Abstract
Set2 is a RNA Polymerase II (RNAPII) associated histone methyltransferase involved in the co-transcriptional methylation of the H3 K36 residue (H3K36me). It is responsible for multiple degrees of methylation (mono-, di-, tri-), each of which has a distinct functional consequence. The extent of methylation and its genomic distribution is determined by different factors that co-ordinate to achieve a functional outcome. In yeast, the Set2-mediated H3K36me is involved in suppressing histone exchange, preventing hyperacetylation and promoting maintenance of well-spaced chromatin structure over the coding regions. In metazoans, separation of this enzymatic activity affords greater functional diversity extending beyond the control of transcription elongation to developmental gene regulation. This review focuses on the molecular aspects of the Set2 distribution and function, and discusses the role played by H3 K36 methyl mark in organismal development.
Introduction
The eukaryotic genome is packaged into a nucleoprotein complex termed chromatin, consisting of repeating units called nucleosomes. Each nucleosome consists of a proteinaceous core composed of four pairs of histone proteins, around which is wrapped ~146 basepairs of DNA. While this arrangement is necessary to accommodate the genome within the confines of the nucleus, it prevents access to the DNA for processes such as transcription and replication. Studies on individual gene loci have shown that positioned nucleosomes at promoters are unfavorable to initiation of transcription. Therefore, transcription initiation requires the disassembly or mobilization of the nucleosome1, followed by post-transcriptional reassembly or replacement of the nucleosome. The cell has evolved specific mechanisms, allowing the transcription machinery access to DNA. This includes histone modifications, histone chaperones and chromatin remodeling complexes, the combined action of which regulates nucleosome stability. In chromatin-based regulation of transcription, histone modifications play a central role as they are used as a signal or a dock for effector proteins modulating downstream events, for as long as they persist.
Several proteins involved in the process of transcription elongation are also known to be necessary for chromatin maintenance during transcription. A key histone modification in this process is the co-transcriptional methylation of H3K36me by Set2. We review the roles of Set2 in the establishment and maintenance of the H3 K36 methyl mark, and its functional consequences to achieve complex and critical outcomes such as organismal development.
Set2-mediated H3 K36 methylation
Transcription over a gene proceeds through three distinct stages of initiation, elongation and termination, with each stage requiring a defined set of factors that modulate each process2, 3. Initiation of transcription on a chromatin template begins with nucleosomal destabilization over the promoter. Nucleosomal depletion over the promoters occurs through a combination of different processes including histone modifications, chromatin remodeling and the presence of DNA-sequences influencing nucleosome stability. These events promote the binding of DNA sequence-specific activator proteins that recruit the RNAPII and general transcription factors to the promoter.
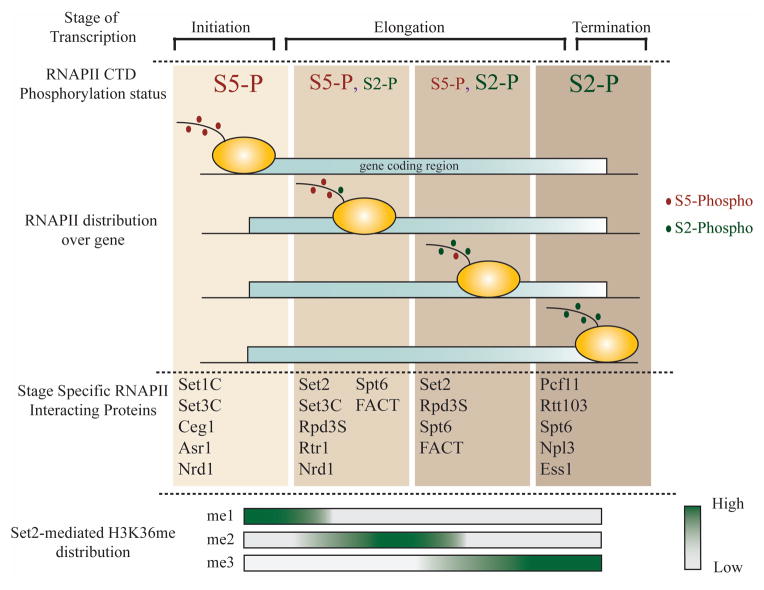
One of the key features of the transcription cycle is the phased manner in which several different proteins are recruited to the gene to aid RNAPII. These dynamic associations coordinate the different events of transcription to achieve proper gene expression. What facilitates these associations is an unusual structural feature of the RNAPII enzyme. The C-terminal domain (CTD) of the largest subunit of RNAPII, Rpb1 consists of tandem heptapeptide repeats (Y1-S2-P3-T4-S5-P6-S7) that are highly conserved across species4,5. This domain forms a platform through which different proteins interact with RNAPII. The dynamism of these interactions is facilitated by the sequential, reversible phosphorylation of S5 and S2 residues of the CTD repeats in a transcription-stage specific manner (Fig. 1). Upon formation of the preinitiation complex (PIC), the kinase activity of TFIIH (Kin28 in yeast and Cdk7 in higher eukaryotes) phosphorylates the S5 residue. This activity recruits different transcription elongation complexes including Set1C, Paf1C, Spt6 (independent of CTD6)and FACT7 (Spt16, Pob3, Nhp6a and b), allowing the RNAPII to proceed into elongation. About 500–750 base pairs (bp) into the gene body, Rtr1, a phosphatase associated with the RNAPII and Ssu72, dephosphorylates the S5 residue8, 9, resulting in the gradual loss of S5 phosphorylation mark towards the 3′ ends of genes. After the RNAPII clears the promoter, the C-terminal domain kinase-1 (CTDK-1)(catalytic subunit Ctk1 in yeast and Cdk9 in higher organisms), phosphorylates the S2 residue, stimulating the recruitment of the Set2 histone lysine methyltransferase10 (KMT). This KMT catalyzes the transfer of multiple methyl groups (mono-, di-, tri-methylation) on the histone H3 K36 residue. The various degrees of methylation are distributed in a graded manner over the coding region (Fig. 1, bottom) with H3 K36 mono-methylation (H3K36me1) over the 5′ ends, di-methylation (H3K36me2) over mid-coding regions, and tri-methylation (H3K36me3) towards the 3′ ends of genes11. This distribution is determined by the association of different regulatory proteins with Set2 (as discussed in the next section) and the transcription status of the gene12 ultimately affecting its function.
Figure 1. RNAPII CTD phosphorylation cycle across the coding region.
Distribution of the RNAPII CTD phosphorylation marks across the coding region of genes. Differential phosphorylation of the S5 or S2 residues in the heptad repeat contributes to the stage-specific association of transcription regulatory proteins. The font size of the CTD phosphorylation at the top of the figure corresponds to the relative abundance of that post-translational CTD modification at a particular stage. The distribution of the Set2-dependent H3K36 methyl marks (me1, me2, me3) across the coding region is denoted as a heat map at the bottom.
The Set2 protein and its KMT activity are conserved across species. However, Set2 orthologs in metazoans (Table 1) have evolved to include several structural and functional features that distinguish them from the yeast counterpart. They show considerable variation in the degree of methylation catalyzed as well as the substrates targeted. One striking feature is the separation of the H3 K36 mono-/di-methylation activity from the tri-methylation activity on different proteins. This alludes to the distinct functions of each methyl mark, and separation of the catalytic activities is necessary for regulation based on their protein interaction profile or tissue-specific expression patterns.
Table 1.
