Abstract
The development of the mammalian kidney has been studied at the genetic, biochemical, and cell biological level for more than 40 years. As such, detailed mechanisms governing early patterning, cell lineages, and inductive interactions are well described. How genes interact to specify the renal epithelial cells of the nephrons and how this specification is relevant to maintaining normal renal function is discussed. Implicit in the development of the kidney are epigenetic mechanisms that mark renal cell types and connect certain developmental regulatory factors to chromatin modifications that control gene expression patterns and cellular physiology. In adults, such regulatory factors and their epigenetic pathways may function in regeneration and may be disturbed in disease processes.
Keywords: Kidney Development, Epigenetics, Intermediate Mesoderm, Nephrogenesis
The term “epigenetics’ refers to heritable traits that are not encoded directly within the DNA of the genome. Heritability, in a conventional sense, refers to the passing of a trait or phenotype from one organism to its offspring. Yet heritability at the cellular level is also a critical issue as specific traits, or gene expression patterns, must be passed on from a mother cell to a daughter cell. Nowhere is this more apparent than in the development of the embryo. Indeed, we know that intrinsic cellular memory is imprinted very early in stem and progenitor cells, long before the eventual cellular phenotype is fully manifested. Yet these stem and progenitor cells often undergo hundreds of rounds of cell division and still remember their eventual fates. The genome and all its associated histone proteins, referred to as chromatin, must contain this epigenetic information that specifies individual cell types. How is this epigenetic memory established and propagated? Genetic screens in fruit flies identified families of genes that modify the inheritance of gene expression patterns at very specific times during development. These modifier genes were sorted into the Polycomb and Trithorax groups based on whether they repressed or activated gene expression respectively.
Although epigenetics had been studied in a variety of model systems over the years, the biochemical functions of many Polycomb and Trithorax family proteins were poorly understood. It was not until the biochemistry of chromatin, its post-translational modifications, and the enzymes responsible for such modifications were revealed that the functions of epigenetic genes became clear. We now know that many Polycomb or Trithorax family genes encode histone methyltransferases or co-factors for histone methyltransferase complexes that modify chromatin and determine chromatin structure. These chromatin modifications are imprinted during development as stem cells lose pluripotency and establish tissue specific cell–lineage pathways. Thus epigenetic imprints are not only established during development but are central to compartmentalizing the genome into accessible and inaccessible domains unique for a particular cell type.
If we are to understand how epigenetics impacts human disease, it becomes necessary to start at the beginning. In Nephrology, we must begin with the development of the kidney, the specification of the cell lineages that constitute the adult kidney, and the genes responsible for imprinting a renal specific fate onto more pluripotent progenitor or stem cells. This review will summarize much of what we know about the genetic control of renal development and also introduce what little we know about the epigenetic components of that developmental program. How the epigenetic programs are disturbed in renal disease can then be addressed.
Early Development of the Kidney and Regionalization
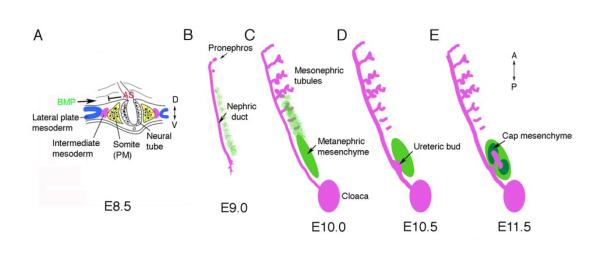
The adult kidney is composed of a large number of specialized epithelial, stromal and endothelial cells. Renal epithelial and stromal cells share a common lineage that is specified early in development, but diverges once kidney development proceeds. In mammals, this lineage is apparent shortly after gastrulation in a region of mesoderm, called the intermediate mesoderm, that lies between the paraxial mesoderm and the lateral plate mesoderm along the medio-lateral axis (Fig. 1) 1, 2. Moreover, the kidney develops along the anterior posterior axis in a temporal sequence. Early anterior kidney structures include the pro- and mesonephros, whose complexity, size, and duration varies greatly among vertebrate species. In the mouse, the pronephros is barely detectable, whereas mesonpehric tubules are well developed with a proximal glomerulus and convoluted tubules that empty into the nephric duct (Fig. 1 B-E). The adult, or metanephric kidney, forms at the posterior end of this intermediate mesoderm. Thus, the intermediate mesoderm requires both medio-lateral patterning and anterior-posterior patterning signals to determine the adult kidney field.
Figure 1. The Intermediate Mesoderm: its Origin and Derivatives.
A) In amniotes, the kidney arises from the intermediate mesoderm, between the paraxial somatic mesoderm (PM) and the lateral plate mesoderm, shown in a cross section through a mouse embryo at embryonic day 8.5 (E8.5) at approximately the 6th somite. Antagonistic signals (AS) from somites may counteract BMP signals from the lateral plate to generate the pronephric field. B-E) The kidneys develop from intermediate mesoderm along the Anterior-Posterior (AP) axis in a specific temporal and spatial order. B) The Wolffian or (pro)nephric duct, is visible at E9.0 in the mouse and grows caudally by proliferation and extension, inducing epithelial tubules from the adjacent mesenchyme. The pronephros is very rudimentary. C) Mesonpehric tubules at E10, as the nephric duct reaches the cloaca, are more developed in the mid-thoracic region, with glomeruli at the proximal end and convoluted tubules draining into the nephric duct. Posterior cells adjacent to the duct form an aggregate called the metanephric mesenchyme. D) By E10.5 an outgrowth of the duct, the ureteric bud, invades the metanephric mesenchyme. E) By E11.5, this ureteric bud has bifurcated and the induced mesenchyme, called the cap mesenchyme, surrounds the tips. Cap mesenchymal cells are the stem cells of the nephrons and generate the podocytes, the parietal epithelium, the proximal tubules, and the distal tubules.
The origin of the intermediate mesoderm (IM) has been explored in a number of experimental organisms. Cell fate mapping studies have identified the odd-skipped related gene (Osr1), which encodes a zinc-finger DNA binding protein, as one of the earliest markers of the lateral plate mesoderm (LPM) and intermediate mesoderm (IM) in chick and mouse embryos 3. Expression of the lim type homeobox gene Lhx1 is evident in the prospective LPM and is another markers for this posterior, lateral mesoderm 4. The expression domains for Osr1 and Lhx1 overlap and encompass prospective IM as well as more LPM; Osr1 is expressed along the entire A-P axis from the first somites. Markers exclusive to the IM, Pax2 and Pax8, are activated within the IM from approximately the 6th somite, in a very narrow stripe of cells just lateral to the paraxial mesoderm, but their expression does not extend into the more lateral plate 5. Shortly thereafter, Lhx1 expression becomes more restricted to the IM and to the nephric duct as it begins to form and extend caudally. Osr1 remains expressed in the mesenchymal cells surrounding the nephric duct and in the more lateral plate mesodermal derivatives but is excluded from the Pax2 positive cells of the nephric duct itself.
Genetic analyses in mice suggest that Lhx1, Osr1, and Pax2/8 have critical roles in early specification of the IM. Lhx1 null mice lack the nephric duct, although Pax2 expression is observed in cells at the boundary between the paraxial and lateral plate mesoderm shortly after gastrulation 4. Pax2 null mutants do develop a nephric duct 6, 7, but the duct is completely absent in a Pax2/8 double mutant, suggesting that these Pax genes function redundantly in this early IM domain 5. Pax2/8 double mutants also do not express Lhx1. Mice homozygous for an Osr1 null allele, the expression of which precedes that of Pax2/8, still exhibit nephric duct formation and Pax2 expression in the anterior IM, yet lack more developed mesonephric tubules and the metanephric mesenchyme in the posterior IM 3, 8. Whether this anterior Pax2 expression in the Osr1 mutants is due to some partial rescue or redundancy by Osr2 or is completely cell autonomous and independent of Osr1 function remains to be determined.
Specifying the IM along the Mediolateral Axis
The activation of the Pax2/8 expression domain might be the first indication that the LPM and IM have assumed separate fates. This activation appears to depend on signals that come from the lateral plate and on opposing signals from the somite. A model for IM fate commitment was first proposed by James and Schultheiss 9. Embryonic manipulation in the chick suggested that low concentrations of bone morphogenetic proteins (BMPs) activate IM specific genes, whereas higher concentrations activate lateral plate markers. Ectopic BMPs can shift the position of the IM, even transforming more paraxial mesoderm into an IM phenotype. These data are consistent with earlier observations that BMPs could replace the overlying surface ectoderm as inducers of the primary nephric duct within the IM 10.
