Abstract
Biomineralization is an extremely efficient biologically guided process towards the advancement of nano-bio integrated materials. As a prime module of the natural world, enzymes are expected to play a major role in biogenic growth of inorganic nanostructures. Although there have been developments in designing enzyme-responsive nanoparticle systems or generation of inorganic nanostructures in an enzyme-stimulated environment, reports regarding action of enzymes as reducing agents themselves for the growth of inorganic nanoparticles still remains elusive. Here we present a mechanistic investigation towards the synthesis of metal and metallic alloy nanoparticles using a commonly investigated enzyme, Jack bean urease (JBU), as a reducing as well as stabilizing agent under physiological conditions. The catalytic functionality of urease was taken advantage of towards the development of metal-ZnO core-shell nanocomposites, making urease an ideal bionanoreactor for synthesizing higher order nanostructures such as alloys and core- shell under ambient conditions.
Inorganic nanoparticles are considered potential structural building blocks for new functional materials1,2,3,4,5. As many of the properties of these materials depend on the size, shape and composition of the building blocks, exploration of facile and efficient methods for design and fabrication of such structures is critically important. Biomolecule-directed growth of inorganic nanoparticles has evolved as an area of intense research owing to the capability of biomolecules in synthesis and assembly of inorganic nanostructures under benign conditions such as room temperature and in aqueous medium. Several biomolecules such as proteins, peptides, DNA and RNA have been used as templates for the growth and design of nanoparticle ensembles6,7,8,9,10,11,12,13,14,15 taking advantage of their various and distinctive molecular structures, specificities, functionalities and versatility in recognition and assembly. In this regard, development of simple preparative protocol through biogenic routes towards the synthesis of higher order nanostructures such as metallic alloys and core-shell compositions is potentially appealing as environmentally friendly alternatives to harsh chemical methods.
Enzymes, a key ingredient of the bio-systems, have been subject of particular attention in nanoparticle-biomolecule interaction studies, where nanoparticles function as enzyme responsive systems. The chemical or electrostatic attachment of enzymes to the nanoparticles has resulted in enhancement, retention or inhibition of catalytic activity of the enzyme16,17,18,19,20,21,22 that inspired the design of enzyme biosensors. On the other hand, the spatially confined environment of enzymes could be anticipated to facilitate the crystallization of inorganic materials with nanometer precisions23,24,25. There have been several reports of enzyme stimulated synthesis of metallic and metal oxide nanoparticles, where the product of an enzyme catalyzed reaction facilitates the formation of nanoparticles23,24,25,26,27,28,29. For instance, oxidases such as glucose-oxidase (GOx) catalyze oxidation of glucose producing H2O2 that acts as a reducing agent for the synthesis of Au nanoparticles26, resulting in the developement of an optical detection path for enzyme activity and sensing of glucose. Similarly, E-coli Glutathione Reductase catalyzes the NADPH-dependent reduction of HAuCl4, leading to the formation of Au nanoparticles at its active site28. However, paucity of literature is evident with respect to studies in which the enzymes can act both as reducing agent as well as stabilizers for metal nanoparticles. Until now, only α-amylase has been demonstrated as a pure enzyme that could generate Au nanoparticles from its corresponding salt30. Understanding the mechanism of the reduction capability of enzymes is not only critical to take full advantage of the nanoscale materials but also in studies related to structural alteration of enzymes that has profound influence on its kinetics.
Herein, we report the biogenic mineralization of metallic nanoparticles such as Au, Ag and Pt and their alloys using urease from jack bean plant (Canavalia ensiformis) as the reducing agent and stabilizer in water under mild conditions. Further the catalytic activity of the enzyme was exploited in order to synthesize metal and ZnO core-shell nanostructures at room temperature (Figure 1). To the best of our knowledge, the present work demonstrates for the first time that enzymes can be envisaged for the synthesis of alloys or core-shell nanostructures. Significantly, this is achieved in water under physiological conditions, thus expanding the current paradigm for biogenic nanoparticle synthesis with precisely controlled size and composition.
Figure 1. Synthesis of metal@ZnO core-shell nanostructures using urease as a template.
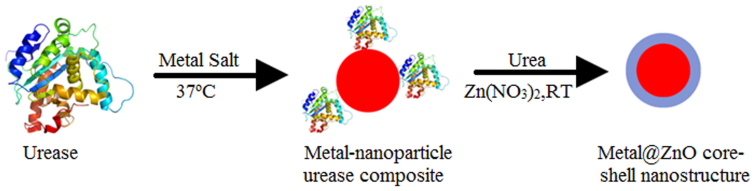
Step 1: Synthesis of metallic nanoparticles using urease both as reducing as well as stabilizing agent. Step 2: hydrolysis of urea by the nanoparticle-enzyme composite led to the increase in pH around the enzyme due to production of ammonia that catalyzed the growth of ZnO shells around the metal nanoparticle-enzyme composite.
