Abstract
Objective
Metaplastic lineages in the oxyntic mucosa of the stomach are critical preneoplastic precursors of gastric cancer. Recent studies have demonstrated that Spasmolytic polypeptide-expressing metaplasia (SPEM) in the mouse oxyntic mucosa arises from transdifferentiation of mature gastric chief cells. Other investigations of intestinal progenitor cells have shown that cells demonstrating transcriptional activity for Lgr5 in the intestine, colon and gastric antrum function as adult stem cells. We have now investigated whether cells demonstrating Lgr5 transcriptional activity in the oxyntic mucosa of mice might be responsible for development of metaplasia.
Design
Lgr5-EGFP-IRES-CreERT2/+;Rosa26R mice were utilized to examine the distribution of Lgr5 transcriptionally active cells in the normal oxyntic mucosa as well as after treatment with DMP-777 or L635 to induce acute SPEM. Lineage mapping was performed to determine if LGR5-expressing cells gave rise to SPEM.
Results
Cells expressing transcriptional activity for Lgr5 in the oxyntic mucosa were present as scattered rare cells only along the lesser curvature of the stomach. These cells also stained for markers of chief cells (intrinsic factor and pepsinogen) but never showed any staining for proliferative markers (Ki-67). In Lgr5-EGFP-IRES-CreERT2/+;Rosa26R mice induced with tamoxifen, treatment with either DMP-777 or L-635 to induce acute oxyntic atrophy caused induction of SPEM, but no lineage mapping into SPEM from Lgr5-expressing cells was observed.
Conclusion
The results indicate that, while chief cells with Lgr5-transcriptional activity are present along the lesser curvature of the gastric oxyntic mucosa, they are not responsible for production of metaplasia.
Keywords: SPEM, chief cell, Lgr5, TFF2, oxyntic atrophy
INTRODUCTION
In the normal acid-secreting (oxyntic) gastric mucosa, cell lineages are derived through orderly differentiation from progenitor cells located in the neck region of gastric glands. Committed progenitor cells give rise to either short-lived surface cells or long-lived cells of the gastric gland including gastric parietal cell, mucous neck cells and zymogen-secreting chief cells.[1, 2] In particular, as mucous neck cells migrate towards the bases of oxyntic glands they undergo redifferentiation into chief cells without undergoing any intervening cell divisions.[1, 3] Thus, in the oxyntic glands, proliferating cells are confined to the upper third of the glands.[4] This pattern contrasts with the organization of the gastric antrum as well as the intestinal crypts, where proliferative cells are located nearer the bases of antral glands and intestinal crypts. Recent studies have noted that cells expressing Lgr5 in the antrum and the intestines can give rise to all of the lineages of these mucosal units, suggesting that Lgr5 cells represent a critical pluripotent progenitor population.[5, 6, 7, 8] In contrast, cells with Lgr5 transcriptional activity were rare in the oxyntic regions of the stomach and only rarely showed any mapping as progenitors of oxyntic mucosal lineages.[5]
Gastric neoplasia is thought to arise in the setting of pre-neoplastic metaplastic cell lineages.[9] Metaplasia develops in the human stomach following the loss of gastric acid-secreting parietal cells (oxyntic atrophy), usually in concert with chronic infection with Helicobacter pylori.[10] In humans, oxyntic atrophy is associated with two types of metaplastic mucous cell lineages: intestinal metaplasia, which resembles intestinal goblet cells,[11] and Spasmolytic Polypeptide Expressing Metaplasia (SPEM), which is phenotypically similar to the deep antral gland cells[12]. In models of acute and chronic induced oxyntic atrophy in mice, only SPEM is observed.[13, 14, 15] While the origin of these metaplastic lesions has been uncertain, recent investigations have demonstrated that, in mice, SPEM is derived from transdifferentiation of mature chief cells into the mucous cell metaplasia.[16, 17] This transdifferentiation causes the recrudescence of proliferative activity in post-mitotic chief cells, which is further augmented in the presence of inflammatory infiltrates.[16]
Given the position of cells with Lgr5 transcriptional activity in the bases of some oxyntic glands, we have sought to determine whether these Lgr5-expressing cells may contribute to the induction of metaplasia in the setting of acute oxyntic atrophy. While we have determined that the cells with Lgr5-transcriptional activity in the oxyntic mucosa are phenotypic chief cells, lineage mapping studies indicate that these cells do not contribute to SPEM. The results therefore indicate the transdifferentiation of chief cells into SPEM is not driven by Lgr5-expressing progenitor cells.
MATERIALS AND METHODS
Mice
Eight to ten week old mice were used for all studies. Generation of Lgr5-EGFP-IRES-CreERT2/+ and Rosa26RLacZ mice has been described previously.[5] Lgr5-EGFP-IRES-CreERT2/+ mice were generated by standard embryonic stem cell targeting in which EGFP was knocked into the Lgr5 coding region along with an internal ribosome re-entry site (IRES) followed by CreERT2. Cre recombinase was activated in Lgr5-EGFP-IRES-CreERT2/+/Rosa26RLacZ mice by intraperitoneal injection of tamoxifen (1 mg/0.1 ml corn oil) for 4 doses every other day. During the experiments, the mice were maintained with regular mouse chow and water ad libitum in a temperature-controlled room under a 12-hour light/dark cycle. The care, maintenance, and treatment of animals in these studies followed protocols approved by the Institutional Animal Care and Use Committee of Vanderbilt University.
Drugs
Preparation and treatment of the DMP-777 have been described previously.[18] L-635 was a gift of Merck & CO., Inc (Rahway, NJ). Mice were administered L-635 orally as a gavage (350 mg/kg) once daily for 3 days.
