Abstract
Engagement of innate viral sensors elicits a robust antiviral program via the induction of type I interferons (IFNs). Innate defense mechanisms against ssDNA viruses are not well defined. Here, we examine type I IFN induction and effectiveness in controlling a ssDNA virus. Using mouse embryonic fibroblasts (MEFs), we found that a murine parvovirus, minute virus of mice (MVMp), induced a delayed but significant IFN response. MEFs deficient in mitochondrial antiviral signaling protein (MAVS) mounted a wild-type IFN response to MVMp infection, indicating that RIG-I-dependent RNA intermediate recognition is not required for innate sensing of this virus. However, MVMp-induced IFNs, as well recombinant type I IFNs, were unable to inhibit viral replication. Finally, MVMp infected cells became unresponsive to Poly (I:C) stimulation. Together, these data suggest that the MVMp efficiently evades antiviral immune mechanisms imposed by type I IFNs, which may in part explain their efficient transmission between mice.
Keywords: Innate immunity, ssDNA viruses, Interferons, RIG-I, Antiviral defense
Introduction
Most cell types in mammalian hosts detect viral infection via cytosolic and nuclear pattern recognition receptors (PRRs), which sense the nucleic acid products of viral replication. The RIG-I-like receptors (RLRs), RIG-I (retinoic acid inducible gene-I) and MDA5 (melanoma differentiation-associated gene 5), are involved in sensing cytosolic RNA species, and activation via the adapter molecule MAVS leads to the production of type I IFNs, via IRF3, and of pro-inflammatory cytokines, via NF-κB (Hornung et al., 2006; Kato et al., 2005, 2006; Pichlmair et al., 2006). Viral DNA species found within the cytosol and nucleus are known to be detected by an ever-increasing group of PRRs, including RNA polymerase (Pol) III (Ablasser et al., 2009; Chiu et al., 2009), DAI (Takaoka et al., 2007), IFI16 (Unterholzner et al., 2010), and LRRFIP1 (Yang et al., 2010), which lead to downstream production of type I IFNs and pro-inflammatory cytokines in a cell- and context-specific manner.
The parvoviruses are a group of non-enveloped, single-stranded DNA viruses that infect a diverse range of species from rodents to humans. Parvoviral infections are responsible for significant clinical burden in many of the affected species. For instance, canine parvovirus and feline panleukopenia virus cause significant mortality in infected dogs (Goddard and Leisewitz, 2010) or cats (Truyen et al., 2009). There are a number of parvoviruses that infect humans, including erythrovirus B19, adeno-associated viruses, and the recently discovered human bocaviruses 1–4 and human parvovirus PARV4 (Brown, 2010). In humans, the best documented clinical manifestations occur with parvovirus B19 infection and include fifth disease, arthropathy, transient aplastic crisis, persistent anemia, and hydrops fetalis (Young and Brown, 2004). Infection with the prototype strain of MVM (MVMp) is asymptomatic in newborn mice, where it can only be detected by seroconversion, and non-pathogenic in adult mice, indicating a high degree of adaptation to its natural host (Kimsey et al., 1986).
Although, MVM has a high, RNA virus-like mutation rate, and exists as multiple in vivo and culture-adapted strains that infect a series of disparate or overlapping differentiated host cell types in vitro and in vivo (Cotmore and Tattersall, 2007), the prototype MVMp strain exhibits a pronounced tropism for fibroblasts. A recent report described activation of the innate immune response in murine embryonic fibroblasts (MEFs) infected with this virus (Grekova et al., 2010). This study found that MEFs from wild-type C57BL/6 and CD1 mice robustly produced type-I IFNs and upregulated anti-viral genes, such as protein kinase R (PKR) and 2′–5′ oligoadenylate synthetase (OAS), in response to MVMp infection (Grekova et al., 2010).
Here, we examined the role of MAVS, which is involved in the detection of RNA Pol III-synthesized RNA intermediates in response to dsDNA viruses (Ablasser et al., 2009; Chiu et al., 2009). We further probed the relevance of type I IFNs in the antiviral protection against MVMp infection. The results of this study suggest that parvovirus MVMp efficiently evades antiviral immune mechanisms imposed by type I IFNs in this cell type.
Results
MVMp infection in murine embryonic fibroblasts leads to delayed and limited activation of the IFN response
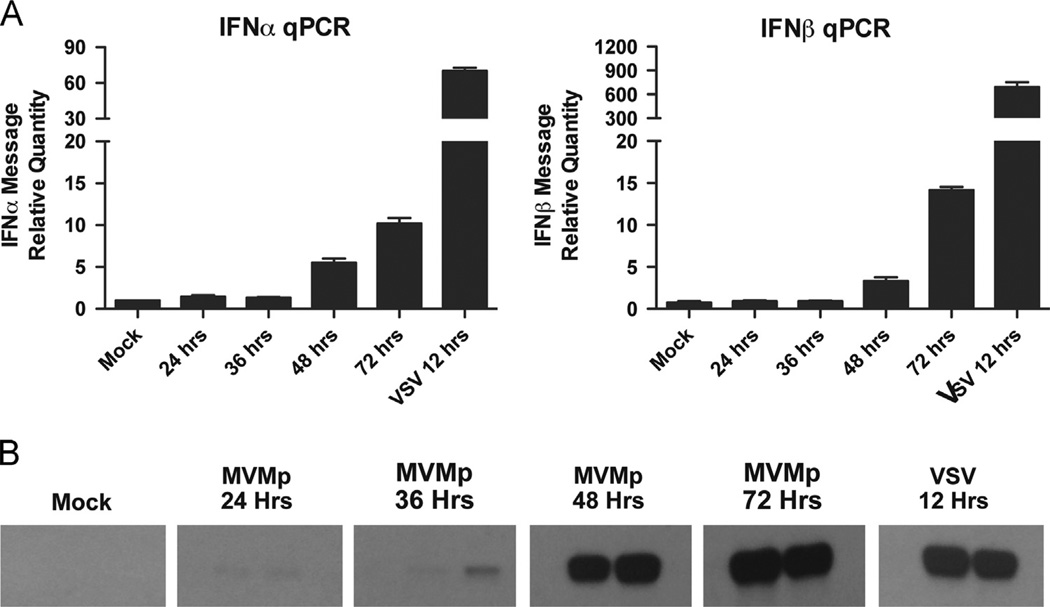
To examine the innate immune response to MVMp, we infected MEFs with MVMp and performed a time course experiment, which measured type I IFN mRNA induction by RT-qPCR during the first 72 h. Typical infection efficiency at this time point was around 30% (data not shown). We found a small, but statistically significant upregulation of IFN-α and IFN-β, especially at time points later than 48 h post infection (Fig. 1A). Consistent with these data, we found activation of STAT1, as measured by its phosphorylation, at later time points in MVMp-infected MEFs (Fig. 1B). The IFN response induced by MVMp infection occurred much later and was smaller in magnitude when compared to that following vesicular stomatitis virus (VSV) infection. These results demonstrated that MVMp infection in MEFs activates a delayed but measurable IFN response.
