Abstract
Topoisomerases are the enzymes responsible for maintaining the supercoiled state of DNA in the cell and also for many other DNA topology-associated reactions. Type IA enzymes alter DNA topology by breaking one DNA strand and passing another strand or strands through the break. Although all type IA topoisomerases are related at the sequence, structure, and mechanism level, different type IA enzymes do not participate in the same cellular processes. We have studied the mechanism of DNA relaxation by E. coli topoisomerases I and III using single-molecule techniques to understand their dissimilarities. Our experiments show important differences at the single molecule-level, while also recovering the results from bulk experiments. Overall, topoisomerase III relaxes DNA using fast processive runs followed by long pauses, whereas topoisomerase I relaxes DNA through slow processive runs followed by short pauses. These two properties combined give rise to the overall relaxation rate, which is faster for topoisomerase I than for topoisomerase III, as expected from many biochemical observations. The results help understand better the role of these two topoisomerases in the cell and also serve to emphasize the power of single-molecule experiments to uncover new functional characteristics of biological molecules.
Introduction
Topoisomerases are the enzymes responsible for altering the topological state of DNA in the cell. They are found in all three phylogenetic kingdoms and in almost every cell type (1,2). In order to change the topology of DNA, topoisomerases transiently break either one or both strands of a DNA duplex to create an opening that permits topological changes. To create each break, all topoisomerases employ a tyrosine to form a transient high energy phosphotyrosine bond which can be reversed to reseal the break in the DNA backbone. Topoisomerases have been implicated in many cellular processes, such as transcription, replication, DNA repair, and recombination (1,2). Although not all topoisomerases are essential for cell viability, their importance has made them ideal targets for chemotherapeutics. There are now several successful antibiotics as well as anti-cancer drugs that target topoisomerases (3). Partly for this reason and also due to their cellular importance and their fascinating mechanism of action, topoisomerases have been the source of many detailed studies on their structure, function and mechanism.
Topoisomerases have been classified into two types based on their mechanism of action. Type I enzymes cleave one DNA strand at a time followed either by passage of one or two DNA strands through the break or swivelling around the intact strand before resealing the broken DNA backbone. Type II topoisomerases cleave both DNA strands in concert and can pass a double strand through the transient break. In general, type I enzymes do not require any high energy cofactor for activity; the reaction is driven by the energy stored in the supercoiled DNA as torsional strain. In cases where the reaction is not energetically favorable, such as during the introduction of positive supercoils, an energy-providing cofactor is needed for activity. In contrast, type II enzymes always require ATP hydrolysis for activity. The energy released by ATP hydrolysis is not used for cleavage/religation of the DNA backbone, but instead it is used to modulate protein conformational changes during the reaction cycle (4).
Type I enzymes are sub-classified into three different sub-types: IA, IB and IC based on sequence and structural similarity as well as their mechanism of action (5,6). All type IA enzymes form a transient phosphotyrosine covalent bond with the 5′ end of the broken DNA strand, whereas type IB and IC enzymes form a transient covalent bond with the 3′ end of the broken strand. Two possible relaxation mechanisms are possible: enzyme-bridged strand passage, where the protein bridges the opening of one strand to allow passage of the other through the break, and swiveling, where one strand rotates or swivels around the other. One important difference between the two mechanisms is that during enzyme-bridged strand-passage it is necessary to hold to both ends of the broken strand, whereas for swiveling one of the ends detaches from the protein and can move freely or almost freely. Type IA enzymes always use an enzyme-bridged strand-passage mechanism whereas enzymes from the other two sub-types, IB and IC, always employ a controlled swiveling mechanism. Overall, the three sub-types perform a very similar overall reaction, but the mechanism employed in each case is substantially different. Each type I sub-type represents a different way to solve the same topological problem.
Type IA topoisomerases
Type IA enzymes are ubiquitous. They are present in bacteria, archaea, and eukarya and are clearly related both at the sequence and structural level (7). In general, type IA topoisomerases relax only negatively supercoiled DNA. An exception is reverse gyrase, which is capable of introducing positive supercoils into DNA by coupling its strand passage activity with a DNA unwinding activity (8). Type IA topoisomerases require single stranded DNA regions for activity (9), either already present in the DNA or created by other proteins. In the case of DNA relaxation, detecting the presence of single stranded regions provides a robust mechanism for sensing the overall topological state of the DNA, as only underwound or negatively supercoiled DNA is likely to present single stranded regions.
