cycling conditions:
 To reduce the risk of plasmid deletions, do not allow the cultures to grow for more than 16 hours. Colonies resuspended in 50 μl of ddH20 should be inoculated into media within 30 min because the cells will lyse in the suspension over time.
To reduce the risk of plasmid deletions, do not allow the cultures to grow for more than 16 hours. Colonies resuspended in 50 μl of ddH20 should be inoculated into media within 30 min because the cells will lyse in the suspension over time. Load and run colony PCR reactions on an agarose gel. Clones that have the correct fragment will have a PCR product of length (N*102) + 538 for TALENs or (N*102) + 278 for TALE-activators, where N stands for the number of repeat arrays in the TALE repeat array-encoding fragment being constructed.
Using a QIAprep Spin Miniprep Kit and following the manufacturer’s instructions, isolate plasmid DNA from the overnight cultures started in Step 6 that have fragments of the right length as determined by colony PCR and analysis of product size performed in Step 7.
-
Use the following sequencing primers to verify the sequence of TALEN expression vectors: 1) oSQT1 for the forward read 2) oSQT3 from the reverse read 3) oJS2980 reverse read that includes the 0.5 domain. Use the following sequencing primers to verify the sequence of TALE-activator expression vectors: 1) oSQT1 for forward read, 2) oSQT38 for reverse read that includes the 0.5 domain, 3) depending on the number of TALE repeats, users may need to design internal primers in order to sequence the entire TALE array. These should be designed to prime off of a unique TALE repeat within the array -- recall that four repeats with different hypervariable residues (NN, NG, NI, or HD) are used and these repeats will be on one of the four frameworks (β, γ, δ, or ε) bearing sequence differences at non-hypervariable residues, providing opportunities for unique repeats to exist within any given array. Users can acquire the expected sequence of the TALE-derived domains either from the CSV filed downloaded from ZiFiT in step 5 of Basic Protocol 1 or directly from ZiFiT (ZiFiTBeta.partners.org) by clicking on the “Generate expected nucleotide sequence” under “TALE Sequence Analysis”.
Sequence verified TALEN or TALE-activator constructs can then be prepped for transfection into cell types in which the CMV promoter or EF1alpha promoter, respectively, are active. The TALEN expression vector also has a T7 promoter that can be used to synthesize TALEN-encoding mRNA for use in downstream applications (Sander et al., 2011).
Commentary
Background Information
Potential users of TALE technology should be aware that various architectures have been described in the literature for both TALENs and TALE-TFs. For TALENs, differences exist in the sequences and numbers of TALE repeats used and the length and amino acid composition of additional TALE-derived domains required for DNA-binding that flank the TALE repeat array. For TALE-TFs, variations exist in the same parameters as for TALENs but also in the identity of the transcriptional regulatory domain used. We have recently validated robust architectures for the design of both TALENs and TALE-TFs that mediate gene activation and have performed large-scale validations of proteins made on these particular frameworks (Reyon et al., 2012b) (Maeder et al., manuscript in preparation).
Any of the three assembly methods described by our lab to date – REAL (Reyon et al., 2012a; Sander et al., 2011), REAL-Fast (Reyon et al., 2012a), and FLASH – can be used to build TALENs or TALE-TFs on our robust, well-validated architectures because all three approaches generate TALE repeat arrays of identical DNA and protein sequences. Therefore, the choice of which platform to use should depend upon the number of TALE proteins to be constructed. In general, we recommend that REAL or REAL-Fast should be used if building 12 or fewer TALEs, manual FLASH should be used if building 12 to 24 TALEs, and FLASH should be used if building 24 or more TALEs.
Critical Parameters and Troubleshooting
It is important to ensure that the sequence for which TALE repeat arrays are being designed is genomic DNA sequence and not cDNA. Users are also encouraged to sequence their region of interest from their specific organism or cell type to ensure there no unexpected polymorphisms are present that might hamper binding by the TALEN or TALE-TF. Another important consideration is to ensure that there are no repetitive sequences within the target binding site. Users are encouraged to use the RepeatMasker webserver (www.RepeatMasker.org) to identify and exclude any such repeats.
Because TALE repeat arrays are very repetitive by nature, users should have a heightened awareness for the possibility of recombination within plasmid constructs when analyzing sequencing results. To reduce the probability of a recombination event, we take the following precautions: (1) we use only recA- strains to propagate plasmids, (2) we take care not to grow cultures of bacteria harboring these plasmids for more than 16 hours, and (3) we re-sequence plasmids following any passage through bacteria (including expansion for preparation of midi- or maxi-preps).