β-galactosidase analysis
Assessment of β-galactosidase activity was used to examine the efficiency of recombination at the Rosa26RLacZ reporter allele after induction of Cre activity. Stomach whole-mounts were prepared, fixed and exposed to 5-bromo-4-chloro-3-indoyl-β-D-galactopyranoside (X-gal) substrate using a method previously reported.[19] The stomachs were opened along the greater curvature and washed with ice-cold fixative containing 1% formaldehyde, 0.2% glutaraldehyde and 0.02% Nonidet P-40 (NP40) in PBS (Ca+ and Mg+ free). The stomachs were immediately incubated for 2 h in a 20-fold volume of ice-cold fixative at 4 °C on a rolling platform. The fixative was removed and the tissues washed twice in PBS for 20 min at room temperature (20 °C) on a rolling platform. The β-galactosidase substrate (5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6·3H2O, 2 mM MgCl2, 0.02% NP40, 0.1% sodium deoxycholate and 1 mg/ml X-gal in PBS) was then added and the tissues incubated in the dark overnight at room temperature. The substrate was removed and the tissues washed twice in PBS for 20 min at room temperature on a rolling platform. The tissues were then fixed overnight in a 20-fold volume of 4% paraformaldehyde in PBS at 4 °C in the dark on a rolling platform. The paraformaldehyde was removed and the tissues washed twice in PBS for 20 min at room temperature on a rolling platform. The stained tissues were transferred to tissue cassettes and paraffin blocks prepared using standard methods. Tissue sections were prepared and counterstained with nuclear fast red (Vector Laboratories, Burlingame, CA). The number of X-gal positive cells was analyzed in all strips of each stomach. Using an Ariol SL-50 slide scanner imaging system (Genetix, San Jose, CA; Vanderbilt Epithelial Biology Center Shared Resource) and light microscopy (200 ×), a mucosal length of 26,000 µm was analyzed to determine positive cells in the oxyntic mucosa.
Immunohistochemistry
For immunohistochemistry deparaffinized sections were rehydrated, and antigens were retrieved using Target Retrieval solution (Dako, Carpinteria, CA) in a pressure cooker for 15 minutes followed by standard detection with 3,3-diaminobenzidine (DAB) or alkaline phosphatase using a kit from Vector Laboratories. Samples were counter-stained with hematoxylin. In some cases, secondary antibodies were conjugated to AlexaFluor 488 (Invitrogen, Camarillo, CA) or Cy-3 and nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI). We used the following primary antibodies : mouse anti-TFF2 (1:100, a gift from Dr Nicholas Wright, Cancer UK, London, England), mouse anti-H+/K+-ATPase (1:2000, a gift from of Dr Adam Smolka, Medical University of South Carolina, Charleston, SC), rabbit anti-intrinsic factor (1:1000, a gift from Dr David Alpers, Washington University, St. Louis, MO), rabbit monoclonal anti-GFP (1:500. Cell Signaling, Danvers, MA), rat IgG anti-Ki-67 (1:50, Dako), sheep IgG anti-pepsinogen II (1:50000 Abcam, Cambridge, MA). The number of GFP positive cells was analyzed in all strips of each stomach. Using the Ariol SL-50 slide scanner and light microscopy (200 X), a mucosal length of 31,000 µm was analyzed to determine immunoreactive positive cells in the oxyntic glands.
For immunofluoresence staining of β-galactosidase, deparaffinized sections were sections were rehydrated, and antigens were retrieved using Target Retrieval solution (Dako) in a pressure cooker for 30 minutes followed by slow cooling without releasing pressure at room temperature for 1 hour, and slides were blocked with protein block serum-free (Dako) and incubated with rabbit anti-β-galactosidase (1:20000, AbD Serotec, Oxford, UK) overnight at 4°C. For double immunofluoresence staining of Mist1 and GFP, sections were incubated with rabbit anti-Mist1 (1:100,000, a gift from Dr Jason Mills, Washington University, St. Louis, MO) overnight at 4°C. Biotinylated secondary antibodies were visualized using streptavidin-conjugated horseradish peroxidase in conjunction with cy3-conjugated Tyramide Signal Amplification kit (PerkinElmer Life Sciences, Boston, MA). For Mist1 immunostaining, biotinylated secondary antibodies were visualized using streptavidin-conjugated horseradish peroxidase in conjunction with Fluorescein-conjugated Tyramide Signal Amplification kit. Sections were mounted using ProLong Gold Antifade Reagent with 4,6-diamino-2-phenylindole (DAPI) (Invitrogen) was used for nuclear counterstain and mounting medium.
Statistics
The data were analyzed with the JMP software package (version 4.0; SAS Institute, Cary, NC). GFP positive cell numbers and X-gal positive cell numbers were compared by analysis of variance followed by post hoc analysis of significant means by the Dunnett test. For all comparisons, P values less than 0.05 were considered statistically significant.