The dorsolateral BMP signals may be opposed by as yet unidentified negative signals emanating from the somites (Fig. 1) 11. Other secreted signals that are known to promote IM marker gene expression and kidney development are activin and retinoic acid. In a study by Preger-Ben Noon et al. 12, Activin also induced Lhx1 expression in the chick embryo along the entire body axis but failed to induce Pax2 expression more anteriorly. These results suggest that signals emanating from the medial tissues, neural tube and somites, compete with signals from the more dorsolateral surface ectoderm, to determine the activation of IM-specific genes, perhaps in a concentration dependent manner. How these signals are integrated and whether they truly oppose each other remains to be determined. However, the mediolateral axis is only one body axis. The developing IM must also be specified along the anterior-posterior (A-P) axis as is evident from the morphological differences between mesonephric and metanephric tissues.
Specification of the IM along the Anterior-Posterior Axis
The mammalian kidney forms along the length of the body axis in a manner reminiscent of its evolutionary history (Fig. 1). Many of the same genes, such as Pax2 (Fig. 2), are expressed along the entire IM; however, the morphological structures derived from different regions along the axis are unique. The Hox genes function in regional specification and patterning of the axial skeleton, and central and peripheral nervous system. Functional redundancy exists among paralogous groups within the four Hox gene clusters. Therefore, most phenotypes are evident only if multiple genes of a group are deleted 13, 14. Compound murine mutants for hoxa11 and hoxd11 exhibit metanephric branching defects and hypoplasia, suggesting a necessity for Hox gene function in the developing metanephros 15. More severe renal defects are evident if all three Hox11 paralogous genes are deleted and include complete agenesis and lack of ureteric bud outgrowth 16. These posterior defects did not affect genes like Pax2 or the Wilms’ tumor suppressor gene, Wt1, whose expression is found along the entire A-P axis within the IM. Rather, Hox11 genes are necessary for the expression of more posterior markers that delineate the metanephric mesenchyme only and include Gdnf (glial derived neurotrophic factor) and six2 (the sina oculis-related homeobox 2 gene), which demarcate the metanephric mesenchyme by E10.5. Since Hox11 genes are important for differentiating the metanephric mesenchyme from more anterior IM derivatives, then altering the pattern of Hox genes might induce a regional shift in identity. Mugford et al. 17 utilized an Osr1 driver to activate hoxd11 in the more anterior IM, encompassing the normal Osr1 domain. This ectopic hoxd11 expression partially transformed mesonephric tubules to a more metanephric phenotype, as based on the expression of marker genes exclusive to metanephric tubules, such as Calbindin3, suggesting that Hox11 genes are necessary for specifying the metanephric identity from more anterior or mesonephric IM.
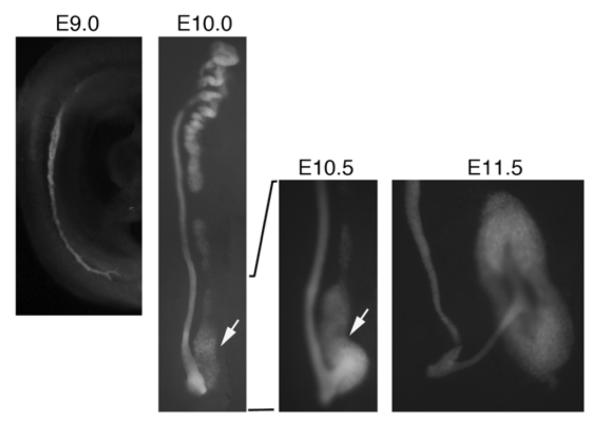
Figure 2. Pax2 Expression Marks the Developing Kidney.
Embryos that carry the enhanced green fluorescent protein inserted into a single allele of Pax2 are show with expression in all tissues that contribute to the kidneys. At E9.0, the nephric duct is visible within the intermediate mesoderm; note the posterior cellular processes that extend as the duct migrates caudally. By E10.0, the nephric duct and mesonephric tubules are well developed; the metanephric mesenchyme (arrow) is adjacent to the posterior duct. By E10.5, the ureteric bud begins to emerge and migrate into the mesenchyme. By E11.5, the ureteric bud has bifurcated and cap mesenchyme is seen aggregating around the tips of the buds. Pax2 expression persists in all of the ureteric bud epithelial and cap mesenchyme derivatives but is down-regulated in more differentiated nephron epithelial cells.
In the most anterior IM and at earlier stages, Hox genes also appear to set the boundary for competence to respond to the mediolateral IM patterning signals described in the previous section. The signals that induce the expression of IM-specific genes, such as Lhx1 and Pax2, are present along the entire body axis, but the IM specific markers are only induced posterior to the sixth somite in the chick embryo, suggesting that only mesoderm formed posterior to this region is competent to respond and make IM 18. This anterior boundary of prospective IM is coincident with the anterior expression boundary of Hox4 paralogues. Remarkably, by using retinoic acid to shift the hoxb4 expression domain more rostrally or by just overexpressing hoxb4 with plasmids in the chick embryo, the boundary for IM competence was shifted more anteriorly 12. These data strongly argue for the existence of an A-P patterning code within the mesoderm that is regulated by specific Hox gene paralogous groups.
How is the A-P patterning code translated into a biological response along the mesoderm? Direct interactions of Hox proteins with other IM specific transcription factors, specifically Eya1 and Pax2, may be necessary for Six2 gene activation in the metanephric mesenchyme, suggesting that regionalization of specific IM compartments depends upon the intersection of expression domains between the mediolateral factors, such as Pax2/8 and Lhx1, and the A-P factors 19. Specific pattern of Hox gene expression may predisposes the mesoderm to respond to IM inductive signals at the anterior boundary, which initiates expression of Lhx1 and Pax2/8 along the entire body axis caudal to the sixth somite, whereas a posterior combination of Hox genes, consisting primarily of the Hox11 paralogous group, are needed to activate genes such as Gdnf and Six2 and distinguish the metanephric mesenchyme from more anterior mesonephric tissue. However the posterior IM is ultimately specified, its development proceeds along a markedly different path compared to more anterior IM as the adult kidney is formed.
The Ureteric Bud and Collecting Duct Lineage
The signals that drive ureteric bud outgrowth have been well studied over the years. The receptor tyrosine kinase RET, the secreted neurotrophin GDNF, and the membrane anchored co-receptor, GFRα1 (GDNF family receptor alpha 1) are critical for initiating a signaling cascade that triggers outgrowth of Ret positive cells from the nehpric duct towards the GDNF signal emanating from the metanephric mesenchyme 20. There are many modifiers of the RET/GDNF signaling pathway that precisely regulate the position and number of buds and the subsequent branching morphogenesis of the ureteric bud epithelia. A complex network of inhibitors restricts GDNF/RET signaling to a region of the nephric duct to prevent ectopic ureter budding. BMP4 expressed in mesenchymal cells surrounding the nephric duct inhibits GDNF/RET signaling. Yet, BMP signaling is blocked by the secreted BMP antagonist Gremlin (Grem1), which is expressed in the metanephric mesenchyme. This allows for precise tuning of the BMP signal to enable bud positioning and branching 21. The Gdnf expression domain is restricted to the more anterior IM by the transmembrane protein Slit2 (slit homolog 2) and its receptor Robo2 (roundabout homolog 2), which are homologus to Drosophila proteins that provide axon guidance cues, and thus repress ectopic ureteric bud outgrowth 22. Within the ureteric bud epithelial cells, the cytoplasmic protein Spry1 (Sprouty homolog 1) limits the intensity or duration of RET signaling, as Spry1 mutants develop ectopic ureter buds, a phenotype that can be suppressed by a reduction of Gdnf gene dosage 23, 24.