Anticipated Results
TALENs and TALE-TF activators built using the FLASH protocol described here have been extensively validated in large-scale experiments and shown to yield proteins that are highly active in human cells (Reyon et al., 2012b) (Maeder et al., manuscript in preparation). TALENs made by FLASH have also been well-validated in zebrafish (Cade et al., 2012; Moore et al., 2012; Sander et al., 2011) and TALENs built on this same architecture have been shown by other groups to be active in C. elegans (Wood et al., 2011), rat (Tesson et al., 2011), and human pluripotent stem cells (Hockemeyer et al., 2011). We emphasize again that the amino acid and DNA sequences of TALE repeat arrays built using FLASH are identical to those made using our previously published REAL (Reyon et al., 2012a; Sander et al., 2011) and REAL-Fast platforms (Reyon et al., 2012a).
Timing Considerations
Once all the required α units, extension units, and termination units have been prepped and digested, users can build and sequence verify TALEN constructs in a week. The timeline can be broken down as follows:
| Time | Procedure |
|---|---|
| Day 1 | Identify target sites using ZiFiT (Basic Protocol 1) |
| Day 1 | Array digested units into plates |
| Day 2 | Build TALE fragment (Basic Protocol 2) |
| Day 3 | Clone TALE fragment (Basic Protocol 3) |
| Day 4 | Start overnights and identify the clones that have fragments of the correct length using colony PCR (Basic Protocol 3) |
| Day 5 | Sequence clones that have the correct length fragments (Basic Protocol 3) |
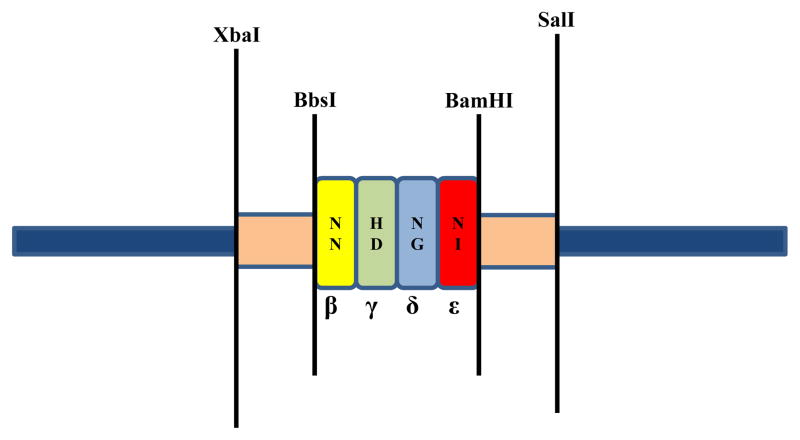
Figure 2. Restriction map of a FLASH plasmid harboring an extension unit.
Extension units are released from their plasmid vectors using a quadruple digest. In the example shown, the extension unit contains coding sequence for four TALE repeats (colored rectangles). Digestion of the plasmid with BbsI and BamHI releases the unit from the plasmid with appropriate overhangs for use in the FLASH assembly method. Digestion of the plasmid with the additional restriction enzymes XbaI and SalI prevents the released unit fragment religating back into the vector, thereby eliminating the need to gel purify the fragment.
Acknowledgments
This work was supported by a National Institutes of Health (NIH) Director’s Pioneer Award DP1 OD006862 (J.K.J.), NIH R01 GM088040 (J.K.J.), the Jim and Ann Orr MGH Research Scholar Award (J.K.J.), NIH T32 CA009216 (J.D.S.), and National Science Foundation DBI-0923827 (D.R. and J.K.J.).
Footnotes
Author Contributions
J.D.S. and J.K.J. conceived of the original FLASH assembly strategy. D.R., M.L.M., C.K., S.Q.T., and J.E.F. experimentally validated and further refined the strategy. D.R. and J.D.S. designed the updated version of the ZiFiT Targeter software. D.R., M.L.M., J.D.S. and J.K.J. wrote the paper.
Competing Financial Interests
J.D.S. and J.K.J. are inventors on a patent application describing the FLASH assembly method. J.K.J. has a financial interest in Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Key References
Reyon et al., 2012. This paper describes the development and validation of the FLASH strategy for assembling TALE repeat arrays and the archive of 376 α units, extension units, and termination units needed to practice the method. It presents large-scale studies demonstrating that FLASH-assembled TALENs have high activities, a robust success rate, and an essentially limitless targeting range in human cells.
Internet Resources
Provides access to the ZiFiT software program for engineering TALENs
http://www.addgene.org/talengineering
TALEN and TALE-activator expression vectors are available through Addgene.