Set2 Orthologs: Targets and functions
| Orthologs | Targets | Function | Reference |
|---|---|---|---|
| Yeast | |||
|
| |||
| Set2 (KMT3) | H3 K36 me1, me2, me3 | Transcription elongation. | 10 |
| Maintains histone stability over the coding regions. | 69 | ||
| Prevents incorporation of acetylated histones. | 69 | ||
| Stimulates deacetylase activity. | 55, 56, 60, 61 | ||
| C. elegans | |||
|
| |||
| MES-4 | H3 K36 me1, me2 | X-chromosome silencing, germ cell viability. | 100, 102, 103 |
| Germ cell viability. | 100 | ||
| MET-1 | H3 K36 me3 | Transcription elongation specific mark. | 105 |
| Vulval development. | 105 | ||
| D. melanogaster | |||
|
| |||
| dMES4 | H3 K36 me1, me2 | H4 K16 acetylation during transcription elongation. | 86 |
| HYPB/dSet2 | H3 K36 me3 | Transcription elongation | 86 |
| Dosage compensation | 84, 85 | ||
| Role in larval-pupal transition. | 108 | ||
| H. sapiens | |||
|
| |||
| SETD2/HYPB/KMT3A | H3 K36 me3 | Splicing. | 93, 95, 96 |
| NSD1/KMT3B | H3 K36 me1, me2, H4 K20 me3 | Post-gastrulation embryonic development. | 107 |
| NSD2/WHSC1 | H3 K36 me2 | Cranio facial development. | 71 |
| Heart development. | 71 | ||
| NSD3/WHSC1L1 | H3 K36 me2 | ||
| ASH1L | H3 K36 me3, H3 K4 me | ||
| SMYD2 | H3 K36 me2 | Regulation of p53 and Rb proteins. | 53 |
| SETMAR | H3 K36 me2 | Non homologous end joining. | 109 |
| A. thaliana | |||
|
| |||
| SDG8/EFS | H3 K36 me2, me3, H3 K9 me3 | Stress response to fungal infections. | 115 |
| Flowering response. | 112, 113 | ||
| Anther and ovule development. | 114 | ||
| SDG26 | H3 K36 me1 | Flowering response. | 112, 113 |
Factors affecting H3K36 methylation by Set2
Set2-mediated H3K36me is modulated by a variety of factors that either associate with the enzyme or belong in the same pathway (Table 2). In this section, we outline the different chromatin related factors that influence the activity of Set2 and thereby the degree and distribution of the H3 K36 methyl mark.
Table 2.
Factors affecting Set2-mediated H3 K36 methylation
| Factor | Effect on H3 K36 methylation | Reference |
|---|---|---|
| RNAPII and elongation factors | ||
|
| ||
| RNAPII CTD Ser-5-P and S-2-P | Affects me2, me3 | 10, 23, 24, 25 |
| CTDK1 (Ctk1, Ctk2, Ctk3) | Affects me1, me2, me3 | 26, 27, 28 |
| Bur1 | Affects me3 | 29, 33 |
| Paf1C (Ctr9 and Paf1) | Affects me3 | 33 |
| Factors affecting nucleosome stability | ||
|
| ||
| Asf1 | Affects me3 | 37 |
| Spt6/Iws1 | Affects me3 | 35, 36 |
| FACT complex | Affects me3 | 40, 41 |
| Histone H4 K44, H2B L116, L117 | Affects me2, me3 | 21, 22 |
| Histone H3 P38 and Fpr4 proline isomerase | Affects me3 | 43 |
| Factors affecting RNA stability | ||
|
| ||
| HnRNP-L (Human) | Affects me3 | 44 |
| Histone H3 K36 Demethylases | ||
|
| ||
| Jhd1 | Affects me1, me2 | 47, 49, 50 |
| Rph1 | Affects me2, me3 | 47, 48, 50 |
Domains within the Set2 protein regulate its lysine methylation activity (Fig. 2). The evolutionarily conserved catalytic site is found within a ~130 amino-acid (a.a) SET domain, named after three Drosophila genes containing this domain, Su(var)3–9, E(z) and trithorax13. This region is flanked on both sides by cysteine-rich regions, the Associated with SET (AWS) and the Post-SET domains that are required for the methyltransferase activity14. These domains are thought to determine the specificity of the lysine residue targeted for methylation15, 16. The active site in the SET domain accommodates the aliphatic side chain and the ε-amino group of the lysine residue and requires a methyl donor, S-adenosyl-L-methionine (AdoMet) as a cofactor. The structure of SET domain allows the substrate and cofactor separate access to the catalytic site such that the cofactor can transfer the methyl group to the active site and leave17. This feature is useful for the SET domain containing proteins to allow multiple degrees of methylation without the substrate having to leave the active site. Interestingly, the presence of a phenylalanine or tyrosine at the catalytic site of SET domains determines the degree to which the target lysine can be methylated. For example mutating F281 to Y in the N. crassa Dim-5 methyltransferase, converts it from an H3K9 trimethylase to a mono-/dimethylase18. Thus, stearic hindrance in the active site is responsible for limiting the degree to which an enzyme can methylate lysine substrates19.
Figure 2. Set2 domain architecture and its effect on H3 K36 methylation.
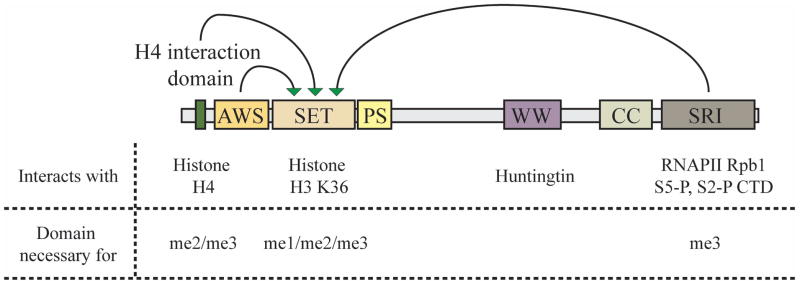
Domain architecture and interactions of the yeast Set2 protein and their effect on H3K36me. The green arrows indicate the necessity of the domain for the methyltransferase activity of the SET domain. AWS: Associated with SET; SET: Su(var), E(z) and Trithorax; PS: Post-SET; WW: Typtophan-rich domain; CC: coiled coil domain; SRI: Set2-Rpb1 interaction domain.
Set2 can methylate both histone octamers and nucleosomes20. Studies with yeast Set2 revealed that histone H4 allosterically activates H3K36me. Further analysis led to the identification of a trans-histone methylation pathway with the H4 K44 residue necessary for positioning the active site of Set2 on H3 K36 on nucleosome templates14. Additionally, residues on H2A and H3 surrounding H4 K44 in nucleosome are also required for H3K36me21. Set2 interacts with this region through a nine-residue domain at its N-terminus (Fig. 2). Loss of this domain reduced Set2 binding to nucleosomes and abrogated di- and tri-methylation of K36, while binding the CTD was unaffected14. While Set2 binds directly to unmodified H3 K36, initiating catalysis, the addition of methyl groups inhibits direct binding. Therefore, the trans-histone methylation pathway provides an anchor for the Set2 enzyme thereby facilitating multiple methylation of H3 K36. Interestingly, the human nuclear receptor SET domain-containing-2 (NSD2) enzyme that catalyzes H3 K36me1 and H3K36me2, also targets H4 K44 when methylating histone octamers but not nucleosomes. However, while H3K36me by NSD2 requires H4 K44, it is independent of its methylation status22.