How RET activation impacts cell movement and proliferation is still not entirely clear, in part due to the large number of tyrosine residues phosphorylated on the RET cytoplasmic domain. In response to chemotactic agents, phosphatidylinositol (PI) 3-kinase is activated at the leading edge of migrating cells to promote lamellipodia formation, extension, and cell movement 25, 26. In contrast, the lipid phosphatase PTEN (phosphatase and tensin homolog), which dephosphorylates the substrates for PI 3-kinase, is located at the trailing edge 27, 28. In kidney organ cultures, inhibition of PI 3-kinase completely blocks ureteric bud outgrowth, suggesting that ureteric bud epithelial cell migration is essential for invasion of the mesenchyme 26. In vivo, deletion of the PTEN phosphatase in the ureteric bud epithelia also leads to abnormal branching and patterning defects, consistent with a role for PI 3-kinase and PTEN in shaping the ureteric bud by counteracting the effects of PI 3-kinase 29. However, cell movement is not enough to drive invasion, as localized proliferation and extension must contribute to the growing bud tip. Other downstream effectors likely to transduce RET signaling include the mitogen activated protein kinases (MAPKs) whose inhibition also leads to branching defects 30, 31. Mutations to specific tyrosine residues in RET isoforms indicate at least two important docking sites for intracellular second messengers, Y1015 and Y1062 which bind Grb2/7 and Shc proteins and activate the PI 3-kinase and the MAPK pathways during kidney development 32, 33. Taken together, the data point to multiple signaling pathways downstream of activated RET that coordinate proliferation and cell migration of UB epithelial cells, yet still maintain the integrity of the bud.
Despite all the complexity, the RET pathway is not the only promoter of ureteric bud outgrowth as a significant portion of RET mutant kidneys still exhibit a rudimentary bud 34. Deletion of the fibroblast growth factor receptors (Fgfr) 1 and 2 demonstrated that Ffgr2 but not Fgfr1 functions in the ureteric bud epithelium to fine tune the pattern of branching morphogenesis and to determine the size of the kidney and the number of nephrons 35. Similarly, deletion of the receptor tyrosine kinase Met, whose ligand is hepatocyte growth factor, also restricted branching morphogenesis, kidney size, and nephron number 36. These results raise the possibility that inhibitors of RET signaling can inhibit other tyrosine kinases important for branching morphogensis, for example the Sprouty1 protein is known to suppress Fgfr signaling 37, further increasing the complexity of signaling in the ureteric bud epithelium and making interpretations at the biochemical level difficult. Surprisingly, when both GDNF and Spry1 are lost, ureteric bud outgrowth and branching are restored but now depend on Fgf10 38.
At this point, we can summarize ureteric bud outgrowth as a complex phenomenon that requires both positive and negative signaling to drive cell movement and proliferation of epithelial cells at a precise position along the nephric duct. The proteins involved include secreted signaling factors and tyrosine kinase receptors, many of which are also utilized for other chemotactic processes such as axon guidance and directed cell migration.
The Metanephric Mesenchyme and Induction
The metanephric mesenchyme is the anlagen of the adult kidney, and as a result has received the most attention over the years. Once the bud invades the metanephric mesenchyme it provides a permissive signal that stimulates the condensation of metanephric mesenchymal cells around the ureteric bud tips. This step begins the polarization of the mesenchyme to generate the epithelial cells of the nephron (Fig. 3). The most compelling data to date indicates that WNT proteins are the primary initiators of condensation. Multiple Wnt genes are expressed in the ureteric bud and the stalk, but genetic ablation experiments show that Wnt9b encodes the only WNT protein that meets all the criteria for the inducer of the metanephric mesenchyme 39. Wnt9b expressing cells can mimic inductive signals and promote mesenchymal aggregation in vitro, whereas loss of Wnt9b in vivo prevents metanephric mesenchymal aggregation but has no effect on initial budding and branching of the ureteric epithelium. The initial WNT inductive signal transduction is canonical, as it can be mimicked by activation of β-catenin, although this must be attenuated in the early condensates as constitutively active β-catenin inhibits mesenchymal aggregates from progressing to polarized epithelia 40.
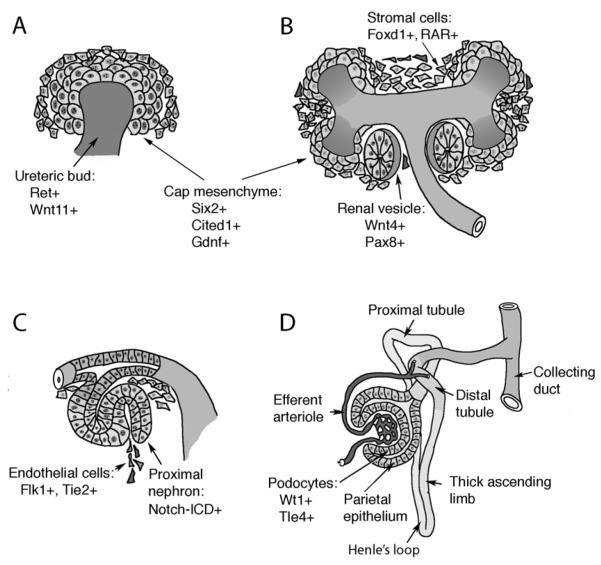
Figure 3. The Maturation of Nephrons.
A) Invasion of the metanephric mesenchyme by the ureteric bud provides inductive signals that promotes mesenchyme cells to condense around the bud tips at E11.5 of mouse development. These so-called cap mesenchymal cells express a unique combination of markers (Six2, Gdnf, Cited1) and define a stem cell population. B) Cap mesenchyme polarizes into a primitive epithelial sphere, the renal vesicle, coincident with the expression of additional markers, such as Wnt4 and Pax8. Cells in the metanephric mesenchyme that do not aggregate at the bud tips express Foxd1and the retinoic acid receptors (RARs) and mark the stromal population. C) The renal vesicle fuses to the ureteric stalk, which forms the collecting ducts, and generates an S-shaped body with a proximal and distal cleft. The more proximal cleft is infiltrated by endothelial cells and forms the glomerular tuft. The proximal portion of the S-shaped body activates the Notch pathway, a seen by the presence of the cleaved Notch intracellular domain (ICD). D) The nephron begins to take shape as glomerular development proceeds and the more proximal tubules elongate and grow towards the medulla to form the descending and ascending limbs of Henle’s loop.
Prior to induction at E10.5, the metanephric mesenchyme expresses a unique set of marker genes, many of which are known to regulate important events in early kidney development. By E11.5, the metanephric mesenchyme has been invaded by the ureteric bud epithelium and condensations of mesenchymal cells around the ureteric bud tips are visible (Fig. 3A). These condensates, now referred to as the cap mesenchyme, are the progenitor cells of the nephron epithelia and are themselves surrounded by stromal cells, which remain mesenchymal and migrate towards the interstitium (Fig. 3 A, B). By E13.5, the S-shaped bodies derived from the cap mesenchyme become infiltrated by endothelial precursors to form the glomerular tuft, which consists of the capillary loops, the mesangium, the glomerular basement membrane and the podocyte cells (Fig 3C).
One question that has plagued investigators is the pluripotency of the metanephric mesenchyme. Are these the stem cells of the kidney or are the metanephric mesenchymal cells a heterogenous mixture of epithelial, stromal, and endothelial precursors? Osr1 positive cells appear capable of making either stromal or epithelial precursors prior to E10.5 41, consistent with the broad expression of pattern Osr1 in the IM and lateral plate mesoderm. Yet once the metanephric mesenchyme is induced, the Osr1 positive population appears to make only epithelial cells, suggesting that stromal progenitor cells have now turned off Osr1 and are a separate lineage. The metanephric stromal cells, which express many genes not found in the cap mesenchyme including foxd1, the retinoic acid receptors, and Transcription factor 21 (Tcf21/Pod1), are necessary for providing signals to promote epithelial cell survival and proliferation 42-45. Cell lineage tracing in the chick embryo 46 suggest that a large population of metanephric stromal cells are derived form paraxial mesoderm and that few arise from the IM. Taken together, the data would suggest that stromal cells and epithelial precursors share a common Osr1 positive lineage before the onset of metanephric induction. However, after E11.5 stromal and cap-mesenchyme lineages are separate. Indeed, it is possible that new stromal precursor cells, which are Osr1 negative and are coming from more paraxial mesoderm, may be migrating into the metanephric mesenchyme after induction.
Cell lineage tracing of the cap mesenchyme reveals a self-renewing population that can be considered the epithelial stem cells of the nephron. Lineage tracing in which the Cre recombinase was driven by either the Cited1 47 or Six2 48 genes clearly demonstrates that cap mesenchyme is pluripotent with respect to epithelial cell types, as its derivatives include glomerular, proximal tubular, and distal tubular epithelia. As such, the cap mesenchyme must proliferate and generate cells of the renal vesicles, the progenitors of the nephrons, while also repopulating the aggregates around the tips of the branching ureteric buds for the next round of nephron formation. The decision of whether to differentiate or self-renew requires Six2, as its loss leads to precocious metanephric mesenchyme differentiation 49. Rather than forming renal vesicles in particular positions under the ureteric bud tips, Six2 mutant embryonic kidneys have epithelial structures all along the T-shaped ureteric bud, which results in exhaustion of the cap mesenchyme population and in rudimentary kidneys. All the cap mesenchyme has presumably seen the inductive Wnt9b signals from the ureteric bud tips, otherwise the cells would not aggregate around the ureteric bud tips. However, cap mesenchyme requires down regulation of canonical, β-catenin mediated, Wnt signaling 50 and the expression of Wnt4 to become polarized into renal vesicles 51. Thus, one function of Six2 could be to suppress the intrinsic Wnt4 signals emanating from induced cap mesenchyme that promote epithelial polarity.