Results
The simple synthetic procedure of incubation of JBU (2 mg/ml) with HAuCl4 (5.5 × 10−4 M) in presence of K2CO3 in water at its optimal pH 7.4 and 37°C yielded a very stable red sol signifying the formation of Au nanoparticles. Similarly when AgNO3 or H2PtCl4 was reacted with urease under similar reaction conditions, Ag and Pt nanoparticles were formed. The Au and Ag nanoparticles exhibited their plasmon resonance bands at 522 nm and 412 nm respectively as shown in Figure 2a. From transmission elctron microscopy (TEM) studies, the average diameter of Au nanoparticles was found to be 8.9 ± 1.6 nm (Figure 2b), while the mean size of Ag and Pt nanoparticles were found to be 4.1 ± 1.2 nm and 2.8 ± 0.8 nm respectively (Supplementary Figures S2 & S3). A careful structural investigation of the Ag nanoparticles by TEM, powder XRD and XPS suggested the formation of a Ag-Ag2O composite nanostructure (Supplementary Figures S2, S4–S6). The average particle sizes of Au and Ag-Ag2O nanoparticles (as calculated from TEM images) correlated well with the optical properties of the nanoparticles as predicted by theoretical calculations and earlier experimental reports31,32,33 (Supplementary Figures S7 & S8). A selected-area electron diffraction (SAED) pattern for the resulting Au nanoparticles (inset 2b) showed the (111), (200), (220) and (311) facets of Au, which was further confirmed through powder XRD studies (Supplementary Figure S9). The high resolution TEM image in Figure 2c revealed the lattice fringe of the (200) face of Au. It is worth mentioning that the nucleation and growth kinetics of the nanoparticles and the position of the plasmon resonance band depended very much on the reaction conditions. For example, while K2CO3 was used to adjust the pH of the medium, the Au nanoparticles were formed within 6 hours and the plasmon resonance peak appeared at 522 nm. On the other hand, when the nanoparticles were synthesized in Phosphate buffer saline (PBS) at pH 7.4, the nanoparticle formation required approximately 48 hours with the appearance of a plasmon resonance peak at 537 nm (Supplementary Figure S10). TEM images of Au nanoparticles in PBS showed substantial agglomeration, (Supplementary Figure S11) justifying the red shift in the plasmon resonance peak compared to those synthesized in presence of K2CO3. The increased ionic strength of the medium might have led to the agglomeration of the Au nanoparticles synthesized in PBS as previously reported34. The sizes of all the metallic nanoparticles (Au, Ag and Pt) synthesized using urease are well in accordance with the sizes of the nanoparticles synthesized using biosynthetic routes involving natural reducing agents. For example, synthesis of Au nanoparticles with an average particle size of 8.3 ± 2.6 nm using (HRE) - Sub E virus, spherical Ag nanoparticles of average diameter 5.3 nm using natural hydrocolloid gum kondagogu and Pt nanoparticles having average size of 2.2 nm using natural wood have been reported35,36,37.
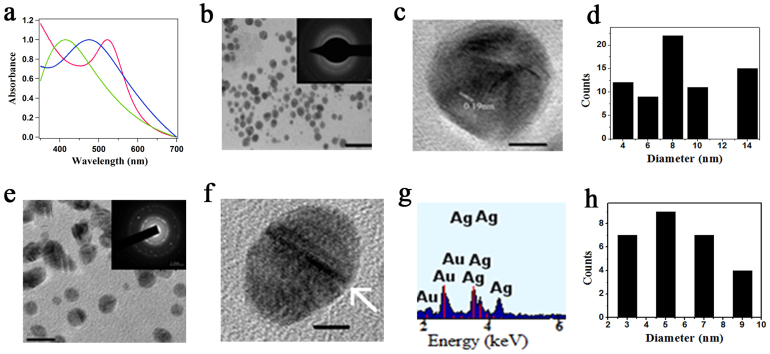
Figure 2.
(a) UV-visible spectrum of Ag (green), Au (red) and AuAg alloy (blue) nanoparticles in presence of K2CO3. (b) TEM image; scale bar 50 nm and SAED pattern (inset) of Au nanoparticles (c) HRTEM image of Au nanoparticles; scale bar 2 nm. (d) Particle size distribution of Au nanoparticles (e) TEM image; scale bar 10 nm, SAED pattern (inset) of AuAg alloy nanoparticles (f) HRTEM image of AuAg alloy nanoparticles; scale bar 2 nm. Arrow signifies the twins faults in the alloy. (g) EDX spectrum of AuAg alloy nanoparticles. (h) Particle size distribution of AuAg alloy nanoparticles.
Next, we explored the possibility of synthesizing metallic alloy nanoparticles using urease as a reducing agent. The incubation of the enzyme with two metal salts such as HAuCl4 and AgNO3 led to the formation of AuAg alloy nanoparticles in water at ambient temperature. When 1:1 molar ratio of HAuCl4 and AgNO3 was added in the reaction medium, the resulting nanoparticles showed a plasmon resonance peak at 476 nm, depicting the formation of Au0.5Ag0.5 nanoparticles38 (Figure 2a). The nanoparticles had an average diameter of 5.6 ± 2.0 nm as calculated from the TEM image (Figure 2e). The HRTEM image (Figure 2f) showed twins and stacking faults in most of the particles, probably due to high internal strain energy existing in these particles39. Energy dispersive X-ray (EDX) analysis confirmed the presence of both Au and Ag in the alloy nanoparticles (Figure 2g). We successfully extended this methodology towards the formation of other metallic alloys such as AgPt and AuPt nanoparticles (Supplementary Figures S13–S15).
The conjugation of nanoparticles with enzyme often leads to the alteration of its activity21. In case of urease, the increased pH of the medium is a good indicator for the measurement of enzyme activity, primarily, due to the liberation of ammonia by the hydrolysis of urea by urease. Ammonia molecules react with water to form ammonium hydroxide that subsequently dissociates to form hydroxide ions resulting in the net increase in the solution pH. As shown in Figure 3a, the decomposition of urea by the native urease led to a pH increase of the medium from 6.9 to 9.0 whereas the Au nanoparticle-urease composite (as synthesized by the reduction of the metal salt with urease) could increase the pH from 6.8 to 7.7, thus indicating a decrease in the enzyme activity. These results were further confirmed using a colorimetric assay, where the solution pH increase was monitored by a pH sensitive dye, bromocresol purple (Supplementary Figure S16). Notably, denatured urease (enzyme heated at 90°C for 30 minutes in water) could not increase the pH of the medium. For understanding the role of Au nanoparticles in modulating enzymatic activity of urease, citrate stabilized Au nanoparticles were functionalized with urease and the activity of the enzyme was monitored by the pH change of the medium while decomposing urea. The pH of the medium increased from 6.9 to 9.1, clearly indicating that mere functionalization of Au nanoparticles separately synthesized had no distinct impact on the enzyme activity. However, during the process of reducing the metal salt to form Au nanoparticles, urease had some loss of activity, although it was not completely denatured. From these results, it was predicted that there could be some conformational changes and chemical modifications in urease during the synthesis of metal nanoparticles resulting in partial inhibition of the enzyme function.
