Abstract
American Tegumentary Leishmaniasis is caused by parasites of the genus Leishmania, and causes significant health problems throughout the Americas. In Panama, Leishmania parasites are endemic, causing thousands of new cases every year, mostly of the cutaneous form. In the last years, the burden of the disease has increased, coincident with increasing disturbances in its natural sylvatic environments. The study of genetic variation in parasites is important for a better understanding of the biology, population genetics, and ultimately the evolution and epidemiology of these organisms. Very few attempts have been made to characterize genetic polymorphisms of parasites isolated from Panamanian patients of cutaneous leishmaniasis. Here we present data on the genetic variability of local isolates of Leishmania, as well as specimens from several other species, by means of Amplified Fragment Length Polymorphisms (AFLP), a technique seldom used to study genetic makeup of parasites. We demonstrate that this technique allows detection of very high levels of genetic variability in local isolates of Leishmania panamensis in a highly reproducible manner. The analysis of AFLP fingerprints generated by unique selective primer combinations in L. panamensis suggests a predominant clonal mode of reproduction. Using fluorescently labeled primers, many taxon-specific fragments were identified which may show potential as species diagnostic fragments. The AFLP permitted a high resolution genetic analysis of the Leishmania genus, clearly separating certain groups among L. panamensis specimens and highly related species such as L. panamensis and L. guyanensis. The phylogenetic networks reconstructed from our AFLP data are congruent with established taxonomy for the genus Leishmania, even when using single selective primer combinations. Results of this study demonstrate that AFLP polymorphisms can be informative for genetic characterization in Leishmania parasites, at both intra and inter-specific levels.
Introduction
Leishmaniasis is a neglected tropical disease caused by protozoa of the genus Leishmania, and has a variety of clinical manifestations, ranging from the mild cutaneous to life threatening visceral leishmaniasis [1]. In Panama, cutaneous and muco-cutaneous leishmaniasis are the main manifestations, caused almost exclusively by Leishmania panamensis [2]–[5]. Although some authors have sporadically reported the presence of other species, their clinical relevance has not been demonstrated [5]–[11]. The burden of the disease in the country is on the rise, as growing urban areas get in closer relationship with sylvatic reservoirs [2], [5], [12].
The study of the genetic diversity in Leishmania is very important in order to unveil key aspects of the population genetics and epidemiology of this parasite. The genetic variability of Leishmania parasites has been studied using various genetic marker systems, including both protein based multilocus enzyme electrophoresis (MLEE, reviewed by [13]), and DNA based polymorphism detection tools (PCR – RFLP or DNA sequence typing). Although MLEE has been very useful in the past to study genetic variation in Leishmania, it has important limitations, leading researchers to explore and evaluate DNA sequence based systems as potentially more user-friendly and efficient. Some of the DNA loci studied in Leishmania include the ribosomal internal transcribed spacer (ITS, [14]), gp63 genes [15], and hsp70 genes [16]. These studies have covered several Leishmania species from both Old and New World territories. However, data on the genetic composition of Leishmania species that cause the disease in Panama are scarce and are necessary for a better understanding of key aspects of the biology, genetics and epidemiology of the parasite. Although some attempts have been made previously to characterize the genetic diversity of the Leishmania species causing leishmaniasis in Panama, particularly using kinetoplast based RFLP [17] and ITS sequencing [18], more data are required for a better understanding of the local parasite populations.
Here we have explored amplified fragment length polymorphisms (AFLP) to characterize genetic variability of Leishmania parasites isolated from Panamanian cutaneous Leishmaniasis patients. AFLP is a very useful technique for rapidly visualizing polymorphic DNA fragments from organisms with no previous sequence information [19]. This technique has been shown to be highly reproducible as it combines the specificity of restriction fragment length polymorphisms (RFLP) with the sensitivity of the polymerase chain reaction (PCR). AFLP has been successfully used to study the biology, genetics, ecology, and phylogeny of many organisms [20]. The use of AFLP in Leishmania parasites has been limited, with only a few reports in members of the Leishmania (Leishmania) subgenus [21]–[24]. Here we probed the reproducibility and suitability of the technique for scanning the genome of Leishmania parasites for polymorphisms. We demonstrate that AFLP is very useful for both intra and inter-species genetic analyses.
Materials and Methods
Ethics Statement
This research was approved by the INDICASAT-AIP institutional review board. Although data were analyzed anonymously, written informed consent was obtained from patients before the samples were taken.
Parasites and Culture
Leishmania isolates were obtained from biopsy samples of cutaneous leishmaniasis patients in Panama City and other parts of Panama Province. Reference strains were obtained from several sources, including cryobanks at INDICASAT-AIP, University of Panama, Instituto Conmemorativo Gorgas de Estudios de la Salud, Walter Reed Army and the Leishmania collection at the Instituto Osvaldo Cruz (CLIOC) (Table 1). Primary parasite isolations were done in NNN biphasic medium [25] at room temperature. Promastigotes were then cultured at 25°C in T25 tissue culture flasks containing 10 ml of Schneider’s insect medium (Sigma, USA) plus gentamycin (50 µg/ml) and 20% (v/v) heat-inactivated fetal calf serum.