Additional secreted signaling molecules that are essential for mesenchyme polarization include Fgf8 and Bmp7. Loss of Fgf8 results in depletion of the cap mesenchyme and significant cell death in the peripheral, nephrogenic zone 52. Using a Pax3-Cre driver to conditionally delete Fgf8 in the metanephric mesenchyme inhibited Wnt4 and Lhx1 expression, but not Pax2 expression. Despite the absence of detectable Wnt4, some cells did progress to the renal vesicle stage at early times after induction, but by approximately E14.5 significant cell death prevented any further development and mesenchymal aggregate formation. Bmp7 deletion also led to developmental arrest, but metanephric mesenchyme cells induced at E11.5 did progress to the polarized epithelial stage and were able to generate tubules 42, 53, 54. It appears that Bmp7 deletion also depleted the cap mesenchyme stem cell population 55. More recently, Fgf9 and Fgf20 were specifically identified as the critical ligands secreted by the ureteric bud for maintaining nephron progenitor cells within the population of cap mesenchyme 56. These data are consistent with a role for Fgfs and Bmp7 in providing survival signals for the metanephric mesenchyme, the progenitors in the cap mesenchyme, and perhaps expansion of the stromal cell population that helps support the cap mesenchyme 57, 58.
Six and Eya proteins are known to interact physically and genetically in other developing tissues 59, yet Eya1 mutations are completely recalcitrant to the inductive signals emanating from the ureteric bud and thus do not mimic the Six2 phenotype 60, 61. Instead, Eya1 appears to interact with Six1, as both genes are essential for early metanephric mesenchyme specification and are associated with Branchio-Oto-Renal (BOR) syndrome in humans carrying one mutant allele of either gene 62. BOR is characterized by unilateral or bilateral renal hypoplasia, dysplasia, or agenesis in addition to cochlear defects and cranio-facial fistulas.
In addition to the stroma and cap mesenchyme, the early metanephros also contains precursors of the vasculature. Angioblasts are integral to the development of the glomerular tuft, which depends on a precise level of vascular endothelial growth factor (VEGF) signaling 63. However, these Flk1 positive angioblasts also signal to other cell types to propagate the inductive signals 64. A recent paper also described crosstalk in ureteric bud cells in culture in which VEGF was able to promote phosphorylation of RET, perhaps accounting for some of the redundancy in the budding mechanism 65.
Clearly induction of the metanephric mesenchyme by the ureteric bud is the critical step in kidney development and leads to all subsequent differentiation. Induction appears to be sequential and requires at least 2 different Wnt proteins to begin the process of epithelia polarization. Many of the genes expressed in the metanephric mesenchyme prior to induction are needed in order to respond to Wnt signals, suggesting that the mesenchyme is already programmed to become renal tissue and just needs a permissive signal, not an instructive signal. Indeed, Saxen 2 made this distinction early on merely by observing that any inducing tissue that could promote epithelial cell polarization of the mesenchyme always resulted in the differentiation of renal epithelia and not any other type of epithelia.
Patterning of the Nephron
The epithelial cells of the nephrons are derived from the cap mesenchyme that makes first the renal vesicle and then the s-shaped body (Fig. 3C). The renal vesicle (Fig. 3B) is a primitive epithelium with basement membrane and lumen that is in close proximity to the ureteric bud stalk. By the S-shaped body stage, the renal vesicle has fused to the ureteric stalk to form a continuous epithelial tubule with common apical lumen. Differential expression of cadherin genes provided some early evidence for patterning of the renal vesicle 66. However, recent large scale expression screens coupled to three-dimensional reconstruction reveal significant differences in gene expression in the renal vesicle, along the proximal-distal axis, with respect to the adjacent ureteric bud stalk, the progenitors of the collecting ducts 67. These differences in gene expression are likely to underlie the regionalization of the vesicle into glomerular, proximal, and distal segments. Strikingly, the distal segments fuse to the prospective collecting ducts by degradation of the epithelial basement membrane and integration of distal renal vesicle cells into the prospective collecting tubules 67.
The definitive proximal-distal axis of the nephron is clear by the S-shaped body stage (Fig. 3C). At the most proximal end are the precursors of the glomerular podocyte cells, the visceral glomerular epithelium. At the distal end, the S-shaped body has fused to the branching ureteric tree to form the collecting tubules. Until recently, little was known regarding the signals that specify this proximal distal axis and the different epithelial cell types that arise along the axis. Thus, one of the most significant discoveries in recent years is the role of the Notch pathway in proximal-distal patterning of the S-shaped body 68. While Notch and its ligands Delta, Jagged, and Serrate are known to specify neural cell fates by lateral inhibition in the fly eye and the mouse immune system, a direct role in regional specification had not been seen before. Kidneys from mice homozygous for a Notch2 null allele have a complete absence of more proximal renal cell types, including the gomeruli and the proximal convoluted tubules 68. These mutants have normal ureteric bud epithelial branching and more distal derivatives form the renal vesicle. Conversely, the expression of an activated Notch intracellular domain in wild-type cap mesenchyme can transform more distal fates to more proximal fates. Similar results were observed in Xenopus with the ectopic expression of the downstream target of Notch, Hairy/Enhancer of Split, which can also instill more proximal fates along the developing pronephros 69. In zebrafish, knockdown of either Notch3, its potential ligand Jagged2, or its downstream effectors has demonstrated a role for Notch in differentially specifying the fates of transport epithelia from multicilliated cells, two different terminal cell types interspersed along the pronephric duct 70. This fate decision is somewhat akin to the Notch mediated lateral inhibition in the fly eye. Interestingly, these specific Notch knockdowns did not affect the single midline glomerulus in the zebrafish larvae pronephros, unlike the loss of Notch signaling in the mouse metanephric kidney. This apparent discrepancy is likely to reflect the unique origin of the zebrafish pronephric glomerulus, which does not require the same genetic components as the metanephric glomerulus. For example, loss of zebrafish Pax2 results in ablation of the pronephric duct and tubules but not the midline glomerulus 71. Most surprisingly, the activation of Notch can bypass the need for Wnt signaling in the induction of metanephric mesenchyme and the conversion to renal epithelia 72. These results further underscore the complexity and potential cross-talk among signaling molecules in the mesenchyme and its derivatives.
At the extreme proximal end of the nephron, there has been significant progress made in understanding the development of the glomerulus and the relationships between podocytes, the endothelial cells, and the mesangial precursor cells of the glomerular tuft (for review see 63). I believe this is due to the clinical relevance of the glomerular filtration barrier and the numerous human genetic mutations that impact podocyte function, the integrity of the slit diaphragm, which forms the filtration barrier, and the stability of the glomerular basement membrane, which contains unique collagen and laminin chains. By contrast, the mechanisms that differentiate the cell types along the axis of the nephron have not been investigated, in large part because molecular markers that distinguish these early decision making events were unavailable. However, this is issue is being addressed through large scale expression screens undertaken by GenitoUrinary Development Molecular Anatomy Project (www.gudmap.org), which has found unique expression signatures for all specific anatomical structures within the developing metanephric kidney 73, 74. Thus, new and better reagents coupled to improved anatomical descriptors are at hand for tackling these outstanding issues of terminal epithelial sub-type specification and function.
