Abstract
Using DNA as programmable, sequence specific ‘glues’, shape-controlled hydrogel units are self-assembled into prescribed structures. Here we report that aggregates are produced using hydrogel cubes with edge length ranging from 30 micrometers to 1 millimeter, demonstrating assembly across scales. In a simple one-pot agitation reaction, 25 dimers are constructed in parallel from 50 distinct hydrogel cube species, demonstrating highly multiplexed assembly. Using hydrogel cuboids displaying face-specific DNA glues, diverse structures are achieved in aqueous and in interfacial agitation systems. These include dimers, extended chains, and open network structures in an aqueous system; and dimers, chains of fixed length, T-junctions, and square shapes in the interfacial system, demonstrating the versatility of the assembly system.
Introduction
Self-assembly is the process by which small components self-organize into larger structures. Initially developed as a concept for engineering molecular complexes1, self-assembly has been applied to construct structures across scales using monomer units ranging from nano-scale to macro-scale dimensions2. Diverse techniques have been developed for mesoscale (micrometer- to millimeter-scale) self-assembly using magnetic force3, hydrophile-lipophile balance4,5, capillary interaction6 and synthetic chemical binding7 to control the assembled architecture.
Increasing the complexity of mesoscale self-assembly faces a crucial challenge, namely the difficulty of engineering a large set of orthogonal specific binding interactions between the monomer units. This challenge can be potentially addressed by using DNA, biology's information carrier, as programmable “glue” to direct the assembly of mesoscale units. DNA contains four different nucleotide bases, each of which forms a base pair with another complementary base according to a set of canonical rules: adenine with thymine, and guanine with cytosine. By simply arranging the sequence of these four nucleotide bases in different DNA strands, a combinatorially large set of binding interactions can be designed as specific hybridizations between complementary DNA strands. DNA hybridization based self-assembly principles have been utilized successfully by the field of DNA nanotechnology to generate diverse complex synthetic DNA/RNA nano-structures8-11 with arbitrarily prescribed geometry12-29 and dynamic functions30-41. Furthermore, DNA strands can be made into hydrogels through covalent42-44 or non-covalent interactions45. DNA has also been used as templates or glues to mediate the self-assembly of fluorophores46, proteins47, inorganic nanoparticles48-51, carbon nanotube52, lipid vesicles53, and even living cells54. Recently, it was reported that short single-stranded DNA probes attached to a glass surface can successfully catch 100 μm size hydrogel microspheres decorated with sequences complementary to the probes55. Building on these previous successes, we address the next challenge here: to fully utilize the versatile programmability of DNA to direct the self-assembly of mesoscale objects into complex higher order structures with precisely prescribed architecture and geometry.
As part of our work to increase the complexity of the architectural and geometrical control of DNA direct mesoscale assembly, here, we report on combining DNA directed assembly principle with microfabrication technology to assemble mesoscale objects using shaped controlled hydrogel units. The central conceptual innovation here is the decoration of DNA glues onto the prescribed surfaces of a non-spherical hydrogel object to produce an asymmetric glue pattern. These new assembling units, by combining the molecular programmability of DNA glue and the shape controllability of microfabrication, will provide a powerful platform to achieve programmable assembly of complex mesoscale structures. To implement this strategy, a crucial technical innovation was necessary: we invent a novel strategy to use in situ rolling circle amplification to produce and attach “giant” DNA glues to the surface of hydrogel cubes. Based on this technical innovation, we demonstrate that giant DNA glue strands, but not short DNA primers, induce the assembly of hydrogel gel cubes with an edge length across scales (30 μm to 1 mm), and that they result in the self-assembly of cube dimers in a highly multiplexed fashion (25 orthogonal dimer pairs from 50 distinct cube species in one pot mixing). We then develop a method to engineer hydrogel cuboids that display giant DNA only on designated faces. Using this technology, we demonstrate the assembly, in aqueous and in interfacial systems, of diverse structures: linear chains with extended or fixed length, open networks, T-junctions, and 2×2 square structures. Thus we establish DNA directed assembly of shape-controlled mesoscale units as a promising route to produce complex structures with sophisticated geometrical and architectural control.
Results
Self-assembly of hydrogels with uniform surface DNA glues
In the first part of our study, we developed a strategy to use complementary DNA molecules as glue to direct the self-assembly of hydrogel cubes with an edge length of 250 μm. Our initial attempt to assemble such hydrogel cubes carrying short complementary DNA strands (36 nt poly-T linker followed by 20 nt complementary sequences) failed to induce hydrogel assembly (Fig. 2c). This failure may be attributed to the rugged surface morphology of the hydrogel, as revealed by scanning electron microscopy (SEM, Fig. 1d), and the relatively small size and weak binding interactions between short complementary DNA strands. To accommodate for this rugged hydrogel surface, we developed a strategy to decorate hydrogel surface with “giant” single-stranded DNA (Fig. 1a). Specifically, in step (1), amine-bearing short DNA strands (brown, 56 nt) were conjugated to PEG-NHS monomers (blue, MW 3500 Da) using a standard protocol56. (2) The DNA-PEG-acrylate was mixed with photocrosslinkable poly(ethylene glycol)-diacrylate (PEG-DA, 4000 MW) and 0.5 wt% photoinitiator, and exposed to UV under a photomask with 250 μm × 250 μm square holes. The height of the cubes was controlled using No. 2 microscope cover glass slides (250 μm in thickness) as spacers. Upon UV exposure, 250 μm × 250 μm × 250 μm hydrogel cubes uniformly modified with short DNA primers were produced. (3) The DNA primers hybridized with complementary, circular DNA templates (produced by circularization of short linear DNA using CircLigase™); through a published procedure known as rolling circle amplification (RCA)57, the primer was amplified to produce a long strand with repeated sequences complementary to the circular template. In this paper, we call this long single-strand DNA product “giant DNA”.
