Abstract
Gastric cancer (GC) remains a major cause of morbidity and mortality worldwide and there is therefore a clear need to search for more sensitive early diagnostic biomarkers. We performed a systematic review of eight published miRNA profiling studies that compared GC tissues with adjacent noncancerous tissues. A miRNA ranking system was used that took the frequency of comparisons, direction of differential expression and total sample size into consideration. We identified five miRNAs that were most consistently reported to be upregulated (miR-21, miR-106b, miR-17, miR-18a and miR-20a) and two miRNAs that were downregulated (miR-378 and miR-638). Six of these were further validated in 32 paired sets of GC and adjacent noncancerous tissue samples using real-time PCR. MiR-21, miR-106b, miR-17, miR-18a and miR-20a were confirmed to be upregulatedin GC tissues, while the expression of miR-378 was decreased. Moreover, we found a significant association between expression levels of miR-21, miR-106b, miR-17, miR-18a and miR-20a and clinicopathological features of GC. These miRNAs may be used for diagnostic and/or prognostic biomarkers for GC and therefore warrant further investigation.
Introduction
Despite a recent decrease in the incidence of gastric cancer (GC) [1], it remains a cause of major morbidity and mortality worldwide, especially in Eastern Asia. A total of one million new cases of GC occurred in 2008, with 738,000 deaths [2]. This accounts for 8% of the total cases of cancer and 10% of total deaths. Although endoscopy can detect the early stages of GC, most cases are still diagnosed at an advanced stage, which results in a poor prognosis [3]. The 5-year survival rate for GC cases with stage II ranges from 30% to 50%, but falls to between 10% and 25% for patients with stage III disease [4]. Although endoscopic techniques are developing rapidly, their value for the early detection of GC is limited due to a lack of sensitivity, high costs and inconvenience. New diagnostic and prognostic biomarkers for GC are therefore urgently required.
MicroRNAs (miRNA) are short noncoding RNA molecules of 19–25 nt. They regulate gene expression at the post-translational level by guiding the RNA-induced silencing complex to miRNA target sites in the 3′ untranslated region of mRNA, leading to mRNA degradation or the inhibition of translation [5]. Previous studies have shown that numerous miRNAs are aberrantly expressed in many kinds of cancers, and miRNA expression profiling has shown certain miRNAs to be associated with tumor development, progression and response to therapy. They are therefore good candidates for using as diagnostic, prognostic and predictive biomarkers [6].
Several studies have been conducted to search for biomarkers by identifying the differential expression of miRNAs between GC tissue samples and corresponding non-tumor gastric tissue from the same patient [7]–[14]. These studies have resulted in the identification of hundreds of differentially expressed miRNAs. However, many of these are likely to be false positives, and only a small fraction could be used as diagnostic or prognostic biomarkers. A logical approach to distinguish important miRNAs from a large number of candidate miRNA lists is to search for the intersection of miRNAs identified in multiple independent studies [15]. Although this method has become increasing popular [15], [16], [17], no published study has identified the intersections of GC-related miRNAs based on a large number of miRNA expression profiling studies.
We conducted this systematic review to identify the most important differentially expressed miRNAs that have been consistently reported in a series of independent miRNA expression profiling studies in GC patients. Moreover, we further validated some of the miRNAs that were most up- or downregulated using real-time PCR in 32 pairs of GC and matched adjacent non-tumor tissue samples.
Materials and Methods
Ethics Statement
The study was approved by the ethics committee of Shanghai Jiaotong University School of Medicine, and written informed consent was obtained from all patients at study entry.
Search Strategy
Potential studies published in English were collected from Medline using the following keywords: ‘miRNA’ OR ‘microRNA’ OR ‘miR’, ‘gastric’ OR ‘stomach’, ‘profiling’ OR ‘microarray’. Lists of references of review articles and original articles were searched manually for additional publications.
Inclusion Criteria of the Literature
For a study to be included in this systematic review, several criteria had to be met: 1) studies had to be miRNA profiling studies in GC patients; 2) studies had to use GC tissues and their corresponding adjacent non-tumor tissues for comparison; 3) methods had to comprise miRNA microarray techniques. Furthermore, only full-text publications in English were included. The profiling studies that used GC cell lines or serum samples from GC patients, those that compared GC biopsies from tumors with different stages of disease, and those that used different miRNA technologies were not included. Review articles were also not included in this systemic review.
Data Extraction and Lists of miRNA
Differentially expressed miRNAs were identified from each included profiling study. Relevant information was determined (i.e., chromosomal location, pre-miRNA length, mature miRNA sequence and potential targets of the miRNAs), and missing information was identified from the miRBase database (www. mirbase.org/) and Pubmed.
Ranking
Each included profiling study [7]–[14] provided a list of differentially expressed miRNAs (Table S1). Griffith and Chan devised a method to rank potential molecular biomarkers for comparison groups [16], [17], which has been used for miRNA profiling studies. For example, Ma et al. [15] identified the intersections of colorectal cancer-related miRNAs based on a large number of miRNA profiling studies. Thus, the criteria for the literature included in this current systematic review were based on those in their reports [15]. MiRNAs were ranked to the criteria in the following order of importance: (i) the miRNA was consistently reported as differentially expressed in a consistent direction of change; (ii) the frequency of the miRNA was reported in the microarray studies; (iii)the total sample size for each consistent reported miRNAs.