Integrating Patterning, Lineage Specification, and Gene Regulation with Epigenetics
The developing kidney is useful as a model system for studying epithelial cell specification, mesenchymal-epithelial interactions, and complex patterning events in three dimensions. We know much about which genes are important for morphogenesis and which genes affect the downstream expression of known markers. However, despite all of this available knowledge, it is still difficult to ascribe biochemical functions to many of the proteins that we know are essential for kidney development. In fact, many of the genes that regulate early development were not identified from their functions in the kidney at all. Some genes such as Osr1, Pax2, Pax8, Eya1, Six1, and Six2 were identified purely by sequence homology to regulatory genes in the fly. Other genes such as Gdnf and Wnt4 were assigned kidney functions once they were mutated in mice and the consequent kidney phenotypes became evident. Particularly for the intrinsic, cell autonomous, nuclear factors, such as the Pax, Eya, and Hox families, how they function to specify early patterns and renal cell lineages remains mysterious. Because many of the early marker genes encode DNA binding proteins, it is assumed that they regulate some aspect of transcription, presumably of other kidney specific genes. How do proteins like Pax2/8, Lhx1, Eya1, and Osr1 talk to the transcription regulatory machinery to establish the early IM and the kidney epithelial lineages? The standard tools of genetics are unlikely to yield all the answers because the biological readouts, i.e. the failure to develop, are not very specific. These problems are not unique to kidney development. To understand the biochemical function of a nuclear protein, its interaction with DNA and with other cellular factors must be defined within the appropriate context. Yet, the context is often inaccessible to standard biochemical purification or manipulation because cells corresponding to that progenitor state, in which these proteins function, are not available as stable lines. However, new ways of thinking about developmental competence are emerging from the rapidly progressing field of epigenetics and chromatin biology, which can impact how we think about early gene regulation and cell lineage restriction.
The restriction of cell fate is a sequential process during development that implies a heritable imprint on the genome of a progenitor population. These types of epigenetic imprints are within the realm of the Polycomb and Trithorax family of genes, whose protein products are involved in establishing and maintaining patterns of histone methylation on chromatin 75-78. The histone octamer is the primary protein component of the nucleosome and its modification dramatically impacts the structure of chromatin. Many of the most interesting developmental regulatory genes in pluripotent embryonic stem (ES) cells have a bivalent pattern of histone modification, with low levels of both positive and negative epigenetic marks, that are then resolved into active or inactive marks upon differentiation along particular lineages 79. These findings imply that there must be cell lineage determining factors that control the locus and tissue specificity of histone modifications during early developmental decision-making. In the developing eye, the Pax6 proteins were proposed to have such early lineage decision-making potential in the fly and the mouse 80. In the intermediate mesoderm, Pax2 expression marks all of the cells fated to become epithelia and is necessary to establish and maintain the epithelial phenotype 7 (Fig. 2). Pax2 mutant mesenchyme cannot respond to inductive signals nor aggregate into early renal vesicles 6.
More recent studies indicate that Pax2 may function as part of an epigenetic network that specifies early renal cell lineages (Fig. 4). Pax2 can provide the DNA binding specificity for an MLL3/4 Trithorax like protein complex that promotes histone H3, lysine 4 methylation (H3K4me) to mark regions of active chromatin 81. Through its interaction with PTIP, Pax2 recruits the MLL proteins such that positive epigenetic marks are established at genes slated for transcription activation. The Pax/PTIP complex can also promote long-distance enhancer and promoter interactions through chromatin looping 82, again suggesting effects on chromatin structure to activate gene expression. However, Pax proteins also interact with other proteins, such as the co-repressor Grg4/Tle4, to recruit Polycomb complexes that establish repressive epigenetic marks on chromatin 83. Thus, this dual potential for activation and repression may be temporally regulated by the availability of co-factors. These data suggest that Pax2 may provide some locus and tissue specificity to imprint a kidney specific epigenetic fate by partitioning the genome of the IM into active and inactive domains unique for the renal lineage (Fig. 4). Whether H3K4 trimethylation promotes gene expression or merely inhibits Polycomb mediated repression still needs to be clarified, as genetic evidence suggests that repression is the default state in the absence of Trithorax mediated derepression 84. In any case, more definitive proof of this concept awaits better technology that could characterize chromatin modifications at single genes in a spatial and temporal manner, in small numbers of cells, during development.
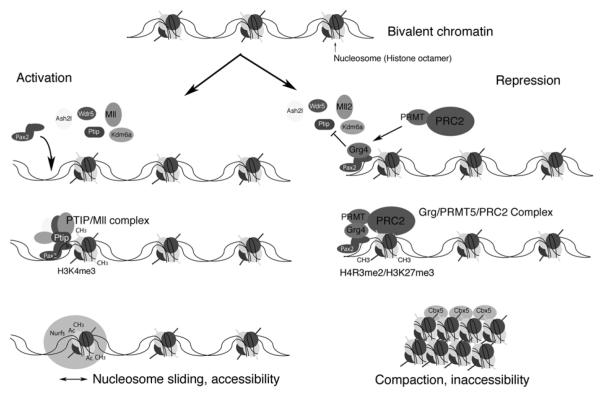
Figure 4. A Chromatin Model for Epigenetic Specification of Cell Lineages.
As cells make lineage decisions, alterations in chromatin structure compartmentalizes the genome into active and inactive domains. In pluripotent embryonic stem (ES) cells, tissue-specific genes and developmental regulators are marked with a bivalent histone code that encompasses low levels of both positive and negative histone methylation marks. As cells become specified and their fates are restricted, cell-type specific DNA binding proteins could provide locus specificity for the modification of chromatin into active or repressed domains. During intermediate mesoderm specification, the Pax2/8 proteins might interact with a histone H3K4 methyltransferase (PTIP/Mll) complex to prevent repression of kidney specific genes by the Polycomb group (PcG) complexes. High levels of H3K4 trimethylation could then recruit nucleosome remodeling factors (Nurfs) that maintain accessibility of genes and facilitate transcription. Conversely, the repressor Grg4 prevents PTIP and Mll from assembling at a Pax2 biding site. Grg4 recruits a histone arginine methyltransferase PRMT and the Polycomb repressor 2 complex (PRC2). PRC2 mediated methylation of histone H3K27 could recruit heterochromatin binding proteins that compact DNA into tightly packaged, silent chromatin. Ash2l, absent small or homeotic like 2; Cbx5, chromobox homolog 5; Kdm6a, 4 lysine (K)-specific demethylase 6A; Mll, mixed-lineage leukemia ; Ptip, Pax trans-activation domain interacting protein; Wdr5, WD repeat domain 5.
Epigenetics and the Control of Phenotypic Stability
Given the critical function of histone methylation in development, one might ask what roles do epigenetic pathways play in adult kidneys? Once cell phenotypes are imprinted and if these imprints are stable in non-dividing cells, then one might assume that histone methylases and associated factors are superfluous. However, both gain and loss of epigenetic modifiers are associated with a variety of disease states, including cancer. Studies in mice have shown that reducing histone H3K4 methyltransfease activity can alter cellular phenotypes. In podocytes, this leads to changes in gene expression patterns and ultimately a glomerular sclerosis phenotype 85. In cardiac myocytes, deletion of PTIP, a co-factor for the histone methyltransferase MLL3/4, alters gene expression and the electrophysiology, ultimately sensitizing cells to arhythmia 86. These data suggest that there is a maintenance function to H3K4 methylation that stabilizes the gene expression patterns in terminally differentiated cells. Thus, disruption of epigenetic pathways could lead to slowly progressing, chronic diseases.
Epigenetic regulation may also be important for regeneration of renal epithelial cells after injury. In humans acute renal failure is a common result of nephrotoxicity or ischemia, however the injured kidneys will recover and repopulate the damaged tubules if the degree of injury is limited. The origin of these regenerating proximal tubule cells has been studied in some detail. Recent cell lineage tracing methods demonstrate that adult regenerating proximal tubule cells are derived primarily from preexisting, surviving proximal tubular epithelia and not from a pre-existing population of renal stem cells 87, 88. What promotes these surviving epithelial cells to enter the mitotic cycle and repopulate the damaged tubules? Reactivation of developmental genes such as Pax2 has been described 89, suggesting that an embryonic program may drive regeneration. The genetics of Polycystic Kidney Disease are particularly illuminating in that regard. In mouse models, loss of PKD1 function in dividing cells leads to rapid and early cystogenesis. However, if PKD1 is deleted in non-dividing cells, cyst formation is rare and focal 90, suggesting a developmental window in which PKD1 is essential. Yet subsequent renal injury can promote cyst formation in adults that have deleted PKD1 after this critical window has passed 91, again suggesting that cell division is necessary for the cystic phenotype. The point is further illustrated in mutations of HNF1β, a critical developmental regulator that activates, among others, the PKD2 gene. If HNF1β is lost in development, PKD2 is not activated and renal cysts form early. However, if HNF1β is lost in adult epithelial cells, PKD2 is unaffected and cyst formation is not observed, suggesting that HNF1β is not necessary to maintain PKD2. Then, if the adult HNF1β mutants are injured, PKD2 is lost again in the newly regenerating epithelial cells 92. These elegant experiments point to an epigenetic memory that maintains PKD2 expression in the absence of the activator HNF1β. Re-entry into the cell cycle must reset the epigenetic switch, now re-establishing the requirement for HNF1β in PKD2 activation. Whether DNA replication and cell division somehow dilute out the epigenetic marks that maintain PKD2 expression remains to be seen. If histone methylation marks are replicated, as DNA and associated methylation at CpG islands, then there would be no need for the activators that reset the marks. However, recent data in Drosophila suggests that it is the proteins and enzymes that set the marks that are bound to chromatin during replication and may be involved in passing the marks onto daughter cells 93.