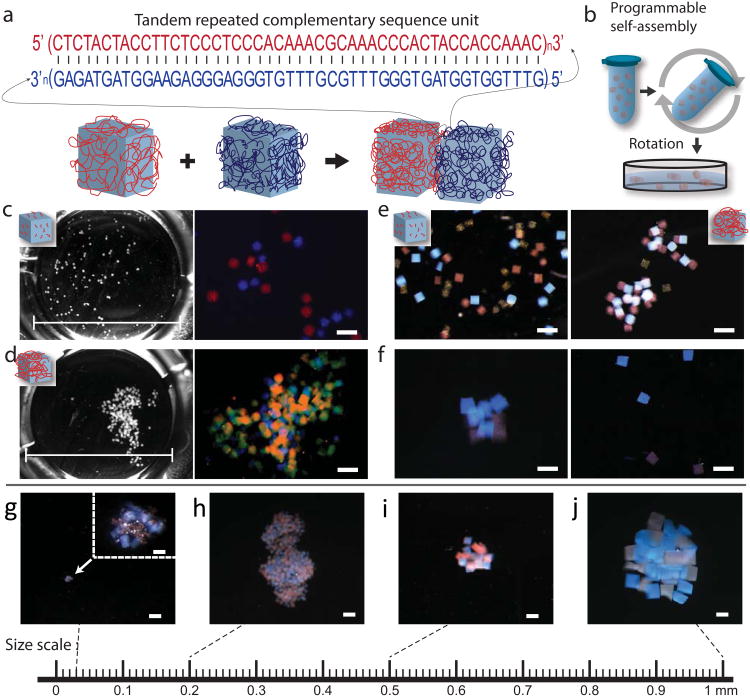
Figure 2. Self-assembly of hydrogel cubes with uniform giant DNA glue modification.
a, Schematic of giant DNA directed hydrogel assembly. Giant DNA containing tandem repeats of complementary 48-nt sequences was uniformly amplified on the surface of red and blue hydrogel cubes. Hybridization between the complementary DNA sequences resulted in assembly of hydrogel cubes. b, Hydrogel cubes were assembled in a 0.5 ml tube filled with assembly buffer under mild rotation, transferred to a petri dish with 1.6 cm diameter, and imaged under microscope. See Method for details. c, d, Phase contrast and fluorescent microscopy images of the post-assembly system, in which hydrogel cubes were modified with short 56-nt (c) or amplified giant (d) DNA strands. Hydrogel cubes carrying complementary DNA a or a* were labeled with red or blue fluorescent microbeads, respectively, and stained with SYBR Gold. e, Red and blue hydrogel cubes carrying complementary short or giant DNA strands failed or succeeded to assemble into aggregates, in the presence of competitive yellow hydrogel cubes that were not modified with DNA. f, Aggregates assembled from red and blue hydrogel cubes carrying complementary giant DNA fell apart after 1 hour Baseline-ZERO™ DNase treatment (left: before DNase treatment; right: after DNase treatment). Scale bar, 500 μm in c, d, g, f. Aggregates were assembled from red and blue hydrogel cubes with various edge lengths, g: 30 μm, h: 200 μm, i: 500 μm and j: 1 mm. Giant DNA glue was uniformly amplified on the hydrogel surface. Scale bar, 1 mm in the main panels; 50 μm in the inset of panel g.
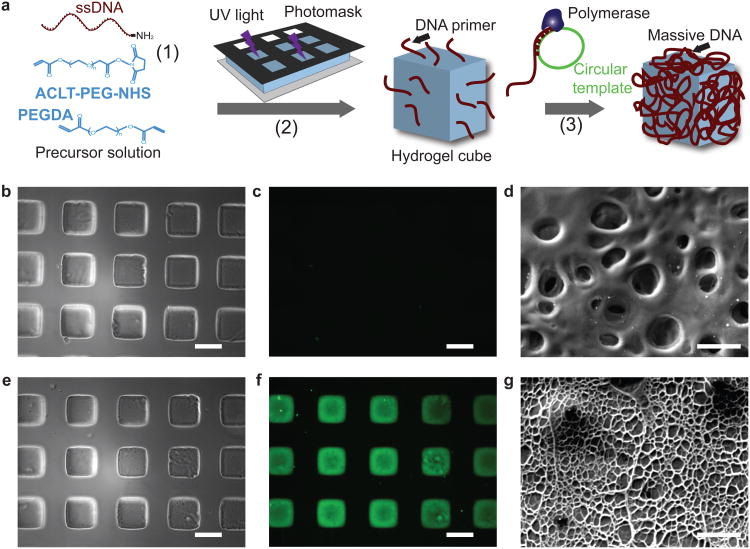
Figure 1. Fabrication of hydrogel cubes with uniform giant DNA glue modification.
a, Schematic of the fabrication process of hydrogel cubes uniformly modified with giant DNA. b-g, Phase contrast (b, e), fluorescent (c, f), and scanning electron microscopy (d, g) images of hydrogels carrying short 56-nt single-stranded DNA primers (b-d) or amplified single-stranded giant DNA (e-g). The gels in c and f were stained with SYBR Gold before imaging. Scale bar, 200 μm in b, c, e, f; scale bar, 10 μm in d, g.