The interaction of Set2 with the phosphorylated CTD of RNAPII targets it from the middle to 3′ end of genes. This association with the S5 and S2 di-phosphorylated CTD occurs through its C-terminal Set2-Rpb1 interaction (SRI) domain23. Optimal binding of SRI to the CTD requires a minimum of two S5/S2 di-phosphorylated heptad repeats24, 25. Loss of the SRI domain abolished di- and tri-methylation of H3 K36 (Fig. 2), although it had no effect on mono-methylation14. Further deletion of the C-terminal domain of Set2 can partially restore di-methylation26. Loss of the CTDK-1 kinase subunit, Ctk1 abrogates methylation of H3 K3627, 28, while deletion of BUR1 (another Cdk9 ortholog in yeast) results in the loss of H3 K36 tri-methylation only29. The Bur1-Bur2 cyclin-dependent kinase is required for the recruitment of the Paf1 complex and subsequent H2B ubiquitylation and H3 K4 methylation30–32. Loss of either Paf1 or its interacting partner Ctr9, results in loss of H3 K36 tri-methylation33 presumably by affecting the transcription elongation pathway. Interestingly, all these mutations also perturb the S2 phosphorylation levels on RNAPII CTD, resulting in destabilization of the Set2 protein34.
The requirement of amino acid residues on H4, H2A and H3 for H3K36me indicates that Set2 needs intact nucleosomes to methylate its target lysine, in vivo. Consistent with this observation, a number of histone chaperones involved in maintenance of nucleosomal structure during elongation are essential for H3K36me3. In humans, the recruitment of the H3 K36 tri-methyltransferase SETD2 has been dissected with respect to Spt6, which is the histone chaperone involved in post-elongation nucleosomal reassembly. It is a phospho-S2 binding protein26, 35 that targets the proximal conserved heptad repeats. Spt6-CTD binding is necessary for the recruitment of Interacts with Spt6-1 (Iws1), which in turn recruits the SETD2 protein36. These observations suggest that the tri-methylation of H3 K36 is coupled with reassembly of the nucleosome after the passage of RNAPII.
Both the FACT complex29 and Asf137, histone chaperones involved in transcription elongation38, 39, are also necessary for maintaining H3 K36 tri-methylation but loss of either factor does not affect mono- or di-methylation. The FACT complex has been shown to facilitate the reassembly of H2A-H2B dimers that are preferentially mono-ubiquitinated on histone H2B40, 41. Consistent with this fact, loss of Large cells 1 (Lge1) a component of the yeast Bre1-ubiquitin ligase complex is associated with a loss of H3 K36 tri-methylation only42.
Other factors that influence H3 K36 tri-methylation include the Fpr4 prolyl isomerase and the RNA-associated protein HnRNPL. Fpr4 mediated prolyl isomerization of the H3 P38 residue renders the conformation of the H3 K36 residue unsuitable for tri-methylation43. HnRNPL associates with the human SETD2 protein and promotes the trimethylation of the target lysine44. Interestingly, HnRNPL has been implicated in regulating splicing, which in turn has been shown to activate H3 K36 tri-methylation45.
In addition to the factors that interact with Set2 and modulate its activity, histone lysine demethylases (KDM) can enzymatically remove H3 K36 methyl groups. Yeast H3 K36 demethylases include Jhd1 and Rph1. Both proteins contain the Jumonji C (JmjC) domain and catalyze the oxidative demethylation of methyl-lysine targets46. Rph1 demethylates both H3 K36 tri- and di- methyl substrates47, 48 while Jhd1 targets di- and mono-methyl H3 K3649, 50. Interestingly, metazoan H3 K36 demethylases bind H3 K4 methylated residues and are targeted to the 5′ ends of genes. This feature may be important to maintain the genomic distribution of the different H3 K36 methylated states (mono-, di-, tri-)51, 52.
Each factor affecting H3 K36 methylation modulates the number of methyl group added. Such a level of cellular control alludes to the functional exclusivity of each degree of methylation. Therefore, the interplay between these factors over chromatin determines the degree of methylation of the target residue as well as its genomic location, creating a variety of functional outcomes. Additionally, human SET and MYND domain-containg protein 2 (SMYD2) has also been shown to methylate p53 and retinoblastoma (Rb) proteins, enhancing the functional diversity of the SET proteins53.
Functional consequences of H3 K36 methylation
Regulation of chromatin processes by histone modifications occurs through several chromatin modifying proteins. Histone modifications can be viewed as signals on histones that are recognized by specific domains on these effector molecules. Methyl-lysine residues are recognized by a number of conserved domains including chromo-, Plant homeo domain (PHD), WD40, Tudor and PWWP domains54. These domains are found in a diverse range of protein complexes affecting chromatin structure, all of which coordinate to achieve specific functional consequences.
Set2/Rpd3S pathway
H3 K36 methylation is a co-transcriptional event that is enriched over the 3′ end of the coding region. Loss of Set2-mediated H3 K36 methylation resulted in hyperacetylation of histones over this region. A similar phenotype was observed on the loss of Eaf3 and Rco1, two components of the Rpd3S histone deacetylase complex55, 56. The Rpd3S complex is recruited to the coding region through its interaction with the S5 and S2 di-phosphorylated CTD of RNAPII57, 58. Eaf3 is a subunit shared with the NuA4 complex59 and contains a chromodomain that binds to methylated H3 K36 and H3 K455, 60. Interestingly, the PHD domain of Rco1 mediates the modification-independent nucleosomal binding of the Rpd3S complex so as to enhance the specificity of Eaf3 towards di- or tri-methylated H3 K3642, 61 and the subsequent deacetylation of histones H3 and H4. Thus, the Set2-mediated H3 K36 methylation coordinates the recruitment and activation of the Rpd3S complex over the coding regions, resulting in the maintenance of a hypo-acetylated state (Fig. 3). Although the H3 K36 methyl mark is present on all active genes, the Set2/Rpd3S pathway affects infrequently transcribed genes and long genes in yeast62. Mammalian cells also possess a complex similar to yeast Rpd3S that is involved in the deacetylation over transcribed loci63. Linking the co-transcriptional H3 K36 methyl mark to the activation of histone deacetylase complex ensures that the coding regions are hypo-acetylated after the passage of RNAPII.
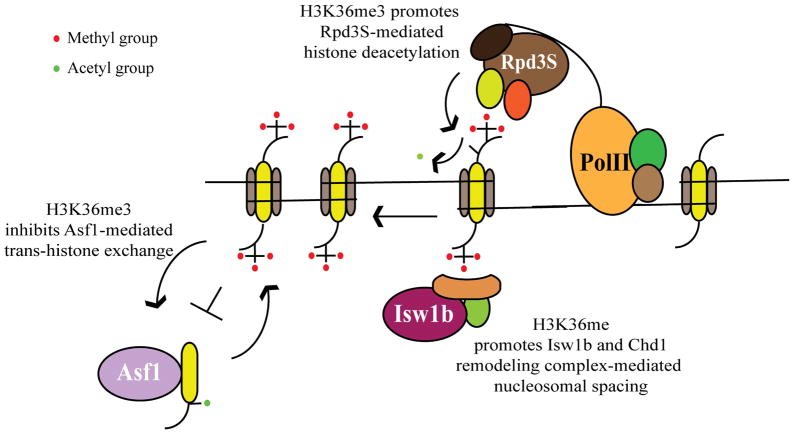
Figure 3. Role of Set2-mediated H3K36 methylation in nucleosomal dynamics.
Set2-mediated K36 methylation regulates nucleosome dynamics through different mechanisms. It suppresses histone exchange over the coding regions by preventing binding of the Asf1 protein to the methylated histones, and promotes the retention of the original nucleosome. This prevents the enrichment of pre-acetylated histones on the coding regions. H3K36me also recruits the Isw1b protein complex to maintain regular nucleosomal spacing after the passage of the elongating polymerase. H3K36me also activates the deacetylase activity of the RNAPII-associated Rpd3S complex, to remove the acetyl marks that have been added by histone acetyltransferase complexes.