Fig. 1.
MVMp activates a delayed and minor interferon response in MEFs. Wild-type MEFs were infected with MVMp (MOI 1) or VSV (MOI 1). (A) Cells were collected at the indicated time points and IFNa or IFNb mRNA levels were measured by RT-qPCR. (B) Cells were collected at the indicated time points, and lysates (in duplicate) were subjected to SDS-PAGE followed by Western blotting for phospho-STAT1.
RNA pol III-dependent stimulation of the MAVS signaling pathway is not involved in the IFN response to MVMp in MEFs
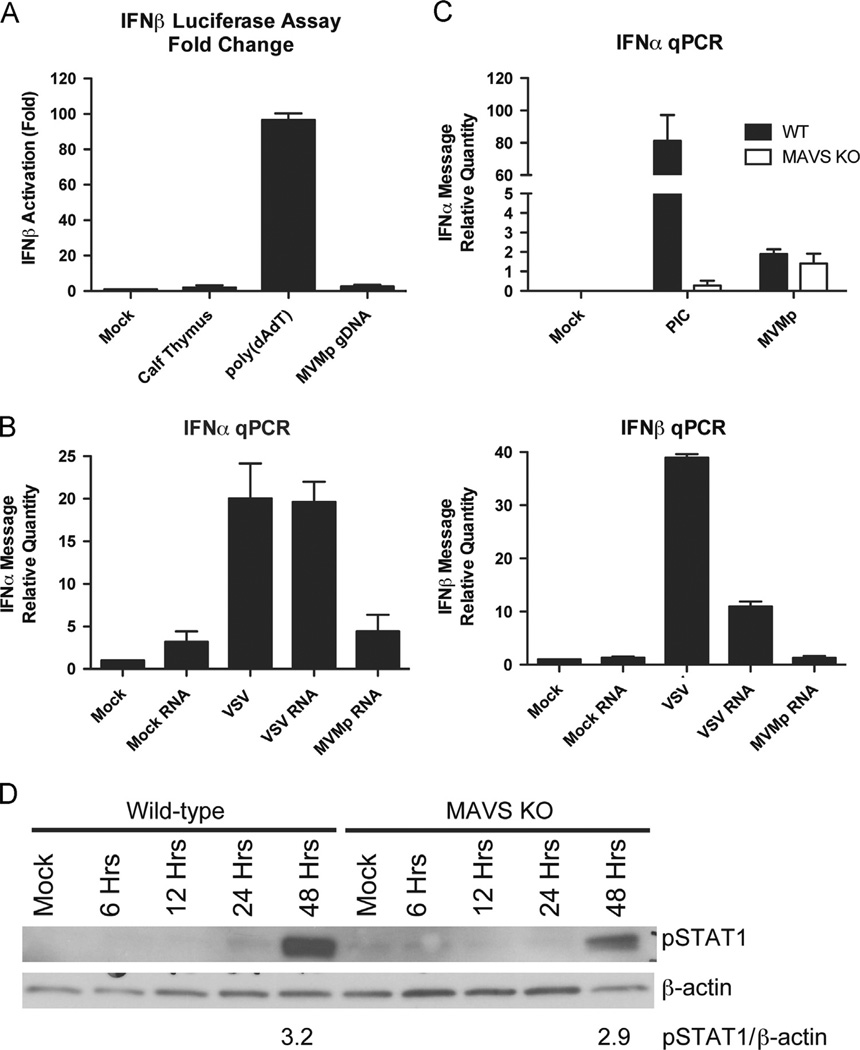
We next tested whether MVMp infection results in the generation of RNA Pol III-dependent RNA recognized by RLRs. AT-rich DNA converted to 5′-triphosphate RNAs by RNA Pol III was used as a positive control. These RNAs activate RIG-I, which results in IFN production through downstream signaling via MAVS (Ablasser et al., 2009; Chiu et al., 2009). To test whether MVMp genomic DNA is able to stimulate the RNA Pol III pathway, we used HEK-293T cells, which are only able to induce IFNs in response to Poly (dA:dT) DNA through the RNA Pol III pathway (Ablasser et al., 2009; Chiu et al., 2009). These cells were transfected with a reporter plasmid that expresses luciferase under control of the IFN-β promoter. The cells were then transfected with genomic DNA purified from MVMp virions. Calf thymus DNA and Poly (dA:dT) DNA were used as negative and positive controls, respectively. When the cells were analyzed for luciferase activity, we found that transfection with MVMp genomic DNA failed to activate the IFN-β promoter in HEK-293T cells, whereas Poly (dA:dT) activated IFN-β robustly in these cells (Fig. 2A). These results indicate that the RNA Pol III-dependent IFN pathway is not activated by MVMp genomic DNA in HEK-293T cells.
Fig. 2.
The RNA Pol III-dependent stimulation of the MAVS signaling pathway is not involved in the IFN response to MVMp in MEFs. (A) HEK-293T cells were transiently transfected with an IFNβ-luciferase reporter plasmid. They were then stimulated with calf thymus DNA, Poly (dA:dT), or MVMp genomic DNA (all at 1 µg/ml). Luciferase levels were measured after 24 hours. The levels are reported as fold change over mock-treated cells. (B) Wild-type MEFs were infected with VSV (MOI 1) or MVMp (MOI 1) or left uninfected. Twenty-four hours later, total cellular RNAs were purified from the cells and then 50 µg of each was transfected into TLR7-deficient bone marrow cells. Cells were collected after eight hours, and IFNα and IFNβ mRNA levels were quantified by RT-qPCR. (C, D) Wild-type or MAVS-deficient MEFs were infected with MVMp (MOI 1) or transfected with Poly (I:C) (PIC) (1 µg/ml). Cells were collected after 48 hours and IFNα mRNA levels were measured by RT-qPCR (C). Cells were collected at the indicated time points, and lysates were subjected to SDS-PAGE followed by Western blotting for phospho-STAT1. (D) The relative intensity of the pSTAT1 band normalized to the β-actin band is indicated.