We first verified the successful production of giant DNA via RCA reaction in free solution and on glass slides(Supplementary Fig. S2a, agarose gel showed the synthesis of high molecular weight product; b, fluorescent microscopy image showed the formation of large membrane-like giant DNA; c, scanning electron microscopy (SEM) image showed the fiber-like giant DNA on glass slides). We then fabricated hydrogel cubes carrying giant DNA as described above and characterized them using fluorescent DNA staining. In comparison with hydrogels carrying short 56-nt DNA primers (Fig. 1b, phase contrast imaging; c, fluorescent imaging, DNA was stained by SYBR Gold), 3 hours of RCA amplification at 37°C resulted in significant DNA staining by SYBR Gold (Fig. 1e, phase contrast imaging; f, fluorescent imaging). As the DNA primer was tethered to the gel precursor, it was anticipated that giant DNA was amplified both inside and on the surface of hydrogel cubes. This is consistent with our experimental observation that only after RCA amplification, the hydrogel cubes stained with fluorescent DNA dye appeared with expected fluorescence under microscope (Fig. 1c,f). We note that as the RCA process is diffusion dependent and it is anticipated that it would be diffusion limited in the regions inside the gels relative to the gel surface. Importantly, SEM imaging of the gel surface before (Fig. 1d) and after (Fig. 1g) the RCA amplification provided direct evidence that giant DNA was amplified on the gel surface. After the amplification, the initially rugged surface of the gel was covered uniformly by fiber-like structures, with reduced surface roughness (SEM image, Fig. 1g).Collectively, the above experiments suggested that the surfaces of hydrogel cubes were decorated with giant DNA produced via RCA, as designed.
Next, we demonstrated the giant DNA directed assembly of hydrogel cubes. Using the procedure described above, we fabricated250 μm × 250 μm × 250 μm hydrogel cubes carrying giant DNA encoding tandem repeated complementary sequence (Fig. 2a) and self-assembly was performed by mixing these hydrogel cubes in a 0.5 ml microtube filled with assembly buffer under mild rotation, using a tube rotator with a fixed speed of 18 rpm (Fig. 2b, see details in Method). All the assembly experiments were performed at room temperature in our paper. To visualize the assembled structure, the two species of hydrogel cubes carrying complementary DNA sequences a and a* were labeled with red (fluoresbrite® microspheres with excitation at 512 nm and emission at 554 nm) or blue (fluoresbrite® microspheres with excitation 360 nm and emission 407 nm) fluorescent microbeads respectively. After the assembly reaction in the tube, the hydrogel cubes were transferred to a 1.6 cm diameter petri dish for imaging (Fig. 2b). Large aggregates were observed for hydrogel cubes carrying complementary giant DNA (Fig. 2d, left, phase contrast imaging; right, fluorescent imaging). In contrast, hydrogel cubes carrying only 56-nt short DNA primers without RCA amplification failed to produce aggregates under the same assembly conditions(Fig. 2c, left, phase contrast imaging; right, fluorescent imaging). Including DNA-free yellow hydrogel cubes (i.e. cubes that contain yellow microbeads) in the reaction system did not change the assembly outcome for either the hydrogel cubes carrying 56-nt short DNA primers (Fig. 2e, left) or hydrogel cubes carrying giant DNA (Fig. 2e, right). Moreover, no yellow hydrogel cubes were observed in the assembled structure, confirming that the assembly was directed by giant DNA on the hydrogel cube surface. This giant-DNA-dependent nature of the assembly was further verified in a DNA degradation experiment (Fig. 2f): assembled hydrogel cubes carrying complementary giant DNA became dispersed after treatment with DNase, which was expected to degrade the giant DNA on the hydrogel cube surface. In previous work, using short DNA strand as glues, researchers were successful in catching small spherical particles (100 μm diameter) on a surface. However, in our system, relatively large cuboid shaped particles (250 μm edge width) needed to assemble in a strong agitation (to disrupt non-specific interactions) solution-based system, and thus much stronger glue interactions were likely required to enable the assembly. Over 70% specific binding was observed by analyzing the binding event between two hydrogel cubes uniformly carrying same or complementary DNA glues. The giant DNA directed hydrogel cube binding was also quantified under different temperatures (4°C, 25°C and 37°C). We observed that the specific assembly yield increased as the temperature decreased (Supplementary Fig. S3). However nonspecific binding also increased significantly at 4°C (data not shown). The assembly experiments in the remainder of the paper were performed under room temperature (Supplementary Fig. S3).
We next demonstrated that the interaction between complementary giant DNA was capable of directing the assembly of hydrogel cubes with a wide range of edge lengths. Cubes carrying complementary DNA a or a* were labeled with red or blue color microbeads respectively. Assembly reaction was performed as described above. Aggregates were observed from hydrogel cubes with edge lengths of 30 μm, 300 μm, 500 μm, and 1000 μm (Fig. 2g-j). These experiments indicate that the hybridization between complementary giant DNA was sufficiently strong to induce the assembly of hydrogel cubes across scales.