Conclusions
The genetic studies of kidney development provide novel insights into complex patterning, epithelial cell specification, branching morphogenesis, and epigenetic imprinting mechanisms. These developmental mechanisms have potential clinical importance in many diseases and in regenerating proximal tubules after injury. The systematic characterization of genes in the kidney and their localized expression patterns (www.gudmap.org) have provided powerful resources for delineating regulatory networks that drive development and impact disease. There are few organ systems for which we have such a sophisticated level of understanding. Nevertheless, as the recent work with Notch/Wnt signaling or GDNF/FGF signaling have shown, the complexities and redundancies inherent in any developing organ system can still surprise us and leave much to be determined.
Acknowledgments
This work was supported by National Institutes of Health grant DK073722 (G.R.D.) and DK082409 (S.R.P.). We thank the members of our labs for discussion and insights.
Financial Support: National Institutes of Health grant DK073722 to G.R.D. National Institutes of Health grant DK082409 to S.R.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement: The authors have no financial interests of conflict to disclose.
References
- 1.Dressler GR. The cellular basis of kidney development. Annu Rev Cell Dev Biol. 2006;22:509–529. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- 2.Saxen L. Organogenesis of the Kidney. In: BARLOW PW, GREEN PB, WHITE CC, editors. Developmental and Cell Biology. 19 ed Cambridge University Press; Cambridge, UK: 1987. (Series 19). [Google Scholar]
- 3.James RG, Kamei CN, Wang Q, Jiang R, Schultheiss TM. Odd-skipped related 1 is required for development of the metanephric kidney and regulates formation and differentiation of kidney precursor cells. Development. 2006;133:2995–3004. doi: 10.1242/dev.02442. [DOI] [PubMed] [Google Scholar]
- 4.Tsang TE, Shawlot W, Kinder SJ, Kobayashi A, Kwan KM, Schughart K, Kania A, Jessell TM, Behringer RR, Tam PP. Lim1 activity is required for intermediate mesoderm differentiation in the mouse embryo. Dev Biol. 2000;223:77–90. doi: 10.1006/dbio.2000.9733. [DOI] [PubMed] [Google Scholar]
- 5.Bouchard M, Souabni A, Mandler M, Neubuser A, Busslinger M. Nephric lineage specification by Pax2 and Pax8. Genes Dev. 2002;16:2958–2970. doi: 10.1101/gad.240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brophy PD, Ostrom L, Lang KM, Dressler GR. Regulation of ureteric bud outgrowth by Pax2-dependent activation of the glial derived neurotrophic factor gene. Development. 2001;128:4747–4756. doi: 10.1242/dev.128.23.4747. [DOI] [PubMed] [Google Scholar]
- 7.Soofi A, Levitan I, Dressler GR. Two novel EGFP insertion alleles reveal unique aspects of Pax2 function in embryonic and adult kidneys. Dev Biol. 2012;365:241–250. doi: 10.1016/j.ydbio.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q, Lan Y, Cho ES, Maltby KM, Jiang R. Odd-skipped related 1 (Odd 1) is an essential regulator of heart and urogenital development. Dev Biol. 2005;288:582–594. doi: 10.1016/j.ydbio.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.James RG, Schultheiss TM. Bmp signaling promotes intermediate mesoderm gene expression in a dose-dependent, cell-autonomous and translation-dependent manner. Dev Biol. 2005;288:113–125. doi: 10.1016/j.ydbio.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 10.Obara-Ishihara T, Kuhlman J, Niswander L, Herzlinger D. The surface ectoderm is essential for nephric duct formation in intermediate mesoderm. Development. 1999;126:1103–1108. doi: 10.1242/dev.126.6.1103. [DOI] [PubMed] [Google Scholar]
- 11.Mauch TJ, Yang G, Wright M, Smith D, Schoenwolf GC. Signals from trunk paraxial mesoderm induce pronephros formation in chick intermediate mesoderm. Dev Biol. 2000;220:62–75. doi: 10.1006/dbio.2000.9623. [DOI] [PubMed] [Google Scholar]
- 12.Preger-Ben Noon E, Barak H, Guttmann-Raviv N, Reshef R. Interplay between activin and Hox genes determines the formation of the kidney morphogenetic field. Development. 2009;136:1995–2004. doi: 10.1242/dev.035592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wellik DM. Hox patterning of the vertebrate axial skeleton. Dev Dyn. 2007;236:2454–2463. doi: 10.1002/dvdy.21286. [DOI] [PubMed] [Google Scholar]
- 14.Zakany J, Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev. 2007;17:359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Patterson LT, Pembaur M, Potter SS. Hoxa11 and Hoxd11 regulate branching morphogenesis of the ureteric bud in the developing kidney. Development. 2001;128:2153–2161. doi: 10.1242/dev.128.11.2153. [DOI] [PubMed] [Google Scholar]
- 16.Wellik DM, Hawkes PJ, Capecchi MR. Hox11 paralogous genes are essential for metanephric kidney induction. Genes Dev. 2002;16:1423–1432. doi: 10.1101/gad.993302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mugford JW, Sipila P, Kobayashi A, Behringer RR, McMahon AP. Hoxd11 specifies a program of metanephric kidney development within the intermediate mesoderm of the mouse embryo. Dev Biol. 2008;319:396–405. doi: 10.1016/j.ydbio.2008.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barak H, Rosenfelder L, Schultheiss TM, Reshef R. Cell fate specification along the anterior-posterior axis of the intermediate mesoderm. Dev Dyn. 2005;232:901–914. doi: 10.1002/dvdy.20263. [DOI] [PubMed] [Google Scholar]
- 19.Gong KQ, Yallowitz AR, Sun H, Dressler GR, Wellik DM. A Hox-Eya-Pax complex regulates early kidney developmental gene expression. Mol Cell Biol. 2007;27:7661–7668. doi: 10.1128/MCB.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costantini F, Kopan R. Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell. 2010;18:698–712. doi: 10.1016/j.devcel.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michos O, Goncalves A, Lopez-Rios J, Tiecke E, Naillat F, Beier K, Galli A, Vainio S, Zeller R. Reduction of BMP4 activity by gremlin 1 enables ureteric bud outgrowth and GDNF/WNT11 feedback signalling during kidney branching morphogenesis. Development. 2007;134:2397–2405. doi: 10.1242/dev.02861. [DOI] [PubMed] [Google Scholar]
- 22.Grieshammer U, Le M, Plump AS, Wang F, Tessier-Lavigne M, Martin GR. SLIT2-mediated ROBO2 signaling restricts kidney induction to a single site. Dev Cell. 2004;6:709–717. doi: 10.1016/s1534-5807(04)00108-x. [DOI] [PubMed] [Google Scholar]
- 23.Basson MA, Akbulut S, Watson-Johnson J, Simon R, Carroll TJ, Shakya R, Gross I, Martin GR, Lufkin T, McMahon AP, Wilson PD, Costantini FD, Mason IJ, Licht JD. Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev Cell. 2005;8:229–239. doi: 10.1016/j.devcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Basson MA, Watson-Johnson J, Shakya R, Akbulut S, Hyink D, Costantini FD, Wilson PD, Mason IJ, Licht JD. Branching morphogenesis of the ureteric epithelium during kidney development is coordinated by the opposing functions of GDNF and Sprouty1. Dev Biol. 2006;299:466–477. doi: 10.1016/j.ydbio.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 25.Besset V, Scott RP, Ibanez CF. Signaling complexes and protein-protein interactions involved in the activation of the Ras and phosphatidylinositol 3-kinase pathways by the c-Ret receptor tyrosine kinase. J Biol Chem. 2000;275:39159–39166. doi: 10.1074/jbc.M006908200. [DOI] [PubMed] [Google Scholar]
- 26.Tang MJ, Cai Y, Tsai SJ, Wang YK, Dressler GR. Ureteric bud outgrowth in response to RET activation is mediated by phosphatidylinositol 3-kinase. Dev Biol. 2002;243:128–136. doi: 10.1006/dbio.2001.0557. [DOI] [PubMed] [Google Scholar]