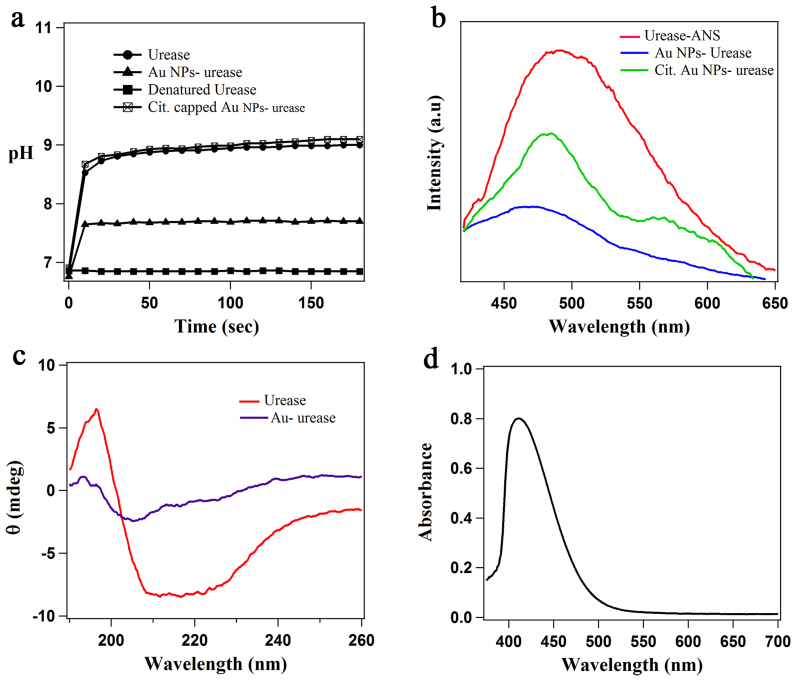
Figure 3.
(a) pH change of the medium by native urease, Au nanoparticle-urease composite (urease reduced Au nanoparticles), denatured urease and functionalized Au nanoparticles with urease (b) Emission spectra of ANS in native urease, Citrate capped Au nanoparticles-urease composite and Au nanoparticle-urease composite (urease reduced Au nanoparticles) depicting the conformational modifications in enzyme structure (λex = 370 nm). (c) Circular dichroism spectrum of native urease and Au nanoparticle urease composite in water. (d) UV-visible spectrum of DTNB treated urease after incubation with HAuCl4 for 48 hours, showing the absence of SPR band of Au nanoparticles.
The conformational studies were performed by fluorescence spectroscopy using 1-anilino-8-naphthalene sulfonate (ANS) as an extrinsic probe, as reported earlier for protein studies40. ANS is highly fluorescent in hydrophobic environment such as interior of an enzyme, but has very low quantum yield in water. Upon unfolding, the intensity will decrease as ANS will be released from the hydrophobic interior of the enzyme into the surrounding medium. Predictibly, as shown in Figure 3b, the emission intensity of ANS at around 490 nm was quenched and blue-shifted to around 470 nm, when urease was involved in the formation of Au nanoparticles. The quenching of ANS emission might be due to two factors: (i) unfolding of urease during reduction and subsequent binding to the Au nanoparticle surface (ii) energy transfer from the ANS donor to the Au nanoparticle acceptor as the emission wavelength of ANS matches with the surface plasmon resonance band of Au nanoparticles. In order to confirm this, we performed a controlled experiment in which urease solution was incubated with ANS and citrate capped Au nanoparticles (maintaining the HAuCl4 concentration same as used in the urease reduction). The fluorescence results (Figure 3b) showed that the emission intensity of ANS was indeed quenched by the Au nanoparticles with a change in line-shape signifying the energy transfer40. However the quenching was not as much as observed for the urease reduced Au nanoparticles, suggesting that both energy transfer as well as partial unfolding of urease contributed to the emission quenching of ANS.
To get a further insight into the conformational change of the enzyme, Circular Dichroism (CD) and FTIR studies were performed. The CD spectrum of native urease and Au NP-urease is shown in Figure 3c. The decrease in the α-helical content of a protein, which indicates the unfolding of the protein, can easily be monitored by tracking the loss in CD signal at 222 nm41. As shown in Figure 3c, the CD signal at 222 nm in case of Au NP-urease composite showed a large decrease in intensity compared to that for the native urease, which is a clear indication of the enzyme being unfolded when it was involved in nanoparticle synthesis. This was further evidenced from the FTIR spectra, where the characteristic amide I band in native urease shifted from 1650 cm−1 to 1635 cm−1 in Au nanoparticle-urease composite (Supplementary Figure S17) signifying substantial perturbation in the enzyme structure42. The second derivative of the amide I band of urease showed peaks at 1656, 1644 and 1636 cm−1 corresponding to α-helix, disordered and β-sheet respectively, whereas in case of the enzyme reduced Au nanoparticles, these peaks were shifted to 1652, 1641 and 1634 cm−1, with a decrease in intensity41,43. Thus from fluorescence, CD and FTIR studies, it was clearly evident that there was a significant change in the structure of urease leading to partial inhibition of the enzyme activity.
Enzymes have a great structural complexity, as they are composed of several amino acid residues. The arrangement of these constituent amino acids into helices or sheets accounts for their structures with specific functionalities. Thus understanding the mechanism towards the growth of nanoparticles and subsequent structural and functional changes in urease is engrossing. It is established that cysteine has reducing as well as stabilizing capability for the generation of metal nanoparticles owing to high affinity of thiols for metals44. In enzymes also, cysteines have been postulated to be preferred sites for metal seeding and nanoparticle conjugation. Thus we assumed that the free and exposed cysteine groups in an enzyme could act as a reducing agent for the synthesis of metal nanoparticles. To support our assumption, the free cysteine groups in JBU were modified by reaction with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) in non-denaturating conditions45. The addition of HAuCl4 to the DTNB modified urease, did not result in the formation of Au nanoparticles, clearly suggesting the involvement of cysteine in the formation of nanoparticles (Figure 3d). The modification of the cysteine groups in urease was also evidenced by using FTIR, while observing the disappearance of a weak band at 2660 cm−1 in the Au nanoparticle-urease composite, attributed to the S-H stretching mode in urease cysteine14 (Supplementary Figure S19). Interestingly, in case of heat-denatured urease, we observed faster growth of Au nanoparticles compared to the native enzyme, clearly indicating that it is not essential for the enzyme to retain its native structure for being used as a reducing agent.