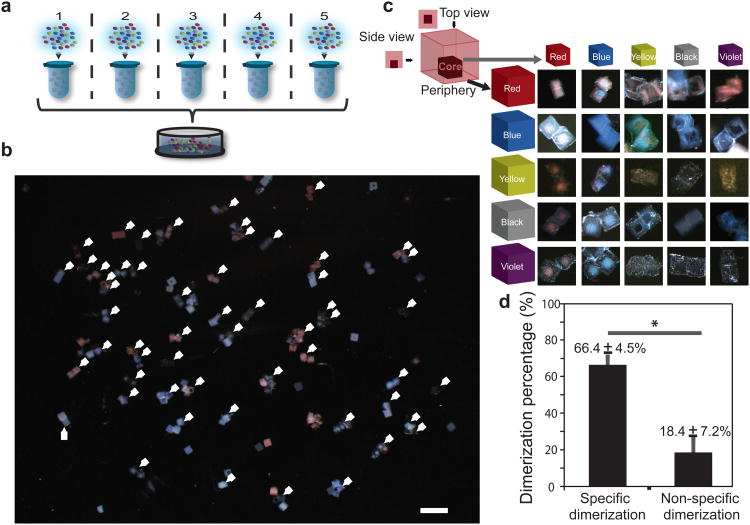
As complementary DNA molecules hybridize with each other in a sequence-dependent fashion, it is possible to generate a large number of specific interactions using giant DNA glues with orthogonal sequences. To test whether such DNA sequence-dependent specificity can be applied in our hydrogel cube self-assembly system, fifty 24-nt DNA sequences were designed to produce 25 pairs of orthogonal specific interactions (Fig. 3a; also see Methods, and Supplementary Table S1 for DNA sequences). To visually differentiate distinct cube species under stereomicroscope, colored microbeads were trapped in the core and periphery parts of the cubic hydrogel respectively (Fig. 3c, top left corner) and pairwise combinations of 5 colors (red, blue, yellow, black, and violet) generated 25 distinct signatures. Following fabrication, giant DNA containing tandem repeated sequence was amplified using RCA reaction on the hydrogel surface as described before. The 25 structures were self-assembled in aqueous assembly system. To avoid forming large aggregate that can trap microgels inside and hence hinder the quantification of specific assembly yields, only one copy for each of the 50 hydrogel species was included in one of five independent experiments (Fig. 3a). The assembly was performed using agitation of repeated mild rotation at a fixed speed of 18 rpm and strong hand shaking every 30 min to disrupt non-specific binding. The assembled structures were then transferred to a petri dish for imaging and quantification. Each of the 25 expected specific dimer structures were all identified (Fig. 3b, c). In total, 83 structures were analyzed in 6 independent experiments: except 15% of non-assembled single hydrogel cubes and hydrogel cubes missed in the operation process putatively due to the nonspecific binding to microtube surface, about 66% specific binding (Fig. 3d). These experiments demonstrated that the interaction between giant DNA glue is sequence-specific and that highly multiplexed assembly can be achieved using our system. To the best of our knowledge, this system includes more specific interactions than all reported mesoscale self-assembly work.
Figure 3. Self-assembly of multiple hydrogel cubes dimers.
a, Schematic for multiplexed self-assembly of 25 orthogonal pairs of dimers in 5 independent experiments. b, The final assembled structure from 5 independent experiments were pooled together into a single petri-dish for imaging, and the assembled specific structure was indicated by a white triangle. Scale bar, 1 mm. c, Schematic (top left corner) describes the double layer structure of hydrogel cube used in the multiplexed self-,assembly of 25 dimers. The core cube was 100 μm × 100 μm × 100 μm and the periphery cube was 300 μm × 300 μm × 300 μm. See Method for fabrication details. The core and periphery hydrogel cubes were labeled with distinct colored microbeads respectively, and pairwise combinations of 5 colors (red, blue, yellow, black and violet) generated 25 distinct signatures. Images of 25 corresponding dimers are shown. d, Hydrogel cube dimers were identified under microscopy and quantified as the specific (66 ± 5%) and non-specific (18 ± 7%) binding events, n = 5, *P < 0.05.
Self-assembly of hydrogels with face-specific DNA glues
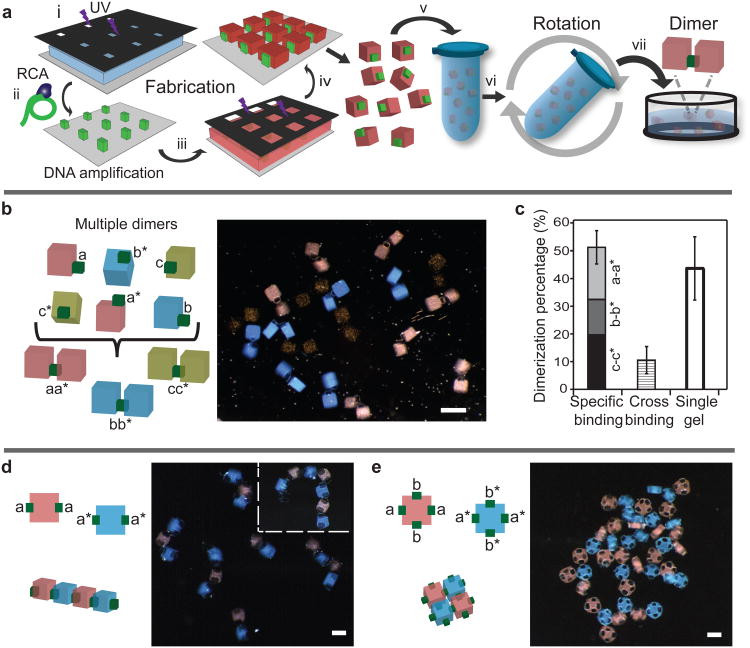
To assemble structures with controlled architecture rather than aggregates, we fabricated hydrogel cuboid units with face-specific giant DNA modification. The procedure is illustrated in Fig. 4a. In Steps 1-4, we describe the procedure to make a two-component cube composite structure where a lager “body-cube” displays smaller DNA-modified “pad-cubes” on its designated sides; in Steps 5 and 6, we describe the agitation system for their assembly. (Step 1) The 150 μm × 150 μm × 150 μm hydrogel pad-cubes were made from a precursor solution that contains 20 wt% PEGDA (4 KDa) and PEG (3.5 KDa) acrylate-ssDNA primers using photolithography. A photomask with 150 μm × 150 μm square holes was used to control the cross-section shape of the pad-cube; No. 1 microscope cover glass slides (150μm in thickness) were used as spacers to control the height of the pad cube. (Step 2) The un-polymerized reagent was washed away and giant DNA was produced via RCA reaction as described before. We now have arrays of 150 μm pad-cubes (colored green in Fig. 4a) with uniform giant DNA modification. (Step 3) To make the larger body-cube, we added a second solution that contained only 20 wt% PEGDA (4 KDa), and covered it with a second photomask with 250 μm × 250 μm square holes. This photomask was aligned carefully with the pad-cubes made in Step 2 such that this photomask covered half of the cross-section area of each pad-cubes (to protect them from subsequent UV exposure). No. 2 microscope cover glass slides (250 μm in thickness) were used as spacers to control the height of the body-cubes. (Step 4) Subsequent UV treatment resulted in the polymerization of the second 250 μm × 250 μm × 250 μm body-cube. Un-polymerized reagent was washed away. At the end of Step 4, we produced an array of cubes: the red 250 μm body-cube covered half of the green 150 μm pad-cubes; only the green 150 μm pad-cubes were modified with giant DNA. As a consequence, the cube composite only had giant DNA modification on designated faces that display the green pads. For exposition simplicity, we refer to this composite structure as a hydrogel cube with surface specific DNA modifications. (Step 5) The hydrogel cubes were collected into a 0.5 ml microtube filled with the assembly buffer. (Step 6) Assembly was performed by rotating the tube. (Step 7) The solution was transferred to a petri dish and imaged under microscope.