Table 1. Leishmania specimens used in this study.
| Species | Code | Source and characteristics |
| L. (V.) panamensis | Ps | Reference strain, Centro de Investigación y Diagnóstico de Enfermedades Parasitarias, Facultad de Medicina, Universidad de Panamá (CIDEP) |
| L. (V.) panamensis | P1 to P26 | Field isolates from Leishmania specimen bank, INDICASAT-AIP |
| L. (L.) chagasi | Cha2 | Reference strain, CIDEP |
| L. (L.) chagasi | Cha5 | Reference strain, CIDEP |
| L. (L.) mexicana | Mex | Leishmania Type Culture Collection, Instituto Oswaldo Cruz, IOC-L 561, International Reference MHOM/BZ/1982/BEL21 |
| L. (L.) aristidesi | Ari | Reference strain, CIDEP |
| L. (L.) major | Maj | Strain donated by Dr. M. Bozza, Federal University of Rio de Janeiro. |
| L. (L.) major | Majw1 | MHOM/SA/1991/WR-1088 (1) |
| L. (V.) lainsoni | Lai | IOC-L 1023, MHOM/BR/1981/M6426 |
| L. (V.) braziliensis | Bra | IOC-L 566, MHOM/BR/1975/M2903 |
| L. (V.) guyanensis | Gc | Reference strain, MHOM/BR/1975/M4147 |
| L. (V.) guyanensis | Gf | Reference strain Dr. M. Bozza, Federal University of Rio de Janeiro, IOC/L 0565 |
| L. (V.) guyanensis | Gw3 | MHOM/GF/2010/WR-3017 (1) |
| L. (L.) donovani | Doni | MHOM/SD/75/1246 Kartown (1) |
| L. (L.) donovani | Donw2 | MHOM/IN/2006/WR-2801 (1) |
| L. (L.) donovani | Donw3 | MHOM/SD/1980/Hansen-WR-378 (1) |
| L. (V.) peruviana | Perw2 | MHOM/PE/2005/WR-2771 (1) |
(1) Kindly provided by Dr. C. Spadafora.
DNA Extractions
High molecular weight DNA was extracted from stationary phase promastigote cultures using a salting out procedure as recommended by the manufacturer (Wizard® Genomic DNA purification kit, Promega, USA). DNA aliquots were checked for integrity, digestibility, and absence of nucleases or PCR inhibitors. Species identities were verified by hsp70 PCR – RFLP, using MluI, RsaI, BccI or HaeIII restriction endonucleases [16], [26].
AFLP
Fluorescent AFLP reactions were done using a commercial kit, as recommended by the manufacturer (AFLP Microbial Fingerprinting Kit, P/N 402948, Applied Biosystems, USA). Based on standard primer combination scanning procedures previously performed on a small number of L. panamensis isolates [27], thirteen selective primer combinations were chosen and used at the selective amplification step. Amplification products were detected on an automated sequencer Genetic Analyzer 3130 using GeneScan ROX 500 as internal size standard (Applied Biosystems, USA). Electropherograms were analyzed using GeneMarker software v2.2.0 (SoftGenetics LLC, USA) using the default parameters recommended as optimal for AFLP markers by the manufacturer (http://www.softgenetics.com/GeneMarker.html). Further details of AFLP protocol and parameters used for allele calling are included in Supporting Information. Peak patterns were converted to dominant presence - absence (1-0) matrices. Measures were taken to minimize scoring errors, which included careful examination of each electropherogram to exclude doubtful peaks, manual checking of bin sets defined by the software, setting minimal threshold at 100 relative fluorescent units, and considering only peaks with sizes between 50 and 500 base pairs. Although some preliminary analyses were done on individual matrices (matrices generated by one selective primer combination), a single concatenated matrix was prepared to evaluate the performance of these markers in all tested Leishmania specimens.
Consistency of AFLP profiles was tested using two approaches. First, the reproducibility of the technique was estimated by performing, at one time point, ten full replicates from a single culture of L. panamensis promastigotes (Ps reference isolate), from DNA extraction to allele calling using one primer combination (EcoRI-0/MseI-G). The error rate was then estimated between every possible pair of replicates, using previously described procedures [28]–[29]. In a second approach we wanted to examine the stability of AFLP profiles during in vitro propagation to check whether this procedure could be responsible for the generation of the genetic variability detected. Promastigotes of the same strain were cultured for one year, under the same culture conditions, subculturing twice a week. Genomic DNA was extracted from samples taken every month and AFLP profiles were generated using ten selective primer combinations. The error rate was then calculated for every possible pairwise comparison between time points using the concatenated dataset. In addition, to check if profiles were accumulating changes over time, pairwise errors were calculated between the dataset generated at the first month and those generated at each subsequent time point. In this time course experiment, the estimation of error rates was done considering two types of band mismatches: “unstable mismatches” (UMM; alleles appearing and disappearing sporadically, possibly representing error of the technique) and “stable mismatches” (SMM; alleles which appear or disappear once and stay that way, representing also putative real new polymorphisms).
Data Analysis
A concatenated presence/absence matrix was used to score polymorphism levels, count group-specific fixed, private or fixed-private alleles, and derive a Jaccard distance (Jaccard distance = 1 −Jaccard similarity; [30]). This distance matrix was used for phylogenetic and ordination analyses. Individual matrices, containing only presence/absence data from individual selective primer combinations were used to calculate primer-specific Jaccard distance matrices. These individual distance matrices were tested for concordance by means of Mantel tests [31], as implemented in PAST v2.17b software [32]. The test statistic R ranges from −1 to 1, and statistical significance was estimated by permutation tests.
Relationships among specimens and taxa were studied using distance based methods, as recommended for dominant, anonymous markers. Phylogenetic relationships were explored using phylogenetic networks [33]. Split graphs depicting phylogenetic relationships among specimens were constructed using Jaccard distance data transformation and the Neighbor-Net method [34], as implemented in SplitsTree 4 v4.12.6 [35]. Robustness of clustering was tested by nonparametric bootstrapping (1000 resamplings). As an additional measure of the consistency of clustering, other clustering methods were applied to the same dataset, namely Bio Neighbor Joining [36] and Unweighted Pair Group Method with Arithmetic mean (UPGMA; [37]) using the same software. Additionally, ordination analyses were performed for a better understanding of the multivariate nature of AFLP data in a lower dimensional space. Principal coordinate analysis (PCoA) was applied to the Jaccard distance matrix using FAMD (Fingerprint Analysis with Missing Data, v 1.25, release May 2010) [38].