Archive of 376 plasmids required to practice FLASH are available by request from the Joung lab
References
- Blount BA, Weenink T, Vasylechko S, Ellis T. Rational diversification of a promoter providing fine-tuned expression and orthogonal regulation for synthetic biology. PLoS One. 2012;7:e33279. doi: 10.1371/journal.pone.0033279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- Briggs AW, Rios X, Chari R, Yang L, Zhang F, Mali P, Church GM. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultmann S, Morbitzer R, Schmidt CS, Thanisch K, Spada F, Elsaesser J, Lahaye T, Leonhardt H. Targeted transcriptional activation of silent oct4 pluripotency gene by combining designer TALEs and inhibition of epigenetic modifiers. Nucleic Acids Res. 2012;40:5368–5377. doi: 10.1093/nar/gks199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, Joung JK, Sander JD, Peterson RT, Yeh JR. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ. Simple Methods for Generating and Detecting Locus-Specific Mutations Induced with TALENs in the Zebrafish Genome. PLoS Genet. 2012;8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A, Lohmueller JJ, Silver PA, Armel TZ. Engineering synthetic TAL effectors with orthogonal target sites. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler R, Scholze H, Hahn S, Streubel J, Bonas U, Behrens SE, Boch J. Transcriptional activators of human genes with programmable DNA-specificity. PLoS One. 2011;6:e19509. doi: 10.1371/journal.pone.0019509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- Kuntz ID, Chen K, Sharp KA, Kollman PA. The maximal affinity of ligands. Proc Natl Acad Sci U S A. 1999;96:9997–10002. doi: 10.1073/pnas.96.18.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011a;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Huang S, Zhao X, Wright DA, Carpenter S, Spalding MH, Weeks DP, Yang B. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011b;39:6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Liu B, Spalding MH, Weeks DP, Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012;30:390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- Liu J, Li C, Yu Z, Huang P, Wu H, Wei C, Zhu N, Shen Y, Chen Y, Zhang B, Deng WM, Jiao R. Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J Genet Genomics. 2012;39:209–215. doi: 10.1016/j.jgg.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Mahfouz MM, Li L, Piatek M, Fang X, Mansour H, Bangarusamy DK, Zhu JK. Targeted transcriptional repression using a chimeric TALE-SRDX repressor protein. Plant Mol Biol. 2012;78:311–321. doi: 10.1007/s11103-011-9866-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz MM, Li L, Shamimuzzaman M, Wibowo A, Fang X, Zhu JK. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci U S A. 2011;108:2623–2628. doi: 10.1073/pnas.1019533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Moore FE, Reyon D, Sander JD, Martinez SA, Blackburn JS, Khayter C, Ramirez CL, Joung JK, Langenau DM. Improved Somatic Mutagenesis in Zebrafish Using Transcription Activator-Like Effector Nucleases (TALENs) PLoS One. 2012;7:e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morbitzer R, Elsaesser J, Hausner J, Lahaye T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 2011;39:5790–5799. doi: 10.1093/nar/gkr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morbitzer R, Romer P, Boch J, Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc Natl Acad Sci U S A. 2010;107:21617–21622. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon D, Khayter C, Regan MR, Joung JK, Sander JD. Engineering Designer Transcription Activator-Like Effector Nucleases (TALENs) Curr Protoc Mol Biol. 2012a doi: 10.1002/0471142727.mb1215s100. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012b;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462–468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholze H, Boch J. TAL effectors are remote controls for gene activation. Curr Opin Microbiol. 2011;14:47–53. doi: 10.1016/j.mib.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Sun N, Liang J, Abil Z, Zhao H. Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Mol Biosyst. 2012;8:1255–1263. doi: 10.1039/c2mb05461b. [DOI] [PubMed] [Google Scholar]
- Tesson L, Usal C, Menoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, Gregory PD, Anegon I, Cost GJ. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- Tremblay JP, Chapdelaine P, Coulombe Z, Rousseau J. TALE proteins induced the expression of the frataxin gene. Hum Gene Ther. 2012 doi: 10.1089/hum.2012.034. [DOI] [PubMed] [Google Scholar]
- Wang Z, Li J, Huang H, Wang G, Jiang M, Yin S, Sun C, Zhang H, Zhuang F, Xi JJ. An Integrated Chip for the High-Throughput Synthesis of Transcription Activator-like Effectors. Angew Chem Int Ed Engl. 2012;51:8505–8508. doi: 10.1002/anie.201203597. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Ochiai H, Sakuma T, Horch HW, Hamaguchi N, Nakamura T, Bando T, Ohuchi H, Yamamoto T, Noji S, Mito T. Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger and TAL effector nucleases. Nat Commun. 2012;3:1017. doi: 10.1038/ncomms2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber E, Gruetzner R, Werner S, Engler C, Marillonnet S. Assembly of designer TAL effectors by Golden Gate cloning. PLoS One. 2011;6:e19722. doi: 10.1371/journal.pone.0019722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Meyer BJ. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]