RESULTS
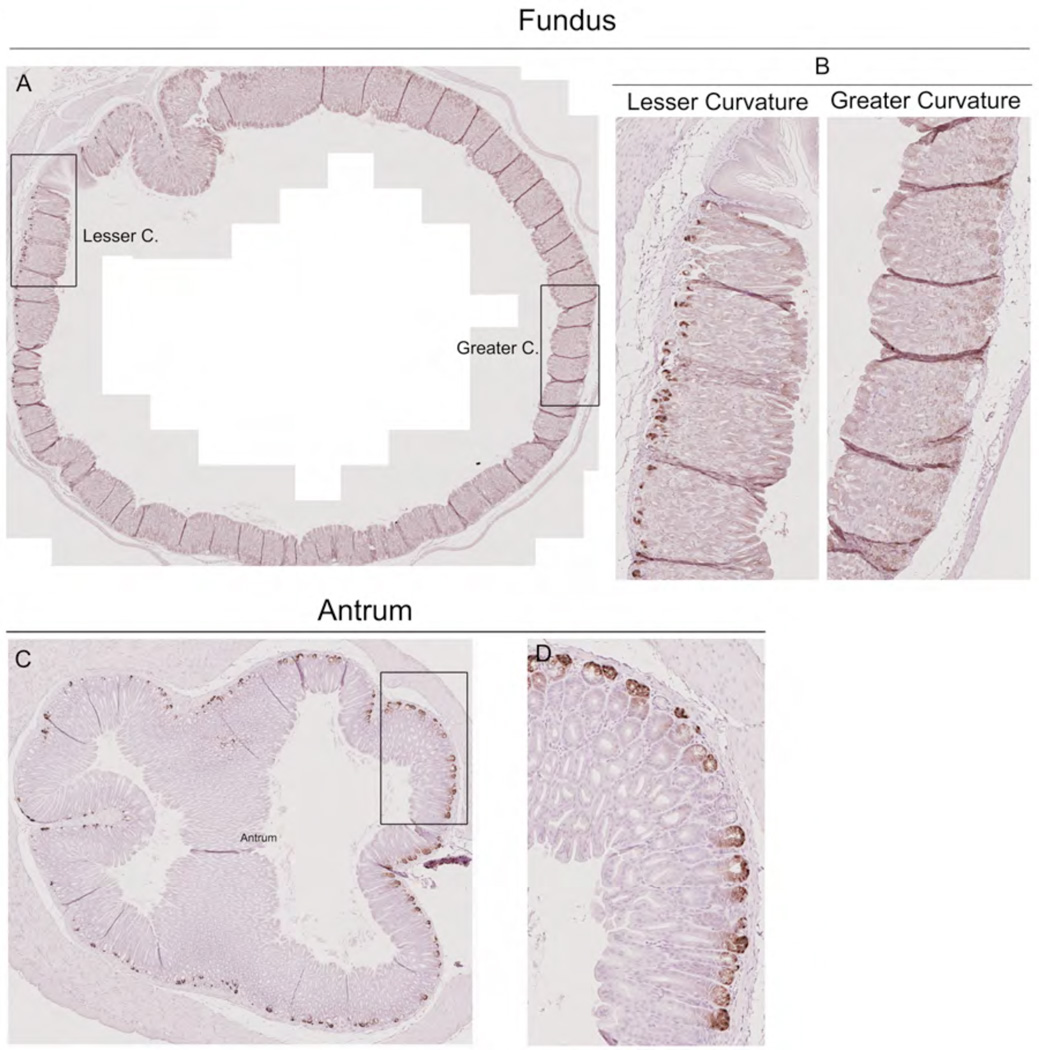
We first re-examined the geographic distribution of cells expressing Lgr5 transcriptional activity in the stomach Lgr5-EGFP-IRES-CreERT2 mice (n=4) that express both tamoxifen-inducible Cre recombinase and GFP under the control of the Lgr5 promoter. Scattered GFP staining cells were observed, sometimes as clusters, at the bases of glands on the lesser curvature of the gastric oxyntic mucosa, but no cells were observed along the greater curvature of the oxyntic region (Figure 1A, B). In contrast, GFP staining cells were observed in the bases of antral glands throughout the entire circumference of the antrum (Figure 1C,D).
Figure 1. Cells with Lgr5 transcriptional activity in the mouse stomach.
Stomachs from Lgr5-EGFP-IRES-CreERT2/+ mice were immunostained for GFP expression. A. Circumferential section of the gastric oxyntic region shows GFP immunostaining cells at the bases of glands only on the lesser curvature. Bar size=700 µm. B. Higher magnification of GFP-staining cells along the lesser curvature of the oxyntic region. Note no GFP-staining cells along the greater curvature of the gastric oxyntic region. Bar size=100 µm. C. Circumferential section of the antrum stained for GFP shows stained cells at the bases of glands throughout the antrum. Bar size=100 µm. D. Higher magnification of GFP-staining cells in the antrum. Bar size=100 µm.
In the intestine and the gastric antrum, Lgr5 transcript expression marks a population of proliferative stem cells with the potential to differentiate into all of the cell lineages of the glands and crypts[5, 19]. However, we did not observe any dividing cells through Ki-67 staining at the bases of oxyntic glands (Supp. Fig. 1). Thus, the position of the Lgr5-expressing cells in the oxyntic mucosa appeared to indicate a distinct population of cells.
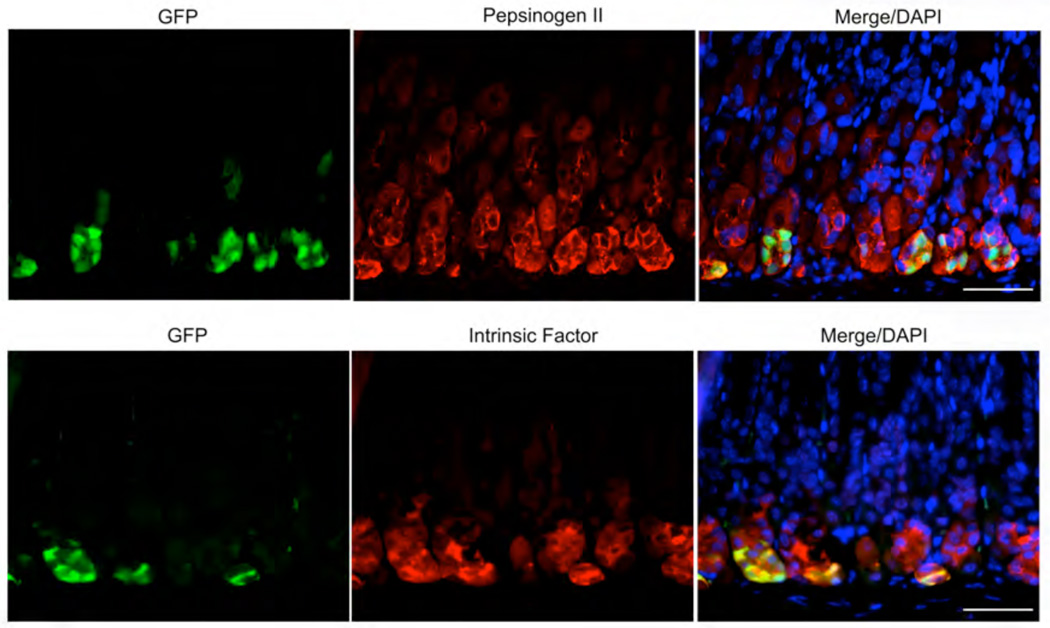
Given the position of the cells in the bases of oxyntic glands, we sought to determine whether these Lgr5-expressing cells might be chief cells. Figure 2 demonstrates that all of the Lgr5-expressing cells in the oxyntic mucosa also stained for pepsinogen II and intrinsic factor. In addition, most of the GFP staining Lgr5-expressing cells stained for Mist1 (data not shown). The staining of Lgr5-expressing cells with pepsinogen and intrinsic factor antibodies suggest that these cells are phenotypic chief cells. However, it is notable that most chief cells on the lesser curvature did not show Lgr5 expression and no chief cells on the greater curvature showed Lgr5 expression. Thus, Lgr5-transcriptional activity is present in a sub-population of phenotypic chief cells on the lesser curvature.