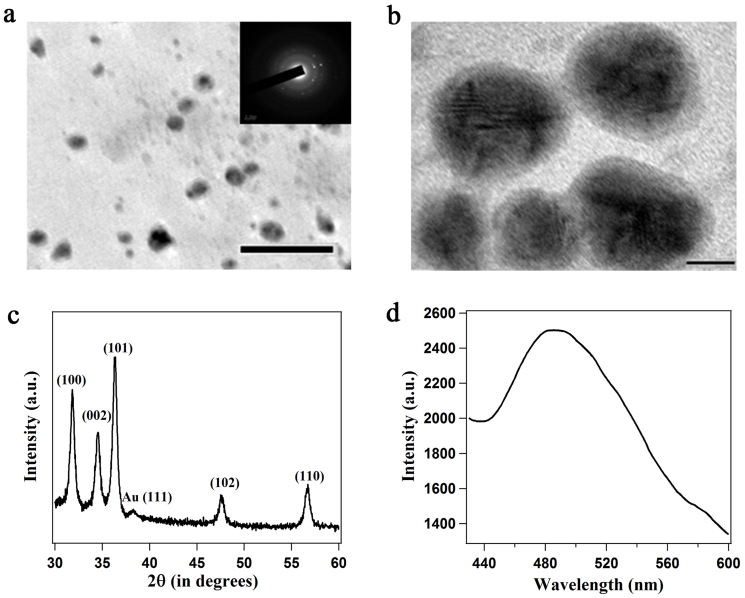
The primary activity of urease is the decomposition of urea, resulting in the liberation of ammonia with a net increase in the solution pH, making the environment suitable for the growth of metal oxide nanoparticles. Hence, we explored the possibility of the production of a semiconductor material such as ZnO as a shell around the Au nanoparticle-enzyme composite taking advantage of the catalytic ability of urease. Under laboratory conditions addition of ammonia into a reaction medium containing metal salts resulted in the synthesis of metal oxide nanoparticles through condensation of the hydrolyzed product46. In the present method, although the urease activity was partially inhibited during nanoparticle synthesis, the hydrolysis of urea by Au nanoparticle-urease composite led to a homogeneous increase of the solution pH to 7.7 that has been exploited to synthesize ZnO nanoparticles. When Au nanoparticle-urease composite was incubated with urea and zinc nitrate hexahydrate in water at room temperature, ZnO nanoshells with an average thickness of 2.1 ± 0.4 nm were formed around the Au nanoparticles (Figure 4a). The ZnO nanoshells showed high crystallinity as evidenced from the SAED pattern. The HRTEM image (Figure 4b) clearly revealed the electron-dense core of Au surrounded by lesser dense shell of ZnO. The UV-visible spectrum of the Au@ZnO core-shell nanoparticles (Supplementary Figure S20) showed two distinct bands centred at 340 nm and 530 nm, characteristic of Au@ZnO core shell nanoparticles47. The intensity of the band at 530 nm decreased significantly and was red shifted by 8 nm compared to pure Au nanoparticles. The X-ray diffraction pattern (Figure 4c) of the Au-ZnO core-shell nanoparticles suggested nanocrystalline structure of the ZnO nanoshells with wurtzite structure (JCPDS card no. 0-3-0888). Scherrer analyses of the (101) and (102) reflections were used to calculate the crystal diameters of ZnO and showed an average size of 11.3 nm, which was consistent with the overall diameter of Au-ZnO core-shell particles as measured by TEM. Along with the characteristic peaks of ZnO, one small reflection at 38.2° was observed, which is characteristic of (111) plane of Au. EDX analysis further confirmed the presence of both Au and Zn in the composite material (Supplementary Figure S21). The ZnO nanoshells also showed their characteristic emissions at 421 nm (λex = 340 nm) (Supplementary Figure S22) and 490 nm (λex = 390 nm) (Figure 4d), originating from the oxygen defects present in the crystal. Elemental analysis of the Au-ZnO nanoparticles acquired by Inductively coupled plasma-atomic emission spectrometry (ICP-AES) measurements showed Au: Zn molar ratio of 55 : 45, which was in close agreement with the theoretical calculations based on Au@ZnO core-shell morphology (details in Supplementary information). It is noteworthy to mention that Au-denatured urease composite (Au nanoparticles reduced by heat-denatured urease) could not form the ZnO nanoshells, as the inherent characteristics of ZnO were not evidenced by XRD and fluorescence experiments.
Figure 4.
(a) TEM image; scale bar 50 nm and SAED pattern (inset) of Au@ZnO core-shell nanoparticles. (b) HRTEM image; scale bar 5 nm. (c) powder XRD pattern of Au-ZnO nanoparticles, the principal Bragg reflections of ZnO are identified. (d) Emission spectrum of Au@ZnO core-shell nanoparticles (λex = 390 nm).