H3K36 methylation and nucleosomal dynamics
Nucleosomal dynamics during transcription elongation is usually a consequence of the passage of RNAPII64. Recent in vitro work has shown that moderate RNAPII elongation occurs over the nucleosome with the loss of a single H2A-H2B dimer, leaving a hexameric nucleosomal complex behind65. In conjunction with this observation, in vivo studies have shown a continuous exchange of the H2A-H2B dimers over the coding regions66. In yeast, H3 turnover over coding regions is very slow for a majority of genes with the exception of highly transcribed genes67, 68. We have recently shown that the H3 K36 methyl mark over the coding region prevents the replacement of histone H3 over the coding region with newly synthesized ones69. Interestingly, this feature also prevents the enrichment of pre-acetylated histones over the coding region, suggesting that co-transcriptional acetylation was partly a consequence of histone exchange69, apart from the recruitment of histone acetyltransferase complexes70. Indeed, targeting SET domain proteins to the promoters results in transcriptional repression20. Wolf-Hirschhorn syndrome candidate 1(WHSC1, formerly known as NSD2) is recruited by the cardiac-specific transcription factor Nkx2-5 and represses the platelet-derived growth factor α gene71. This observation can be attributed to the ability of H3K36me to suppress histone exchange and thereby promoter acetylation. This suggests that histone exchange over promoters could be involved in the maintenance of histone acetylation. In a recent study, depletion of one copy of H3 results in the loss of nucleosomes over the promoter regions while the coding regions were enriched for H3 K36 methylated nucleosomes72, suggesting that this histone mark is required for nucleosomal retention during transcription elongation.
The suppression of histone exchange over the coding regions is achieved by H3 K36 methylation through two distinct mechanisms. H3 K36 di- and tri-methylation impedes the interaction of Asf1– a histone chaperone involved in histone exchange during transcription elongation– to histones over the coding regions69. At the same time, this methyl mark recruits the imitation switch 1b (Isw1b) chromatin-remodeling complex over the coding regions, which acts along with chromodomain-helicase-DNA binding-1 (Chd1) to re-establish chromatin integrity following the passage of RNAPII73–75 (Fig. 3). These mechanisms ensure that while H2A-H2B dimer exchange occurs unimpeded, the H3-H4 tetramer core remains intact. This results in the persistence of the H3 K36 methyl mark over the coding region following transcription elongation. Recently, the common core of Rpd3S and the promoter-specific Rpd3L deacetylase complexes composed of three subunits– Rpd3, Sin3 and Ume155–was shown to possess a histone modification-independent histone chaperone activity76. The core subunits prevent nucleosome eviction but not the remodeling activity of the RSC complex, indicating the possible involvement of another H3K36me-recruited complex in the maintenance of chromatin integrity. Histone exchange in metazoans accompanies transcription, resulting in the replacement of the replication-specific histone H3.1 with the transcription-specific H3.377, 78. Interestingly, the latter histone variant is enriched for the H3 K36 methyl mark, added following the process of transcription elongation and histone replacement79. The H3K36me on H3.3 may prevent it from undergoing further exchange, while facilitating further rounds of transcription.
Set2/Rpd3S pathway and cryptic transcript initiation
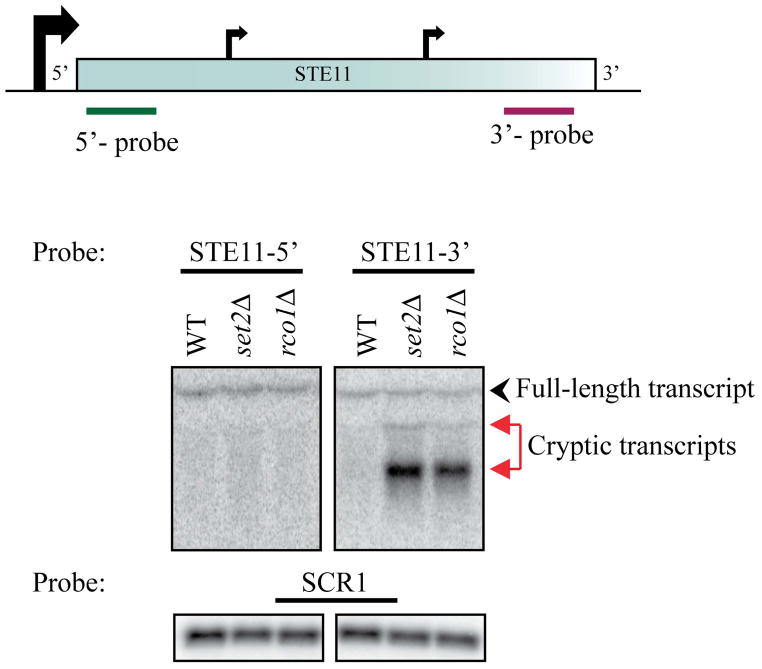
Transcription initiation at sites other than the 5′ end of the gene is a widespread phenomenon in Drosophila and humans80. The use of multiple start sites coupled with antisense and intergenic transcription indicates the complexity of the transcriptome. Most of these transcripts are tightly regulated, alluding to their function in critical cellular events. In yeast, loss of the Set2/Rpd3S pathway results in the widespread initiation of transcription from within the coding regions (Fig. 4), resulting in both sense and antisense transcripts62. Several chromatin-associated proteins regulate cryptic transcript initiation, suggesting the importance of chromatin structure in suppressing this phenotype81, 82. Interesting, many of these proteins are involved in the maintenance of H3 K36 methylation. Recent studies have shown that cryptic transcription is activated in response to cellular stress, and that some of these transcripts are translated, although no function has been assigned to these proteins81.
Figure 4. Cryptic transcription upon loss of Set2/Rpd3S pathway.
Total RNA was prepared from the wild type, set2Δ and rco1Δ strains and subjected to Northern blot analysis. The probes used in these assays were from the 5′ and 3′ ends of the STE11 gene (top). An SCR1 probe was used as a loading control. The full-length transcript and the short cryptic transcripts are indicated. The approximate start sites for the cryptic transcripts from within the coding regions are indicated.
H3 K36 tri-methylation and dosage compensation
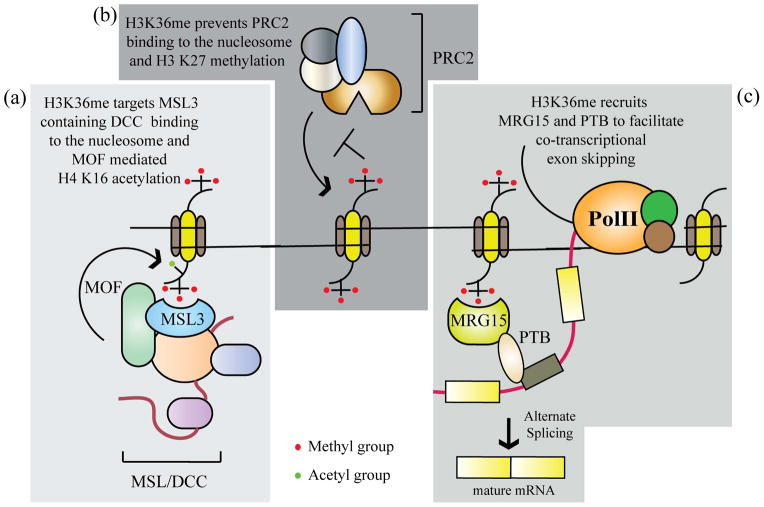
Dosage compensation is a process that ensures gene expression is normalized between the autosomes and the sex chromosome83. In male flies it involves an ~ 2-fold increase in transcription from the single X chromosomes to equal that from diploid autosomes. The Male Specific Lethal (MSL) complex in male flies is targeted to the X chromosomes in a DNA sequence specific manner, followed by their spread over the chromosomes. This ectopic spreading is facilitated by H3 K36 tri-methylation by recruiting the chromodomain-containing MSL3 subunit of the MSL complex84 to the 3′ ends of genes. The males absent on the first (MOF) subunit of the MSL complex catalyzes the acetylation of H4 K16 residue, which is involved in the maintenance of active chromatin by preventing compaction of chromatin85 (Fig. 5a). The exact role of the MSL complex in upregulating transcription is yet to be deciphered. Interestingly, the loss of Drosophila Set2 results in an increase in H4 K16 acetylation over the coding regions86.