Results
Here we show results of studying the genetic variation in Leishmania parasites circulating in Panama as depicted by the anonymous, multilocus fingerprinting technique AFLP. We analyzed samples of parasites isolated from local patients suffering cutaneous leishmaniasis as well as several reference strains covering both subgenera, Leishmania (Viannia) and Leishmania (Leishmania). All tested Leishmania specimens, which included specimens of L. panamensis, L. guyanensis, L. braziliensis, L. peruviana, L. lainsoni, L. chagasi, L. major, L. mexicana, L. aristidesi and L. donovani (Table 1), were successfully typed using hsp70 gene PCR – RFLP (data not shown). All Leishmania specimens isolated from local patients turned out to be L. panamensis.
The implementation of the AFLP, using a rather conservative procedure for allele calling, allowed the generation of a significant number of fragments. In total, 2457 loci were scored from the 42 specimens and 13 selective primers. Out of these 2457 loci, 2455 were polymorphic. Many fixed, private and fixed private fragments were observed for different groups of specimens. The group corresponding to L. panamensis specimens presented 1104 fragments, of which 781 were polymorphic, 323 fixed, 294 private and 37 fixed private fragments (Table 2). Although numbers may not be representative due to the small number of specimens tested, the closely related species L. guyanensis showed 739 fragments, of which 375 were polymorphic, 364 were fixed, 103 were private, and 21 were fixed private alleles.
Table 2. Total number of fragments, polymorphisms and taxon-specific fragments detected by AFLP analysis of all Leishmania specimens tested.
| Group specific fixed private alleles (bp) | ||||||
| Code | Selective primer combination (1) | Bands detected inL. panamensis | Polymorphism inL. panamensis (%) | L. panamensis | Subgenus Viannia | Subgenus Leishmania |
| R10 | EcoRI-0/MseI-A | 99 | 89 | 159 | – | – |
| R11 | EcoRI-0/MseI-C | 97 | 59 | 180, 312 | 64, 66, 116, 136, 141, 219, 270, 351, 399 | – |
| R12 | EcoRI-0/MseI-G | 85 | 63 | 65, 183, 200, 341, 355, 373 | 66, 92, 136, 226, 250 | 120 |
| R13 | EcoRI-0/MseI-T | 102 | 74 | 156, 356, 389 | 88, 205, 265 | - |
| S9 | EcoRI-A/MseI-0 | 113 | 73 | 367 | 116, 216, 341 | – |
| S12 | EcoRI-A/MseI-G | 79 | 64 | 67, 141, 154, 239 | 92, 104, 160, 165 | – |
| S13 | EcoRI-A/MseI-T | 78 | 79 | 377, | 216 | 103 |
| T9 | EcoRI-C/MseI-0 | 107 | 56 | 101, 183, 411, | 198, 219, 351 | 121 |
| U9 | EcoRI-G/MseI-0 | 105 | 77 | 74, 85, 133, 134, 252 | 66, 135, 245 | 131, 170 |
| V9 | EcoRI-T/MseI-0 | 75 | 49 | 88, 136, 144, 153, 210, 330 | 76, 99, 106, 162, 180 | – |
| V13 | EcoRI-T/MseI-T | 71 | 78 | 155, 197, 208 | 100, 180 | – |
| W13 | EcoRI-AA/MseI-T | 52 | 86 | – | 87, 128, 203 | – |
| Z12 | EcoRI-AT/MseI-G | 41 | 70 | 170, 211 | 165 | – |
| Total | 1104 | 70 | ||||
(1) EcoRI: 5′-GACTGCGTACCAATTC-3′; MseI: 5′-GATGAGTCCTGAGTAA-3′.
The analysis of the L. panamensis group revealed varied proportions of polymorphisms, ranging from 49% (primer combination V9) to 89% (primer combination R10, Table 2), and 70% when considering all primers. All L. panamensis isolates showed different fingerprints. Several fragments were detected which appeared to be specific for the groups L. (Viannia), L. (Leishmania) and L. panamensis (Table 2).
The pairwise error values obtained from the reproducibility experiment ranged from 0% (identical profiles) to 6.3% (most divergent profiles), with a mean value of 3.1%. When we checked the stability of AFLP profiles in time, the mean estimated pairwise errors were 2.5%, 0.27%, and 2.2% when considering all mismatches, only “stable mismatches”, or only “unstable mismatches”, respectively (Figure S1, panel A). These figures suggest that most of the mismatches observed are probably due to errors of the procedure, rather than generation of genuine new polymorphisms. When we compared the AFLP profiles from the first month against profiles generated during each subsequent month, the error rate values did not show a significant correlation with time, either when considering “stable” or “unstable” alleles (Figure S1, panels B and C). As error rates did not show a tendency to increase with time (Spearman correlation P>0.05 in both cases), it seems that at least in this isolate, and under our experimental conditions, the in vitro propagation required for the AFLP procedure does not seem to significantly contribute to the variability observed in this study.
As an additional assessment of the properties of AFLP markers for these species, we evaluated the congruency of distance matrices generated from each particular selective primer combination, using all specimens. Each possible pair of Jaccard distance matrices was compared by means of Mantel tests, which showed that all datasets were strongly and significantly correlated (Mantel R values ranged from 0.88 to 0.98, all highly significant at P<0.001; Table S1).