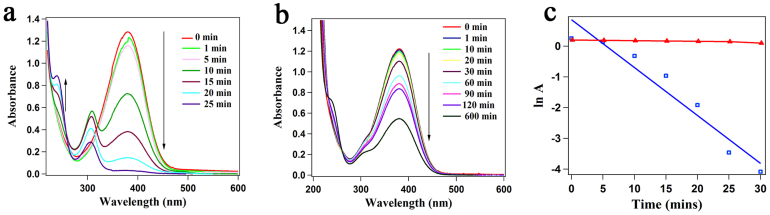
In order to have further evidence of Au-ZnO composite material with core-shell morphology, we performed the reduction of p-nitroaniline with NaBH4 using Au-urease and Au@ZnO-urease as catalysts. It is well known that Au nanoparticles can be used as an efficient heterogeneous catalyst for the reduction of p-nitroaniline to 1,4-diaminobenzene in presence of NaBH448. It was found that the reaction was completed within 25 minutes when urease reduced Au nanoparticles were used as catalyst, as evidenced from the UV-visible spectrum (Figure 5a). When the reduction was carried out in presence of Au@ZnO core shell nanoparticles synthesized by the present method, as catalyst, the reaction did not proceed substantially even after 10 hours (Figure 5b). The results implied that due to the presence of ZnO shell on Au, the nanoparticles could not function as an efficient catalyst for the reduction reaction. A plot of logarithm of absorbance (ln A) for p-nitroaniline at 380 nm showed a linear decrease with time, confirming the reduction of p-nitroaniline, whereas in case of Au@ZnO core shell nanoparticles as catalyst, the plot of ln A versus time (Figure 5c) showed that the reaction proceeded very slowly, which further confirmed that the Au nanoparticles were indeed coated with ZnO layer.
Figure 5.
(a) Time-dependent UV-visible spectrum showing the reduction of p-nitroaniline to 1,4-diaminobenzene with NaBH4 in presence of urease reduced Au nanoparticles as catalyst. (b) Time-dependent UV-visible spectrum for the reduction of p-nitroaniline to 1,4-diaminobenzene with NaBH4 catalyzed by Au@ZnO core-shell nanoparticles synthesized using urease. (c) Plot of ln (A) versus time for the reduction of p-nitroaniline to 1,4-diaminobenzene with NaBH4 using urease reduced Au nanoparticles as catalyst (blue line and markers) and Au@ZnO core shell nanoparticles as catalyst (red line and markers).
Discussion
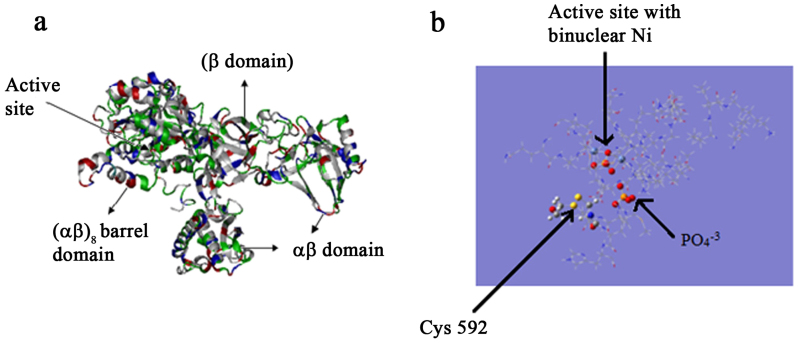
In our synthetic methodology, the involvement of urease in reducing the metal salts and subsequent binding to the nanoparticle surface led to conformational changes in the enzyme. The result was the partial inhibition of urease activity due to which hydrolysis of urea by the nanoparticle-urease conjugate led to pH enhancement only to a slightly basic 7.7, whereas in case of native urease the solution pH increased to 9.0. To have an insight into the mechanism of synthesis of nanoparticles by urease, the reported crystal structure of the JBU was examined. Jack Bean Urease was the first enzyme to be crystallized and the first example of a nickel metalloenzyme49. Spectroscopic, crystallographic and theoretical studies50,51,52,53 have illustrated not only the molecular architecture and amino acid sequences in urease, but also the mechanism of enzymatic activity. It has been suggested that the essential cysteine residues serve as acid catalysts in the mechanism of action of urease54. According to earlier reports, of the 36 cysteines present in JBU, only 3 of them (Cys59, Cys207 and Cys592) are the exposed ones and chemical modification of these groups impairs its activity51 (protein data bank entry 3LA4). Out of these, Cys592 is located on the mobile flap adjacent to the active site (Figure 6b) and plays a critical role in catalysis50. In the present study, we hypothesized that the involvement of Cys592 in the formation of metallic nanoparticles followed by formation of disulfide bonds resulted in the structural changes in the mobile flap. As the mobile flap is involved in the regulation of access to the active site containing the nickel centre, the structural modification of the Cys592 and subsequent attachment to the metal nanostructures through the Metal-S bond greatly influenced the flap loosing its mobility. In case of denatured urease, five other buried cysteine residues per subunit become exposed and more reactive55, accounting for the faster growth of metallic nanoparticles by denatured urease.
Figure 6.
(a) Overall structure of the Jack bean urease monomer (b) stereo diagram of the active site architecture containing a binuclear nickel centre and mobile flap of urease. For clarity, only cys592 has been highlighted along with Ni ions and PO43− residues present in the active site.
Although the conformational changes in the enzyme structure did not have any impact on the metallic nanoparticle synthesis, they had a definitive role in the formation of ZnO nanostructures. In case of native urease, Zn2+ binds on the negatively charged enzyme surface through electrostatic interaction at around pH 9. Previous reports have suggested the formation of zinc hydroxide intermediate under the basic conditions and further dehydration of these intermediates yield ZnO on the enzyme surface propelled by the "salting out" effect56,57. In our case, the decomposition of urea by Au nanoparticle-urease composite led to an increase of solution pH to 7.7. Even at this near neutral conditions, the entropy enhancement due to the disruption of the hydration layer around urease (because of Zn2+ binding to the enzyme surface) was enough to convert Zn(OH)2 to ZnO nanoshells.
In summary, we have demonstrated the use of urease as an effective biomolecular reactor towards the growth of metallic nanoparticles, metallic alloys and metal-metal oxide core-shell nanostructures under ambient conditions. The exposed cysteine residues in the enzyme were found to be responsible for the generation of metal and metallic alloy nanoparticles. Although there were conformational changes leading to partial inhibition of the enzyme activity during the nanoparticle synthesis, the activity of urease could still be employed for the synthesis of metal-ZnO core-shell nanoparticles. The use of urease as a reducing and stabilizing agent for the synthesis of nanoparticles demonstrates the practicability of this catalytic nanoreactor system as an alternative to the current environmentally harsh and energy-exhaustive methods for material synthesis. Further these studies will give a mechanistic intimation towards the capability of microorganisms such as fungus, virus and bacteria in synthesizing nanoparticles58,59,60. The proposed methodology can be extended easily towards the generation of a range of alloys and core-shell nanostructures involving metal and metal oxides. Further the immobilization and growth of these nanoparticle-enzyme composites on various substrates will afford opportunities for the development of technologically relevant systems.