Next, we examined whether MVMp-infected MEFs generate an RNA species capable of stimulating IFN through the MAVS signaling pathway. To this end, we infected wild-type MEFs with MVMp and purified total RNA from cellular extracts after 48 h. The purified RNAs were then used to transfect TLR7-deficient bone marrow cells. These cells were chosen to avoid contributions from TLR7 while probing the ability of cytosolic RNA sensors to detect purified RNA from MVMp infected cells. We found that TLR7-deficient bone marrow cells did not produce IFN-α (Fig. 2B) or IFN-β (Fig. 2C) in response to RNAs isolated by MVMp-infected cells beyond the level seen from RNAs isolated from uninfected cells. In contrast, the RNAs isolated from VSV-infected cells induced robust production of IFN-α and IFN-β in transfected TLR7-deficient bone marrow cells, as previously described (Kato et al., 2008). These results suggest that immunostimulatory RNAs are not produced in MVMp-infected MEF.
It remained possible that immunostimulatory RNAs are produced in MVMp-infected MEFs but remain undetectable in our assays. To further probe whether immunostimulatory RNAs stimulate type I IFNs in MVMp-infected MEFs, we examined MAVS-deficient MEFs. MAVS-deficient MEFs infected with MVMp generated IFN-α mRNA levels equivalent to wild-type MEFs, as measured by RT-qPCR 48 h post-infection (Fig. 2D). In addition, STAT1 phosphorylation in MAVS-deficient MEFs upon infection with MVMp was comparable to that seen upon infection of wild-type MEFs, further demonstrating that MAVS is not involved in the innate immune response to MVMp in MEFs (Fig. 2e). Taken together, these data suggest that the RNA-sensing pathways are not activated upon MVMp infection in MEFs.
MVMp infection is not blocked by type I IFNs
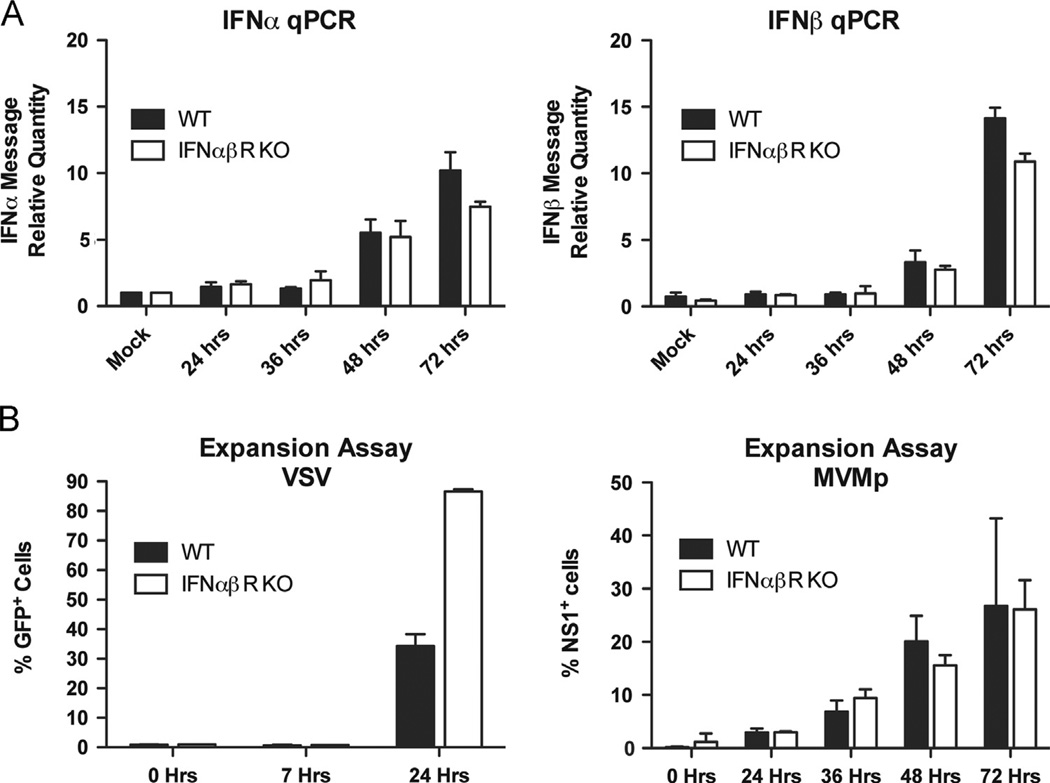
Next, we examined whether the low levels of secreted IFNs can actively inhibit MVMp infection of MEFs. First we examined IFN synthesis upon MVMp infection in IFN-αβR-deficient MEFs. Since IFN-α production is controlled by a positive feedback loop through the IFN-αβR (Honda et al., 2006), the levels of IFN-α produced in the IFN-αβR-deficient MEFs would be reduced compared to wild-type MEFs if they were responding to even low levels of secreted IFNs. However, we found that the low levels of both IFN-α and IFN-β mRNA produced by MEFs of either genotype were comparable for the first 48 h of infection (Fig. 3A). Although by 72 h there was a slight decrease in the levels of IFN mRNAs produced in the IFN-αβR-deficient MEFs compared to the wild-type cells, the levels of IFN-α and IFN-β remained below the limit of detection by ELISA throughout infection (data not shown). These results suggest that there is minimal feed-forward signaling through the IFNαβR, and this only occurs at a very late time point, likely reflecting the very low levels of IFN being produced by infected cells.
Fig. 3.
Signaling through the IFNαβR does not affect MVMp replication in MEFs. (A) Wild-type or IFNαβR-deficient MEFs were infected with MVMp (MOI 1) and IFNα and IFNβ mRNA levels were quantified after eight hours by RT-qPCR. (B) Wild-type or IFNαβR-deficient MEFs were infected with MVMp (MOI 1) or VSV-GFP (MOI 0.1) and were collected at the indicated time points. The percentage of infected cells in the culture was measured by flow cytometry.