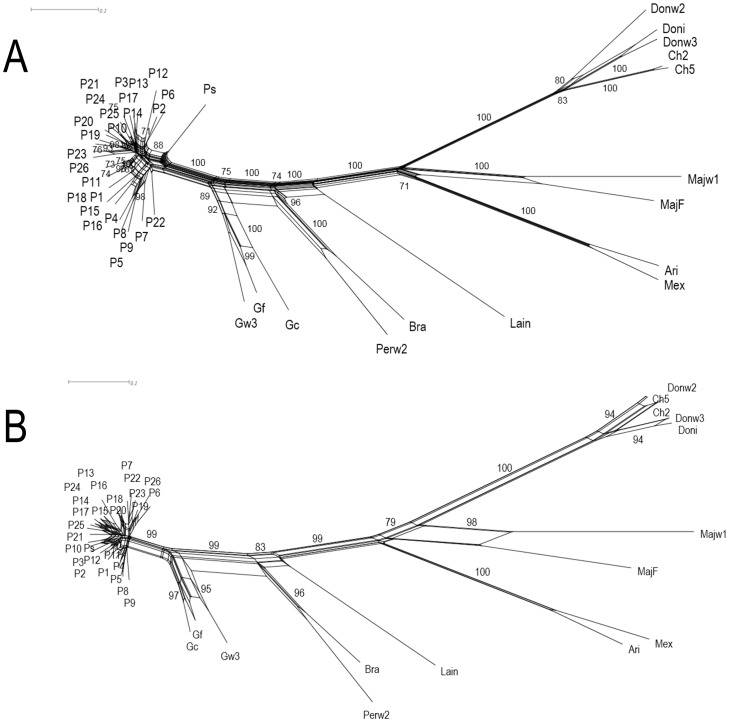
Cluster and ordination analyses were done on concatenated presence – absence matrix to explore the relationships among all tested species of Leishmania. The Neighbor-Net split graph obtained after Jaccard transformation showed very good species definition, with very high bootstrap support (Figure 1A). Within the L. panamensis group, some groups could also be well defined with significant statistical support. Trees generated using other methods, namely BioNJ and UPGMA, showed the same overall topologies (Figure S2). The Neighbor-Net phylogenetic network displayed perfect definition of species into both subgenera, Leishmania (Viannia) and Leishmania (Leishmania). The genetic relationships among the Leishmania (Viannia) specimens showed clear distinction of all species tested, and even closely related species like L. panamensis and L. guyanensis could be well separated with strong statistical support. This split graph also showed a significant number of incompatible splits, particularly among L. panamensis and L. guyanensis isolates. As also shown by several authors from the analyses of other marker systems, L. peruviana and L. braziliensis occupy closely related but clearly distinct positions, while L. lainsoni is represented as the most divergent species within the L. (Viannia) subgenus. The L. (Leishmania) subgenus group was well defined with robust clusters for the included species. We tested the ability of each individual matrix to allow reconstruction of phylogenetic relationships among all specimens. We detected some selective primer combinations, particularly combination V9 (Table 2), that were able to generate trees with almost similar resolution and topology as the concatenated dataset (Figure 1B). However, using the full concatenated matrix was required to achieve better intra-specific node resolution in L. panamensis (Table S2).
Figure 1. Split graph showing the results of Neighbor-Net analysis obtained on Jaccard distances among Leishmania species tested.
Specimen names according to Table 1. Bootstrap values over 70% are shown. Panel A: split graph generated using the concatenated matrix (fit: 96%). Panel B: split graph generated using matrix corresponding to selective primer V9 (fit: 97%).
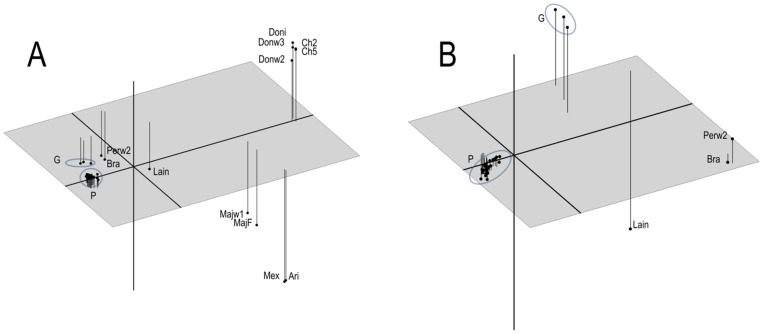
The principal coordinate analysis confirmed the results of the clustering analyses, showing a clear separation between specimens of the Leishmania (Viannia) and Leishmania (Leishmania) subgenera, the latter being the most diverse (Figure 2A). When the PCoA plot was done including only the Leishmania (Viannia) species, more resolution was observed in three dimensions (Figure 2B), confirming the clear separation between L. panamensis and L. guyanensis, the L. peruviana – L. braziliensis relationship, and the divergent position of L. lainsoni.
Figure 2. Principal coordinate analysis (PCoA) plots depicting genetic relationships among Leishmania species tested.
Panel A: PCoA plot including all specimens tested. P, L. panamensis strains; G, L. guyanensis strains. Other specimen names according to Table 1. The variance explained is 64%. Panel B: PCoA plot including only species from Leishmania (Viannia) subgenus. The variance explained is 63%.
Discussion
This study presents a genetic analysis of Leishmania parasites circulating in Panama as well as several reference strains covering both subgenera, Leishmania (Viannia) and Leishmania (Leishmania), using AFLP markers. The AFLP system has been applied frequently in other taxa, and is convenient because of its reproducibility and sequence information independence. Once optimized for a specific taxonomic group, AFLP allows easy and rapid genome screening for polymorphisms, and the detection of hundreds to thousands of variable sites.
Prior to AFLP typing, we tested species identity of all specimens by using hsp70 PCR-RFLP. All isolates from cutaneous leishmaniasis patients were identified as L. panamensis, in agreement with a previous report in Panama [17]. Other species have been sporadically reported in Panama, but their clinical relevance is still to be defined [18].