Methods
Synthesis of metallic nanoparticles
Metallic nanoparticles were synthesized by incubating a solution containing 2 mg/ml enzyme solution, K2CO3 and the appropriate metal salt (HAuCl4, AgNO3 or K2PtCl4) at 37°C with mild stirring for various times (6 hours for Au and Ag nanoparticles and 36 hours for Pt nanoparticles). The morphology of the nanoparticles and reaction time depended on the method of synthesis of nanoparticles. For example, when PBS buffer was used to maintain the pH of the medium to 7.4, instead of K2CO3 the formation of Au nanoparticles required 48 hours.
Synthesis of alloy nanoparticles
The synthesis of all the alloy nanoparticles was carried out in 2.2 ml of 2 mg/ml enzyme solution containing 2.5 mg of K2CO3. In all the three cases, metal salts were added such that their individual concentration in the final solution was 2.7 × 10−4 M, and the resulting solution was stirred at 37°C for 24 hrs.
Synthesis of Au@ZnO Core-shell nanoparticles
The Au nanoparticle-urease composites (by reduction of HAuCl4 with urease) were centrifuged and washed with water several times to remove any free enzyme unbound to the Au nanoparticle surface. The obtained pellet was re-dispersed in 0.1 M NaNO3. To 2 mL of the Au nanoparticle-urease composite solution, was added 10 mg of urea and 50 μL of 0.02 M Zn(NO3)2·6H2O and the reaction mixture was stirred at room temperature for 12 hrs.
Details of synthesis for individual metal nanoparticles are given in the supplementary information.
Characterization of nanoparticles
A Varian Cary 100 Bio spectrophotometer was used for UV-visible measurements. Emission spectra were recorded using a fluoromax-4p fluorometer from Horiba (Model: FM-100). Powder X-ray diffraction patterns (XRD) were recorded on a Bruker D8 Advance diffractometer with Cu Kα source (wavelength of X-rays was 0.154 nm). Transmission electron microscopy (TEM) images were recorded using a Philips CM 200 microscope and High resolution transmission electron microscope images were recorded using a JEOL JEM-2100 microscope at an operating voltage of 200 kv. FTIR spectra were recorded in KBr pellet using a Bruker Tensor 27 instrument. Circular Dichroism (CD) studies were performed using a JASCO J-815 spectropolarimeter. ICP-AES measurements were performed using instrument from M/s. Spectro, Germany (Model: Arcos). XPS spectra were recorded using an ESCA instrument: VSW of UK make.
Author Contributions
B.S. performed majority of the experiments and contributed towards the manuscript preparation. S.M. performed several of the spectroscopic and microscopic investigations and interpreted the data. T.K.S. conceived and supervised the project, interpreted the data and wrote the manuscript. All authors discussed the results.
Supplementary Material
Supplementary Information
Acknowledgments
This work is supported by SERB, DST, India research project No. SR/S1/PC-32/2010. B. S. and S. M. acknowledge the research fellowships from UGC (India) and CSIR (India) respectively. We thank SAIF, NEHU, Shillong for TEM facility, SAIF, IIT Bombay for TEM and ICP-AES facilities and UGC-DAE consortium for scientific research, Indore for powder XRD as well as XPS facilities. Helpful discussions with Dr. Deepa Dey is highly acknowledged.
References
- Daniel M. C. & Astruc D. Gold Nanoparticles: Assembly, Supramolecular Chemistry, Quantum-Size-Related Properties, and Applications toward Biology, Catalysis, and Nanotechnology. Chem. Rev. 104, 293–346 (2004). [DOI] [PubMed] [Google Scholar]
- Goesmann H. & Feldmann C. Nanoparticulate Functional Materials. Angew. Chem. Int. Ed. 49, 1362–1395 (2010). [DOI] [PubMed] [Google Scholar]
- Costi R., Saunders A. E. & Banin U. Colloidal Hybrid Nanostructures: A New Type of Functional Materials. Angew. Chem. Int. Ed. 49, 4878–4897 (2010). [DOI] [PubMed] [Google Scholar]
- Talapin D. V., Lee J.-S., Kovalenko M. V. & Shevchenko E. V. Prospects of Colloidal Nanocrystals for Electronic and Optoelectronic Applications. Chem. Rev. 110, 389–458 (2010). [DOI] [PubMed] [Google Scholar]
- Stark W. J. Nanoparticles in Biological Systems. Angew. Chem. Int. Ed. 50, 1242–1258 (2011). [DOI] [PubMed] [Google Scholar]
- Berti L. & Burley G. A. Nucleic acid and nucleotide-mediated synthesis of inorganic nanoparticles. Nat. Nanotechnol. 3, 81–87 (2008). [DOI] [PubMed] [Google Scholar]
- Ma N., Sargent E. H. & Kelley S. O. One-step DNA-programmed growth of luminescent and biofunctionalized nanocrystals. Nat. Nanotechnol. 4, 121–125 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H. et al. Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme. Nat. Nanotechnol. 6, 93–97 (2011). [DOI] [PubMed] [Google Scholar]
- Niemeyer C. M. Nanoparticles, Proteins, and Nucleic Acids: Biotechnology Meets Materials Science. Angew. Chem. Int. Ed. 40, 4128–4158 (2001). [DOI] [PubMed] [Google Scholar]
- Gugliotti L. A., Feldheim D. L. & Eaton B. E. RNA-Mediated Metal-Metal Bond Formation in the Synthesis of Hexagonal Palladium Nanoparticles. Science 304, 850–852 (2004). [DOI] [PubMed] [Google Scholar]