All type IA topoisomerases have a common structural core that is responsible for the DNA strand passage reaction. This core, of around 67 kDa in Escherichia coli topoisomerase I, is supplemented by other domains at the N- or C- terminus of the protein. The strand passage domain has a characteristic fold formed by the convergence of four domains (Figure 1) that create a toroidal shape with a hole in the center large enough to accommodate double stranded DNA (10). The active site is found where two of the domains come together, just outside the central hole. Separation of the domain is required for entrance and exit to the central hole and this involves a large conformational change of the protein. Sequence comparison of the C-terminal region extending from the core domain of the type IA enzymes shows that this region does not present the same level of conservation as the core. For example, E. coli topoisomerases I and III have C-terminal domains of ≈30 kDa and 14 kDa in size, respectively, and with different properties (11–13).
Figure 1. Structure of E. coli topoisomerase III, a member of the type IA family of topoisomerases.
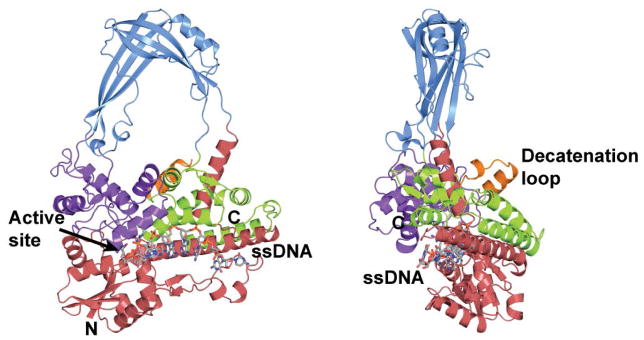
The diagram shows the structure of the intact E. coli topoisomerase III in complex with single stranded DNA (15). The enzyme is formed by four domains (red, blue, purple, and green) that form a toroidal shape with a central hole. The active site is found at the interface of two of the domains. Single stranded DNA binds along a groove before it enters the active site. Topoisomerase III has a 17 amino acid loop, the decatenation loop, which is needed for decatenation activity (32). Topoisomerase I does not have this loop.
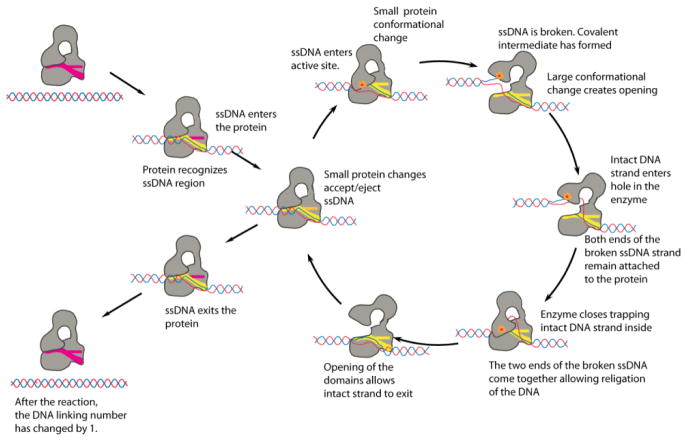
Based on the structure of E. coli topoisomerase I, a detailed mechanism for type IA enzymes was proposed (10,14) (Figure 2). The proposed mechanism of action involves several steps (Figure 2), and crystal structures for many of these intermediate steps have been obtained (14–18). All these structures provide pictures of some of the probably stable intermediates, but structures of the other stages, putatively the most unstable ones, are still unknown and the exact conformation of them remain largely hypothetical. Single molecule experiments (19,20) have supported the general features of the proposed mechanism and confirmed that type IA enzymes change the linking number in steps of one, as is mandated in an enzyme-bridged strand passage mechanism that transports a single DNA strand through a single stranded DNA break.
Figure 2. Proposed mechanism of DNA relaxation catalyzed by type IA topoisomerases.
The diagram shows the proposed mechanism employed by type IA enzymes to relax DNA (10,14). The conformations of many of the different intermediates are based on structures, but some intermediates have not been observed and remain hypothetical. Many of the features of the proposed mechanism have been confirmed through biochemical, structural, and single molecule experiments.