Figure 5. H3 K36 methylation in transcription regulation.
H3K36me regulates transcription rates by a number of methods. (a) In dosage compensation in flies, it recruits the MSL/DCC to the coding regions of the X-chromosome, stimulating the H4 K16 acetylation by the MOF subunit. This increases the rate of transcription over the X-chromosome to equal that of the autosomes. (b) H3K36me prevents the binding and subsequent methylation of the H3K27 residue by the repressive polycomb PRC2 complexes. (c) H3K36me regulates alternate splicing of the transcript in a co-transcriptional manner. The MRG15 (yeast Eaf3) protein binds the H3K36me mark through its chromodomain. PTB binds MRG15 and facilitates exon skipping.
H3 K36 methylation and Polycomb silencing
Polycomb group (PcG)-mediated silencing is involved in dosage compensation through X-inactivation, the developmental regulation of homeotic (Hox) genes and cancers87, 88. Two main PcG complexes have been identified that work in conjunction with each other, although some exceptions exist. Polycomb repressive complex 2 (PRC2) complex contains a histone methyltransferase targeting the H3 K27 residue. This methylation is thought to recruit the PRC1 complex resulting in silencing of the locus. PRC2 itself recognizes H3 K27 methylation, activating the methyltransferase allosterically, resulting in the spreading of the mark, and creating a silenced genomic region. Active marks, predominantly H3 K4 and K36 methylation, restrict this spreading activity of PRC289. H3 K36 di- and tri-methylation prevent PRC2 complex binding and its activity89, 90 (Fig. 5b). It has been recently shown that the PRC2 enzyme is strongly activated by dense nucleosome packing. Interestingly, amino acids 35–42 on histone H3 of the neighboring nucleosome bound an acidic region of the Su(z)12 subunit of PRC2 through electrostatic interactions91. Knockdown of the human SETD2 protein results in global up-regulation of H3 K27 tri-methylation levels36. Interestingly, in Drosophila, the antagonistic trithorax group protein Absent, small or homeotic-1-like (ASH1L) has been shown to be a H3 K36 di-methylase90 in vitro. While H3K36me inhibits PRC2-mediated H3 K27 methylation, Drosophila Setd2 has been shown to act on H3 K27 methylated templates, suggesting that Polycomb-mediated silencing can be reversed by active transcription.
H3K36 methylation and Alternative splicing
Alternative splicing (AS) of precursor mRNA is a co-transcriptional process, in which differentially selected splice sites result in the production of functionally distinct mRNA and protein isoforms92. AS is a tightly regulated process requiring the recruitment of a vast array of proteins that catalyze the splicing event. A key event in this coordinated process involves the recognition of the splice site by proteins that either promote or suppress splicing, which results in inclusion or exclusion of the exon, respectively. Polypyrimidine-tract binding protein 1 (PTBP1) protein is a splice junction-binding protein that suppresses splicing resulting in exon exclusion (Fig. 5c). PTBP1 is recruited on nascent RNA by MORF4 related gene on chromosome 15 (MRG15, a homolog of yeast Eaf3) bound to methylated H3 K3693. The selection of fibroblast growth factor receptor 2 (FGFR2) exon IIIc over exon IIIb in mesenchymal cells is due to the distribution of H3 K36 tri-methylation over exon IIIb93, resulting in PTBP1-mediated exon exclusion. However, in epithelial cells, exon IIIb is included due to the epithelial splicing regulatory protein 1 (ESRP1)94 and is not enriched for the H3 K36 methyl mark. Indeed, the epithelial-to-mesenchymal transition (EMT), a hallmark of metastasis, involves a switch from FGFR2-IIIb to FGFR2-IIIc94, indicating a role of tissue-specific H3K36me distribution in carcinogenesis. Similarly, mouse PC4 and SF2 interacting protein 1 (Psip1) binds H3 K36 tri-methylated nucleosomes directly through a PWWP domain and regulates AS, usually at downstream exons95. In agreement with a role in regulating exon selection, H3K36me3 is differentially distributed between included exons and introns96. These observations point to the role of H3 K36 methylation in alternate splicing. The chromatin remodeler Chd1, involved in the Set2-dependent maintenance of chromatin integrity after RNAPII elongation73, also plays a role in alternate splicing97.
H3K36 methylation and Development
The sequential activation and regulation of gene networks is a hallmark of developmental processes resulting in morphological outcomes. Developmental regulation serves to generate a spatio-temporal pattern of gene expression that not only specifies the exact time, but also the cell in which the gene must be expressed. One point to note here is that these networks not only regulate the activation but also coordinate the repression of genes to specify cell fates. Additional regulation at RNA and protein levels, determines the precise outcome. Any perturbation in this network would lead to developmental defects and/or carcinogenesis98. In this section, we enumerate the developmental effects of the H3 K36 methylation pathway.
MES-4 and epigenetic germline maintenance
Divergent somatic and germline lineages are established during early division cycles of metazoan embryonic development and are maintained throughout organismal development99. The germ cell lineage is characterized by unique chromatin architecture, and the factors involved in the maintenance of this lineage are mostly chromatin-associated proteins. Maintenance of germ cell lineage requires a balance between gene expression and overall repression to ensure genomic integrity99. In particular, two classes of genes involved in chromatin state specification in C. elegans highlight this disparity. The maternal-effect sterile (MES) genes are maternally supplied factors that are crucial for the development of the germ cell lineage. Loss of any one of the four MES factors results in the deterioration of the primordial germ cells in the zygote, leading to the production of sterile worms100. MES-2, MES-3 and MES-6 form a repressive complex similar to the PRC2 complexes in other metazoans species, and are involved in silencing the X-chromosomes during germ cell development101, 102. MES-4 is a histone methyltransferase involved in H3 K36 methylation (Table 1). MES-4 and the H3 K36 di-methyl mark are found enriched over the autosomes, but not over the X-chromosome (except the tip of the left arm) in the germline tissue103, 104. This exclusion of MES-4 over the X-chromosome is thought to be due to the MES-2,-3,-6 complex. Interestingly, loss of MES-4 results in the de-silencing of the X-chromosome.
MES-4-mediated H3 K36 methylation has some notable differences compared to the yeast Set2 protein. Although MES-4 is found over the coding regions, it does not require RNAPII for its activity or localization. Indeed, the distribution of H3 K36 di-and tri-methylation over germline-specific genes is enriched towards the 5′ end rather than the 3′ end of the gene, as expected from studies in yeast104. MES-4 is the predominant H3 K36 methyltransferase in the germ line cells. Methyltransferase-1 (MET-1), the SETD2 homolog catalyzing the co-transcriptional H3 K36 tri-methylation is not expressed until late embryonic stages (100-cell stage) and is not involved in germline specification or maintenance. Interestingly, MET-1 mutants in association with other specific mutations alter the somatic cell lineage to give rise to the vulva, by failing to repress the epithelial growth factor (EGF) gene lin-3 in worms105. Loss of Setd2 in mice resulted in severe vascular defects106.