- Whaley S. R., English D. S., Hu E. L., Barbara P. F. & Belcher A. M. Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly. Nature 405, 665–668 (2000). [DOI] [PubMed] [Google Scholar]
- Dickerson M. B., Sandhage K. H. & Naik R. R. Protein- and Peptide-Directed Syntheses of Inorganic Materials. Chem. Rev. 108, 4935–4978 (2008). [DOI] [PubMed] [Google Scholar]
- Moyano D. F. & Rotello V. M. Nano Meets Biology: Structure and Function at the Nanoparticle Interface. Langmuir 27, 10376–10385 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M. et al. Protein-Assisted One-Pot Synthesis and Biofunctionalization of Spherical Gold Nanoparticles for Selective Targeting of Cancer Cells. Angew. Chem. Int. Ed. 51, 9272–9275 (2012). [DOI] [PubMed] [Google Scholar]
- Chen C. L., Zhang P. & Rosi N. L. A New Peptide-Based Method for the Design and Synthesis of Nanoparticle Superstructures: Construction of Highly Ordered Gold Nanoparticle Double Helices. J. Am. Chem. Soc. 130, 13555–13557 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadiali J. E. & Stevens M. M. Enzyme-Responsive Nanoparticle Systems. Adv. Mater. 20, 4359–4363 (2008). [Google Scholar]
- Wang Z., Levy R., Fernig D. G. & Brust M. Kinase-Catalyzed Modification of Gold Nanoparticles: A New Approach to Colorimetric Kinase Activity Screening. J. Am. Chem. Soc 128, 2214–2215 (2006). [DOI] [PubMed] [Google Scholar]
- Choi Y., Ho N. H. & Tung C. H. Sensing Phosphatase Activity by Using Gold Nanoparticles. Angew. Chem. Int. Ed. 46, 707–709 (2007). [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lorenzo L., de la Rica R., Alvarez-Puebla R. A., Liz-Marzan L. M. & Stevens M. M. Plasmonic nanosensors with inverse sensitivity by means of enzyme-guided crystal growth. Nat. Mater. 11, 604–607 (2012). [DOI] [PubMed] [Google Scholar]
- You C. C., Agasti S. S., De M., Knapp M. J. & Rotello V. M. Modulation of the Catalytic Behavior of α-Chymotrypsin at Monolayer-Protected Nanoparticle Surfaces. J. Am. Chem. Soc. 128, 14612–14618 (2006). [DOI] [PubMed] [Google Scholar]
- Deka J., Paul A. & Chattopadhyay A. Modulating enzymatic activity in the presence of gold nanoparticles. RSC Adv. 2, 4736–4745 (2012). [Google Scholar]
- Bonomi R., Caccolaro A., Sansone A., Scrimin P. & Prins L. J. Detection of Enzyme Activity through Catalytic Signal Amplification with Functionalized Gold Nanoparticles. Angew. Chem. Int. Ed. 50, 2307–2312 (2011). [DOI] [PubMed] [Google Scholar]
- Shenton W., Mann S., Colfen H., Bacher A. & Fischer M. Synthesis of Nanophase Iron Oxide in Lumazine Synthase Capsids. Angew. Chem. Int. Ed. 40, 442–445 (2001). [DOI] [PubMed] [Google Scholar]
- de la Rica R. & Matsui H. Urease as a Nanoreactor for Growing Crystalline ZnO Nanoshells at Room Temperature. Angew. Chem. Int. Ed. 47, 5415–5417 (2008). [DOI] [PubMed] [Google Scholar]
- Johnson J. M., Sinsinger N., Sun C., Li D. & Kisailus D. Urease-Mediated Room-Temperature Synthesis of Nanocrystalline Titanium Dioxide. J. Am. Chem. Soc. 134, 13974–13977 (2012). [DOI] [PubMed] [Google Scholar]
- Willner I., Baron R. & Willner B. Growing Metal Nanoparticles by Enzymes. Adv. Mater. 18, 1109–1120 (2006). [Google Scholar]
- Virkutyte J. & Varma R. S. Green synthesis of metal nanoparticles: Biodegradable polymers and enzymes in stabilization and surface functionalization. Chem. Sci. 2, 837–846 (2011). [Google Scholar]
- Scott D., Toney M. & Muzikar M. Harnessing the Mechanism of Glutathione Reductase for Synthesis of Active Site Bound Metallic Nanoparticles and Electrical Connection to Electrodes. J. Am. Chem. Soc. 130, 865–874 (2008). [DOI] [PubMed] [Google Scholar]
- Xiao Y., Patolsky F., Katz E., Hainfeld J. F. & Willner I. "Plugging into Enzymes": Nanowiring of Redox Enzymes by a Gold Nanoparticle. Science 299, 1877–1881 (2003). [DOI] [PubMed] [Google Scholar]
- Rangnekar A. et al. Retention of Enzymatic Activity of α-Amylase in the Reductive Synthesis of Gold Nanoparticles. Langmuir 23, 5700–5706 (2007). [DOI] [PubMed] [Google Scholar]
- Link S. & El-Sayed M. A. Size and Temperature Dependence of the Plasmon Absorption of Colloidal Gold Nanoparticles. J. Phys. Chem. B 103, 4212–4217 (1999). [Google Scholar]
- Haiss W., Thanh T. K. N., Aveyard J. & Fernig D. G. Determination of Size and Concentration of Gold Nanoparticles from UV-Vis Spectra. Anal. Chem. 79, 4215–4221 (2007). [DOI] [PubMed] [Google Scholar]
- Lu H., Yu L., Liu Q. & Du J. Ultrafine silver nanoparticles with excellent antibacterial efficacy prepared by a handover of vesicle templating to micelle stabilization. Polym. Chem. 4, 3448–3452 (2013). [Google Scholar]
- Du S. et al. Aggregation and adhesion of gold nanoparticles in phosphate buffered saline. J. Nanopart. Res. 14, 758 (2012). [Google Scholar]
- Slocik J. M., Naik R. R., Stone M. O. & Wright D. W. Viral templates for gold nanoparticle synthesis. J. Mater. Chem. 15, 749–753 (2005). [Google Scholar]