Validation of the miRNAs Using Real Time PCR
To validate the profiling results, 32 fresh GC tissues and their paired non-tumor gastric tissues were obtained from the Renji Hospital, affiliated to the Shanghai Jiaotong University School of Medicine. Total RNA was extracted from 32 pairs of matched human GC specimens (including cancer and adjacent noncancerous tissues) using TRIzol reagent (Invitrogen). The RNA concentration and purity was measured using Nanodrop ND-2000, and the ultraviolet absorption measurement method was applied to detect the purity of the RNA, only those A260/A280 located between 1.80–2.00, and A260/A230>1.7, were used for the final experiment, otherwise the RNA must be re- extracted. Reverse transcription from 3 µg RNA was done usingAll-in-OneTM First-Strand cDNA Synthesis Kit(Genecopoeia, Guangzhou, China), according to the manufacturer’s protocol. In brief, the prepared RNA reverse transcription reaction solution was incubated at 37°C for 60 minutes and terminated at 85°C for 5 minutes, and then stored at −20°C for further analysis. Quantitative PCR (qPCR) was performed using an ABI Prism 7900HT Sequence Detection System (Applied Biosystems, USA) with SYBR Premix Ex Taq II (Takara). The primers (miR-21-5p, miR-106b-5p, miR-17-5p, miR-18a-5p, miR-20a-5p and miR-378-5p) including U6 were obtained from Genecopoeia (Guangzhou, China). Quantification was calculated using the 2 −ΔΔCT method and is presented as normalized pattern.
Statistical Analysis
The results were analyzed using SAS 9.2 software (SAS Institute Inc. USA). Data are presented as means ± SD. Student’s t-test was used to compare values between two independent groups.
Results
Literature Selection and Study Characteristics
A total of 104 studies were searched in Pubmed using our search strategy, 73 of which were excluded after screening the titles and abstracts. 23 studies were excluded after reading the full text. Only eight studies were finally included in this systematic review. The detailed study selection was shown in Figure 1. The detailed characteristics of each study are given in Table 1.
Figure 1. Flowchart of the study selection.
Table 1. The characterestics of the eight microRNA profiling studies included in this systemic review.
| Study | Year | Platform | Total samplesize | No. of differentialmiRNAs | Up-regulatedmiRNAs in GC | Down-regulatedmiRNAs in GC |
| Saito et al [7] | 2012 | Toray Industries | 20 | 10 | NR | 10 |
| Wang et al [8] | 2012 | miRCURY LNA microarray platform | 6 | 18 | NR | 18 |
| (Exiqon, Denmark) | ||||||
| Oh et al [9] | 2011 | Agilent Human miRNA microarray(V2) | 80 | 80 | 40 | 40 |
| Ding et al [10] | 2010 | µParaflo™ microfluidic chip | 12 | 15 | 8 | 7 |
| (LC Sciences, TX, USA) | ||||||
| Tchernitsa et al [11] | 2010 | NCODE muti-species microarray probe set | 12 | 22 | 20 | 2 |
| (Invitrogen, CA, USA) | ||||||
| Ueda et al [12] | 2010 | Ohio State University custom microRNA | 320 | 35 | 22 | 13 |
| microarray chip | ||||||
| Yao et al [13] | 2009 | miRCURY LNA array (V 11.0) | 6 | 24 | 22 | 2 |
| Guo et al [14] | 2008 | µParaflo™ microfluidic chip | 6 | 19 | 12 | 7 |
| (LC Sciences, TX, USA) |
Differentially Expressed miRNAs
A total of 223 differentially expressed miRNAs were reported in the eight microarray studies (Differentially expressed miRNAs in each study were detailed in Table S1); 124 were upregulated in GC, and 99 were downregulated. Among the 223 differentially expressed miRNAs, 48 were reported in at least two studies; 39 (81.3%) had a consistent direction and nine (18.7%) had an inconsistent direction of altered expression. Among the former 39, 20 were upregulated in GC, and 19 were downregulated. Three of these miRNAs were reported in five microarray studies (miR-21, miR-106b and miR-378), four were reported in four studies (miR-17, miR-18a, miR-20a and miR-638), and seven were reported in three studies (miR-19a, miR-20b, miR-25, miR-30d, miR-923, miR-375, and miR-148a). Their chromosomal locations, pre-miRNA lengths, mature sequences and the potential targets are listed in Tables 2–4.