Figure 4. Self-assembly of hydrogel cubes with face-specific DNA glue modifications.
a, Hydrogel cubes carrying giant DNA glue on designated faces were fabricated in steps 1-4, collected in a 0.5 ml microtube in step 5, mixed and agitated in step 6, and transferred to a petri-dish for imaging in step 7. b, Hydrogel cubes displaying face-specific giant DNA with tandem repeats a/a* (with the body cube labeled red), b/b* (blue body cube), or c/c* (yellow body cube). The pad cube carrying giant DNA glue is colored green. Letters x and x* denote complementary DNA sequences (see SuSupplementary Table S1 for sequence details). Scale bar, 500 μm. c, Quantification of specific assembly, non-nonspecific assembly, and un-assembled single cubes. *P < 0.05. d, Linear chain structures were assembled from the red and blue gels. Left, schematic. Right, microscopy images. The inset shows a chain containing 7 cubes. Scale bar, 200 μm. e, Net-like structures were self-assembled from the red and blue cubes. Scale bar, 200 μm.
Using the above strategy, we demonstrated the multiplexed assembly of three hydrogel cube dimer species (Fig. 4b, c). In this experiment, six hydrogel cube species were manufactured (Fig. 4b, left). The first species is a red hydrogel cube (i.e. the body cube contained red microbeads) that displays giant DNA with tandem repeating sequence “a”. We call this cube the red cube a. The other five species are red cube a* (where sequence a* is complementary to sequence a), blue cube b, blue cube b*, yellow cube c, and yellow cube c*. Multiple copies of each of the six cube species were made separately and then mixed at room temperature in the same tube for assembly (see Methods for details). After 6 h rotation, the solution was imaged (Fig. 4b, right).Three populations of the structures were observed and quantified (Fig. 4c). In total 77 assembled structures were analyzed in 6 independent experiments:46% were the expected, specific dimers that formed between two same-color cubes that presumably carried complementary sequences; 10% were the dimers that formed between two different-color cubes that carried non-complementary sequences; 44% were the un-assembled single cubes. The specificity of dimer formation was quantified as 82% by dividing the number of specific dimers over the total number of dimers.
Using hydrogel cubes that display giant DNA on multiple designated faces, we next constructed linear chain structures and net-like structures. To make the chain structures, two cube species were made: a red cube that displays giant DNA a on two opposite faces, and a blue cube that displays giant DNA a* on two opposite faces (Fig. 4d, left). The assembly of these two species produced chain structures that contained alternating red and blue cubes, as expected (Fig. 4d, right). The longest chain observed contained 7 cubes (Fig. 4d, top-right corner). We then made a red cube species that displays giant DNA a on two opposite faces, and giant DNA b on another two opposite faces; and a blue cube species that displays pairs of a* and b* on opposite faces (Fig. 4e, left). The assembly of these two species resulted in the formation of net-like structures with alternating red and blue cubes that were connected via DNA modified sides (Fig. 4e, right). The linear and net-like structures were each assembled in 3 independent experiments, where each experiment used 40 red and 40 blue complementary cubes. In this experimental system, 39% of the gel cubes remained as un-assembled, whereas 58% were assembled into a linear structure (due to their small size, 3% of cubes were lost during the assembly and quantification process). Among the 58% assembled cubes, 42% were only connected to a different color cube and hence called specifically assembled cubes; the remaining 16% were connected non-specifically to at least one same color cube, and were called non-specifically assembled cubes. For the net-like structures, the specifically assembled cubes, non-specifically assembled cubes, and un-assembled cube monomers were respectively 56%, 30% and 14%.