Although all type IA enzymes are structurally related, mechanistically there are important differences amongst them. In addition, the presence of more than one type I enzyme in most organisms suggests that different enzymes, even if they are of the same sub-type, play different roles in the cell. For these reasons, discerning the mechanistic differences amongst members of the same sub-type is important to understand their functional role in the cell. E. coli is an excellent example of a case where two closely related type IA enzymes, topoisomerases I and III, are involved in different cellular processes. In vivo a major function of topoisomerase I, together with two type II enzymes (topoisomerase IV and gyrase), is to maintain the supercoiled state of DNA in the cell (21). The function of topoisomerase III, the other type IA enzyme, is to resolve single-stranded DNA recombination and replication intermediates (22–27), and in some instances double-stranded intermediates (28). These differences in functions are reflected in their in vitro characteristics: topoisomerase I is a better DNA relaxation enzyme than topoisomerase III (22), whereas topoisomerase III is a better decatenating enzyme (29,30). From a structural perspective the two enzymes are also different in subtle ways. The major difference between the two is the presence of a much larger C-terminal domain in topoisomerase I that is involved in DNA binding and is essential for DNA relaxation activity (11,12), whereas the C-terminal domain of topoisomerase III also binds DNA but is smaller and dispensable (31). Another small but significant difference is the presence in topoisomerase III of a 17 amino acid region, termed the decatenation loop, that resides at the base of the central hole and whose removal decreases decatenation activity markedly (32).
Single-molecule studies of type IA topoisomerases
Biochemical experiments have been instrumental to study topoisomerases and to understand their differences and similarities. Nevertheless, experiments in bulk can hide some details by the inevitable averaging that occurs when looking at billions of molecules at a time. For this reason, it is important to look at the action of topoisomerases at the single-molecule level. Single-molecule studies of type I topoisomerases have been very informative and helped uncover new aspects of the mechanism of these enzymes (33). Type IA enzymes were amongst the first topoisomerases studied at the single-molecule level and these studies confirmed the enzyme-bridged strand passage mechanism (19). More recently, studies of eukaryotic type IB topoisomerases revealed some of the constraints imposed by the protein on the swiveling of the DNA strands and also helped understand the role of some topoisomerase poisons (34–36). Finally, single-molecule studies were essential to clarify the mechanism of topoisomerase V, the sole member of the type IC group (37).
In order to understand the mechanistic difference between the two E. coli type IA topoisomerases, we analyzed DNA relaxation by the two enzymes at the single-molecule level using a magnetic tweezers setup (38). In this type of experiments, one end of a long single DNA molecule is attached to a functionalized glass slide while the other end is attached to a small (≈1 – 3μm) paramagnetic bead. By using a rotating magnet, it is possible to introduce supercoils into the DNA at will while monitoring the position of the bead through a microscope. Rotating the magnet causes the DNA to over- or underwind and eventually plectonemes are formed in the single DNA molecule (39). This is always the case for positively supercoiled or overwound DNA, but it only happens at low forces (< 0.7 pN) for negatively supercoiled or underwound DNA as in the latter case DNA eventually melts and no plectonemes are formed. In the experimental setup, plectoneme appearance results in DNA length shortening and hence there is a straightforward relationship between the length of the DNA molecule stretched by the magnet and the supercoiled state of the DNA. Thus, by monitoring the position of the bead it is possible to measure accurately the supercoiled state of DNA while following DNA relaxation events in real time and under well-defined conditions.