Table 2. Up-regulated miRNAs (n = 20) reported in at least two expression profiling studies.
| miRNAsname | Chromosomallocalization | Pre-miRNAlength | Mature sequence | Studies(reference) | Total samplesizes | Potential Target |
| miR-21 | 17q23.1 | 72 | 8-uagcuuaucagacugauguuga - 29 | 5 [9], [10], [11], [12], [14] | 430 | PTEN, VEGF,STAT3,MMP-2 |
| miR-106b | 7q22.1 | 81 | 13-aaaagugcuuacagugcagguag-35 | 5 [10], [11], [12], [13], [14] | 356 | RB1, CDKN1A,IL-10,RUNX1 |
| miR-17 | 13q31.3 | 71 | 14-caaagugcuuacagugcagguag-36 | 4 [9], [12], [13], [14] | 412 | STAT3,ZNFX1,EIF5A2,SMAD12 |
| miR-18a | 13q31.3 | 71 | 6-uaaggugcaucuagugcagauag-28 | 4 [9], [12], [13], [14] | 412 | DICER1,Myc,HIF1A,Eralpha |
| miR-20a | 13q31.3 | 71 | 8-uaaagugcuuauagugcagguag-30 | 4 [9], [10], [12], [14] | 418 | SERF1A,PKD2,SMAD12,TRPV6 |
| miR-19a | 13q31.3 | 71 | 14-aguuuugcauaguugcacuaca-35 | 3 [9], [12], [14] | 406 | TNF-α,SOCS1,CCND1,TGFBR2 |
| miR-20b | Xq26.2 | 71 | 6-caaagugcucauagugcagguag-28 | 3 [10], [12], [14] | 338 | CDKN1A,MYLIP,ESR1,STAT3 |
| miR-25 | 7q22.1 | 84 | 14-aggcggagacuugggcaauug-34 | 3 [10], [11], [12] | 344 | F8A3,FAM47C,FBXO21,LY9 |
| miR-18b | Xq26.2 | 71 | 6-uaaggugcaucuagugcaguuag-28 | 2 [9], [14] | 86 | Eralpha, ESR1 |
| miR-340 | 5q35.3 | 95 | 16-uuauaaagcaaugagacugauu-37 | 2 [13], [14] | 12 | PELI1,PTPDC1,RNF11,SOX4 |
| miR-214 | 1q24.3 | 110 | 30-ugccugucuacacuugcugugc -51 | 2 [9], [11] | 92 | BUB3,RFX3,KLF12,ARHGAP12 |
| miR-224 | Xq28 | 81 | 8-caagucacuagugguuccguu-28 | 2 [9], [12] | 400 | API5L1,RAB9B,CXCR4,CD40 |
| miR-135b | 1q32.1 | 97 | 16-uauggcuuuucauuccuauguga-38 | 2 [9], [12] | 400 | BGLAP,APC,RUNX2,KLF4 |
| miR-34a | 1p36.22 | 110 | 22-uggcagugucuuagcugguugu-43 | 2 [9], [13] | 86 | SIRT1,MYC,HDAC1,LEF1 |
| miR-125b | 11q24.1 | 88 | 15-ucccugagacccuaacuuguga-36 | 2 [9], [11] | 92 | p53,MUC1,VDR,IL-6 |
| miR-27a | 19p13.13 | 78 | 10-agggcuuagcugcuugugagca-31 | 2 [9], [11] | 92 | SGOL1,ZNF135,NPM1,DLEU1 |
| miR-103 | 5q34 | 78 | 48-agcagcauuguacagggcuauga-70 | 2 [9], [11] | 92 | DICER1,AXIN2,HRB,ARIH2 |
| miR-223 | Xq12 | 110 | 26-cguguauuugacaagcugaguu-47 | 2 [9], [13] | 86 | GABRB3,SEMA3A,TRAPPC10 |
| miR-181a-2 | 9q33.3 | 110 | 39-aacauucaacgcugucggugagu-61 | 2 [12], [13] | 326 | p27,CDX2,BCL2,K-RAS,GATA6 |
| miR-92 | 13q31.3 | 78 | 11-agguugggaucgguugcaaugcu-33 | 2 [11], [12] | 332 | PARP2,CXCL9,SIX3,NRP2 |
Table 4. The differentially expressed miRNAs (n = 9) with an inconsistent direction between studies.
| miRNAsname | Chromosomallocalization | Pre-miRNAlength | Mature sequence | Studies(reference) | Direction ofexpression | Totalsamplesizes | Potential Target |
| miR-106a | Xq26.2 | 70 | 13-aaaagugcuuacagugcagguag-35 | 14 | ↑ | 6 | RB1, CDKN1A, E2F1, VEGFA |
| 9 | ↓ | 80 | |||||
| miR-27a | 19p13.13 | 70 | 10 -agggcuuagcugcuugugagca-31 | 9 | ↑ | 80 | CUX1, SMAD12, PIGK, PCDHB4 |
| 8 | ↓ | 6 | |||||
| miR-107 | 10q23.31 | 70 | 50 -agcagcauuguacagggcuauca -72 | 9 | ↑ | 80 | PLAG1, CDK6, BACE1, CRKL |
| 8 | ↓ | 6 | |||||
| miR-200b | 1p36.33 | 94 | 21-caucuuacugggcagcauugga-42 | 9 | ↑ | 80 | ZNF396, RAB22A, FZD1, FLRT3 |
| 8 | ↓ | 6 | |||||
| miR-181c | 19p13.13 | 70 | 27 -aacauucaaccugucggugagu-48 | 12 | ↑ | 320 | IL-2, BCL-2, CDX2, GATA6 |
| 9 | ↓ | 80 | |||||
| miR-345 | 14q32.2 | 98 | 18 -gcugacuccuaguccagggcuc-39 | 12 | ↑ | 320 | ABCC1, NTRK3, CDKN1A |
| 9 | ↓ | 80 | |||||
| miR-29b | 7q32.3 | 80 | 10-gcugguuucauauggugguuuaga-33 | 12 | ↑ | 320 | USP28, LIN9, WDR26, PROS1 |
| 9 | ↓ | 80 | |||||
| miR-451 | 17q11.2 | 72 | 17-aaaccguuaccauuacugaguu-38 | 12 | ↑ | 320 | CXCL16, CDKN2B, PSMB8, MBP |
| 9 | ↓ | 80 | |||||
| miR-222 | Xp11.3 | 110 | 31-cucaguagccaguguagauccu-52 | 11 | ↑ | 12 | PTPN3, BCAR3, ADAM22,CD2AP |
| 9 | ↓ | 80 |
Table 3. Down-regulated miRNAs (n = 20) reported in at least two expression profiling studies.