Figure 2. Cells with Lgr5 transcriptional activity in the oxyntic mucosa are phenotypic chief cells.
Sections of stomach from Lgr5-EGFP-IRES-CreERT2/+ mice were dual immunostained for either (A) GFP and pepsinogen II, (B) GFP and intrinsic factor or (C) GFP and Mist1. The immunostaining shows that the GFP-expressing cells all also labeled with pepsinogen, intrinsic factor and Mist1. Bar size=50 µm.
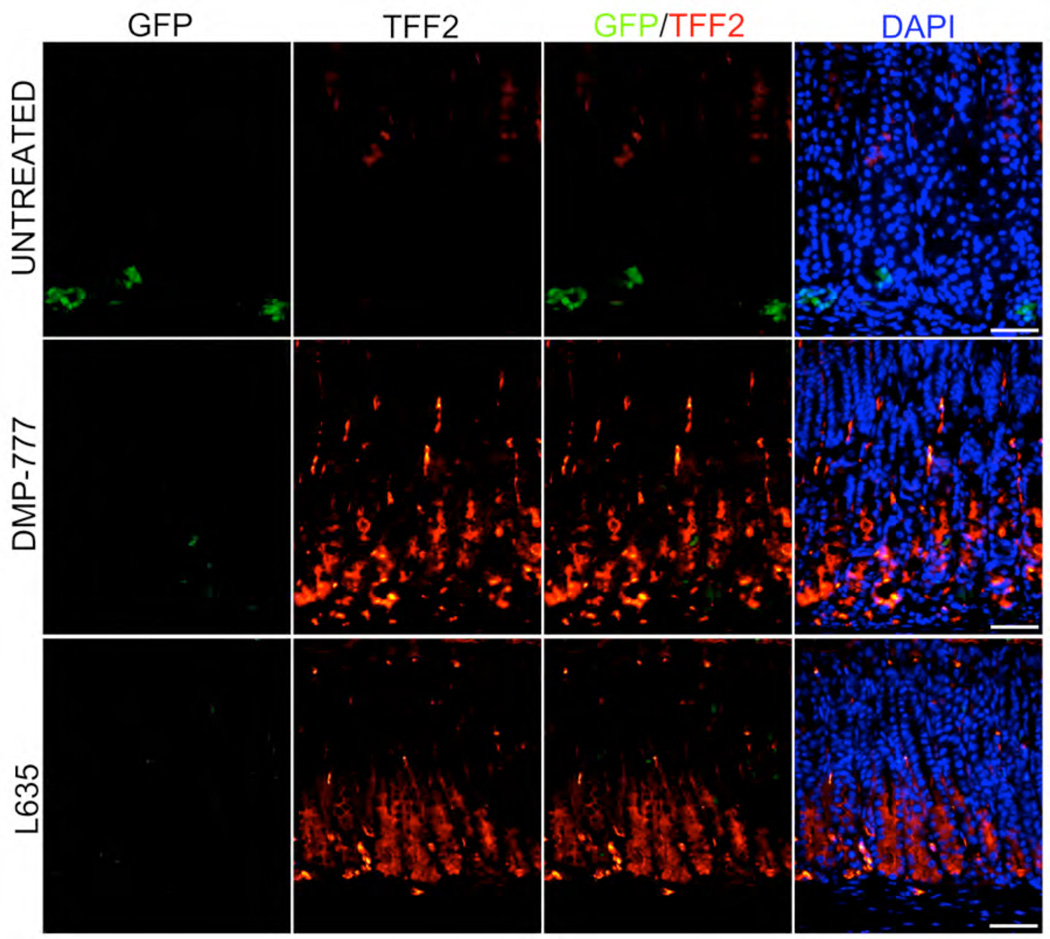
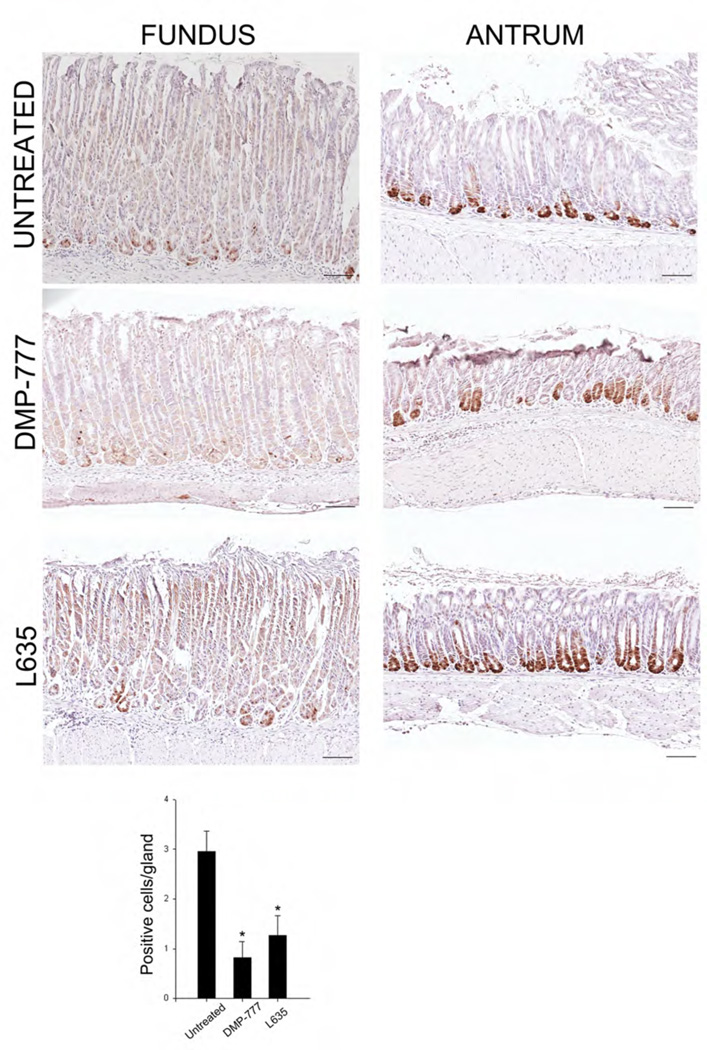
We have recently demonstrated that loss of parietal cells leads to induction of metaplasia in mice through the transdifferentiation of chief cells into SPEM.[16] We therefore sought to test whether Lgr5-expressing chief cells may contribute to SPEM. In DMP-777 treatment (n=4), SPEM develops after 14 days of drug treatment in the absence of inflammation,[20] while with L-635 SPEM develops after just 3 days of treatment (n=4) in association with a prominent inflammatory infiltrate.[16] In both acute oxyntic atrophy models, we saw induction of SPEM (as demonstrated by TFF2-expressing cells at the bases of gastric glands)(Figure 3) with a concomitant reduction in the numbers of GFP staining cells at the bases of oxyntic glands (Figure 4). However, no significant changes in the numbers or distribution of Lgr5-expressing, GFP-staining cells were observed in the antrum in either model (Supp. Figure 2). Similar results were obtained when sections were stained with immunohistochemistry for GFP (Figure4).