Interfacial self-assembly of hydrogel to complex structures
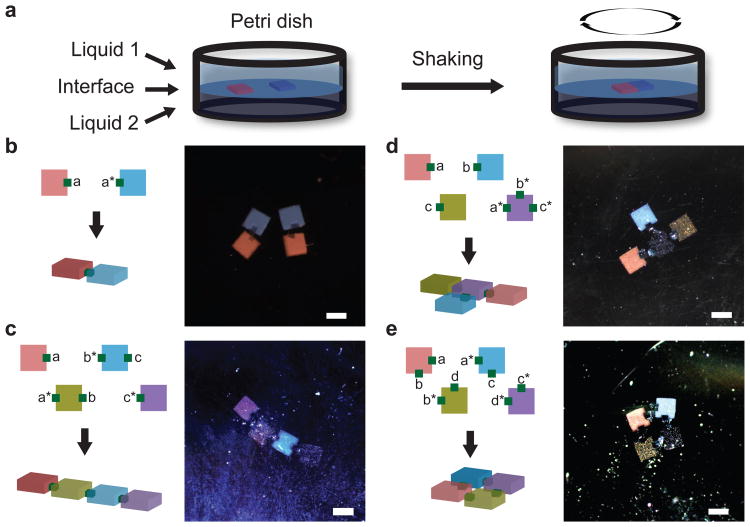
We next programmed the self-assembly of hydrogel units into prescribed finite structures. To avoid the rotation of the hydrogel cubes in the vertical direction during assembly, flat hydrogel cuboids were fabricated and floated on a liquid/liquid interface between aqueous PBS and Fluorinert® FC-40 liquid; horizontal shaking was applied to facilitate assembly (Fig. 5a). The assembled structures were directly imaged in this interfacial system. We analyzed the binding between two hydrogel cubes carrying uniform DNA modification in the interfacial system. Over 70% stable specific binding was observed only between hydrogel cubes carrying complimentary DNA, which is consistent with what we observed in the aqueous liquid. Using similar fabrication strategy as in Fig. 4a, we made a two-component gel composite: the body is a 1 mm (length) × 1 mm (width) × 0.3 mm (height) flat cuboid and the pads are DNA-modified 250 μm × 250 μm × 250 μm cubes. Note that the length ratio between the body and the pad cubes increased to 4:1 (as compared to the 2.5:1 in Fig. 4). Using the interfacial system and the flat cuboids that carry relatively smaller pads, we were able to assemble dimers (Fig. 5b), linear chains with finite length (Fig. 5c), a T-junction (Fig. 5d), and a square structure (Fig. 5e). Note that the linear chain, the T junction, and the square are all composed of 4 distinct cuboid species; by simply changing the pad (and hence surface giant DNA) modification pattern, we were able to change the assembled structure from a chain to a T-junction, and to a square. The assembly process details are captured in time-lapse movies. Supplementary Movie 1 shows two copies of dimers self-assembly (Fig. 5b). Supplementary Movie 2 shows the chain structure self-assembly (Fig. 5c). Supplementary Movie 3 shows the T-junction self-assembly (Fig. 5d). Supplementary Movie 4 shows the self-assembly of the square structures (Fig. 5e).
Figure 5. Self-assembly of hydrogel cuboids at the liquid-liquid interface.
a, Schematic of hydrogel self-assembly at the liquid-liquid interface. Hydrogel cuboids were floated at the interface formed between upper aqueous PBS liquid and lower Fluorinert® FC-40 liquid, and agitated with rotary shaking. b, Dimers were formed from the red and blue gels. See SuSupplementary Movie 1 for the assembly process. c-e, Schematic and color stereomicroscopy images of four hydrogel cuboids (labeled as red, blue, yellow and violet) self-assembled into chain (c), T-junction (d) or square shape (e), based on their surface DNA glue pattern. For the assembly process, see Supplementary Movies 2, 3, and 4 respectively. Scale bar, 1 mm.
For the dimer experiments (Fig. 5b), two copies of each component were included; for the linear (Fig. 5c), T-junction (Fig. 5d) and square (Fig. 5e) structures, only one copy of each component was included. We tested the assembly of each system for three times or more, and all together more than 20 experiments were performed for these structures. Out of these more than 20 experiments, the intended structure always formed as desired. However, it is important to note that such systems (involving only one copy of each component) are significantly simpler than systems that involve multiple copies of the same components. We have performed an experiment to form two copies of the T-junction. In this experiment (Supplementary Fig.S4), we started with 14 hydrogel cubes: two copies of center hydrogel cube (purple) and four copies of each of the three side hydrogel cubes (red, blue and yellow). We attempted the experiments three times, and only successfully assembled two copies of T-junctions in one of the three experiments. It thus remains a challenge to assemble complex structures using multiple copies for each component in the interfacial systems.
Discussion
Although short DNA primer has been reported to assemble nano-particles and microscale hydrogels, we demonstrate, for the first time, the DNA-directed self-assembly of shape-controlled hydrogel modules to build complex structures in a programmable fashion. Acting like sequence specific glue and tethered onto a microgel surface, giant single-stranded DNA exhibits a significant capability for binding objects across scales, with sizes ranging from 30 micrometers to a millimeter. Additionally, giant DNA glues offer significant diversity over current mesoscale self-assembly systems: fifty DNA sequences were designed to generate 25 orthogonal pairs of specific interactions. This is the largest of number of orthogonal binding interactions that have been used simultaneously in the same reaction system for mesoscale assembly. The designable DNA glues thus provide much richer options for programming mesoscale self-assembly.
For self-assembling complex structures, the unit fabrication is crucial. As cube units uniformly carrying DNA glues assembled only into aggregates lack of architecture control, we developed a precisely controlled fabrication technique by which specific giant DNA glue is decorated on a prescribed face of a hydrogel cuboid. By changing the position of DNA glues, various structures including dimers, linear chains, and open networks were assembled. In an interfacial system, we further demonstrated that hydrogel cuboids can be fabricated with 4 different DNA glues on 4 designated faces, and by simply changing the surface DNA decoration pattern, we assembled discrete structures including dimers, T-junctions, linear chains with fixed length, and squares. We have successfully introduced programmability into self-assembling mesoscale structure.