| miRNAsname | Chromosomallocalization | Pre-miRNAlength | Mature sequence | Studies(reference) | Total samplesizes | Potential Target |
| miR-378 | 5q32 | 66 | 5 -cuccugacuccagguccugugu-26 | 5 [7], [8], [9], [13], [14] | 118 | CDK6, VEGF,POLH,DFFA |
| miR-638 | 19p13.2 | 100 | 16-agggaucgcgggcggguggcggccu-40 | 4 [8], [9], [10], [13] | 104 | NKX2,CCNG2,WDR47,GPR116 |
| miR-30d | 8q24.22 | 70 | 6 -uguaaacauccccgacuggaag-27 | 3 [9], [10], [12] | 412 | RUNX2, TP53,GNAI2 |
| miR-923* | 3 [7], [9], [10] | 112 | ||||
| miR-375 | 2q35 | 64 | 40 -uuuguucguucggcucgcguga-61 | 3 [9], [10], [12] | 412 | JAK2,HuD, N-Cadherin,CASP3 |
| miR-148a | 7p15.2 | 68 | 6 -aaaguucugagacacuccgacu-27 | 3 [9], [11], [12] | 412 | POU2F1,BRWD1,VASH2,GSR |
| miR-31 | 9p21.3 | 70 | 8 -aggcaagaugcuggcauagcu- 28 | 2 [8], [14] | 12 | RHOA,FOXP3,ARPC5,CASR |
| miR-133b | 6p12.2 | 119 | 66-uuugguccccuucaaccagcu -87 | 2 [7], [14] | 26 | CASP9,BCL2L2,IGF1R,PITX3 |
| miR-139 | 11q13.4 | 68 | 7-ucuacagugcacgugucuccagu-29 | 2 [9], [14] | 86 | TMF1,FMR1,EIF4,EBF1 |
| miR-768* | 2 [7], [14] | 26 | ||||
| miR-141 | 12p13.31 | 95 | 17- caucuuccaguacaguguugga-38 | 2 [8], [10] | 18 | CD46,EGFR,DLC1,MYOCD |
| miR-663 | 17q23.2 | 93 | 15-aggcggggcgccgcgggaccgc -36 | 2 [8], [10] | 18 | ZFR2,GRIN2D,PRRT1,SHOX |
| miR-494 | 14q32.31 | 81 | 48-ugaaacauacacgggaaaccuc -69 | 2 [8], [9] | 88 | PTEN,CDK6, |
| miR-623 | 13q32.3 | 98 | 16- aucccuugcaggggcuguugggu-38 | 2 [8], [9] | 88 | IL6ST,AR,ZNF197,ACSM2A |
| miR-30c | 6q13 | 70 | 17-uguaaacauccuacacucucagc -39 | 2 [10], [12] | 332 | MUC17,UBE2I |
| miR-193b | 16p13.12 | 83 | 14 -cgggguuuugagggcgagauga -35 | 2 [7], [9] | 100 | ARL8B,CACS3,PCDH17,CCR1 |
| miR-939 | 8q24.3 | 82 | 15 -uggggagcugaggcucugggggug-38 | 2 [7], [9] | 100 | IL-6, TNF-α |
| miR-29c | 1q32.2 | 88 | 16 -ugaccgauuucuccugguguuc-37 | 2 [7], [12] | 340 | TCEA3,PPP2R5E,HEBP2,MAFA |
| miR-30a | 6q13 | 70 | 6 -uguaaacauccucgacuggaag-27 | 2 [9], [12] | 400 | RUNX2,MET,CDH1,WNT5A |
Human mir-923 appears to be a frgament of the 28S rRNA, so is removed from the microRNA database;
Human mir-768 overlaps an annotated snoRNA, HBII-239. Phylogenetic analysis in all vertebrates supports the snoRNA annotation, with poor conservation of the reported mature miRNA sequence,it is therefore removed from the microRNA database.
Validation of the Selected miRNAs in GC Patients
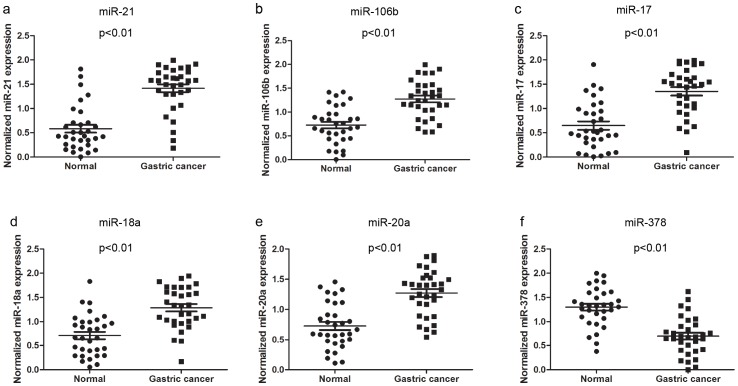
To validate the expression of the six most consistently reported miRNAs (miR-21, miR-106b, miR-17, miR-18a, miR-20a and miR-378), the expression of these miRNAs in GC biopsies and adjacent noncancerous tissues were compared in 32 cases of GC using real-time PCR. The raw Ct values of the six miRNAs were shown in Table S2.The results showed that miR-378 was downregulated in GC tissues, whereas the other five miRNAs (miR-21, miR-106b, miR-17, miR-18a and miR-20a) were upregulated in GC (Figure 2). Our results were consistent with those of the original profiling studies. Furthermore, we explored the relationship between the expression of these miRNAs with the clinical and pathological features of GC. We found that the expression of miR-21 was significantly higher in cases of GC cases with larger tumor sizes (≥8 cm), poor differentiation and metastasis with lymph node involvement and later stage disease. MiR-106b, miR-17 and miR-18a levels were significantly higher in poorly differentiated GC, cases with lymph node involvement, or late stage disease, while miR-20a levels were significantly higher in cases of GC with lymph node involvement. However, no relationship was found between the expression of miR-378 and the clinicopathological features of GC. These results are detailed in Table 5.
Figure 2. Expression levels of miR-21, miR-106b, miR-17, miR-18a, miR-20a and miR-378 in GC and adjacent noncancerous tissue samples.
Using U6 as a normalization control, the expression of miR-21, miR-106b, miR-17, miR-18a and miR-20a was significantly higher in GC tissues, while the expression of miR-378 was significantly lower.
Table 5. The relationship between the six selected microRNAs and gastric cancer characteristics.