When we analyzed our specimens with the AFLP procedure, large numbers of fragments could be detected in all cases. Of particular interest were the fixed private alleles which, if validated with more isolates, may represent diagnostic fragments useful for rapid molecular species discrimination in a clinical setting. These fixed private alleles are particularly interesting because the Leishmania species that have been sequenced so far have a very high level of synteny sharing and a very low number of species-specific genes [39].
In terms of the number of fragments, our findings differ from those reported by Kumar and coworkers in Leishmania donovani [21]. Employing the same restriction enzyme system (Tru9I – EcoRI, and +3 primers), they found high number of fragments, at levels we observed in L. panamensis only when using lower stringency primers (+0, +1, +2). Although not directly comparable to those of Kumar et al., our results are congruent with the fact that the Leishmania genome is GC rich. Therefore, the use of these restriction enzymes should generate fewer fragments in the useful range of 50 to 500 base pairs [27].
The results of the AFLP typing revealed very high levels of polymorphisms, especially in L. panamensis. Odiwuor and colleagues [23] employed a different combination of enzymes (TaqI – PstI) to analyze the genetic diversity in L. archibaldi, L. donovani, L. chagasi and L. infantum. They found that up to 52% of alleles were polymorphic, a level of variability that is high enough to describe the genetic variability of the tested species and strains by means of clustering and ordination techniques. In a more recent report, these authors used the same AFLP system to analyze some species of Leishmania (Viannia) [24]. The proportions of polymorphisms that we observed in L. panamensis are even higher than those reported for other Leishmania species in these studies. This might be because more strains were typed in our study, and/or because some of the selective primer combinations that we used might probe regions of the L. panamensis genome that are more polymorphic.
Several molecular events may result in an AFLP fragment polymorphism, including mutations at any of the restriction sites, deletions, insertions, translocations, or variation in the size of repetitive sequences. Genomic DNA rearrangements at repetitive sequences have been shown to be rather common in protozoa, contributing to their high genetic variability [40]–[41]. It has been shown that Leishmania genomic DNA may contain up to 25% diverse repetitive sequences, as judged from DNA reassociation kinetics in L. donovani [42]. More recently, genome sequencing has shown that one member of the Leishmania (Viannia) subgenus, L. braziliensis, contains a higher number of repetitive sequences than either L. major or L. infantum, as well as more divergence at the level of DNA and protein sequences [43]. As some of these repetitive sequences are intrinsically polymorphic, they may be contributing to the AFLP polymorphisms that we observed in L. panamensis.
Although AFLP is considered to be a very reproducible fingerprinting technique [44], it is not free of potential errors, which in turn can have a significant effect on further analyses [28]. In our experimental conditions, AFLP had good reproducibility, with error rates roughly similar to values reported by other authors in other organisms [28]. The time stability experiment showed that, at least under our experimental conditions and using this strain, AFLP profiles appear to be stable in time. This is in agreement with the general belief that Leishmania karyotypes are stable in vitro [45]. Our result, however, might be species and/or strain specific, as it has been reported that some strains of L. peruviana may undergo significant karyotype changes during in vitro propagation [46].
The AFLP data presented here also provide some insight into the reproductive mode of these parasites. The Jaccard distance matrices generated by each individual selective primer pair combination (which may be regarded as independent markers), were strongly correlated when all Leishmania specimens were analyzed together. This suggests some degree of linkage disequilibrium, and is consistent with a predominantly clonal mode of propagation. Similar levels of correlation between independent molecular markers have been observed in Trypanosoma cruzi, and interpreted as evidence of a predominantly clonal mode of reproduction [47]. In this regard, our data are consistent with previously reported evidence for the genus [48]. However, this mode of reproduction would also lead one to predict the presence of frequent, repeated fingerprint profiles in the parasite populations. This prediction was not confirmed in our study, as all isolates of L. panamensis had different AFLP profiles. Possible causes for this discrepancy may include: an insufficient sample size, sporadic but significant sexual reproduction, a more heterogeneous range of sand fly vectors and/or hosts than previously suspected for this geographical area, and/or more complex transmission cycles. Studies in L. braziliensis using other markers have found higher molecular diversity in areas with more sylvatic associated transmission cycles [49]. In Panama, accelerated urbanization frequently disrupts natural environments, possibly allowing for more sylvatic related transmission cycles, and therefore higher molecular diversity.
The AFLP data obtained from all tested parasite species allowed us to reconstruct phylogenetic networks that were congruent with the accepted taxonomy for the genus [50]–[53]. Within the L. panamensis group, split graphs showed a considerable amount of reticulation, possibly indicating sampling artifacts or some levels of genetic recombination. Interestingly, some individual selective primer combinations were able to produce networks which robustly reproduced the accepted taxonomy for all specimens. This result is important because performing AFLP using single selective primer combinations should be more affordable and convenient.
The clear separation observed between the closely related L. panamensis and L. guyanensis (particularly in the principal coordinate plot), is not consistent with previous reports questioning the validity of the separation of these two species based on MLEE and RAPD data [54]–[55]. The higher resolution observed in our results might be due to the fact that AFLP allows simultaneous examination of thousands of loci in the Leishmania genome, most of them polymorphic. Although the trees generated with our data show topologies concordant with previous phylogenetic studies for the genus Leishmania, confirmation of the usefulness of AFLP data may require inclusion of more strains, especially for species represented here by a single specimen.