In our experiments, we were able to detect real-time relaxation of a single DNA molecule by both E. coli topoisomerases I and III (40). In the single molecule experiments topoisomerase I has a faster total relaxation rate (0.9 supercoils/second) than topoisomerase III (0.2 supercoils/second), comparable to the ~5 fold difference observed in bulk experiments (see Figure 1 in (40)). Careful analysis of the single-molecule results shows some unexpected differences between the two enzymes that are not evident from bulk experiments. Figure 3 shows examples of single-molecule relaxation events, where the length of the DNA molecule is monitored as a topoisomerase acts on it. It is clear that the enzymes relax DNA by processively removing a few supercoils followed by a pause before starting another relaxation run. DNA relaxation traces by the two topoisomerases show the same characteristics: a pause of variable length before a short run of relaxation events. However, the experiments also show significant differences. For all the substrates studied, topoisomerase III relaxes DNA using very fast relaxation runs, on average as fast as ≈130 supercoils/second for negatively supercoiled DNA. In contrast, topoisomerase I relaxes DNA much more slowly, on average about 3 supercoils/second for negatively supercoiled DNA. When the length of the initial pauses are compared, the two enzymes behave exactly the opposite way: topoisomerase I pauses briefly before relaxation runs, for roughly 6 seconds for negatively supercoiled DNA, whereas topoisomerase III pauses for an extended period of time, about 130 seconds on average. Thus, the single-molecule experiments reveal unexpected differences between these two closely related type IA enzymes: both enzymes can relax DNA by removing a large number of supercoils with high processivity. However, the relaxation velocity is faster for topoisomerase III relaxation runs than for topoisomerase I and the time lags are shorter for topoisomerase I than for topoisomerase III. The combination of these two characteristics results in an overall faster total relaxation rate for topoisomerase I.
Figure 3. Single molecule relaxation by E. coli topoisomerases I and III.
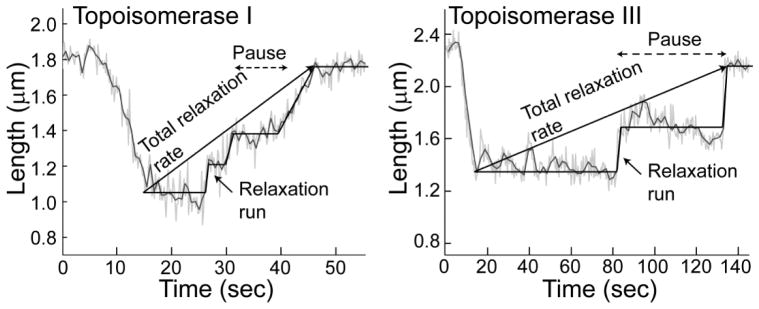
The diagrams show typical relaxation experiments by either topoisomerase I or III (40). In the experiments, the length of the DNA molecule is directly related to the superhelical state of the DNA. In both cases, the enzymes relax DNA using a combination of relaxation runs preceded by pauses. In the case of topoisomerase I, the pauses are short and the relaxation runs are slow. Topoisomerase III behaves the opposite way, the pauses are long and the relaxation runs are fast. The combination of these two general features results in topoisomerase I having a faster overall relaxation rate than topoisomerase III, consistent with bulk experiments. Note the different time scales in the two diagrams. The straight lines overlaying the events serve as guides and do not represent actual fits.
These new observations allow us to understand better the mechanistic differences between the two enzymes. Clearly the two enzymes work by the same overall mechanism, but the mechanism has been fine-tuned to operate differently, probably reflecting differences in their target substrates in the cell. The main cellular role of topoisomerase I is to maintain the topological state of DNA (21) and this is consistent with the characteristics observed in the single-molecule measurements. In our experiments, topoisomerase I relaxes DNA steadily and consistently without pausing for long periods of time. In contrast, topoisomerase III exhibits fast bursts of relaxation activity preceded by long pauses. It appears that topoisomerase I has been optimized as a house-keeping enzyme that constantly monitors the topological state of DNA and helps maintain it at a steady level. It is an impulsive enzyme; it does not wait to act. As soon as the substrate is present, it relaxes it slowly but steadily. On the other hand, topoisomerase III works through fast bursts of activity followed by long pauses, more consistently with an enzyme that prefers not to act on ordinary supercoiled DNA but that can process other substrates very efficiently, perhaps recombination intermediates. Additional experiments are needed to characterize more fully the action of these two enzymes on other substrates, such as experiments on catenated single or double stranded DNA molecules. However, the present experiments already show that despite their many overall similarities, the two enzymes act differently at the single molecule level and that these differences are likely a reflection of their different cellular roles.