| miR-21 | miR-106b | miR-17 | miR-18a | miR-20a | miR-378 | |||||||
| ΔCt | p value | ΔCt | p value | ΔCt | p value | ΔCt | p value | ΔCt | p value | ΔCt | p value | |
| Tumor size | ||||||||||||
| <8 cm | −12.07±0.62 | 0.02 | −1.56±0.40 | 0.48 | −4.67±0.70 | 0.48 | 0.99±0.43 | 0.01 | −4.46±0.59 | 0.41 | 4.06±0.31 | 0.84 |
| ≥8 cm | −14.32±0.63 | −1.94±0.33 | −4.07±0.39 | −0.48±0.34 | −5.09±0.47 | 3.85±0.75 | ||||||
| Location | ||||||||||||
| Cardia | −13.61±0.66 | 0.57 | −0.97±0.50 | 0.18 | −2.68±0.61 | 0.04 | 0.80±0.72 | 0.47 | −4.11±0.66 | 0.42 | 5.22±1.01 | 0.23 |
| Nocardia | −12.89±0.57 | −1.90±0.29 | −4.77±0.45 | 0.21±0.34 | −4.92±0.43 | 3.66±0.57 | ||||||
| Lauren’s classification | ||||||||||||
| Diffuse | −13.37±0.70 | 0.43 | −1.91±0.36 | 0.45 | −4.64±0.50 | 0.49 | 0.11±0.43 | 0.47 | −4.83±0.38 | 0.86 | 3.89±0.70 | 0.58 |
| Intestinal | −12.61±0.65 | −1.50±0.38 | −4.09±0.67 | 0.57±0.46 | −4.69±0.71 | 4.06±0.77 | ||||||
| Differentiation | ||||||||||||
| Well | −11.68±0.68 | 0.02 | −0.84±0.42 | 0.01 | −3.15±0.56 | 0.03 | 1.29±0.38 | 0.02 | −4.12±0.61 | 0.27 | 4.78±0.87 | 0.28 |
| Poor | −13.89±0.59 | −2.18±0.29 | −5.01±0.49 | −0.16±0.39 | −5.05±0.47 | 3.63±0.57 | ||||||
| Metastasis with lymph node | ||||||||||||
| No | −11.18±1.00 | 0.02 | −0.80±0.19 | 0.04 | −5.37±1.30 | <0.001 | 1.14±0.15 | 0.17 | −3.17±0.60 | 0.03 | 5.62±0.98 | 0.07 |
| Yes | −13.72±0.48 | −2.03±0.25 | 0.10±0.31 | 0.11±0.31 | −5.21±0.40 | 3.29±0.55 | ||||||
| pTNM Stage | ||||||||||||
| Early(I+II) | −11.62±0.75 | 0.02 | −0.97±0.45 | 0.03 | 1.35±0.55 | 0.02 | 1.200.49 | 0.03 | −4.22±0.56 | 0.31 | 4.31±0.74 | 0.63 |
| Later(IIII+IV) | −13.82±0.56 | −2.17±0.29 | −0.19±0.33 | −0.18±0.36 | −5.04±0.49 | 3.78±0.89 | ||||||
Discussion
MiRNA microarray studies provide amounts of information, but a common drawback is the lack of consistency among different studies. According to the reports of Griffith et al. and Chan et al. [16], [17], a logical solution to this problem would be to determine the consistency between different studies used different microarray platforms. Several systematic reviews [15]–[17] have used this method to determine differentially expressed genes or miRNAs in various disease states. Applying a similar method, we observed that a total of 48 differentially expressed miRNAs were reported in at least two independent studies among eight GC miRNA profiling studies [7]–[14]. Among these, 39 miRNAs were reported to be altered in a consistent direction, while the findings for nine were inconsistent. Among the 39 miRNAs that had consistent changes, 20 miRNAs were consistently upregulated in GC compared with noncancerous gastric tissue, and 19 were consistently downregulated in GC. We identified the five miRNAs that were most consistently upregulated (miR-21, miR-106a, miR-17, miR-18a and miR-20a) and two most consistently downregulated (miR-378 and miR-638) in at least four profiling studies. Then, we validated these findings using real-time PCR, which further supported the findings of this systematic review. We also determined that the expression of these miRNAs correlated with the clinicopathological features of GC, which suggested that these miRNAs may be useful as biomarkers for GC.
One of the most consistently reported upregulated miRNA in our systematic review was miR-21, which has altered expression and oncogenic activity in different human cancers. Cui et al. [18] showed that the expression of miR-21 was significantly higher in GC tissue compared with adjacent normal tissue. The expression of miR-21 has also been found to be higher in patients with GC with lymph node metastasis than those without, and was also significantly correlated with the histological tumor type and pTNM stage [19], which was validated by our study. Moreover, higher expression levels of miR-21 predicted poor survival in patients with GC [19]. Other studies have found that miR-21 may promote tumor proliferation and invasion in GC by suppressing the expression of PTEN or PDCD4 [20], [21]. Additionally, previous studies have also revealed oncogenic activity of miR-21 in colorectal cancer [22], breast cancer [23] and esophageal cancer [24].
MiR-106b was also consistently reported as an upregulated miRNA in GC tissue by this and previous studies [14], [25]. The high expression of miR-106b has been previously associated with lymph node metastasis [25], [26], and this was validated in our study. Kim et al. [27] found that miR-106b may exert its oncogenic activity by suppressing p21 expression in GC. MiR-106b could induce epithelial-to-mesenchymal transition (EMT) and a tumor initiating cell phenotype in breast cancer by targeting Smad7 and Six1 and activating TGF-β signaling [28]. It may also promote cell proliferation in human hepatocellular carcinoma by downregulating the expression of APC [29].
MiR-17 has known oncogenic activity in humans, and was found to be upregulated in 77.2% of tissue samples of GC compared with adjacent normal gastric tissue. It promotes cell cycle progression and inhibit apoptosis in GC by targeting p21 and p53INP1 (tumor protein p53-induced nuclear protein 1) [30]. Circulating levels of miR-17 are elevated in GC patients, and the concentration of miR-17 is significantly associated with the TNM stage and grade of GC [31]. However, our study found that the expression of miR-17a was higher in the cases of GC without lymph node metastasis than those with lymph node involvement, which may have been a consequence of the small sample size in our study. The high expression of miR-17 is significantly correlated with poor survival outcomes [31]. Previous studies have also found that miR-17 has oncogenic activity in colorectal cancer [32], breast cancer [33] and pancreatic cancer [34].
MiR-18a was found to be upregulated in four studies in this systematic review, and is known to have oncogenic activity in humans. Wu et al. [35] revealed that the expression of miR-18a was significantly upregulated in GC tissue compared with normal gastric tissue, and could directly target PIAS3 (protein inhibitor of activated signal transducer and activator of transcription 3) and was positively correlated with levels of Survivin, Bcl-xl and c-myc. Moreover, the upregulation of miR-18a has been reported in nasopharyngeal carcinoma [36], pancreatic cancer [37], hepatocellular carcinoma [38] and breast cancer [39].