- 27.Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;109:611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- 28.Kolsch V, Charest PG, Firtel RA. The regulation of cell motility and chemotaxis by phospholipid signaling. J Cell Sci. 2008;121:551–559. doi: 10.1242/jcs.023333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D, Dressler GR. PTEN modulates GDNF/RET mediated chemotaxis and branching morphogenesis in the developing kidney. Dev Biol. 2007;307:290–299. doi: 10.1016/j.ydbio.2007.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher CE, Michael L, Barnett MW, Davies JA. Erk MAP kinase regulates branching morphogenesis in the developing mouse kidney. Development. 2001;128:4329–4338. doi: 10.1242/dev.128.21.4329. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe T, Costantini F. Real-time analysis of ureteric bud branching morphogenesis in vitro. Dev Biol. 2004;271:98–108. doi: 10.1016/j.ydbio.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 32.Jain S, Encinas M, Johnson EM, Jr., Milbrandt J. Critical and distinct roles for key RET tyrosine docking sites in renal development. Genes Dev. 2006;20:321–333. doi: 10.1101/gad.1387206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong A, Bogni S, Kotka P, de Graaff E, D’Agati V, Costantini F, Pachnis V. Phosphotyrosine 1062 is critical for the in vivo activity of the Ret9 receptor tyrosine kinase isoform. Mol Cell Biol. 2005;25:9661–9673. doi: 10.1128/MCB.25.21.9661-9673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuchardt A, D’Agati V, Pachnis V, Costantini F. Renal agenesis and hypodysplasia in ret-k-mutant mice result from defects in ureteric bud development. Development. 1996;122:1919–1929. doi: 10.1242/dev.122.6.1919. [DOI] [PubMed] [Google Scholar]
- 35.Zhao H, Kegg H, Grady S, Truong HT, Robinson ML, Baum M, Bates CM. Role of fibroblast growth factor receptors 1 and 2 in the ureteric bud. Dev Biol. 2004;276:403–415. doi: 10.1016/j.ydbio.2004.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishibe S, Karihaloo A, Ma H, Zhang J, Marlier A, Mitobe M, Togawa A, Schmitt R, Czyczk J, Kashgarian M, Geller DS, Thorgeirsson SS, Cantley LG. Met and the epidermal growth factor receptor act cooperatively to regulate final nephron number and maintain collecting duct morphology. Development. 2009;136:337–345. doi: 10.1242/dev.024463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aranda S, Alvarez M, Turro S, Laguna A, de la Luna S. Sprouty2-mediated inhibition of fibroblast growth factor signaling is modulated by the protein kinase DYRK1A. Mol Cell Biol. 2008;28:5899–5911. doi: 10.1128/MCB.00394-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michos O, Cebrian C, Hyink D, Grieshammer U, Williams L, D’Agati V, Licht JD, Martin GR, Costantini F. Kidney development in the absence of Gdnf and Spry1 requires Fgf10. PLoS Genet. 2010;6:e1000809. doi: 10.1371/journal.pgen.1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carroll TJ, Park JS, Hayashi S, Majumdar A, McMahon AP. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell. 2005;9:283–292. doi: 10.1016/j.devcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 40.Park JS, Valerius MT, McMahon AP. Wnt/beta-catenin signaling regulates nephron induction during mouse kidney development. Development. 2007;134:2533–2539. doi: 10.1242/dev.006155. [DOI] [PubMed] [Google Scholar]
- 41.Mugford JW, Sipila P, McMahon JA, McMahon AP. Osr1 expression demarcates a multipotent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev Biol. 2008;324:88–98. doi: 10.1016/j.ydbio.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dudley AT, Godin RE, Robertson EJ. Interaction between FGF and BMP signaling pathways regulates development of metanephric mesenchyme. Genes Dev. 1999;13:1601–1613. doi: 10.1101/gad.13.12.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hatini V, Huh SO, Herzlinger D, Soares VC, Lai E. Essential role of stromal mesenchyme in kidney morphogenesis revealed by targeted disruption of Winged Helix transcription factor BF-2. Genes Dev. 1996;10:1467–1478. doi: 10.1101/gad.10.12.1467. [DOI] [PubMed] [Google Scholar]
- 44.Levinson RS, Batourina E, Choi C, Vorontchikhina M, Kitajewski J, Mendelsohn CL. Foxd1-dependent signals control cellularity in the renal capsule, a structure required for normal renal development. Development. 2005;132:529–539. doi: 10.1242/dev.01604. [DOI] [PubMed] [Google Scholar]
- 45.Mendelsohn C, Batourina E, Fung S, Gilbert T, Dodd J. Stromal cells mediate retinoiddependent functions essential for renal development. Development. 1999;126:1139–1148. doi: 10.1242/dev.126.6.1139. [DOI] [PubMed] [Google Scholar]
- 46.Guillaume R, Bressan M, Herzlinger D. Paraxial mesoderm contributes stromal cells to the developing kidney. Dev Biol. 2009;329:169–175. doi: 10.1016/j.ydbio.2009.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boyle S, Misfeldt A, Chandler KJ, Deal KK, Southard-Smith EM, Mortlock DP, Baldwin HS, de Caestecker M. Fate mapping using Cited1-CreERT2 mice demonstrates that the cap mesenchyme contains self-renewing progenitor cells and gives rise exclusively to nephronic epithelia. Dev Biol. 2008;313:234–245. doi: 10.1016/j.ydbio.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, McMahon AP. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell. 2008;3:169–181. doi: 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Self M, Lagutin OV, Bowling B, Hendrix J, Cai Y, Dressler GR, Oliver G. Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J. 2006;25:5214–5228. doi: 10.1038/sj.emboj.7601381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marose TD, Merkel CE, McMahon AP, Carroll TJ. Beta-catenin is necessary to keep cells of ureteric bud/Wolffian duct epithelium in a precursor state. Dev Biol. 2008;314:112–126. doi: 10.1016/j.ydbio.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kispert A, Vainio S, McMahon AP. Wnt-4 is a mesenchymal signal for epithelial transformation of metanephric mesenchyme in the developing kidney. Development. 1998;125:4225–4234. doi: 10.1242/dev.125.21.4225. [DOI] [PubMed] [Google Scholar]
- 52.Grieshammer U, Cebrian C, Ilagan R, Meyers E, Herzlinger D, Martin GR. FGF8 is required for cell survival at distinct stages of nephrogenesis and for regulation of gene expression in nascent nephrons. Development. 2005;132:3847–3857. doi: 10.1242/dev.01944. [DOI] [PubMed] [Google Scholar]
- 53.Dudley AT, Lyons KM, Robertson EJ. A requirement for bone morphogenetic protein-7 during development of the mammalian kidney and eye. Genes Dev. 1995;9:2795–2807. doi: 10.1101/gad.9.22.2795. [DOI] [PubMed] [Google Scholar]
- 54.Luo G, Hofmann C, Bronckers ALJJ, Sohocki M, Bradley A, Karsenty G. BMP-7 is an inducer of nephrogenesis, and is also required for eye development and skeletal patterning. Genes Dev. 1995;9:2808–2820. doi: 10.1101/gad.9.22.2808. [DOI] [PubMed] [Google Scholar]
- 55.Blank U, Brown A, Adams DC, Karolak MJ, Oxburgh L. BMP7 promotes proliferation of nephron progenitor cells via a JNK-dependent mechanism. Development. 2009;136:3557–3566. doi: 10.1242/dev.036335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barak H, Huh SH, Chen S, Jeanpierre C, Martinovic J, Parisot M, Bole-Feysot C, Nitschke P, Salomon R, Antignac C, Ornitz DM, Kopan R. FGF9 and FGF20 maintain the stemness of nephron progenitors in mice and man. Dev Cell. 2012;22:1191–1207. doi: 10.1016/j.devcel.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown AC, Adams D, de Caestecker M, Yang X, Friesel R, Oxburgh L. FGF/EGF signaling regulates the renewal of early nephron progenitors during embryonic development. Development. 2011;138:5099–5112. doi: 10.1242/dev.065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oxburgh L, Brown AC, Fetting J, Hill B. BMP signaling in the nephron progenitor niche. Pediatr Nephrol. 2011;26:1491–1497. doi: 10.1007/s00467-011-1819-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar JP. The molecular circuitry governing retinal determination. Biochim Biophys Acta. 2009;1789:306–314. doi: 10.1016/j.bbagrm.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sajithlal G, Zou D, Silvius D, Xu PX. Eya 1 acts as a critical regulator for specifying the metanephric mesenchyme. Dev Biol. 2005;284:323–336. doi: 10.1016/j.ydbio.2005.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu PX, Zheng W, Huang L, Maire P, Laclef C, Silvius D. Six1 is required for the early organogenesis of mammalian kidney. Development. 2003;130:3085–3094. doi: 10.1242/dev.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, Bitner-Glindzicz M, Francis M, Lacombe D, Vigneron J, Charachon R, Boven K, Bedbeder P, Van Regemorter N, Weissenbach J, Petit C. A human homologue of the Drosophila eyes absent gene underlies branchio- oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet. 1997;15:157–164. doi: 10.1038/ng0297-157. [DOI] [PubMed] [Google Scholar]