References
- 1.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 2.Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol. 2011;12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bates AD, Berger JM, Maxwell A. The ancestral role of ATP hydrolysis in type II topoisomerases: prevention of DNA double-strand breaks. Nucleic Acids Res. 2011;39:6327–6339. doi: 10.1093/nar/gkr258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forterre P. DNA topoisomerase V: a new fold of mysterious origin. Trends Biotechnol. 2006;24:245–247. doi: 10.1016/j.tibtech.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Forterre P, Gribaldo S, Gadelle D, Serre MC. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 7.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu Rev Biophys Biomol Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 8.Kikuchi A, Asai K. Reverse gyrase--a topoisomerase which introduces positive superhelical turns into DNA. Nature. 1984;309:677–681. doi: 10.1038/309677a0. [DOI] [PubMed] [Google Scholar]
- 9.Kirkegaard K, Wang JC. Bacterial DNA topoisomerase I can relax positively supercoiled DNA containing a single stranded loop. J Mol Biol. 1985;185:625–637. doi: 10.1016/0022-2836(85)90075-0. [DOI] [PubMed] [Google Scholar]
- 10.Lima CD, Wang JC, Mondragon A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature. 1994;367:138–146. doi: 10.1038/367138a0. [DOI] [PubMed] [Google Scholar]
- 11.Ahumada A, Tse-Dinh YC. The Zn(II) binding motifs of E. coli DNA topoisomerase I is part of a high-affinity DNA binding domain. Biochem Biophys Res Commun. 1998;251:509–514. doi: 10.1006/bbrc.1998.9500. [DOI] [PubMed] [Google Scholar]
- 12.Beran-Steed RK, Tse-Dinh YC. The carboxy terminal domain of Escherichia coli DNA topoisomerase I confers higher affinity to DNA. Proteins. 1989;6:249–258. doi: 10.1002/prot.340060307. [DOI] [PubMed] [Google Scholar]
- 13.Zhang HL, DiGate RJ. The carboxyl-terminal residues of Escherichia coli DNA topoisomerase III are involved in substrate binding. J Biol Chem. 1994;269:9052–9059. [PubMed] [Google Scholar]
- 14.Changela A, DiGate RJ, Mondragon A. Structural studies of E. coli topoisomerase III-DNA complexes reveal a novel type IA topoisomerase-DNA conformational intermediate. J Mol Biol. 2007;368:105–118. doi: 10.1016/j.jmb.2007.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Changela A, DiGate RJ, Mondragon A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature. 2001;411:1077–1081. doi: 10.1038/35082615. [DOI] [PubMed] [Google Scholar]
- 16.Feinberg H, Lima CD, Mondragon A. Conformational changes in E. coli DNA topoisomerase I. Nature Struct Biol. 1999;6:918–922. doi: 10.1038/13283. [DOI] [PubMed] [Google Scholar]
- 17.Perry K, Mondragon A. Structure of a complex between E. coli DNA topoisomerase I and single-stranded DNA. Structure (Camb) 2003;11:1349–1358. doi: 10.1016/j.str.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, Cheng B, Tse-Dinh YC. Crystal structure of a covalent intermediate in DNA cleavage and rejoining by Escherichia coli DNA topoisomerase I. Proc Natl Acad Sci USA. 2011;108:6939–6944. doi: 10.1073/pnas.1100300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dekker NH, Rybenkov VV, Duguet M, Crisona NJ, Cozzarelli NR, Bensimon D, Croquette V. The mechanism of type IA topoisomerases. Proc Natl Acad Sci USA. 2002;99:12126–12131. doi: 10.1073/pnas.132378799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dekker NH, Viard T, de La Tour CB, Duguet M, Bensimon D, Croquette V. Thermophilic topoisomerase I on a single DNA molecule. J Mol Biol. 2003;329:271–282. doi: 10.1016/s0022-2836(03)00320-6. [DOI] [PubMed] [Google Scholar]
- 21.Zechiedrich EL, Khodursky AB, Bachellier S, Schneider R, Chen D, Lilley DM, Cozzarelli NR. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J Biol Chem. 2000;275:8103–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]