Mir-20a is another miRNA with oncogenic activity, and was found to be upregulated in four studies in this literature. It has been demonstrated that the circulating level of miR-20a is significantly elevated in GC patients compared to healthy controls, and this is significantly associated with the stage and grade of the tumor [31], [40]. Our study also found that miR-20a was significantly elevated in GC tissues and was significantly associated with lymph node metastasis. Moreover, the upregulation of miR-20a has previously been found in cervical cancer, prostate cancer and ovarian cancer, and this miRNA could promote the cell proliferation or invasion of these cancers [41]–[43].
The most consistently downregulated miRNA in this systematic review was miR-378, which was found to be downregulated in five studies. MiR-378 has been demonstrated to have anti-oncogenic activity in humans [44]. The exogenous expression of miR-378 markedly suppresses the proliferation of GC cells by suppressing CDK6 and VEGF signaling [44]. In our study, although we found that the expression of miR-378 was downregulated in GC tissues, no relationship was found between the expression of miR-378 and the clinicopathological features of GC. This may have been due to the small sample size of this study. It is also reported that miR-378 is significantly downregulated in colorectal cancer, and may play an important tumor suppressor role in this cancer [45]. However, other studies have found that miR-378 may have oncogenic activity in other cancer types [46]–[49]. Therefore, the exact role of miR-378 in carcinogenesis needs to be further elucidated.
Furthermore, we also found that some of the candidate miRNAs identified in our study were slso identified as serum biomarkers in various cancers. For example, serum miR-21 was significantly elevated in perioperative serum from adenomas and colorectal cancer (CRC), and was an independent prognostic marker for CRC [50], [51]; Plasma miR-106b, together with miR-20a and miR-221 have the potential as novel biomarkers for early detection of gastric cancer [40]; Circulating miR-17 may used as a novel noninvasive biomarker for nasopharyngeal carcinoma [52], gastric cancer [53] and CRC [54]; Serum miR-18a may be used as a novel biomarker in breast cancer [55], colorectal cancer [56], hepatocellular carcinoma [57], and pancreatic cancer [58]; Circulating miR-378 may be used as a biomarker in renal cell carcinoma [59] and gastric cancer [60]. These studies further confirmed the importance of the indentified miRNAs, and may expand the application scope of these miRNAs.
In conclusion, our systemic review identified five upregulated miRNAs (miR-21, miR-106b, miR-17, miR-18a and miR-20a) and one downregulated miRNA (miR-378) that are potential novel biomarkers for GC. These miRNAs have been shown to have diagnostic and/or prognostic potential for this cancer and warrant further investigation. Further studies that focus on these miRNAs will help to determine a panel of diagnostic and prognostic GC biomarkers with appropriate levels of sensitivity and specificity.
Supporting Information
Differentially expressed miRNAs identified in each included study.
(XLS)
Raw Ct value of the selected miRNAs.
(XLS)
PRISMA Checklist.
(DOC)
Acknowledgments
We thank Miss Wei-Feng Qu for her excellent editorial work.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of Key Program (No. 30830055), the Ministry of Public Health, China (No. 200802094), the Ministry of Education (No. 20090073110077) to Fang JY, and the Doctor Innovation Foundation of Shanghai Jiao-Tong University School Of Medicine (No. BXJ201219) to Wang JL. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bertuccio P, Chatenoud L, Levi F, Praud D, Ferlay J, et al. (2009) Recent patterns in gastric cancer: a global overview. Int J Cancer. 125: 666–673. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Bray F (2011) Center MM, Ferlay J, Ward E, et al (2011) Global cancer statistics. CA Cancer J Clin. 61(2): 69–90. [DOI] [PubMed] [Google Scholar]
- 3. Ang TL, Khor CJ, Gotoda T (2010) Diagnosis and endoscopic resection of early gastric cancer. Singapore Med J 51: 93–100. [PubMed] [Google Scholar]
- 4. Wöhrer SS, Raderer M, Hejna M (2004) Palliative chemotherapy for advanced GC. Ann Oncol 15: 1585–95. [DOI] [PubMed] [Google Scholar]
- 5. Gartel AL, Kandel ES (2008) miRNAs: Little known mediators of oncogenesis. Semin Cancer Biol 18: 103–10. [DOI] [PubMed] [Google Scholar]
- 6. Iorio MV, Croce CM (2012) MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 4(3): 143–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saito Y, Suzuki H, Imaeda H, Matsuzaki J, Hirata K, et al. (2013) The tumor suppressor microRNA-29c is downregulated and restored by celecoxib in human GC cells. Int J Cancer. 132(8): 1751–60. [DOI] [PubMed] [Google Scholar]
- 8. Wang J, Zhang J, Wu J, Luo D, Su K, et al. (2012) MicroRNA-610 inhibits the migration and invasion of GC cells by suppressing the expression of vasodilator-stimulated phosphoprotein. Eur J Cancer. 48(12): 1904–13. [DOI] [PubMed] [Google Scholar]
- 9. Oh HK, Tan AL, Das K, Ooi CH, Deng NT, et al. (2011) Genomic loss of miR-486 regulates tumor progression and the OLFM4 antiapoptotic factor in GC. Clin Cancer Res. 17(9): 2657–67. [DOI] [PubMed] [Google Scholar]
- 10. Ding L, Xu Y, Zhang W, Deng Y, Si M, et al. (2010) MiR-375 frequently downregulated in GC inhibits cell proliferation by targeting JAK2. Cell Res. 20(7): 784–93. [DOI] [PubMed] [Google Scholar]