We believe that there is still room for improvement in our self-assembly system by increasing the resolution of module fabrication. As demonstrated in this study, hydrogel cuboids with a larger width ratio between the body part and the DNA pad align better with each other than cuboids with a smaller width ratio. It is also possible to improve the assembly by tuning the strength of DNA glue through controlling the DNA density and length on the gel surface. Additionally, better face-to-face alignment could be achieved by improving the gel fabrication method, for example, by minimizing the aspect ratio between the DNA carrying pads and the gel body (note that the assembly in Fig. 5 demonstrated improved face-to-face alignment over Fig. 4). Furthermore, advances in the mixing regimes that better regulate the hydrodynamic forces involved in the assembly process may be used to further enhance the assembly process.
By coupling novel in situ DNA amplification methods and microfabrication techniques, we successfully introduced the diversity and specificity of biomolecular interaction to mesoscale assembly. DNA-directed self-assembly of shape-controlled hydrogel modules proved to be highly programmable and controllable, and will open new doors to address the challenge of building complex self-assembled 3D structures for diverse applications in materials science and especially in biomaterials. One particularly promising direction is to develop tissue engineering application, as recreating the highly defined complicate structure of tissue is a pressing challenge. For example, by encapsulating specific cells inside the hydrogel cubes, the self-assembled structures could be used to build the basic architectures of native tissues.
Methods
Materials
The pre-polymer solution of PEG-DA with average molecular weight 4000 Da was prepared by diluting PEG (Monomer-Polymer&Dajac Labs) in DPBS (Gibco) to a final concentration of 20 wt% with 1 wt% photo-initiator, 1-(4-(2-Hydroxyethoxy)-phenyl)-2-hydroxy-2-methyl-1-propane-1-one (IRGACURE 2959 Ciba) for hydrogel modules fabrication. A circular DNA template was produced by ligating 5′ and 3′ ends of DNA (Invitrogen) with 5′end phosphate modification using a CircLigase ssDNA ligase (EPICENTRE Biotechnology). DNA sequence for DNA glue synthesis was designed using software NUPACK (www.nupack.org) to minimize mis-hybridization between orthogonal sequences. DNA was ordered from Invitrogen. Baseline-ZERO™ DNase was obtained from Epicentre Biotechnology and used with final concentration of 1 U/ml.
Sequence design
We designed 25 orthogonal sequence pairs using a modified version of the Domain Design (DD) software described by Zhang et al58. We first designed 25 domains of 24 bases each, and then concatenated these domains together into 25 domains of 48 bases each. Sequences were designed using a 3-letter alphabet to reduce spurious hybridization. In order to reduce long regions of repeated bases (e.g. poly-A, poly-G, etc.), we assign a higher reward to sequences with a higher Shannon entropy. Using NUPACK thermodynamic analysis package, unintended interactions between the concatenated products were calculated to be ∼108 times less favorable than the intended interactions59.
Fabrication of PEG hydrogels carrying DNA glues
We followed a standard protocol to synthesize acrylate-PEG-DNA by adopting NHS chemistry to conjugate a 5′ amine modified the DNA strand (containing a poly(T-36) linker segment followed by a primer segment) to acrylate-PEG-NHS (Jenkem Technology). Shape-controlled PEG hydrogel was fabricated by following a general photolithograph process as described in our previous work60, in which the photomask was designed using AutoCAD software with 20,0000 dpi resolution (CAD/Art Services; Bandon, OR). Prior to DNA amplification, hydrogel was washed with PBS and DNA amplification buffer thoroughly (40 mM Tris-HCl pH 7.5, 50 mM KCl, 10 mM MgCl2, 5 mM (NH4)2SO4, and 4 mM DTT). Giant DNA was amplified by soaking hydrogel in reaction solution containing 50 μM circular DNA template and 5 U/ul of Phi29 DNA polymerase (EPICENTRE Biotechnology) in 1× DNA amplification buffer at 37°C overnight according to the manufacturer's instructions. Hydrogel carrying patterned DNA glue was fabricated in a two-step fabrication process. In step (1), DNA glue was amplified on a small hydrogel cube with edge with of 150 - 300 μm, which was fabricated by photolithography as described above. In step (2), DNA gel was washed with 1× assembly buffer (0.5 M NaCl, 0.5 mM EDTA, and 0.05% Tween-20 in 1× PBS) thoroughly. Then 20 wt% pre-polymer PEG solution containing 1 v/v% color microbead was added. The final shape of hydrogel was controlled by a secondary photomask which was aligned under microscope with the hydrogel to produce the desired DNA pattern.
Self-assembly of hydrogels in aqueous solution
Hydrogels carrying specific DNA glue was collected in a 0.5 ml microtube filled with assembly buffer containing 0.5 M NaCl, 0.5 mM EDTA, and 0.05% Tween-20 in 1× PBS buffer. To prevent nonspecific binding between hydrogel and microtube, the inside surface of microtube was treated with a corona treater (BD-20AC from Electro-Technic Products Inc.) and then coated with 10% PEGDA (MW 1000) beforehand. To achieve DNA-directed hydrogel assembly, microtube was subjected to agitation of continuous 360-degree upright rotation on a VWR Multimix tube rotator with a fixed speed of 18 rpm and intermittent soft vortex or hand shaking every 30-60 minutes to disrupt non-specific binding or aggregates at room temperature. After assembly, hydrogel was transferred to a petri dish filled with a solution of 20 wt% PEG (MW 3350) in 1× assembly buffer and subjected to further horizontal shaking at a speed of about 60 rpm on a VWR standard orbital shaker. Assembled structures were identified, quantified and imaged using a Stereo Microscope.