Figure 3. Induction of SPEM in treatment of Lgr5-EGFP-IRES-CreERT2/+ mice with DMP-777 or L635.
Sections of the gastric oxyntic region from untreated mice, mice treated with DMP-777 for 14 days or mice treated with L635 for 3 days were dual immunostained for GFP and TFF2. Dual labeling overlays of GFP and TFF2 staining are shown in the third column. Triple labeling overlays with GFP, TFF2 and DAPI are shown in the fourth column. The staining demonstrates that treatment with either drug led to a decrease in immunostaining for GFP. Also none of the GFP staining cells stained for TFF2 in the untreated stomach or in sections with SPEM. Bar size=50 µm.
Figure 4. GFP immunohistochemical staining in oxyntic region and antrum.
GFP staining of cells expressing Lgr5 transcriptional activity in sections of oxyntic and antral mucosa was performed using peroxidase-labeled secondary antibodies and DAB chromagen. Sections of oxyntic mucosa and antrum were stained from untreated Lgr5-EGFP-IRES-CreERT2/+ mice and Lgr5-EGFP-IRES-CreERT2/+ mice treated with either DMP-777 for 14 days or L-635 for three days. The quantitation of GFP positive cells in sections from the oxyntic mucosa is shown at the bottom (*p<0.01 compared with untreated mice). Bar size=100 µm.