Next we asked if viral replication is affected by signaling though the IFN-αβR. Signaling though the IFN-αβR leads to expression of hundreds of interferon-stimulated genes (ISGs), inducing an antiviral state in neighboring cells (Sen, 2001). As expected, replication of VSV, a virus that is sensitive to type I IFNs (Ito and Montagnier, 1977), was enhanced in the IFN-αβR-deficient MEFs (Fig. 3b). In contrast, MVMp replicated approximately equally well in both wild-type and IFN-αβR-deficient MEFs. These results suggest that the low levels of type I IFNs produced during MVMp infection of MEFs are not sufficient to induce an antiviral state in the cell culture.
Exogenous IFN-β has minimal effect on MVMp infection
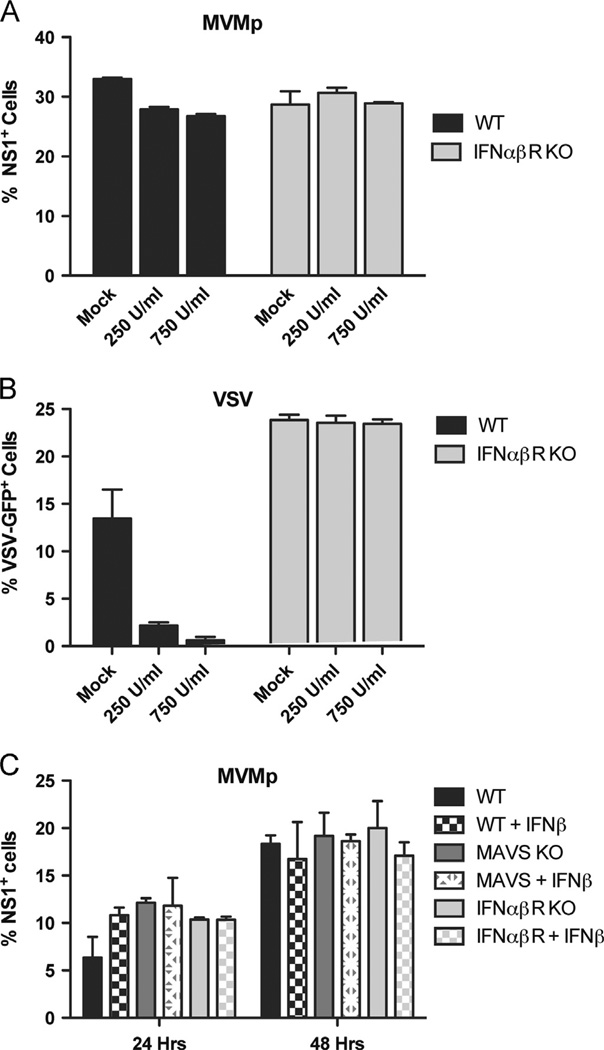
In vivo, many cell types other than fibroblasts are capable of secreting large amounts of type I IFNs in response to virus infections. To test whether exogenous type I IFNs suppress MVMp replication in MEFs, we pretreated wild-type and IFN-αβR-deficient MEFs for 18 hours with large amounts of recombinant IFN-β before infecting them with MVMp. We found that initiation of infection by MVMp was only minimally affected by the addition of IFN-β in wild-type cells (Fig. 4A). This is in contrast to the results we see upon infection with VSV, where IFN-β pretreatment of wild-type, but not IFN-αβR-deficient, cells leads to the almost complete absence of VSV infection in the cell culture (Fig. 4B). In order to confirm these findings, we extended the duration of infection up to 48-hours and measured the extent of infection in target cells derived from wild-type, MAVS KO, and IFN-αβR KO mice pretreated with recombinant IFN-β (Fig. 4C). These data consistently demonstrated that exogenous type I IFN fails to protect MEFs from MVMp infection, and that deficiencies in MAVS or IFN-αβR does not impact MVMp replication. Thus, these data demonstrate that exogenous IFN is not able to prevent MVMp infection, and suggest that MVMp may have a mechanism to counteract IFN-induced host antiviral effector mechanisms.
Fig. 4.
Pretreatment of MEFs with exogenous IFNβ minimally inhibits MVMp replication. (A) Wild-type or IFNαβR-deficient MEFs were pretreated with the indicated amounts of recombinant IFNβ for 18 hours before infection with MVMp (MOI 1). The percentage of infected cells in the culture was measured after 24 h by flow cytometry after intracellular staining for NS1. (B) Wild-type or IFNαβR-deficient MEFs were pretreated with the indicated amounts of recombinant IFNβ for 18 h before infection with VSV-GFP (MOI 0.1). The percentage of infected cells in the culture was measured after 12 hours by flow cytometry after intracellular staining for NS1. (C) Wild-type, MAVS-deficient, and IFNαβR-deficient MEFs were pretreated for 24 h with 250 U/ml of recombinant IFNβ and then infected with MVMp (MOI 1). The percentage of infected cells in the culture was measured at the indicated points by flow cytometry after intracellular staining for NS1.