Loss of MES-4 results in sterility of the progeny of homozygous mutant hermaphrodite worms. This observation indicates that normal development of germ cell in the homozygous mutant worms is due to factors transferred from the heterozygous mother. Interestingly, MES-4 and the associated H3 K36 methyl mark are distributed over the germline-expressed genes in early embryos, even in the absence of transcription. This – coupled with the observation that embryonic expression of catalytically dead mes-4 mutant does not rescue the sterile phenotype – led to the hypothesis that MES-4 mediated H3 K36 methyl mark is an epigenetic signal necessary for the proper development of primordial germ cells104. Inheritance of the epigenetic marks across mitosis (and meiosis) requires the nucleosomes to be stable across the cell cycle, which is not the case due to replication dependent histone turnover. DNA methylation as well as H3 K9 and H3 K27 methylation can undergo trans-generational transfer of epigenetic information. A key event in this transfer is the ability of these marks to recruit or allosterically activate the enzymes that add them. Such an activity has not been described for the Set2 homologs as yet. How the MES-4 protein recognizes and maintains genomic regions with H3 K36 methyl mark in early embryos remains to be determined.
Mammalian homologs of the MES-4 protein include the NSD family of H3 K36 methylases (Table 1). Loss of these proteins displays a wide range of developmental defects. Homozygous mutant embryos of NSD1 die during gastrulation due to excessive apoptosis107. Hemi-zygous mutants of WHSC1 (formerly known as NSD2) are associated with Wolf-Hirschhorn syndrome, which is characterized by craniofacial defects and growth delays71, suggesting a critical role of this family of proteins in embryonic development. Incidentally, Drosophila dSet2 is involved in larval to pupal transition108. Additionally, H3K36 di-methylation by the human SET domain and mariner transposase fusion gene-containing protein (SETMAR), is necessary for non-homologous end joining (NHEJ)109, thereby maintaining genomic integrity.
Flowering control in Arabidopsis
Flowering in plants requires a change in the characteristics of totipotent apical meristem (tissue consisting of undifferentiated cells) from a vegetative (generates shoots and leaves) to a floral (generates flowers) state. In plants, activation of the sexual reproduction pathway is usually in response to environmental stimuli like day length (photoperiod) or temperature changes110. The response to these signals, even if they are transient, induces changes in the gene expression pattern leading to floral commitment. One of the well-studied pathways in flowering control is the regulation of the MADS-box transcription factor, FLOWERING LOCUS C (FLC), which acts as a flowering repressor111. Upon receiving the necessary environmental cue, the floral transition is initiated leading to the repression of FLC. Loss of EARLY FLOWERING IN SHORT DAYS (EFS)/SET DOMAIN GROUP 8 (SDG8), the SET domain containing H3 K36 di- and tri-methylase, results in reduced FLC expression, causing early flowering112. Interestingly, loss of SET DOMAIN GROUP 26 (SDG26), an H3 K36 mono-methylase, causes late flowering, presumably by alleviating repression of FLC113. Interestingly, loss of the Arabidopsis PAF1 complex components cause early flowering and a loss of H3 K36 di- and tri-methylation110, in conjunction with studies in yeast. Repression of FLC involves the PRC2 complex-mediated silencing of chromatin111. Therefore, activation of FLC locus can be explained in terms of SDG8-mediated eviction of the polycomb complex.
Interestingly, EFS is required for anther and ovule development in Arabidopsis, and its loss causes severe defects in the development of the reproductive organs114. EFS is similar to C. elegans MES-4, which is also involved in germline maintenance and in stress response to fungal infections.115
Conclusions
Set2-mediated H3K36me prevents co-transcriptional acetylation by suppressing histone exchange over the coding regions. It also recruits chromatin-remodeling complexes to the coding regions to maintain chromatin structure and nucleosomal spacing. At the same time, it activates the deacetylase activity of the elongating RNAPII-specific Rpd3S complex, removing histone acetyl marks placed by histone acetyltransferases. On the basis of its distribution, H3K36me carries out these functions towards the 3′ ends of genes. Recent studies have demonstrated similar functions for the 5′-specific H3 K4 methyl mark, although its role in affecting histone exchange has not been analyzed yet116. Thus, both of these methyl marks coordinate to maintain chromatin integrity over the coding regions of genes.
Associating the Set2/Rpd3S pathway with the active transcription machinery ensures efficient maintenance of chromatin integrity over the coding regions. It also affords tighter regulation at the level of each transcription cycle, including co-transcriptional events like RNA splicing. Although H3K36me serves to suppress spurious initiation of transcripts and maintain the coding regions in a hypoacetylated state, it does not impede transcription. Thus, it is not a repressive mark, but a regulatory one, which maintains chromatin structure following elongation in a state that allows further rounds of transcription through it while preventing internal initiation.
By suppressing histone exchange, H3K36me ensures its persistence across different rounds of transcription, ensuring transcriptional memory with respect to its genomic position and transcriptional status. This memory is lost upon DNA replication-associated histone exchange on chromatin. Recent studies have suggested that active chromatin is partitioned as dimers, allowing for the marks to be distributed to the daughter strands in the vicinity of the original genomic locus117. This observation, coupled with the trans-generational maintenance of H3K36me by C. elegans MES-4, truly qualifies it as an epigenetic mark.
Acknowledgments
This work is supported by NIH grant R01GM047867 to JLW and the Stowers Institute for Medical Research. We would like to thank Joanne Chatfield for proofreading the manuscript.