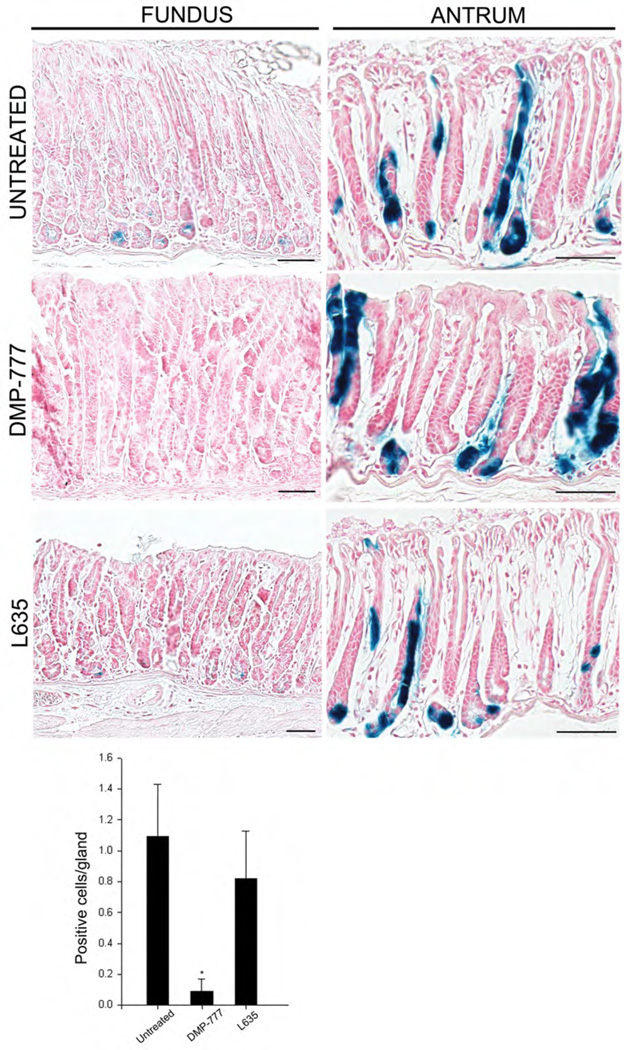
We next sought to examine whether cells with Lgr5 transcriptional activity give rise to SPEM following induction of acute oxyntic atrophy using in vivo lineage tracing. Lgr5-EGFP-IRES-CreERT2/+;Rosa26RLacZ mice were treated with four doses of tamoxifen and following 10 days without treatment were subsequently treated with either 14 days of DMP-777 (n=5) or 3 days of L-635 (n=5). Figure 5 demonstrates that isolated blue X-gal stained β-galactosidase-expressing cells were observed in the bases of oxyntic glands along the lesser curvature, indicative of cells with Lgr5 transcriptional activity. While both DMP-777 and L-635 treatments led to induction of SPEM, we did not observe any X-gal staining in these cells, so they were not derived from previously labeled Lgr5-expressing cells (Figure 5). In contrast, the entire epithelium of multiple antral gastric units was clonally X-gal positive, confirming that Lgr5 positive cells at the base of the antral glands were multipotent stem cells responsible for maintaining epithelial renewal under normal homeostatic conditions as well as oxyntic atrophy (Figure 5). We confirmed the presence of Lgr5 positive stem cell in this region by X-gal staining as well as immunohistochemistry against β-galactosidase protein (Figure 6). These findings indicate that SPEM was not derived from cells with Lgr5 transcriptional activity.
Figure 5. Lgr5-transcriptional activity does not map into SPEM.
Lgr5-EGFP-IRES-CreERT2/+; Rosa26RLacZ mice were treated with 4 doses of tamoxifen and then following a 10 day hiatus mice were either maintained without further treatment, treated with 14 days of DMP-777 or treated with 3 days of L-635. At sacrifice, stomachs were fixed in glutaraldehyde and then incubated with X-gal as whole mounts. The sections demonstrate that only scattered cells were observed at the bases of oxyntic glands even after treatment and no X-gal staining was observed in SPEM. In antrum, multiple clonal X-gal-positive antral glandular lineages were visible in untreated Lgr5-EGFP-IRES-CreERT2/+ mice and Lgr5-EGFP-IRES-CreERT2/+ mice treated with either DMP-777 for 14 days or L-635 for three days. The quantitation of X-gal positive cells in sections from the oxyntic mucosa is shown at the bottom (*p<0.01 compared with untreated mice). Bar size=100µm
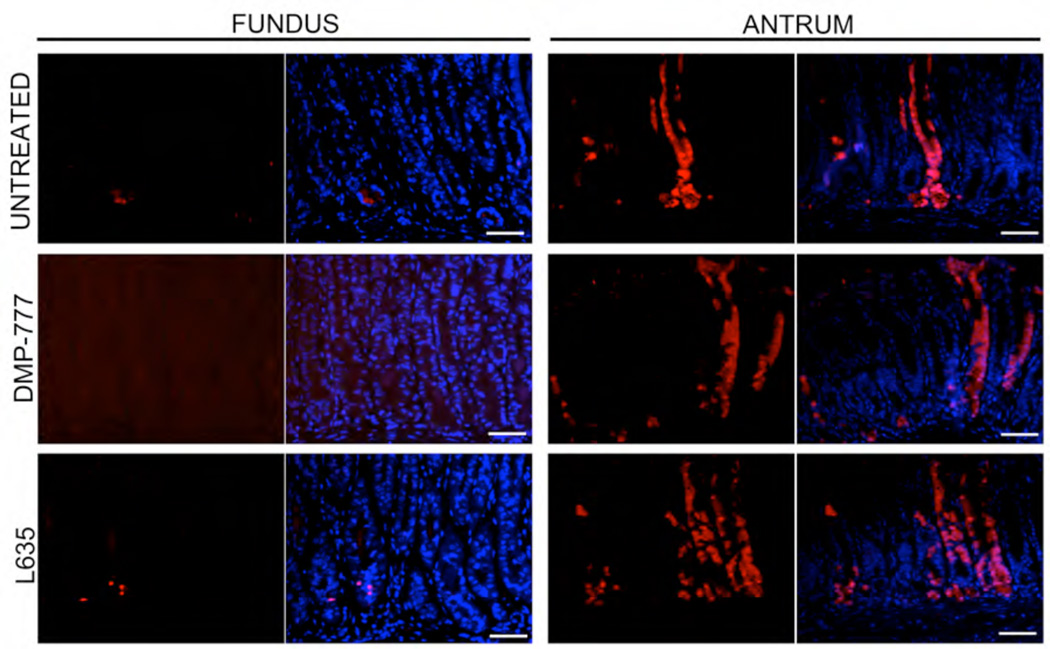
Figure 6. Bacterial β-galactosidase immunohistochemical staining in the oxyntic mucosa and antrum of Lgr5-EGFP-IRES-CreERT2/+ mice.