- 63.Quaggin SE, Kreidberg JA. Development of the renal glomerulus: good neighbors and good fences. Development. 2008;135:609–620. doi: 10.1242/dev.001081. [DOI] [PubMed] [Google Scholar]
- 64.Gao X, Chen X, Taglienti M, Rumballe B, Little MH, Kreidberg JA. Angioblast-mesenchyme induction of early kidney development is mediated by Wt1 and Vegfa. Development. 2005;132:5437–5449. doi: 10.1242/dev.02095. [DOI] [PubMed] [Google Scholar]
- 65.Tufro A, Teichman J, Banu N, Villegas G. Crosstalk between VEGF-A/VEGFR2 and GDNF/RET signaling pathways. Biochem Biophys Res Commun. 2007;358:410–416. doi: 10.1016/j.bbrc.2007.04.146. [DOI] [PubMed] [Google Scholar]
- 66.Cho EA, Patterson LT, Brookhiser WT, Mah S, Kintner C, Dressler GR. Differential expression and function of cadherin-6 during renal epithelium development. Development. 1998;125:4806–4815. doi: 10.1242/dev.125.5.803. [DOI] [PubMed] [Google Scholar]
- 67.Georgas K, Rumballe B, Valerius MT, Chiu HS, Thiagarajan RD, Lesieur E, Aronow BJ, Brunskill EW, Combes AN, Tang D, Taylor D, Grimmond SM, Potter SS, McMahon AP, Little MH. Analysis of early nephron patterning reveals a role for distal RV proliferation in fusion to the ureteric tip via a cap mesenchyme-derived connecting segment. Dev Biol. 2009;332:273–286. doi: 10.1016/j.ydbio.2009.05.578. [DOI] [PubMed] [Google Scholar]
- 68.Cheng HT, Kim M, Valerius MT, Surendran K, Schuster-Gossler K, Gossler A, McMahon AP, Kopan R. Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development. 2007;134:801–811. doi: 10.1242/dev.02773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taelman V, Van Campenhout C, Solter M, Pieler T, Bellefroid EJ. The Notch-effector HRT1 gene plays a role in glomerular development and patterning of the Xenopus pronephros anlagen. Development. 2006;133:2961–2971. doi: 10.1242/dev.02458. [DOI] [PubMed] [Google Scholar]
- 70.Liu Y, Pathak N, Kramer-Zucker A, Drummond IA. Notch signaling controls the differentiation of transporting epithelia and multiciliated cells in the zebrafish pronephros. Development. 2007;134:1111–1122. doi: 10.1242/dev.02806. [DOI] [PubMed] [Google Scholar]
- 71.Majumdar A, Lun K, Brand M, Drummond IA. Zebrafish no isthmus reveals a role for pax2.1 in tubule differentiation and patterning events in the pronephric primordia. Development. 2000;127:2089–2098. doi: 10.1242/dev.127.10.2089. [DOI] [PubMed] [Google Scholar]
- 72.Boyle SC, Kim M, Valerius MT, McMahon AP, Kopan R. Notch pathway activation can replace the requirement for Wnt4 and Wnt9b in mesenchymal-to-epithelial transition of nephron stem cells. Development. 2011;138:4245–4254. doi: 10.1242/dev.070433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brunskill EW, Aronow BJ, Georgas K, Rumballe B, Valerius MT, Aronow J, Kaimal V, Jegga AG, Yu J, Grimmond S, McMahon AP, Patterson LT, Little MH, Potter SS. Atlas of gene expression in the developing kidney at microanatomic resolution. Dev Cell. 2008;15:781–791. doi: 10.1016/j.devcel.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McMahon AP, Aronow BJ, Davidson DR, Davies JA, Gaido KW, Grimmond S, Lessard JL, Little MH, Potter SS, Wilder EL, Zhang P. GUDMAP: the genitourinary developmental molecular anatomy project. J Am Soc Nephrol. 2008;19:667–671. doi: 10.1681/ASN.2007101078. [DOI] [PubMed] [Google Scholar]
- 75.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 76.Rando OJ. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr Opin Genet Dev. 2012;22:148–155. doi: 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 78.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 80.Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 81.Patel SR, Kim D, Levitan I, Dressler GR. The BRCT-Domain Containing Protein PTIP Links PAX2 to a Histone H3, Lysine 4 Methyltransferase Complex. Dev Cell. 2007;13:580–592. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schwab KR, Patel SR, Dressler GR. Role of PTIP in class switch recombination and longrange chromatin interactions at the immunoglobulin heavy chain locus. Mol Cell Biol. 2011;31:1503–1511. doi: 10.1128/MCB.00990-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Patel SR, Bhumbra SS, Paknikar RS, Dressler GR. Epigenetic Mechanisms of Groucho/Grg/TLE Mediated Transcriptional Repression. Mol Cell. 2012;45:185–195. doi: 10.1016/j.molcel.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klymenko T, Muller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 2004;5:373–377. doi: 10.1038/sj.embor.7400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lefevre GM, Patel SR, Kim D, Tessarollo L, Dressler GR. Altering a histone H3K4 methylation pathway in glomerular podocytes promotes a chronic disease phenotype. PLoS Genet. 2010;6:e1001142. doi: 10.1371/journal.pgen.1001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stein AB, Jones TA, Herron TJ, Patel SR, Day SM, Noujaim SF, Milstein ML, Klos M, Furspan PB, Jalife J, Dressler GR. Loss of H3K4 methylation destabilizes gene expression patterns and physiological functions in adult murine cardiomyocytes. J Clin Invest. 2011;121:2641–2650. doi: 10.1172/JCI44641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Humphreys BD, Czerniak S, DiRocco DP, Hasnain W, Cheema R, Bonventre JV. Repair of injured proximal tubule does not involve specialized progenitors. Proc Natl Acad Sci U S A. 2011;108:9226–9231. doi: 10.1073/pnas.1100629108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Humphreys BD, Valerius MT, Kobayashi A, Mugford JW, Soeung S, Duffield JS, McMahon AP, Bonventre JV. Intrinsic epithelial cells repair the kidney after injury. Cell Stem Cell. 2008;2:284–291. doi: 10.1016/j.stem.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 89.Imgrund M, Grone E, Grone HJ, Kretzler M, Holzman L, Schlondorff D, Rothenpieler UW. Re-expression of the developmental gene Pax-2 during experimental acute tubular necrosis in mice 1. Kidney Int. 1999;56:1423–1431. doi: 10.1046/j.1523-1755.1999.00663.x. [DOI] [PubMed] [Google Scholar]
- 90.Piontek K, Menezes LF, Garcia-Gonzalez MA, Huso DL, Germino GG. A critical developmental switch defines the kinetics of kidney cyst formation after loss of Pkd1. Nat Med. 2007;13:1490–1495. doi: 10.1038/nm1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Takakura A, Contrino L, Zhou X, Bonventre JV, Sun Y, Humphreys BD, Zhou J. Renal injury is a third hit promoting rapid development of adult polycystic kidney disease. Hum Mol Genet. 2009;18:2523–2531. doi: 10.1093/hmg/ddp147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Verdeguer F, Le Corre S, Fischer E, Callens C, Garbay S, Doyen A, Igarashi P, Terzi F, Pontoglio M. A mitotic transcriptional switch in polycystic kidney disease. Nat Med. 2009 doi: 10.1038/nm.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Petruk S, Sedkov Y, Johnston DM, Hodgson JW, Black KL, Kovermann SK, Beck S, Canaani E, Brock HW, Mazo A. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell. 2012;150:922–933. doi: 10.1016/j.cell.2012.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]