- Vinod V. T. P., Saravanan P., Sreedhar B., Devi D. K. & Sashidhar R. B. A facile synthesis and characterization of Ag, Au and Pt nanoparticles using a natural hydrocolloid gum kondagogu (Cochlospermum gossypium). Colloids Surf. B 83, 291–298 (2011). [DOI] [PubMed] [Google Scholar]
- Lin X. et al. Platinum nanoparticles using wood nanomaterials: eco-friendly synthesis, shape control and catalytic activity for p-nitrophenol reduction. Green Chem. 13, 283–287 (2011). [Google Scholar]
- Burda C., Chen X. B., Narayanan R. & El-Sayed M. A. Chemistry and Properties of Nanocrystals of Different Shapes. Chem. Rev. 105, 1025–1102 (2005). [DOI] [PubMed] [Google Scholar]
- Rao C. N. R., Matte H. S. S. R., Voggu R. & Govindaraj A. Recent progress in the synthesis of inorganic nanoparticles. Dalton Trans. 41, 5089–5120 (2012). [DOI] [PubMed] [Google Scholar]
- Lystvet S. M., Volden S., Halskau O. & Glomm W. R. Immobilization onto gold nanoparticles alters α-lactalbumin interaction with pure and mixed phospholipid monolayers. Soft Matter 7, 11501–11509 (2011). [Google Scholar]
- Cruz J. C., Pfromm P. H., Tomich J. M. & Rezac M. E. Conformational changes and catalytic competency of hydrolases adsorbing on fumed silica nanoparticles: II. Secondary structure. Colloids Surf. B 81, 1–10 (2010). [DOI] [PubMed] [Google Scholar]
- Kumar C. V. & Chaudhari A. Chemical Chaperones: influence of carboxylate orientation in the refolding of glucose oxidase. Microporous and Mesoporous Materials. 47, 407–410 (2001). [Google Scholar]
- Jiang X., Jiang J., Jin J., Wang E. & Dong S. Effect of Colloidal Gold Size on the conformational Changes of Adsorbed Cytochrome c: Probing by Circular Dichroism, UV-Visible, and Infrared Spectroscopy. Biomacromolecules 6, 46–53 (2005). [DOI] [PubMed] [Google Scholar]
- Ma Z. & Han H. One-step synthesis of cysteine-coated gold nanoparticles in aqueous solution. Colloids Surf. A 317, 229–233 (2008). [Google Scholar]
- Ellman G. L. Tissue Sulfhydryl Groups. Arch. Biochem. Biophys. 82, 70–77 (1959). [DOI] [PubMed] [Google Scholar]
- Kisailus D., Schwenzer B., Gomm J., Weaver J. C. & Morse D. E. Kinetically Controlled Catalytic Formation of Zinc Oxide Thin Films at Low Temperature. J. Am. Chem. Soc. 128, 10276–10280 (2006). [DOI] [PubMed] [Google Scholar]
- Haldar K. K., Sen T. & Patra A. Au@ZnO Core-Shell Nanoparticles Are Efficient Energy Acceptors with Organic Dye Donors. J. Phys. Chem. C 112, 11650–11656 (2008). [Google Scholar]
- Yao Y. et al. A new water-soluble pillar[5]arene: synthesis and application in the preparation of gold nanoparticles. Chem. Commun. 48, 6505–6507 (2012). [DOI] [PubMed] [Google Scholar]
- Sumner J. B. The isolation and crystallization of the enzyme urease. J. Biol. Chem. 69, 435–441 (1926). [Google Scholar]
- Roberts B. P., Miller B. R. III, Roitberg A. E. & Merz K. M. Jr Wide-Open Flaps Are Key to Urease Activity. J. Am. Chem. Soc. 134, 9934–9937 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian A. & Ponnuraj K. Crystal Structure of the First Plant Urease from Jack Bean: 83 Years of Journey from Its First Crystal to Molecular Structure. J. Mol. Biol. 400, 274–283 (2010). [DOI] [PubMed] [Google Scholar]
- Krajewska B. & Zaborska W. Jack bean urease: The effect of active-site binding inhibitors on the reactivity of enzyme thiol groups. Bioorg. Chem. 35, 355–365 (2007). [DOI] [PubMed] [Google Scholar]
- Takishima K., Suga T. & Mamiya G. The structure of jack bean urease- The complete amino acid sequence, limited proteolysis and reactive cysteine residues. Eur. J. Biochem. 175, 151–165 (1988). [DOI] [PubMed] [Google Scholar]
- Dixon N. E., Riddles P. W., Gazzola C., Blakeley R. L. & Zerner B. Jack bean urease (EC 3.5.1.5). V. On the mechanism of action of urease on urea, formamide, acetamide, N-methylurea, and related compounds. Can. J. Biochem 58, 1335–1344 (1980). [DOI] [PubMed] [Google Scholar]
- Jabri E. & Karplus P. A. Structures of the Klebsiella aerogenes Urease Apoenzyme and Two Active-Site Mutants. Biochemistry 35, 10616–10626 (1996). [DOI] [PubMed] [Google Scholar]
- Garczarek F. & Gerwert K. Functional waters in intraprotein proton transfer monitored by FTIR difference spectroscopy. Nature 439, 109–112 (2006). [DOI] [PubMed] [Google Scholar]
- Makarov V., Pettitt M. & Feig M. Solvation and Hydration of Proteins and Nucleic Acids: A Theoretical View of Simulation and Experiment. Acc. Chem. Res. 35, 376–384 (2002). [DOI] [PubMed] [Google Scholar]
- Mann S. Molecular tectonics in biomineralization and biomimetic materials chemistry. Nature 365, 499–505 (1993). [Google Scholar]
- Escosura A., de la Nolte R. J. M. & Cornelissen J. J. L. M. Viruses and protein cages as nanocontainers and nanoreactors. J. Mater. Chem. 19, 2274–2278 (2009). [Google Scholar]
- Ahmad A. et al. Enzyme mediated extracellular synthesis of CdS nanoparticles by the fungus Fusarium Oxysporum. J. Am. Chem. Soc. 124, 12108–12109 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information