Lgr5-EGFP-IRES-CreERT2/+; Rosa26RLacZ mice were treated with 4 doses of tamoxifen and then following a 10 day hiatus mice were either maintained without further treatment, treated with 14 days of DMP-777 or treated with 3 days of L-635. The sections demonstrate that only scattered β-galactosidase (red) positive cells were observed at the bases of oxyntic glands even after treatment and no β-galactosidase staining was observed in SPEM. However, β-galactosidase (red) positive antral glandular lineages were visible in untreated Lgr5-EGFP-IRES-CreERT2/+ mice and Lgr5-EGFP-IRES-CreERT2/+ mice treated with either DMP-777 for 14 days or L-635 for three days. Bar size=50µm
DISCUSSION
Oxyntic atrophy, the loss of parietal cells in the gastric oxyntic mucosa, is requisite for the development of preneoplastic metaplasias.[21] Indeed, in humans oxyntic atrophy is the strongest correlate with risk for gastric cancer.[22] While the association of metaplasia in the stomach with the development of gastric cancer was first proposed over 20 years ago,[11] the origin of metaplasia has been controversial. Recent investigations have established that SPEM lineages in mice are predominantly derived from transdifferentiation of mature chief cells, especially in the presence of significant inflammatory infiltrates as observed in H. felis-infected mice or mice treated with L-635.[16] While those studies demonstrated that metaplasia derives from endogenous mucosal progenitors, it also indicates that metaplasia does not evolve from the normal oxyntic mucosal progenitors.[17] Rather, metaplasia develops from transdifferentiation of mature chief cells and recrudescence of proliferative capacity in the presence of inflammatory infiltrates.
In the intestine, crypt base cells that express Lgr5 can give rise to all of the normal lineages of the intestinal crypt and villus structures and can also give rise to neoplastic lineages.[19] Similarly, Lgr5-expressing cells also exist in the gastric antrum that can give rise to antral glandular lineages.[5] In contrast, few Lgr5-expressing cells exist in the oxyntic mucosa and those are located exclusively along the lesser curvature in mice. The results presented here indicate that the oxyntic mucosal cells with Lgr5 transcriptional activity are phenotypic chief cells, which express both intrinsic factor and pepsinogen. Nevertheless, following treatment with either oxyntic atrophy-inducing drug, we failed to observe any evidence for lineage mapping of the cells with Lgr5 transcriptional activity into SPEM lineages. Thus, in this case, cells with Lgr5 transcriptional activity in the gastric oxyntic mucosa do not represent progenitors for metaplasia. Indeed, in contrast with the Lgr5-expressing cells in the intestinal crypts and antral glands, we have found no evidence for proliferative capacity (as indicated by staining with Ki-67) in any Lgr5-expressing cells in the oxyntic mucosa in either untreated or treated mice.
The significance of Lgr5-transcriptional activity in a small population of cells along the lesser curvature of the stomach remains unclear. Since there presently is no antibody that can reliably detect Lgr5 immunostaining in mice, we cannot assess at this time whether these cells are actually making Lgr5 protein. Thus, the transcripts may be made in these cells without translation. In addition, it is important to note that, while Lgr5-expressing cells in the antrum and the intestines also show proliferative marker staining, no Ki-67 staining is ever seen in the chief cells of the normal stomach. Previous investigations have demonstrated that Olfactomedin 4 (OLFM4) co-labels Lgr5-expressing cells in the intestine[23]. However, we have previously reported that OLFM4 expression is markedly increased in SPEM and intestinal metaplasia in the human stomach[24]. Nevertheless, none of our studies have found up-regulation of Lgr5 expression in metaplasia either in humans or mice[18, 24, 25]. All of these results indicate that expression of lineage markers may vary both between gastrointestinal tissues and among pathological lineages.
While our recent studies on the origin of SPEM have indicated that SPEM originates from mature Mist1-expressing chief cell lineages,[5] it remains unclear whether chief cells, as a general population, can transdifferentiate into SPEM or whether SPEM arises from a subset of chief cells. Interestingly, the present data identify a population of chief cells on the lesser curvature with Lgr5 transcriptional activity, which do not appear to have the capacity to transdifferentiate into SPEM. The data therefore do indicate that subpopulations of chief cells with differing capacities for transdifferentiation do exist. More importantly, however, the results support the concept that SPEM may arise from a general capacity of many chief cells to transdifferentiate into mucous cell metaplasia.
Supplementary Material
SUMMARY BOX.
What is already known about this subject:
In humans, intestinal metaplasia and spasmolytic polypeptide-expressing metaplasia (SPEM) are associated with the development of gastric cancer.
Acute or chronic loss of parietal cells in mouse leads to the emergence of SPEM through transdifferentiation of chief cells into mucous cell metaplasia.
Lgr5-expressing cells in the intestine and gastric antrum are cycling pluripotent progenitor cells that can give rise to all lineages in the mucosa.
What are the new findings:
A small number of cells with Lgr5 transcriptional activity are present in the lesser curvature of oxyntic mucosa of the mouse stomach and these cells are phenotypic chief cells.
Induction of acute parietal cell loss with either DMP-777 or L635 leads to induction of SPEM but none of the mucous cell metaplasia is derived from Lgr5-expresing cells.
Lgr5-transcriptionally active cells are not pluripotent progenitor cells in the gastric acid-secreting mucosa and SPEM is not derived from Lgr5 expressing cells.
How might it impact on clinical practice in the foreseeable future?
Identification of SPEM may be a critical early biomarker for risk of gastric cancer.
ACKNOWLEDGMENTS
We thank Drs. Nicholas Wright, Jason Mills and David Alpers for the gifts of antibodies. These studies were supported by grants to J.R.G. from a Department of Veterans Affairs Merit Review Award 1I01BX000930, RO1 DK071590, and the AGA Funderburg Award in Gastric Biology Related to Cancer. This work was supported by core resources of the Vanderbilt Digestive Disease Center (P30 DK058404), and the Vanderbilt-Ingram Cancer Center.
Abbreviations
- SPEM
spasmolytic polypeptide expressing metaplasia
- TFF2
trefoil factor family 2
Footnotes
None of the authors have any conflicts of interest in the pursuit of this work.
Author contributions: Nam designed and performed experiments and wrote the manuscript; O’Neal performed experiments and edited manuscript; Finke produced L635 and edited manuscript; Coffey designed experiments and edited manuscript; Barker designed experiments and edited manuscript; Goldenring designed experiments and edited manuscript.