References
- 1.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nature genetics. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 2.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes & development. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 3.Sims RJ, 3rd, Mandal SS, Reinberg D. Recent highlights of RNA-polymerase-II-mediated transcription. Current opinion in cell biology. 2004;16:263–271. doi: 10.1016/j.ceb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Bartkowiak B, Mackellar AL, Greenleaf AL. Updating the CTD Story: From Tail to Epic. Genetics research international. 2011;2011:623718. doi: 10.4061/2011/623718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes & development. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nature structural & molecular biology. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 7.Kwon SH, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. Heterochromatin protein 1 (HP1) connects the FACT histone chaperone complex to the phosphorylated CTD of RNA polymerase II. Genes Dev. 2010;24:2133–2145. doi: 10.1101/gad.1959110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosley AL, Pattenden SG, Carey M, Venkatesh S, Gilmore JM, Florens L, Workman JL, Washburn MP. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Molecular cell. 2009;34:168–178. doi: 10.1016/j.molcel.2009.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bataille AR, Jeronimo C, Jacques PE, Laramee L, Fortin ME, Forest A, Bergeron M, Hanes SD, Robert F. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Molecular cell. 2012;45:158–170. doi: 10.1016/j.molcel.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 10.Li B, Howe L, Anderson S, Yates JR, 3rd, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. The Journal of biological chemistry. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
- 11.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 12.Rao B, Shibata Y, Strahl BD, Lieb JD. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Molecular and cellular biology. 2005;25:9447–9459. doi: 10.1128/MCB.25.21.9447-9459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alvarez-Venegas R, Avramova Z. SET-domain proteins of the Su(var)3–9, E(z) and trithorax families. Gene. 2002;285:25–37. doi: 10.1016/s0378-1119(02)00401-8. [DOI] [PubMed] [Google Scholar]
- 14.Du HN, Fingerman IM, Briggs SD. Histone H3 K36 methylation is mediated by a trans-histone methylation pathway involving an interaction between Set2 and histone H4. Genes & development. 2008;22:2786–2798. doi: 10.1101/gad.1700008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lachner M, Jenuwein T. The many faces of histone lysine methylation. Current opinion in cell biology. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- 16.Kouzarides T. Histone methylation in transcriptional control. Current opinion in genetics & development. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 17.Trievel RC, Beach BM, Dirk LM, Houtz RL, Hurley JH. Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell. 2002;111:91–103. doi: 10.1016/s0092-8674(02)01000-0. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, Selker EU, Cheng X. Structural basis for the product specificity of histone lysine methyltransferases. Molecular cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng X, Collins RE, Zhang X. Structural and sequence motifs of protein (histone) methylation enzymes. Annual review of biophysics and biomolecular structure. 2005;34:267–294. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strahl BD, Grant PA, Briggs SD, Sun ZW, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Molecular and cellular biology. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du HN, Briggs SD. A nucleosome surface formed by histone H4, H2A, and H3 residues is needed for proper histone H3 Lys36 methylation, histone acetylation, and repression of cryptic transcription. The Journal of biological chemistry. 2010;285:11704–11713. doi: 10.1074/jbc.M109.085043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Trojer P, Xu CF, Cheung P, Kuo A, Drury WJ, 3rd, Qiao Q, Neubert TA, Xu RM, Gozani O, et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. The Journal of biological chemistry. 2009;284:34283–34295. doi: 10.1074/jbc.M109.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Molecular and cellular biology. 2005;25:3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Phatnani HP, Guan Z, Sage H, Greenleaf AL, Zhou P. Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-terminal domain of Rpb1. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17636–17641. doi: 10.1073/pnas.0506350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vojnic E, Simon B, Strahl BD, Sattler M, Cramer P. Structure and carboxyl-terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. The Journal of biological chemistry. 2006;281:13–15. doi: 10.1074/jbc.C500423200. [DOI] [PubMed] [Google Scholar]
- 26.Youdell ML, Kizer KO, Kisseleva-Romanova E, Fuchs SM, Duro E, Strahl BD, Mellor J. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Molecular and cellular biology. 2008;28:4915–4926. doi: 10.1128/MCB.00001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Molecular and cellular biology. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes & development. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu Y, Sutton A, Sternglanz R, Prelich G. The BUR1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by SET2. Molecular and cellular biology. 2006;26:3029–3038. doi: 10.1128/MCB.26.8.3029-3038.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu H, Hu C, Wong CM, Hinnebusch AG. The Spt4p subunit of yeast DSIF stimulates association of the Paf1 complex with elongating RNA polymerase II. Molecular and cellular biology. 2006;26:3135–3148. doi: 10.1128/MCB.26.8.3135-3148.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Molecular cell. 2005;20:589–599. doi: 10.1016/j.molcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 32.Laribee RN, Krogan NJ, Xiao T, Shibata Y, Hughes TR, Greenblatt JF, Strahl BD. BUR kinase selectively regulates H3 K4 trimethylation and H2B ubiquitylation through recruitment of the PAF elongation complex. Current biology : CB. 2005;15:1487–1493. doi: 10.1016/j.cub.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 33.Chu Y, Simic R, Warner MH, Arndt KM, Prelich G. Regulation of histone modification and cryptic transcription by the Bur1 and Paf1 complexes. The EMBO journal. 2007;26:4646–4656. doi: 10.1038/sj.emboj.7601887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuchs SM, Kizer KO, Braberg H, Krogan NJ, Strahl BD. RNA polymerase II carboxyl-terminal domain phosphorylation regulates protein stability of the Set2 methyltransferase and histone H3 di- and trimethylation at lysine 36. The Journal of biological chemistry. 2012;287:3249–3256. doi: 10.1074/jbc.M111.273953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes & development. 2007;21:160–174. doi: 10.1101/gad.1503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoh SM, Lucas JS, Jones KA. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes & development. 2008;22:3422–3434. doi: 10.1101/gad.1720008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin LJ, Minard LV, Johnston GC, Singer RA, Schultz MC. Asf1 can promote trimethylation of H3 K36 by Set2. Molecular and cellular biology. 2010;30:1116–1129. doi: 10.1128/MCB.01229-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Molecular cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 39.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 40.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 41.Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Molecular cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 42.Li B, Jackson J, Simon MD, Fleharty B, Gogol M, Seidel C, Workman JL, Shilatifard A. Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. The Journal of biological chemistry. 2009;284:7970–7976. doi: 10.1074/jbc.M808220200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson CJ, Santos-Rosa H, Kouzarides T. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell. 2006;126:905–916. doi: 10.1016/j.cell.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 44.Yuan W, Xie J, Long C, Erdjument-Bromage H, Ding X, Zheng Y, Tempst P, Chen S, Zhu B, Reinberg D. Heterogeneous nuclear ribonucleoprotein L Is a subunit of human KMT3a/Set2 complex required for H3 Lys-36 trimethylation activity in vivo. The Journal of biological chemistry. 2009;284:15701–15707. doi: 10.1074/jbc.M808431200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nature structural & molecular biology. 2011;18:977–983. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- 46.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Molecular cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 47.Kim T, Buratowski S. Two Saccharomyces cerevisiae JmjC domain proteins demethylate histone H3 Lys36 in transcribed regions to promote elongation. The Journal of biological chemistry. 2007;282:20827–20835. doi: 10.1074/jbc.M703034200. [DOI] [PubMed] [Google Scholar]
- 48.Klose RJ, Gardner KE, Liang G, Erdjument-Bromage H, Tempst P, Zhang Y. Demethylation of histone H3K36 and H3K9 by Rph1: a vestige of an H3K9 methylation system in Saccharomyces cerevisiae? Molecular and cellular biology. 2007;27:3951–3961. doi: 10.1128/MCB.02180-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang J, Hogan GJ, Liang G, Lieb JD, Zhang Y. The Saccharomyces cerevisiae histone demethylase Jhd1 fine-tunes the distribution of H3K36me2. Molecular and cellular biology. 2007;27:5055–5065. doi: 10.1128/MCB.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 51.Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, Chan SM, Martin DG, Fingerman IM, Briggs SD, et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. The Journal of biological chemistry. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nature reviews Genetics. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 53.Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 54.Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell research. 2011;21:564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 56.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 57.Drouin S, Laramee L, Jacques PE, Forest A, Bergeron M, Robert F. DSIF and RNA polymerase II CTD phosphorylation coordinate the recruitment of Rpd3S to actively transcribed genes. PLoS genetics. 2010;6:e1001173. doi: 10.1371/journal.pgen.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Molecular cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eisen A, Utley RT, Nourani A, Allard S, Schmidt P, Lane WS, Lucchesi JC, Cote J. The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. The Journal of biological chemistry. 2001;276:3484–3491. doi: 10.1074/jbc.M008159200. [DOI] [PubMed] [Google Scholar]