- 11. Tchernitsa O, Kasajima A, Schäfer R, Kuban RJ, Ungethüm U, et al. (2010) Systematic evaluation of the miRNA-ome and its downstream effects on mRNA expression identifies GC progression. J Pathol. 222(3): 310–9. [DOI] [PubMed] [Google Scholar]
- 12. Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, et al. (2010) Relation between microRNA expression and progression and prognosis of GC: a microRNA expression analysis. Lancet Oncol. 11(2): 136–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yao Y, Suo AL, Li ZF, Liu LY, Tian T, et al. (2009) MicroRNA profiling of human GC. Mol Med Report. 2(6): 963–70. [DOI] [PubMed] [Google Scholar]
- 14. Guo J, Miao Y, Xiao B, Huan R, Jiang Z, et al. (2009) Differential expression of microRNA species in human GC versus non-tumorous tissues. J Gastroenterol Hepatol. 24(4): 652–7. [DOI] [PubMed] [Google Scholar]
- 15. Ma Y, Zhang P, Yang J, Liu Z, Yang Z, et al. (2012) Candidate microRNA biomarkers in human colorectal cancer: systematic review profiling studies and experimental validation. Int J Cancer. 130(9): 2077–87. [DOI] [PubMed] [Google Scholar]
- 16. Chan SK, Griffith OL, Tai IT, Jones SJ (2008) Meta-analysis of colorectal cancer gene expression profiling studies identifies consistently reported candidate biomarkers. Cancer Epidemiol Biomarkers Prev 17: 543–52. [DOI] [PubMed] [Google Scholar]
- 17. Griffith OL, Melck A, Jones SJ, Wiseman SM (2006) Meta-analysis and meta-review of thyroid cancer gene expression profiling studies identifies important diagnostic biomarkers. J Clin Oncol 24: 5043–51. [DOI] [PubMed] [Google Scholar]
- 18.Cui L, Zhang X, Ye G, Zheng T, Song H, et al.. (2013)Gastric juice MicroRNAs as potential biomarkers for the screening of GC. Cancer. doi: 10. 1002/cncr. 27903. PMID: 23335180. [DOI] [PubMed]
- 19. Xu Y, Sun J, Xu J, Li Q, Guo Y, Zhang Q (2012) miR-21 Is a Promising Novel Biomarker for Lymph Node Metastasis in Patients with GC. Gastroenterol Res Pract. 2012: 640168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang BG, Li JF, Yu BQ, Zhu ZG, Liu BY, et al. (2012) microRNA-21 promotes tumor proliferation and invasion in GC by targeting PTEN. Oncol Rep. 27(4): 1019–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cao Z, Yoon JH, Nam SW, Lee JY, Park WS (2012) PDCD4 expression inversely correlated with miR-21 levels in GCs. J Cancer Res Clin Oncol. 138(4): 611–9. [DOI] [PubMed] [Google Scholar]
- 22. Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K (2010) STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 39(4): 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han M, Wang Y, Liu M, Bi X, Bao J, et al. (2012) MiR-21 regulates epithelial-mesenchymal transition phenotype and hypoxia-inducible factor-1α expression in third-sphere forming breast cancer stem cell-like cells. Cancer Sci. 103 (6): 1058–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alder H, Taccioli C, Chen H, Jiang Y, Smalley KJ, et al. (2012) Dysregulation of miR-31 and miR-21 induced by zinc deficiency promotes esophageal cancer. Carcinogenesis. 33(9): 1736–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim BH, Hong SW, Kim A, Choi SH, Yoon SO (2012) Prognostic implications for high expression of oncogenic microRNAs in advanced gastric carcinoma. J Surg Oncol. doi: 10.1002/jso.23271. [DOI] [PubMed]
- 26. Tchernitsa O, Kasajima A, Schäfer R, Kuban RJ, Ungethüm U, et al. (2010) Systematic evaluation of the miRNA-ome and its downstream effects on mRNA expression identifiesGC progression. J Pathol. 222(3): 310–9. [DOI] [PubMed] [Google Scholar]
- 27. Kim YK, Yu J, Han TS, Park SY, Namkoong B, et al. (2009) Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in GC. Nucleic Acids Res. 37(5): 1672–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smith AL, Iwanaga R, Drasin DJ, Micalizzi DS, Vartuli RL, et al. (2012) The miR-106b-25 cluster targets Smad7, activates TGF-β signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 31 (50): 5162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shen G, Jia H, Tai Q, Li Y, Chen D (2013) miR-106b downregulates adenomatous polyposis coli and promotes cell proliferation in human hepatocellular carcinoma. Carcinogenesis. 34(1): 211–9. [DOI] [PubMed] [Google Scholar]
- 30.Wang M, Gu H, Qian H, Zhu W, Zhao C, et al.. (2013) miR-17–5p/20a are important markers for GC and murine double minute 2 participates in their functional regulation. Eur J Cancer. doi:pii: S0959–8049(12)01013–1. [DOI] [PubMed]
- 31. Wang M, Gu H, Wang S, Qian H, Zhu W, et al. (2012) Circulating miR-17–5p and miR-20a: molecular markers for GC. Mol Med Report. 5(6): 1514–20. [DOI] [PubMed] [Google Scholar]
- 32. Ma Y, Zhang P, Wang F, Zhang H, Yang Y, et al. (2012) Elevated oncofoetal miR-17–5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat Commun. 3: 1291. [DOI] [PubMed] [Google Scholar]
- 33. Li H, Bian C, Liao L, Li J, Zhao RC (2011) miR-17–5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res Treat. 126(3): 565–75. [DOI] [PubMed] [Google Scholar]
- 34. Yu J, Ohuchida K, Mizumoto K, Fujita H, Nakata K, et al. (2010) MicroRNA miR-17–5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasion. Cancer Biol Ther. 10(8): 748–57. [DOI] [PubMed] [Google Scholar]
- 35.Wu W, Takanashi M, Borjigin N, Ohno SI, Fujita K, et al.. (2013)MicroRNA-18a modulates STAT3 activity through negative regulation of PIAS3 during gastric adenocarcinoge nesis. Br J Cancer. doi: 10.1038/bjc. 2012. 587. PMID: 23322197. [DOI] [PMC free article] [PubMed]
- 36. Luo Z, Dai Y, Zhang L, Jiang C, Li Z, Yang J, et al. (2013) miR-18a promotes malignant progression by impairing microRNA biogenesis in nasopharyngeal carcinoma. Carcinogenesis. 34(2): 415–25. [DOI] [PubMed] [Google Scholar]