The results of our phylogenetic and ordination analyses indicate that AFLP markers are a useful tool for studying the genetic diversity of the Leishmania genus at a higher resolution than was possible with previously used markers. Additionally, AFLP scanning of Leishmania genomes should allow for the rapid identification of polymorphisms associated with clinically relevant traits, such as drug resistance or clinical presentation. Rapid conversion of those polymorphisms into dominant markers would have an immediate application to the clinical practice.
Although the AFLP approach has some limitations associated with the dominant mode of inheritance and the requirement of purified DNA, the possibility of simultaneously examining hundreds or even thousands of sites in the genome is an attractive opportunity to study genetic variation at depths only possible today with next generation sequencing, but at much higher costs.
Here we have demonstrated that AFLP markers allow high resolution genetic analysis of parasites of the Leishmania genus, particularly of the isolates circulating in Panama. We have uncovered a high number of polymorphisms in the main species causing cutaneous leishmaniasis, L. panamensis. This discovery opens up new and exciting possibilities for the generation of knowledge in the fields of molecular epidemiology and taxonomy, and confirms the notion that AFLP is a very promising tool for studying the genetic diversity of these parasites.
Supporting Information
Characterization of mismatches (error rate) detected in AFLP profiles during in vitro cultivation of promastigotes of the Leishmania panamensis Ps isolate during one year. Panel A: box plot showing distribution of pairwise errors (see Materials and Methods for details) considering all mismatches (allMM), stable mismatches (SMM) or unstable mismatches (UMM). Panels B and C: pairwise error rates (considering all mismatches or stable mismatches, respectively) between profiles generated at each time point and the one obtained at first month.
(TIF)
Split graphs obtained from Jaccard distance transformations of the concatenated AFLP matrix for all Leishmania specimens tested, following the algorithms Bio Neighbor Joining (A) and UPGMA (B).
(TIF)
Pairwise Mantel test for Jaccard distance matrices generated from each selective primer combination.
(DOCX)
Number of significant nodes and bootstrap values of UPGMA trees generated from datasets obtained from each selective primer combination. Bootstrap values were calculated after 10 000 resamplings, and only values over 70% are showed.
(DOCX)
Details of AFLP data analysis in GeneMarker v 2.2.0 (SoftGenetics LLC, USA).
(DOCX)
Acknowledgments
We are very grateful to Dr. Gert Van der Auwera for helpful discussion of the results; to Dr. Marcelo Bozza and Dr. Carmenza Spadafora for kind gift of Leishmania strains; to Mrs. Laura Pineda for assistance with parasite cultures; and to Dr. Gabrielle Britton for reviewing the manuscript.
Funding Statement
This work was partially supported by funds from grant FID08-104 from Secretaría Nacional de Ciencia, Tecnología e Innovación (SENACYT, Panamá). No additional external funding was received for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Desjeux P (2004) Leishmaniasis: current situation and new perspectives. Comp Immunol Microbiol Infect Dis 27: 305–318. [DOI] [PubMed] [Google Scholar]
- 2.Vásquez A, Paz H, Alvar J, Perez D, Hernandez C (1998) Informe Final: Estudios Sobre la Epidemiología de la Leishmaniasis en la parte Occidental de la República de Panamá. Ministerio de Salud, República de Panamá.
- 3. Christensen HA, Herrer A, Telford SR Jr (1972) Enzootic cutaneous leishmaniasis in eastern Panama. II. Entomological investigations. Ann Trop Med Parasitol 66: 55–66. [DOI] [PubMed] [Google Scholar]
- 4. Christensen HA, Fairchild GB, Herrer A, Johnson CM, Young DG, et al. (1983) The ecology of cutaneous leishmaniasis in the Republic of Panama. J Med Entomol 20: 463–484. [DOI] [PubMed] [Google Scholar]
- 5. Christensen HA, de Vasquez AM, Petersen JL (1999) Short report: epidemiologic studies on cutaneous leishmaniasis in eastern Panama. Am J Trop Med Hyg 60: 54–57. [DOI] [PubMed] [Google Scholar]
- 6. Takafuji ET, Hendricks LD, Daubek JL, McNeil KM, Scagliola HM, et al. (1980) Cutaneous leishmaniasis associated with jungle training. Am J Trop Med Hyg 29: 516–520. [DOI] [PubMed] [Google Scholar]
- 7. Petersen J, Johnson CM, de Vásquez AM, Sáenz R (1987) Cutaneous leishmaniasis caused by Leishmania mexicana amazonensis in Panama. Rev Med Panama 12: 158–164. [PubMed] [Google Scholar]
- 8. de Vásquez AM, Saenz RE, Petersen JL, Christensen HA, Johnson CM (1990) Leishmania mexicana complex: human infections in the Republic of Panama. Am J Trop Med Hyg 43: 619–622. [DOI] [PubMed] [Google Scholar]
- 9. Kreutzer RD, Corredor A, Grimaldi G Jr, Grogl M, Rowton ED, et al. (1991) Characterization of Leishmania colombiensis sp. n (Kinetoplastida:Trypanosomatidae), a new parasite infecting humans, animals, and phlebotomine sand flies in Colombia and Panama. Am J Trop Med Hyg 44: 662–675. [DOI] [PubMed] [Google Scholar]
- 10. Herrer A (1971) Leishmania hertigi sp. n., from the tropical porcupine, Coendou rothschildi . J Parasitol 57: 626–629. [PubMed] [Google Scholar]