- 22.DiGate RJ, Marians KJ. Identification of a potent decatenating enzyme from Escherichia coli. J Biol Chem. 1988;263:13366–13373. [PubMed] [Google Scholar]
- 23.Wallis JW, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyperrecombination mutation in S cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- 24.Lopez CR, Yang S, Deibler RW, Ray SA, Pennington JM, Digate RJ, Hastings PJ, Rosenberg SM, Zechiedrich EL. A role for topoisomerase III in a recombination pathway alternative to RuvABC. Mol Microbiol. 2005;58:80–101. doi: 10.1111/j.1365-2958.2005.04812.x. [DOI] [PubMed] [Google Scholar]
- 25.Harmon FG, DiGate RJ, Kowalczykowski SC. RecQ helicase and topoisomerase III comprise a novel DNA strand passage function: a conserved mechanism for control of DNA recombination. Mol Cell. 1999;3:611–620. doi: 10.1016/s1097-2765(00)80354-8. [DOI] [PubMed] [Google Scholar]
- 26.Suski C, Marians KJ. Resolution of converging replication forks by RecQ and topoisomerase III. Mol Cell. 2008;30:779–789. doi: 10.1016/j.molcel.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiasa H, Marians KJ. Topoisomerase IV can support oriC DNA replication in vitro. J Biol Chem. 1994;269:16371–16375. [PubMed] [Google Scholar]
- 28.Perez-Cheeks BA, Lee C, Hayama R, Marians KJ. A role for topoisomerase III in Escherichia coli chromosome segregation. Mol Microbiol. 2012;86:1007–1022. doi: 10.1111/mmi.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiasa H, DiGate RJ, Marians KJ. Decatenating activity of Escherichia coli DNA gyrase and topoisomerases I and III during oriC and pBR322 DNA replication in vitro. J Biol Chem. 1994;269:2093–2099. [PubMed] [Google Scholar]
- 30.Hiasa H, Marians KJ. Topoisomerase III, but not topoisomerase I, can support nascent chain elongation during theta-type DNA replication. J Biol Chem. 1994;269:32655–32659. [PubMed] [Google Scholar]
- 31.Zhang HL, Malpure S, Li Z, Hiasa H, DiGate RJ. The role of the carboxyl-terminal amino acid residues in Escherichia coli DNA topoisomerase III-mediated catalysis. J Biol Chem. 1996;271:9039–9045. doi: 10.1074/jbc.271.15.9039. [DOI] [PubMed] [Google Scholar]
- 32.Li Z, Mondragon A, Hiasa H, Marians KJ, DiGate RJ. Identification of a unique domain essential for Escherichia coli DNA topoisomerase III-catalysed decatenation of replication intermediates. Mol Microbiol. 2000;35:888–895. doi: 10.1046/j.1365-2958.2000.01763.x. [DOI] [PubMed] [Google Scholar]
- 33.Koster DA, Crut A, Shuman S, Bjornsti MA, Dekker NH. Cellular strategies for regulating DNA supercoiling: a single-molecule perspective. Cell. 2010;142:519–530. doi: 10.1016/j.cell.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 35.Koster DA, Czerwinski F, Halby L, Crut A, Vekhoff P, Palle K, Arimondo PB, Dekker NH. Single-molecule observations of topotecan-mediated TopIB activity at a unique DNA sequence. Nucleic Acids Res. 2008;36:2301–2310. doi: 10.1093/nar/gkn035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 37.Taneja B, Schnurr B, Slesarev A, Marko JF, Mondragon A. Topoisomerase V relaxes supercoiled DNA by a constrained swiveling mechanism. Proc Natl Acad Sci USA. 2007;104:14670–14675. doi: 10.1073/pnas.0701989104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skoko D, Wong B, Johnson RC, Marko JF. Micromechanical analysis of the binding of DNA-bending proteins HMGB1, NHP6A, and HU reveals their ability to form highly stable DNA-protein complexes. Biochemistry. 2004;43:13867–13874. doi: 10.1021/bi048428o. [DOI] [PubMed] [Google Scholar]
- 39.Marko JF. Torque and dynamics of linking number relaxation in stretched supercoiled DNA. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;76:021926. doi: 10.1103/PhysRevE.76.021926. [DOI] [PubMed] [Google Scholar]
- 40.Terekhova K, Gunn KH, Marko JF, Mondragon A. Bacterial topoisomerase I and topoisomerase III relax supercoiled DNA via distinct pathways. Nucleic Acids Res. 2012;40:10432–10440. doi: 10.1093/nar/gks780. [DOI] [PMC free article] [PubMed] [Google Scholar]