- 37. Morimura R, Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, et al. (2011) Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br J Cancer. 105 (11): 1733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li L, Guo Z, Wang J, Mao Y, Gao Q (2012) Serum miR-18a: a potential marker for hepatitis B virus-related hepatocellular carcinoma screening. Dig Dis Sci. 57(11): 2910–6. [DOI] [PubMed] [Google Scholar]
- 39.Guo L, Zhao Y, Yang S, Cai M, Wu Q, et al.. (2012)Genome-wide screen for aberrantly expressed miRNAs reveals miRNA profile signature in breastcancer. Mol Biol Rep. PMID: 23196705. [DOI] [PubMed]
- 40. Cai H, Yuan Y, Hao YF, Guo TK, Wei X, et al. (2013) Plasma microRNAs serve as novel potential biomarkers for early detection of GC. Med Oncol. 30(1): 452. [DOI] [PubMed] [Google Scholar]
- 41. Kang HW, Wang F, Wei Q, Zhao YF, Liu M, et al. (2012) miR-20a promotes migration and invasion by regulating TNKS2 in human cervical cancer cells. FEBS Lett. 586(6): 897–904. [DOI] [PubMed] [Google Scholar]
- 42. Li X, Pan JH, Song B, Xiong EQ, Chen ZW, et al. (2012) Suppression of CX43 expression by miR-20a in the progression of human prostate cancer. Cancer Biol Ther. 13 (10): 890–8. [DOI] [PubMed] [Google Scholar]
- 43. Fan X, Liu Y, Jiang J, Ma Z, Wu H, et al. (2010) miR-20a promotes proliferation and invasion by targeting APP in human ovarian cancer cells. Acta Biochim Biophys Sin (Shanghai). 42(5): 318–24. [DOI] [PubMed] [Google Scholar]
- 44.Deng H, Guo Y, Song H, Xiao B, Sun W, et al. (2013) MicroRNA-195 and microRNA-378 mediate tumor growth suppression by epigenetical regulation in GC. Gene. doi:pii: S0378–1119(13)00041–3. 10.1016. [DOI] [PubMed]
- 45. Faltejskova P, Svoboda M, Srutova K, Mlcochova J, Besse A, et al. (2012) Identification and functional screening of microRNAs highly deregulated in colorectal cancer. J Cell Mol Med. 16 (11): 2655–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu QP, Xie YZ, Deng Z, Li XM, Yang W, et al. (2012) Ergosterol peroxide isolated from Ganoderma lucidum abolishes microRNA miR-378-mediated tumor cells on chemoresistance. PLoS One. 7(8): e44579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nguyen HC, Xie W, Yang M, Hsieh CL, Drouin S, et al. (2013) Expression differences of circulating microRNAs in metastatic castration resistant prostate cancerand low-risk, localized prostate cancer. Prostate. 73(4): 346–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen LT, Xu SD, Xu H, Zhang JF, Ning JF, et al. (2012) MicroRNA-378 is associated with non-small cell lung cancer brain metastasis by promoting cell migration, invasion and tumor angiogenesis. Med Oncol. 29(3): 1673–80. [DOI] [PubMed] [Google Scholar]
- 49. Eichner LJ, Perry MC, Dufour CR, Bertos N, Park M, et al. (2010) miR-378 mediates metabolic shift in breast cancer cells via the PGC-1β/ERRγ transcriptional pathway. Cell Metab. 12(4): 352–61. [DOI] [PubMed] [Google Scholar]
- 50. Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, et al. (2013) Serum miR-21 as a Diagnostic and Prognostic Biomarker in Colorectal Cancer. J Natl Cancer Inst. 105(12): 849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu GH, Zhou ZG, Chen R, Wang MJ, Zhou B, et al. (2013) Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol. 34(4): 2175–81. [DOI] [PubMed] [Google Scholar]
- 52. Zeng X, Xiang J, Wu M, Xiong W, Tang H, et al. (2012) Circulating miR-17, miR-20a, miR-29c, and miR-223 combined as non-invasive biomarkers in nasopharyngeal carcinoma. PLoS One. 7(10): e46367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang M, Gu H, Wang S, Qian H, Zhu W, et al. (2012) Circulating miR-17–5p and miR-20a: molecular markers for gastric cancer. Mol Med Rep. 5(6): 1514–20. [DOI] [PubMed] [Google Scholar]
- 54. Faltejskova P, Bocanek O, Sachlova M, Svoboda M, Kiss I, et al. (2012) Circulating miR-17–3p, miR-29a, miR-92a and miR-135b in serum: Evidence against their usage as biomarkers in colorectal cancer. Cancer Biomark. 12(4): 199–204. [DOI] [PubMed] [Google Scholar]
- 55. Godfrey AC, Xu Z, Weinberg CR, Getts RC, Wade PA, et al. (2013) Serum microRNA expression as an early marker for breast cancer risk in prospectively collected samples from the Sister Study cohort. Breast Cancer Res. 15(3): R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brunet Vega A, Pericay C, Moya I, Ferrer A, Dotor E, et al. (2013) microRNA expression profile in stage III colorectal cancer: Circulating miR-18a and miR-29a as promising biomarkers. Oncol Rep. 30(1): 320–6. [DOI] [PubMed] [Google Scholar]
- 57. Li L, Guo Z, Wang J, Mao Y, Gao Q (2012) Serum miR-18a: a potential marker for hepatitis B virus-related hepatocellular carcinoma screening. Dig Dis Sci. 57(11): 2910–6. [DOI] [PubMed] [Google Scholar]
- 58. Morimura R, Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, et al. (2011) Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br J Cancer. 105(11): 1733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Redova M, Poprach A, Nekvindova J, Iliev R, Radova L, et al. (2012) Circulating miR-378 and miR-451 in serum are potential biomarkers for renal cell carcinoma. J Transl Med. 10: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu H, Zhu L, Liu B, Yang L, Meng X, et al. (2012) Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett. 316(2): 196–203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially expressed miRNAs identified in each included study.
(XLS)
Raw Ct value of the selected miRNAs.
(XLS)
PRISMA Checklist.
(DOC)