- 11. Herrer A, Telford SR Jr, Christensen HA (1971) Enzootic cutaneous leishmaniasis in Eastern Panama. I. Investigation of the infection among forest mammals. Ann Trop Med Parasitol 65: 349–358. [DOI] [PubMed] [Google Scholar]
- 12. Romero LI, Paz HM, Ortega-Barría E, Bayard V, Hochberg LP, et al. (2004) Evaluation of serological assays based on a novel excreted antigen preparation for the diagnosis of cutaneous leishmaniasis in Panama. J Microbiol Methods 57: 391–397. [DOI] [PubMed] [Google Scholar]
- 13. Schönian G, Mauricio I, Gramiccia M, Canavate C, Boelaert M, et al. (2008) Leishmaniases in the Mediterranean in the era of molecular epidemiology. Trends Parasitol 24: 135–142. [DOI] [PubMed] [Google Scholar]
- 14. Cupolillo E, Grimaldi G Jr, Momen H, Beverley SM (1995) Intergenic region typing (IRT): a rapid molecular approach to the characterization and evolution of Leishmania . Mol Biochem Parasitol 73: 145–155. [DOI] [PubMed] [Google Scholar]
- 15. Victoir K, Banuls AL, Arevalo J, Llanos-Cuentas A, Hamers R, et al. (1998) The gp63 gene locus, a target for genetic characterization of Leishmania belonging to subgenus Viannia . Parasitology 117: 1–13. [PubMed] [Google Scholar]
- 16. Garcia L, Kindt A, Bermudez H, Llanos-Cuentas A, De Doncker S, et al. (2004) Culture-independent species typing of neotropical Leishmania for clinical validation of a PCR-based assay targeting heat shock protein 70 genes. J Clin Microbiol 42: 2294–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miranda A, Carrasco R, Paz H, Pascale JM, Samudio F, et al. (2009) Molecular epidemiology of American tegumentary leishmaniasis in Panama. Am J Trop Med Hyg 81: 565–571. [DOI] [PubMed] [Google Scholar]
- 18. Azpurua J, De La Cruz D, Valderama A, Windsor D (2010) Lutzomyia sand fly diversity and rates of infection by Wolbachia and an exotic Leishmania species on Barro Colorado Island, Panama. PLoS Negl Trop Dis 4(3): e627 doi:10.1371/journal.pntd.0000627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, et al. (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bensch S, Åkesson M (2005) Ten years of AFLP in ecology and evolution: why so few animals? Mol Ecol 14: 2899–2914. [DOI] [PubMed] [Google Scholar]
- 21. Kumar A, Boggula VR, Sundar S, Shasany AK, Dube A (2009) Identification of genetic markers in Sodium Antimony Gluconate (SAG) sensitive and resistant Indian clinical isolates of Leishmania donovani through amplified fragment length polymorphism (AFLP). Acta Trop 110: 80–85. [DOI] [PubMed] [Google Scholar]
- 22. Kumar A, Boggula VR, Misra P, Sundar S, Shasany AK, et al. (2010) Amplified fragment length polymorphism (AFLP) analysis is useful for distinguishing Leishmania species of visceral and cutaneous forms. Acta Trop 113: 202–206. [DOI] [PubMed] [Google Scholar]
- 23. Odiwuor S, Vuylsteke M, De Doncker S, Maes I, Mbuchi M, et al. (2011) Leishmania AFLP: paving the way towards improved molecular assays and markers of diversity. Infect Genet Evol 11: 960–967. [DOI] [PubMed] [Google Scholar]
- 24. Odiwuor S, Veland N, Maes I, Arévalo J, Dujardin JC, et al. (2012) Evolution of the Leishmania braziliensis species complex from amplified fragment length polymorphisms, and clinical implications. Infect Genet Evol 12(8): 1994–2002. [DOI] [PubMed] [Google Scholar]
- 25.Evans DA (1987) Leishmania In: A. E. R. Taylor AER, Baker JR, editors. In vitro methods for parasite cultivation. Academic Press, New York, N.Y. 52–75.
- 26. Montalvo AM, Fraga J, Monzote L, Montano I, De Doncker S, et al. (2010) Heat-shock protein 70 PCR-RFLP: a universal simple tool for Leishmania species discrimination in the New and Old World. Parasitology 137: 1159–68. [DOI] [PubMed] [Google Scholar]
- 27. Restrepo CM, Perez Lao E, De La Guardia C, Sousa O, Calzada JE, et al. (2011) Amplified Fragment Length Polymorphisms Reveals High Intraspecific Variability in Field Isolates of Leishmania panamensis . Curr Trends Biotechnol Pharm 5: 1183–1192. [Google Scholar]
- 28. Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, et al. (2004) How to track and assess genotyping errors in population genetics studies. Mol Ecol 13: 3261–3273. [DOI] [PubMed] [Google Scholar]
- 29. Holland B, Clarke A, Meudt H (2008) Optimizing Automated AFLP Scoring Parameters to Improve Phylogenetic Resolution. Syst Biol 57: 347–366. [DOI] [PubMed] [Google Scholar]
- 30. Jaccard P (1912) The distribution of the flora in the alpine zone. New Phytol 11: 37–50. [Google Scholar]
- 31. Mantel NA (1967) The detection of disease clustering and a generalized regression approach. Cancer Res. 27: 209–220. [PubMed] [Google Scholar]
- 32. Hammer O, Harper DAT, Ryan PD (2001) PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol Electronica 4: 9pp. [Google Scholar]
- 33. Fitch W (1997) Networks and viral evolution. J Mol Evol 44: S65–S75. [DOI] [PubMed] [Google Scholar]
- 34. Bryant D, Moulton V (2004) Neighbor-Net: An Agglomerative Method for the Construction of Phylogenetic Networks. Mol Biol Evol 21: 255–265. [DOI] [PubMed] [Google Scholar]
- 35. Huson DH, Bryant D (2006) Application of Phylogenetic Networks in Evolutionary Studies. Mol Biol Evol 23: 254–267. [DOI] [PubMed] [Google Scholar]
- 36. Gascuel O (1997) BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol 14: 685–695. [DOI] [PubMed] [Google Scholar]