- 60.Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Molecular cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 61.Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 62.Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes & development. 2007;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jelinic P, Pellegrino J, David G. A novel mammalian complex containing Sin3B mitigates histone acetylation and RNA polymerase II progression within transcribed loci. Molecular and cellular biology. 2011;31:54–62. doi: 10.1128/MCB.00840-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Studitsky VM, Walter W, Kireeva M, Kashlev M, Felsenfeld G. Chromatin remodeling by RNA polymerases. Trends in biochemical sciences. 2004;29:127–135. doi: 10.1016/j.tibs.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 65.Kulaeva OI, Gaykalova DA, Pestov NA, Golovastov VV, Vassylyev DG, Artsimovitch I, Studitsky VM. Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nature structural & molecular biology. 2009;16:1272–1278. doi: 10.1038/nsmb.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Molecular cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 67.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 68.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Molecular cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 69.Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, Workman JL. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 70.Ginsburg DS, Govind CK, Hinnebusch AG. NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Molecular and cellular biology. 2009;29:6473–6487. doi: 10.1128/MCB.01033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, Kaneda Y. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 72.Gossett AJ, Lieb JD. In vivo effects of histone H3 depletion on nucleosome occupancy and position in Saccharomyces cerevisiae. PLoS genetics. 2012;8:e1002771. doi: 10.1371/journal.pgen.1002771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, Washburn MP, Workman JL. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nature structural & molecular biology. 2012;19:884–892. doi: 10.1038/nsmb.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maltby VE, Martin BJ, Schulze JM, Johnson I, Hentrich T, Sharma A, Kobor MS, Howe L. Histone H3 lysine 36 methylation targets the Isw1b remodeling complex to chromatin. Molecular and cellular biology. 2012;32:3479–3485. doi: 10.1128/MCB.00389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Radman-Livaja M, Quan TK, Valenzuela L, Armstrong JA, van Welsem T, Kim T, Lee LJ, Buratowski S, van Leeuwen F, Rando OJ, et al. A key role for Chd1 in histone H3 dynamics at the 3′ ends of long genes in yeast. PLoS genetics. 2012;8:e1002811. doi: 10.1371/journal.pgen.1002811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen XF, Kuryan B, Kitada T, Tran N, Li JY, Kurdistani S, Grunstein M, Li B, Carey M. The Rpd3 core complex is a chromatin stabilization module. Current biology : CB. 2012;22:56–63. doi: 10.1016/j.cub.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3. 3 replacement patterns. Nature genetics. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 79.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3. 3 is enriched in covalent modifications associated with active chromatin. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nature reviews Genetics. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 81.Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS biology. 2008;6:e277. doi: 10.1371/journal.pbio.0060277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Silva AC, Xu X, Kim HS, Fillingham J, Kislinger T, Mennella TA, Keogh MC. The replication-independent histone H3-H4 chaperones HIR, ASF1, and RTT106 co-operate to maintain promoter fidelity. The Journal of biological chemistry. 2012;287:1709–1718. doi: 10.1074/jbc.M111.316489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Conrad T, Akhtar A. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nature reviews Genetics. 2011;13:123–134. doi: 10.1038/nrg3124. [DOI] [PubMed] [Google Scholar]
- 84.Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Molecular cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 85.Gelbart ME, Larschan E, Peng S, Park PJ, Kuroda MI. Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nature structural & molecular biology. 2009;16:825–832. doi: 10.1038/nsmb.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bell O, Wirbelauer C, Hild M, Scharf AN, Schwaiger M, MacAlpine DM, Zilbermann F, van Leeuwen F, Bell SP, Imhof A, et al. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. The EMBO journal. 2007;26:4974–4984. doi: 10.1038/sj.emboj.7601926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nature reviews Molecular cell biology. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 88.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Molecular cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 90.Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. The Journal of biological chemistry. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N, Li X, Xu M, Zhang Z, Niu T, et al. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012;337:971–975. doi: 10.1126/science.1225237. [DOI] [PubMed] [Google Scholar]
- 92.Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends in biochemical sciences. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 93.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Molecular cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS genetics. 2012;8:e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nature genetics. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Molecular cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nature reviews Molecular cell biology. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nakamura A, Seydoux G. Less is more: specification of the germline by transcriptional repression. Development. 2008;135:3817–3827. doi: 10.1242/dev.022434. [DOI] [PubMed] [Google Scholar]
- 100.Capowski EE, Martin P, Garvin C, Strome S. Identification of grandchildless loci whose products are required for normal germ-line development in the nematode Caenorhabditis elegans. Genetics. 1991;129:1061–1072. doi: 10.1093/genetics/129.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bender LB, Cao R, Zhang Y, Strome S. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Current biology : CB. 2004;14:1639–1643. doi: 10.1016/j.cub.2004.08.062. [DOI] [PubMed] [Google Scholar]
- 102.Fong Y, Bender L, Wang W, Strome S. Regulation of the different chromatin states of autosomes and X chromosomes in the germ line of C. elegans. Science. 2002;296:2235–2238. doi: 10.1126/science.1070790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bender LB, Suh J, Carroll CR, Fong Y, Fingerman IM, Briggs SD, Cao R, Zhang Y, Reinke V, Strome S. MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development. 2006;133:3907–3917. doi: 10.1242/dev.02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rechtsteiner A, Ercan S, Takasaki T, Phippen TM, Egelhofer TA, Wang W, Kimura H, Lieb JD, Strome S. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS genetics. 2010;6:e1001091. doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Andersen EC, Horvitz HR. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development. 2007;134:2991–2999. doi: 10.1242/dev.009373. [DOI] [PubMed] [Google Scholar]
- 106.Hu M, Sun XJ, Zhang YL, Kuang Y, Hu CQ, Wu WL, Shen SH, Du TT, Li H, He F, et al. Histone H3 lysine 36 methyltransferase Hypb/Setd2 is required for embryonic vascular remodeling. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2956–2961. doi: 10.1073/pnas.0915033107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rayasam GV, Wendling O, Angrand PO, Mark M, Niederreither K, Song L, Lerouge T, Hager GL, Chambon P, Losson R. NSD1 is essential for early post-implantation development and has a catalytically active SET domain. The EMBO journal. 2003;22:3153–3163. doi: 10.1093/emboj/cdg288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stabell M, Larsson J, Aalen RB, Lambertsson A. Drosophila dSet2 functions in H3-K36 methylation and is required for development. Biochemical and biophysical research communications. 2007;359:784–789. doi: 10.1016/j.bbrc.2007.05.189. [DOI] [PubMed] [Google Scholar]
- 109.Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, Radhakrishnan K, Lee SH, Nickoloff JA, Hromas R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci U S A. 2011;108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Adrian J, Torti S, Turck F. From decision to commitment: the molecular memory of flowering. Molecular plant. 2009;2:628–642. doi: 10.1093/mp/ssp031. [DOI] [PubMed] [Google Scholar]
- 111.He Y. Control of the transition to flowering by chromatin modifications. Molecular plant. 2009;2:554–564. doi: 10.1093/mp/ssp005. [DOI] [PubMed] [Google Scholar]
- 112.Zhao Z, Yu Y, Meyer D, Wu C, Shen WH. Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nature cell biology. 2005;7:1256–1260. doi: 10.1038/ncb1329. [DOI] [PubMed] [Google Scholar]
- 113.Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, Shen WH. Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Molecular and cellular biology. 2008;28:1348–1360. doi: 10.1128/MCB.01607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Grini PE, Thorstensen T, Alm V, Vizcay-Barrena G, Windju SS, Jorstad TS, Wilson ZA, Aalen RB. The ASH1 HOMOLOG 2 (ASHH2) histone H3 methyltransferase is required for ovule and anther development in Arabidopsis. PLoS One. 2009;4:e7817. doi: 10.1371/journal.pone.0007817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Berr A, McCallum EJ, Alioua A, Heintz D, Heitz T, Shen WH. Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant physiology. 2010;154:1403–1414. doi: 10.1104/pp.110.161497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Buratowski S, Kim T. The role of cotranscriptional histone methylations. Cold Spring Harbor symposia on quantitative biology. 2010;75:95–102. doi: 10.1101/sqb.2010.75.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]