MVMp infection blocks cytokine production by poly (I:C)
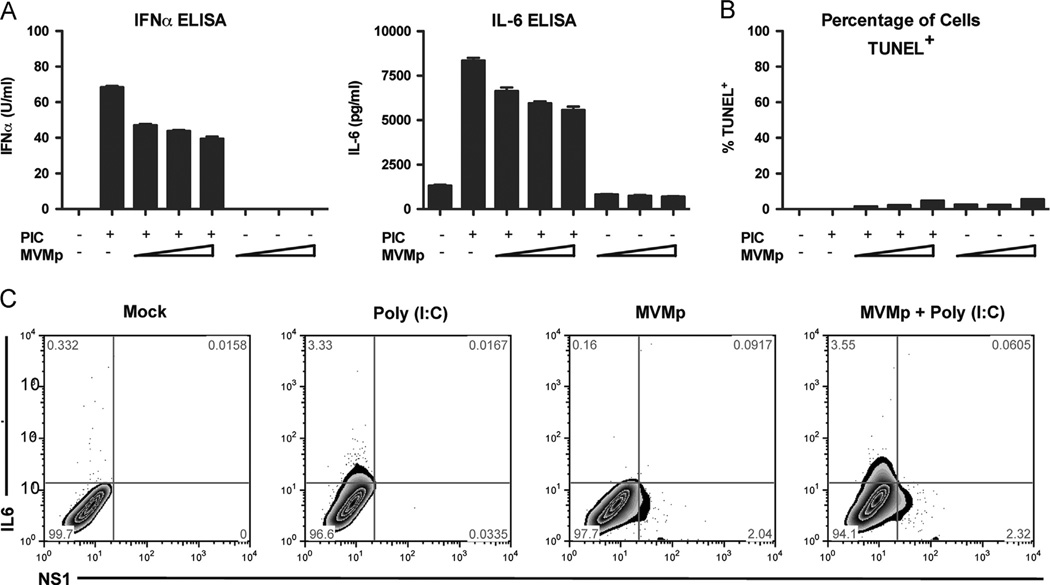
Thus far, our results indicated that MVMp blocks IFN-induced host antiviral defense mechanisms. However, it remains unclear if MVMp is also capable of blocking the RLR signaling pathway in infected cells. To test this hypothesis, we first infected wild-type MEF cells with two-fold dilutions of MVMp for 24 h and then stimulated the cells for an additional 24 h by transfecting them with Poly (I:C), a ligand for MDA5 (Gitlin et al., 2006; Kato et al., 2006). Cell supernatants were collected and the levels of secreted IFN-α and IL-6 were measured by ELISA (Fig. 5A). We found a significant, dose-dependent reduction in the amount of IFN and cytokine secreted by the cells that had been pre-exposed to MVMp compared to those that were uninfected before Poly (I:C) stimulation. To determine whether the decrease in Poly (I:C) response is due to apoptosis of MVMp-infected cells (Mincberg et al., 2011), we performed a TUNEL staining assay on the cells after stimulation. We found that approximately five percent of the cells were TUNEL positive after the maximum dose of MVMp (Fig. 5B). However, this level of cell death in the MVMp-infected cultures is unlikely to be solely responsible for the reduction in the amount of cytokines secreted (approximately 40% reduction at the highest MOI). We next used flow cytometry to examine whether MVMp infection can block cytokine production in response to Poly (I:C) stimulation in a cell-autonomous manner. In this assay, cells were first infected for 24 h with MVMp. They were then transfected with Poly (I:C) for eight hours, before being stained for intracellular IL-6 and MVMp NS1 and analyzed by flow cytometry. Notably, we found that IL-6 expression in response to Poly (I:C) stimulation was excluded from the NS1+ MVMp-infected population (Fig. 5C). These data show that MVMp-infected cells are unable to produce IL-6, suggesting that MVMp is able to block activation of the innate response in a cell-autonomous manner.
Fig. 5.
MVMp infection blocks interferon and cytokine production by Poly(I:C) stimulation. (A, B) Wild-type MEF cells were infected with MVMp (MOI 0.1, 0.3, or 1) for 24 h before stimulation with Poly(I:C) (1 µg/ml). (A) Cell supernatants were collected after 24 h and the levels of secreted IFNα and IL-6 were measured by ELISA. (B) Cells were collected after 24 h and the percentage of TUNEL+ apoptotic cells in the cultures was measured by flow cytometry. Wild-type MEFs were infected with MVMp (MOI 1) for 24 h before stimulation with Poly (I:C) (1 µg/ml). Cells were analyzed for intracellular levels of MVMp NS1 (x-axis) and IL-6 (y-axis) by flow cytometry. (C) Wild-type MEFs were infected with MVMp (MOI 1) for 24 h before stimulation with Poly (I:C)(1 µg/ml). Cells were analyzed for intracellular levels of MVMp NS1 (x-axis) and IL-6 (y-axis) by flow cytometry.
Discussion
Fibroblasts, like most cells, use cytosolic and nuclear PRRs to detect direct viral infection via the sensing of viral nucleic acid replication intermediates. We found that MVMp induces low levels of type I IFNs in MEFs with delayed kinetics, consistent with previous reports (Grekova et al., 2010; Schlehofer et al., 1992). However, no evidence for RNA Pol III-mediated synthesis of RNA intermediates that trigger the MAVS-dependent pathway was found in MVMp-infected cells. Further, IFN induction in MVMp-infected cells occurred independently of the presence of the MAVS gene. Even though MVMp induces type I IFNs in infected cells, we show that the low levels of type I IFNs generated late after infection by MEFs are not sufficient to induce an effective antiviral state. This was demonstrated by the fact that MVMp replicated in IFNαβR-deficient MEFs at levels equivalent to wild-type cells, while even pretreatment with high levels of exogenous IFNβ was not able to significantly block MVMp infection in wild-type MEFs. Further, we provide evidence that MVMp is able to block the innate immune response to another infection-mimicking stimulus applied to MVM- infected cells. Accordingly, MVMp infection reduced secretion of IFN and IL-6 from Poly (I:C)-stimulated MEFs in a dose-dependent manner, and flow cytometric analysis showed that MVMp-infected MEFs were unable to produce IL-6 in response to Poly (I:C) stimulation in a cell autonomous manner. Overall, our results indicate that MVMp elicits delayed and low levels of type I IFNs through a MAVS-independent mechanism, but the virus is not susceptible to the effects of type I IFNs, and that MVMp suppresses the cellular response to an exogenous Poly (I:C) stimulus in a cell autonomous manner.
These results are somewhat at variance with those of Ventoso and colleagues (Ventoso et al., 2010), who found that MVMp infection of NIH3T3 cells and of primary MEFs prepared from the 129Sv strain of mice, was restricted primarily through the activation of the cytosolic sensor double-stranded RNA-dependent protein kinase R (PKR), and that resistance to infection correlated with phosphorylation of eukaryotic translation factor 2α (eIF2 α). These authors also showed that RNA transcribed in vitro from genomic MVM DNA would stimulate PKR-dependent phosphorylation of eIF2α. At present, we do not understand the basis of the apparent discrepancies between our results and those of Ventoso et al. (2010) but it should be noted that here we used exclusively MEFs derived from C57BL/6 mice, and significant differences in susceptibility of different mouse strains, including C57BL/6 and 129Sv, to MVMp have been reported previously (Brownstein et al., 1991).