- 37. Michener CD, Sokal RR (1957) A quantitative approach to a problem of classification. Evolution 11: 490–499. [Google Scholar]
- 38. Schlüter PM, Harris SA (2006) Analysis of multilocus fingerprinting data sets containing missing data. Mol Ecol Notes 6: 569–572. [Google Scholar]
- 39. Rogers MB, Hilley JD, Dickens NJ, Wilkes J, Bates PA, et al. (2011) Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania . Genome Res 12: 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lanzer M, de Bruin D, Wertheimer SP, Ravetch JV (1994) Organization of chromosomes in Plasmodium falciparum: a model for generating karyotypic diversity. Parasitol Today 10: 114–117. [DOI] [PubMed] [Google Scholar]
- 41. Kebede A, De Doncker S, Arevalo J, Le Ray D, Dujardin JC (1999) Size-polymorphism of mini-exon gene-bearing chromosomes among natural populations of Leishmania, subgenus Viannia . Int J Parasitol 29: 549–557. [DOI] [PubMed] [Google Scholar]
- 42. Leon W, Fouts DL, Manning J (1978) Sequence arrangement of the 16S and 26S rRNA genes in the pathogenic haemoflagellate Leishmania donovani . Nucleic Acids Res 5: 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, et al. (2007) Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet 39: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Meudt HM, Clarke AC (2007) Almost forgotten or latest practice? AFLP applications, analyses, and advances. Trends Plant Sci 12: 106–117. [DOI] [PubMed] [Google Scholar]
- 45. Giannini SH, Curry SS, Tesh RB, Van der Ploeg LH (1990) Size-conserved chromosomes and stability of molecular karyotype in cloned stocks of Leishmania major . Mol Biochem Parasitol 39: 9–21. [DOI] [PubMed] [Google Scholar]
- 46. Dujardin JC, De Doncker S, Jacquet D, Bañuls AL, Balavoine M, et al. (2007) Clonal propagation and the fast generation of karyotype diversity: An in vitro Leishmania model. Parasitology 134: 33–9. [DOI] [PubMed] [Google Scholar]
- 47. Telleria J, Barnabé C, Hide M, Bañuls AL, Tibayrenc M (2004) Predominant clonal evolution leads to a close parity between gene expression profiles and subspecific phylogeny in Trypanosoma cruzi . Mol Biochem Parasitol 137: 133–141. [DOI] [PubMed] [Google Scholar]
- 48. Tibayrenc M, Ayala FJ (1999) Evolutionary genetics of Trypanosoma and Leishmania . Microbes Infect 1: 465–472. [DOI] [PubMed] [Google Scholar]
- 49. Cupolillo E, Brahim LR, Toaldo CB, de Oliveira-Neto MP, de Brito ME, et al. (2003) Genetic polymorphism and molecular epidemiology of Leishmania (Viannia) braziliensis from different hosts and geographic areas in Brazil. J Clin Microbiol 41: 3126–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cupolillo E, Grimaldi Jr G, Momen H (1994) A general classification of New World Leishmania using numerical zymotaxonomy. Am J Trop Med Hyg 50: 296–311. [DOI] [PubMed] [Google Scholar]
- 51.Bañuls AL (1998) Apport de la génétique évolutive à l’épidémiologie et à la taxonomie du genre “Leishmania” PHD Thesis, Université des Sciences et Techniques du Languedoc, Montpellier, France, 196 pp.
- 52. Fraga J, Montalvo AM, De Doncker S, Dujardin JC, Van der Auwera G (2010) Phylogeny of Leishmania species based on the heat-shock protein 70 gene. Infect Genet Evol 10: 238–245. [DOI] [PubMed] [Google Scholar]
- 53. Boité MC, Mauricio IL, Miles MA, Cupolillo E (2012) New Insights on Taxonomy, Phylogeny and Population Genetics of Leishmania (Viannia) Parasites Based on Multilocus Sequence Analysis. PLoS Negl Trop Dis 6(11): e1888 doi:10.1371/journal.pntd.0001888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Saravia NG, Segura I, Holguin AF, Santrich C, Valderrama L, et al. (1998) Epidemiologic, genetic, and clinical associations among phenotypically distinct populations of Leishmania (Viannia) in Colombia. Am J Trop Med Hyg 59: 86–94. [DOI] [PubMed] [Google Scholar]
- 55. Bañuls AL, Jonquieres R, Guerrini F, Le Pont F, Barrera C, et al. (1999) Genetic analysis of Leishmania parasites in Ecuador: are Leishmania (Viannia) panamensis and Leishmania (V.) guyanensis distinct taxa? Am J Trop Med Hyg 61: 838–845. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of mismatches (error rate) detected in AFLP profiles during in vitro cultivation of promastigotes of the Leishmania panamensis Ps isolate during one year. Panel A: box plot showing distribution of pairwise errors (see Materials and Methods for details) considering all mismatches (allMM), stable mismatches (SMM) or unstable mismatches (UMM). Panels B and C: pairwise error rates (considering all mismatches or stable mismatches, respectively) between profiles generated at each time point and the one obtained at first month.
(TIF)
Split graphs obtained from Jaccard distance transformations of the concatenated AFLP matrix for all Leishmania specimens tested, following the algorithms Bio Neighbor Joining (A) and UPGMA (B).
(TIF)
Pairwise Mantel test for Jaccard distance matrices generated from each selective primer combination.
(DOCX)
Number of significant nodes and bootstrap values of UPGMA trees generated from datasets obtained from each selective primer combination. Bootstrap values were calculated after 10 000 resamplings, and only values over 70% are showed.
(DOCX)
Details of AFLP data analysis in GeneMarker v 2.2.0 (SoftGenetics LLC, USA).
(DOCX)