REFERENCES
- 1.Karam SM, Leblond CP. Dynamics of epithelial cells in the corpus of the mouse stomach. III. Inward migration of neck cells followed by progressive transformation into zymogenic cells. Anat Rec. 1993;236:297–313. doi: 10.1002/ar.1092360204. [DOI] [PubMed] [Google Scholar]
- 2.Karam SM. Dynamics of epithelial cells in the corpus of the mouse stomach. IV. Bidirectional migration of parietal cells ending in their gradual degeneration and loss. Anat Rec. 1993;236:314–332. doi: 10.1002/ar.1092360205. [DOI] [PubMed] [Google Scholar]
- 3.Ramsey VG, Doherty JM, Chen CC, et al. The maturation of mucus-secreting gastric epithelial progenitors into digestive-enzyme secreting zymogenic cells requires Mist1. Development. 2007;134:211–222. doi: 10.1242/dev.02700. [DOI] [PubMed] [Google Scholar]
- 4.Karam SM, Leblond CP. Dynamics of epithelial cells in the corpus of the mouse stomach. I. Identification of proliferative cell types and pinpointing of the stem cells. AnatRec. 1993;236:259–279. doi: 10.1002/ar.1092360202. [DOI] [PubMed] [Google Scholar]
- 5.Barker N, Huch M, Kujala P, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 6.Jaks V, Barker N, Kasper M, et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 7.Barker N, van de Wetering M, Clevers H. The intestinal stem cell. Genes Dev. 2008;22:1856–1864. doi: 10.1101/gad.1674008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 9.Correa P, Piazuelo MB, Wilson KT. Pathology of gastric intestinal metaplasia: clinical implications. Am J Gastroenterol. 2010;105:493–498. doi: 10.1038/ajg.2009.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filipe MI, Munoz N, Matko I, et al. Intestinal metaplasia types and the risk of gastric cancer: a cohort study in Slovenia. Int J Cancer. 1994;57:324–329. doi: 10.1002/ijc.2910570306. [DOI] [PubMed] [Google Scholar]
- 11.Correa P. A human model of gastric carcinogenesis. Cancer Res. 1988;48:3554–3560. [PubMed] [Google Scholar]
- 12.Schmidt PH, Lee JR, Joshi V, et al. Identification of a metaplastic cell lineage associated with human gastric adenocarcinoma. Lab Invest. 1999;79:639–646. [PubMed] [Google Scholar]
- 13.Fox JG, Li X, Cahill RJ, et al. Hypertrophic gastropathy in Helicobacter felis-Infected wild type C57BL/6 mice and p53 hemizygous transgenic mice. Gastroenterology. 1996;110:155–166. doi: 10.1053/gast.1996.v110.pm8536852. [DOI] [PubMed] [Google Scholar]
- 14.Wang TC, Goldenring JR, Dangler C, et al. Mice lacking secretory phospholipase A2 show altered apoptosis and differentiation with Helicobacter felis infection. Gastroenterology. 1998;114:675–689. doi: 10.1016/s0016-5085(98)70581-5. [DOI] [PubMed] [Google Scholar]
- 15.Goldenring JR, Ray GS, Coffey RJ, et al. Reversible drug-induced oxyntic atrophy in rats. Gastroenterology. 2000;118:1080–1093. doi: 10.1016/s0016-5085(00)70361-1. [DOI] [PubMed] [Google Scholar]
- 16.Nam KT, Lee H-J, Sousa JF, et al. Mature chief cells are cryptic progenitors for metaplasia in the stomach. Gastroenterology. 2010;139:2028–2037. doi: 10.1053/j.gastro.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quante M, Marrache F, Goldenring JR, et al. TFF2 mRNA transcript expression marks a gland progenitor cell of the gastric oxyntic mucosa. Gastroenterology. 2010 doi: 10.1053/j.gastro.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nomura S, Yamaguchi H, Ogawa M, et al. Alterations in gastric mucosal lineages induced by acute oxyntic atrophy in wild-type and gastrin-deficient mice. Am J Physiol Gastrointest Liver Physiol. 2005;288:G362–G375. doi: 10.1152/ajpgi.00160.2004. [DOI] [PubMed] [Google Scholar]
- 19.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 20.Nomura S, Yamaguchi H, Wang TC, et al. Alterations in gastric mucosal lineages induced by acute oxyntic atrophy in wild type and gastrin deficient mice. Amer J Physiol. 2004;288:G362–G375. doi: 10.1152/ajpgi.00160.2004. [DOI] [PubMed] [Google Scholar]
- 21.Meining A, Morgner A, Miehlke S, et al. Atrophy-metaplasia-dysplasia-carcinoma sequence in the stomach: a reality or merely a hypothesis? Best Prac Res Clin Gastroenterol. 2001;15:983–998. doi: 10.1053/bega.2001.0253. [DOI] [PubMed] [Google Scholar]
- 22.El-Zimaity HMT, Ota H, Graham DY, et al. Patterns of gastric atrophy in intestinal type gastric carcinoma. Cancer. 2002;94:1428–1436. doi: 10.1002/cncr.10375. [DOI] [PubMed] [Google Scholar]
- 23.van der Flier LG, Haregebarth A, Strange DE, et al. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 24.Lee HJ, Nam KT, Park HS, et al. Gene expression profiling of metaplastic lineages identifies CDH17 as a prognostic marker in early stage gastric cancer. Gastroenterology. 2010;139:213–225. doi: 10.1053/j.gastro.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nozaki K, Weis V, Wang TC, et al. Altered gastric chief cell lineage differentiation in histamine-deficient mice. Am J Physiol Gastrointest Liver Physiol. 2009;296:G1211–G1220. doi: 10.1152/ajpgi.90643.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.