An obvious question that arises from the present report concerns the nature of the innate sensor(s) that recognize MVMp within infected C57BL/6 embryonic fibroblasts. Since the MAVS-dependent pathway is dispensable for MVMp recognition in MEFs, cytosolic or nuclear sensors of viral DNA, rather than viral mRNA, are likely involved. Current evidence suggests that during productive infection the MVM genome transits the endosomal and cytosolic compartment securely sequestered within the capsid, and is uncoated in the nucleus upon the host cell's entry into S-phase (Cotmore and Tattersall, 2007). However, if some MVM genomes are exposed during transit of the cytosol, DNA sensors that could detect them in this locale include DAI, LRRFIP1, IFI16, or the putative ISD sensor (Stetson and Medzhitov, 2006). Engagement of LRRFIP1 with dsDNA induces type I IFNs, but whether the ssDNA viral genome would stimulate this molecule is unknown (Yang et al., 2010). DAI is not activated by ssDNA (Stetson and Medzhitov, 2006), and ISD is only known to be activated by dsDNA (Stetson and Medzhitov, 2006). However, the MVMp genome does have small regions of dsDNA that form its 3′ and 5′ hairpin telomeres, making it possible that either DAI or ISD sensors might be engaged during viral entry.
Perhaps more significantly, this virus replicates in the cell nucleus via a unidirectional strand displacement mechanism that generates linear duplex replicative forms of the viral DNA, as well as replicative intermediates containing extensive regions of single-stranded DNA (Cotmore and Tattersall, 2012). Based on the delayed IFN production in MVMp-infected cells, which mirrors the viral replication kinetics, we suspect that viral ligands recognized by the DNA sensor are likely derived from this amplified viral replicative-form DNA. These unique DNA molecules may present potential targets for intranuclear DNA sensor such as IFI16, which is now known to detect DNA molecules in both the cytoplasm and nucleus, having recently been shown to detect Kaposi Sarcoma-Associated Herpesvirus (KSHV) DNA specifically in the nuclear compartment (Kerur et al., 2011). However, a previous report suggests that one MVMp sensing pathway is only active in non-transformed cells (Grekova et al., 2010), which might suggest involvement of the ISD sensor in its innate recognition (Stetson and Medzhitov, 2006). It will be of considerable interest in the future to determine which, if any, of these sensors are involved in the innate recognition of nuclear viral DNA in parvovirus-infected cells.
Conflicting results have been reported regarding the sensitivity of MVMp to IFNs. Some reports found that MVMp is sensitive to exogenous IFNs (Grekova et al., 2010; Harris et al., 1974), while others showed no effect of IFN treatment on MVMp replication (Engers et al., 1981). We found a very moderate effect of recombinant IFN-β on MVMp infection, even at very large doses (Fig. 4). In addition, IFN induced by MVMp infection was completely ineffective in preventing viral replication (Fig. 3). The reasons for these differences are not known, but may be due to differences in the mouse strains or cell types examined, the time course of IFN treatment and infection, or the MVM strain used. We specifically used primary early passage MEFs (between passage 2 and 5) to avoid an impaired type I IFN response reported in transformed cells in response to MVMp infection (Grekova et al., 2010). We suspect that the extent of the block in IFN responses we observed with MEFs is therefore further amplified in transformed cells. Notwithstanding these differences, our data showed that MVMp can evade antiviral activities imposed by type I IFNs. Since STAT-1 phosphorylation still occurs in MVMp-infected cells (Fig. 1), we suspect that MVMp interferes with the activity of ISGs that otherwise restrict viral replication.
Our data indicated that poly (I:C)-induced MDA5 signaling is blocked in MVMp-infected cells, although the mechanism of this blockade is unknown. Somewhat similarly, a recent study showed that the NS2 protein of porcine parvovirus is able to block poly I:C-induced IFN responses (Lin et al., 2013). The molecular size and expression levels of the MAVS protein remain intact following MVMp infection (unpublished observations), suggesting that the blockade occurs downstream of MAVS. Based on the fact that MVMp fails to elicit a rapid and effective type I IFN program in MEFs and blocks further response to a secondary stimulus, we propose that this immune evasion strategy may contribute to the replication and spread of rodent autonomous parvoviruses in vivo. In addition, MVMp infected cells may be particularly susceptible to co-infection with RNA viruses, as the RLR signaling pathway is clearly impaired. In addition to its well-known roles in Fifth disease, arthropathy, transient aplastic crisis, persistent anemia, and hydrops fetalis, human parvovirus B19 has been implicated in a variety of human autoimmune diseases, including Hashimoto's thyroidosis and rheumatoid arthritis (Lunardi et al., 2008). Given the importance of parvovirus infection in humans and animals, future elucidation of the viral evasion mechanisms may shed light on proper treatment options for parvovirus-associated diseases.
Materials and methods
Mice
Six- to seven-week old female C57BL/6 mice were obtained from the National Cancer Institute. The MAVS-deficient mice were a kind gift from Zhijian Chen (UT Southwestern, Dallas, TX) (Sun et al., 2006). TLR7-deficient (Lund et al., 2004), IFNαβR-deficient (Hwang et al., 1995) and MAVS-deficient (Sun et al., 2006) mice were bred and housed in the animal facilities at Yale University. MAVS-deficient mice were maintained as heterozygotes to obtain wild-type littermate controls. All procedures used in this study complied with federal guidelines and institutional policies of the Yale Animal Care and Use Committee.
Viruses
Wild-type MVMp virions were generated by transfecting an infectious plasmid clone into A9 cells, or co-transfecting into HEK-293T cells with pXX6–80, a helper plasmid encoding the VA, E2A and E4 regions of adenovirus (Xiao et al., 1998). Transfected and infected cells were grown for 72 h, and virus then released by subjecting the cells to three freeze–thaw cycles in 50 mM Tris–HCl, 0.5 mM EDTA, pH 8.7 (TE 8.7) containing protease inhibitors. The cellular extract was cleared by centrifugation and purified on iodixanol step gradients as previously described (Cotmore and Tattersall, 2012; Farr et al., 2006). Virus titer (PFU/ml) was quantified by plaque assay in A9 cells. Recombinant-derived wild-type VSV and VSV-GFP was kindly provided by John Rose (Yale University, New Haven, CT). VSV was propagated in BHK cells as previously described (Lund et al., 2004).
Cells
Murine embryonic fibroblasts (MEFs) were prepared from day 14.5 embryos from C57BL/6, IFNαβR-deficient mice, and MAVS-deficient and WT littermate control mice (Sun et al., 2006). MEFs were used at passage 2–5 and were maintained in complete DMEM medium (10% FBS, 10 mM HEPES, 100 units/ml penicillin and 100 µg/ml streptomycin). Single-cell bone marrow suspensions were prepared from cells isolated from the femurs and tibias of mice and were maintained in complete RPMI medium. HEK-293T cells were purchased from ATCC and were maintained in complete DMEM medium. A9 ouabr11 cells were maintained in complete DMEM medium (Tattersall and Bratton, 1983).
Cell stimulation
MEFs were plated at 8 × 104 cells per well in 6-well tissue culture plates in 2 ml of total medium. Cells were stimulated with Poly (I:C) (1 µg/ml, Invivogen) or Poly (dA:dT) (1 µg/ml, Invivogen) or genomic DNA extracted from purified MVMp virions (1 µg/ml) complexed with Lipofectamine 2000 (Invitrogen) in OPTIMEM. The transfection medium was replaced with fresh complete DMEM after one hour. Cells were infected with recombinant wild-type VSV or VSV-GFP (MOI 0.1–5) or infected with MVMp (MOI 0.1–1). In some instances, cells were pretreated with recombinant IFN-β (250 or 750 U/ml) (PBL IFN Source) for 18 hours before infection. Cells were collected after 6–12 h for analysis by RT-qPCR and supernatants were collected after 18–24 h for analysis by ELISA. Cells used for intracellular cytokine staining were exposed to Brefeldin A (Sigma-Aldrich) after 8 h of Poly (I:C) stimulation, and harvested after a further ten hours.
Quantitative real time PCR
Total RNA was isolated from cells using an RNeasy Mini kit (QIAGEN, Inc.) according to the manufacturer's protocol. RNA was reverse transcribed using Superscript II and Oligo dT primers (Invitrogen), and the resulting cDNA quantified on an MX3000p instrument (Stratagene (Aligent Technologies, Inc.) using SYBR Green-based assays (QIAGEN, Inc.) and primers listed in Table 1.
Table 1.
Primers used for quantitative RT-PCR.
| IFNα4 forward | CTGCTACTTGGAATGCAACTC |
| IFNα4 reverse | CAGTCTTGCCAGCAAGTTGG |
| IFNb forward | GCACTGGGTGGAATGAGACTATTG |
| IFNb reverse | TTCTGAGGCATCAACTGACAGGTC |
Western Blots
MEFs were trypsinized on ice, spun down, and resuspended in RIPA lysis buffer (1% NP-40, 20 mM Tris, pH 7.5, 150 mM NaCl, 1× protease inhibitor cocktail (Roche)). Proteins were separated by SDS-PAGE on 4–20% precast gels (Bio-Rad Laboratories) and transferred to a nitrocellulose membrane. The membranes were blocked in 5% nonfat dry milk in a buffer containing 20 mM Tris, pH 7.5, 500 mM NaCl, and 0.05% Tween 20, and incubated in anti-phospho-STAT1 antibody (1:1000, Tyr701, Cell Signaling Technology) or anti-β-actin antibody (1:5000, Sigma-Aldrich) diluted in 0.01 M Tris HCl, 0.15 M NaCl, 0.05% Tween-20 (TBST) containing 5% bovine serum albumin. Membranes were washed, incubated with goat anti-rabbit-HRP (for phosphoStat1, 1:50,000, Santa Cruz Biotechnology) or goat anti-mouse-HRP (for β-actin, 1:10,000) diluted in TBST plus 5% nonfat dry milk. Blots were developed using SuperSignal West Femto enhanced chemiluminescent substrate (Thermo Fisher Scientific). The levels of phosphoSTAT1 were normalized to β-actin levels by measuring band intensity using the program “Fiji” (Schindelin et al., 2012).
IFN-β luciferase assay
HEK-293T cells were transiently transfected with an IFN-β-luciferase reporter (300 ng/ml) using Lipofectamine 2000 (Invitrogen). Twenty-four hours later, cells were stimulated with calf thymus DNA, Poly (dA:dT), or MVMp genomic DNA (all at 1 µg/ml) using Lipofectamine 2000. Twenty-four hours later, cells were washed once in PBS, and processed using a Dual-Luciferase Reporter Assay (Promega) according to the manufacturer's instructions. Samples were read on a Lumat LB 9507 luminometer.
ELISA
Levels of murine IFN-α were measured using ELISA as previously described (Lund et al., 2003). Briefly, 96-well Maxisorp plates (Nunc) were coated with rat anti-mouse IFN-α (F18 antibody, Hycult Biotech, Inc.) at a 1:100 dilution in carbonate buffer overnight at 4 °C. After non-specific binding was blocked using blocking buffer (5% FBS in PBS), samples were loaded at 1:2 in blocking buffer. Recombinant IFN-α standards (Hycult Biotech, Inc.) were also diluted in blocking buffer. After incubating samples overnight at 4 °C or 4 h at room temperature, plates were incubated in polyclonal rabbit anti-mouse IFN-α (PBL IFN Source) at a 1:500 dilution in blocking buffer for 2 h at room temperature. Plates were then incubated with horseradish peroxidase-conjugated donkey anti–rabbit F(ab′)2 (Jackson ImmunoResearch Laboratories) at 1:10,000 dilution in blocking buffer for one hour at room temperature. The IL-6 ELISA was performed using a kit according to the manufacturer's instructions (BioLegend). All ELISA plates were developed with the addition of TMB (eBioscience, Inc.) and the reaction was stopped by addition of 2N•H2SO4. The optical density at 450 nm was measured using a Model 680 Microplate reader (Bio-Rad Laboratories) and the concentration of cytokines calculated from the appropriate standard curve.
Flow cytometric analysis
MVMp-infected MEFs were trypsinized on ice, spun down, and fixed in FAC-Fix buffer (1% FBS and 1% paraformaldehyde in PBS) for 20–30 min on ice. Cells were washed twice with FACS buffer (1% FBS in PBS) and permeablized by the addition of intracellular FACS (iFACS) buffer (1% FBS and 0.1% saponin in PBS). Cells were stained in 96-well U-bottom tissue culture plates with anti-NS1 (1:50, CE10B10, (Yeung et al., 1991)) and/or anti-IL-6-PE (1:200, MP5-20F3, eBioscience, Inc.) antibodies in a 50 µl volume of iFACS buffer for 30 min on ice, before being washed twice with iFACS buffer and then incubated with an anti-mouse secondary antibody conjugated to Alexa Fluor 488 (1:500, A11001, Invitrogen). After washing twice with iFACS buffer and once with FACS buffer, cells were resuspended in FACS-Fix buffer. MEFs that had been infected with VSV-GFP were trypsinized on ice, spun down, and then resuspended in FACS-Fix buffer. Apoptotic cells were stained using an In Situ Cell Death Detection Kit according to the manufacturer's instructions (Roche). All cells were processed on a FACSCalibur (BD Biosciences) and the results were analyzed using FlowJo (TreeStar, Inc.)
Acknowledgments
This work was supported by the Public Health Service grants, AI 081884 and AI 064705 (to AI) and CA 029303 (to PT) from the National Institutes of Health. LM is a recipient of the American Heart Association Predoctoral Fellowship.
References
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat. Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KE. The expanding range of parvoviruses which infect humans. Rev. Med. Virol. 2010;20:231–244. doi: 10.1002/rmv.648. [DOI] [PubMed] [Google Scholar]
- Brownstein DG, Smith AL, Jacoby RO, Johnson EA, Hansen G, Tattersall P. Pathogenesis of infection with a virulent allotropic variant of minute virus of mice and regulation by host genotype laboratory investigation. J. Tech. Methods Pathol. 1991;65:357–364. [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore SF, Tattersall P. Mutations at the base of the icosahedral five-fold cylinders of minute virus of mice induce 3′-to-5′ genome uncoating and critically impair entry functions. J. Virol. 2012;86:69–80. doi: 10.1128/JVI.06119-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore SF, Tattersall P. Parvoviral host range and cell entry mechanisms. Adv. Virus Res. 2007;70:183–232. doi: 10.1016/S0065-3527(07)70005-2. [DOI] [PubMed] [Google Scholar]
- Engers HD, Louis JA, Zubler RH, Hirt B. Inhibition of T cell-mediated functions by MVM(i), a parvovirus closely related to minute virus of mice. J. Immunol. 1981;127:2280–2285. [PubMed] [Google Scholar]
- Farr GA, Cotmore SF, Tattersall P. VP2 cleavage and the leucine ring at the base of the fivefold cylinder control pH-dependent externalization of both the VP1 N terminus and the genome of minute virus of mice. J. Virol. 2006;80:161–171. doi: 10.1128/JVI.80.1.161-171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard A, Leisewitz AL. Canine parvovirus. Vet. Clin. North Am. Small Anim. Pract. 2010;40:1041–1053. doi: 10.1016/j.cvsm.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Grekova S, Zawatzky R, Horlein R, Cziepluch C, Mincberg M, Davis C, Rommelaere J, Daeffler L. Activation of an antiviral response in normal but not transformed mouse cells: a new determinant of minute virus of mice oncotropism. J. Virol. 2010;84:516–531. doi: 10.1128/JVI.01618-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RE, Coleman PH, Morahan PS. Erythrocyte association and interferon production by minute virus of mice. Proc. Soc. Exp. Biol. Med. 1974;145:1288–1292. doi: 10.3181/00379727-145-37998. [DOI] [PubMed] [Google Scholar]
- Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- Hwang SY, Hertzog PJ, Holland KA, Sumarsono SH, Tymms MJ, Hamilton JA, Whitty G, Bertoncello I, Kola I. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons alpha and beta and alters macrophage responses. Proc. Natl. Acad. Sci. USA. 1995;92:11284–11288. doi: 10.1073/pnas.92.24.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Montagnier L. Heterogeneity of the sensitivity of vesicular stomatitis virus to interferons. Infect. Immun. 1977;18:23–27. doi: 10.1128/iai.18.1.23-27.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimsey PB, Engers HD, Hirt B, Jongeneel CV. Pathogenicity of fibroblast- and lymphocyte-specific variants of minute virus of mice. J. Virol. 1986;59:8–13. doi: 10.1128/jvi.59.1.8-13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Qiu Z, Liu Q, Cui S. Interferon induction and suppression in swine testicle cells by porcine parvovirus and its proteins. Vet. Microbiol. 2013;163:157–161. doi: 10.1016/j.vetmic.2012.11.032. [DOI] [PubMed] [Google Scholar]
- Lunardi C, Tinazzi E, Bason C, Dolcino M, Corrocher R, Puccetti A. Human parvovirus B19 infection and autoimmunity. Autoimmun. Rev. 2008;8:116–120. doi: 10.1016/j.autrev.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. USA. 2004;101:5598–5603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mincberg M, Gopas J, Tal J. Minute virus of mice (MVMp) infection and NS1 expression induce p53 independent apoptosis in transformed rat fibroblast cells. Virology. 2011;412:233–243. doi: 10.1016/j.virol.2010.12.035. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlehofer JR, Rentrop M, Mannel DN. Parvoviruses are inefficient in inducing interferon-beta, tumor necrosis factor-alpha, or interleukin-6 in mammalian cells. Med. Microbiol. Immunol. 1992;181:153–164. doi: 10.1007/BF00202055. [DOI] [PubMed] [Google Scholar]
- Sen GC. Viruses and interferons. Ann. Rev. Microbiol. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Tattersall P, Bratton J. Reciprocal productive and restrictive virus-cell interactions of immunosuppressive and prototype strains of minute virus of mice. J. Virol. 1983;46:944–955. doi: 10.1128/jvi.46.3.944-955.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truyen U, Addie D, Belak S, Boucraut-Baralon C, Egberink H, Frymus T, Gruffydd-Jones T, Hartmann K, Hosie MJ, Lloret A, Lutz H, Marsilio F, Pennisi MG, Radford AD, Thiry E, Horzinek MC. Feline panleukopenia. ABCD guidelines on prevention and management. J. Feline Med. Surg. 2009;11:538–546. doi: 10.1016/j.jfms.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventoso I, Berlanga JJ, Almendral JM. Translation control by protein kinase R restricts minute virus of mice infection: role in parvovirus oncolysis. J. Virol. 2010;84:5043–5051. doi: 10.1128/JVI.02188-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Li J, Samulski RJ. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat. Immunol. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- Yeung DE, Brown GW, Tam P, Russnak RH, Wilson G, Clark-Lewis I, Astell CR. Monoclonal antibodies to the major nonstructural nuclear protein of minute virus of mice. Virology. 1991;181:35–45. doi: 10.1016/0042-6822(91)90467-p. [DOI] [PubMed] [Google Scholar]
- Young NS, Brown KE. Parvovirus B19. New Engl. J. Med. 2004;350:586–597. doi: 10.1056/NEJMra030840. [DOI] [PubMed] [Google Scholar]