Liquid/liquid interface self-assembly
Liquid/liquid interface was generated between Fluorinert® electronic liquid FC-40 (bottom liquid, 3M™ Chemicals) and aqueous assembly buffer (top liquid) in a petri dish. Hydrogel units were floated on the interface and subjected to agitation of continuous horizontal shaking at a low speed of 60 rpm on a VWR standard orbital shaker (Model 1000, VWR) and intermittent 120 rpm shaking or strong hand shaking every 30-60 minutes to break undesired, spurious aggregates at room temperature. Assembly process was recorded using an image recording software, HyperCam Version 2, under a Stereo Microscope.
Quantification of DNA directed hydrogel assembly
To quantify the specificity of dimer formation, hydrogel cubes with an edge width of 250 μm were fabricated. A gel is uniformly decorated with a particular giant DNA glue species. In total, three different complementary pairs of DNA (a/a*, red; b/b*, blue; c/c*, yellow) were used to generate giant DNA glues. Ten copies of hydrogels of each pair were placed in an Eppendorf tube (n=6) filled with assembly buffer and rotated for over 3 hours. Rotating the hydrogels generated dimers with all different combinations of the three colors. Self-assembly was performed in aqueous liquid system as described above, and the total specific binding among the pairs was identified under a Stereo Microscope. A total of 77 assembled structures were analyzed in 6 independent experiments with 82% of specific binding while non-specific binding was 19%, which confirmed the specificity of DNA interactions. Quantification analysis was performed using STDEV function and TDIST function for standard deviation and P-value respectively in software Excel (Microsoft™).
Giant DNA analysis
Giant DNA was amplified by rolling circle amplification (RCA) using Phi29 DNA polymerase (EPICENTRE Biotechnology) as described earlier and analyzed on a 1% agarose gel with SYBR Green staining. The morphology of giant DNA on surface of microgel or glass slide was analyzed using a scanning electron microscopy (Zeiess EVO SEM). After amplification, microgel was rinsed with PBS (GIBCO, DPBS) thoroughly and the liquid around the gel was dried up using Kimwipes. Following washing, microgel was frozen in -80°C freezer for 3 hours and transferred to a freeze drier for 2 days before scanning electron microscopy imaging.
Assembly of 25 orthogonal pairs of hydrogel dimers
DNA strands, D001, D001*, D002, D002* …… D025, D025* (Supplementary Table S1:DNA sequences), and poly(T-36) with a 5′ end amine modification were purchased from Integrated DNA Technology (IDT) and dissolved in water upon arrival.
DNA oligo phosphorylation
One phosphate group was added to the 3′ end of DNA strands by T4 Polynucleotide kinase (PNK, EPICENTRE Biotechnology) before ligation with poly(T-36) or circularization. Phosphorylation master reaction containing 33 mM Tris-HCl (pH 7.5), 66 mM potassium acetate, 10 mM magnesium acetate, 0.5 mM DTT, 5 mM ATP, 100 μM DNA oligo, and 10 U/μl PNK enzyme, was incubate at 37°C for 3 hours and then PNK enzyme was inactivated by incubation at 70°C for 30minutes.
Circular DNA template preparation
Circular DNA template was prepared by circulating phosphate modified DNA strand using Cirligase II ssDNA ligase (EPICENTRE Biotechnology). Circularization reaction was performed by supplementing 2.5 mM manganese chloride, 1 M Betaine, and 5 U/μl ligase to phosphate modified DNA strand solution and then incubated at 60°C for 6 hours. Then the ligase was inactivated at 80°C for 10 minutes. Then exonuclease I (EPICENTRE Biotechnology) was added to remove linear DNA at 37°C for 1 hour and was inactivated at 80°C for 30 minutes.
Conjugation of acrylate-PEG-DNA primer
Firstly, phosphorylated DNA oligo was ligated to amine-poly(T-36). Ligation reaction was performed with supplementing 20 U/μl T4 RNA ligase (New England Biolab) and 100 μM amine-poly(T-36) in phosphate modified DNA oligo solution and incubated at room temperature (25°C) overnight. Following ligation, amine-poly(T-36)-DNA oligo was conjugated with acrylate-PEG-NHS (Jenkem Technology) as described earlier. Cubic PEG microgel with size of 250 μm × 250 μm × 250 μm carrying DNA primer was fabricated and giant DNA was amplified as described earlier. Pairwise combination of the core and periphery gels that contain one of the 5 color microbeads (polybead Red Dyed 1.0 μm microspheres, polybead Blue Dyed 0.5 μm microspheres, Polybead Yellow Dyed 3.0 μm microspheres, polybead Violet Dyed 1.0 μm microsphere and polybead Black Dyed 10.0 μm microspheres; Polysciences) generated 25 distinct labeling. Fabrication process is similar to that of microgel for dimer assembly as previously described except for the following modification. During the secondary photolithography, the second microgel (periphery) was fabricated to completely wrap the first microgel (core) to form the final structure. To decrease assembly time and avoid aggregation, a single copy of each microgel uniformly carrying DNA D001, D001*, D002, D002* …… D025, D025* was collected in 1.5 ml Eppendorf microtubes filled with 1× assembly buffer. Subsequent self-assembly was performed in aqueous liquid as described earlier. The assembled structure was identified under a Stereo Microscope. A total of 83 specific assembled structures were analyzed in 6 independent experiments and the specificity of assembly was quantified using TDIST function of software Excel (Microsoft) for P-value calculation (n=6).
Supplementary Material
Acknowledgments
The authors thank Wei Sun and Tom Schaus for discussions. This work is funded by the NIH (EB008392; DE019024; HL099073; AR057837; HL092836), NSF CAREER Award DMR0847287, and ONR grants to A.K. and by ONR Young Investigator Program Award N000141110914, ONR Grant N000141010827, NSF CAREER Award CCF1054898, NIH Director's New Innovator Award 1DP2OD007292, and Wyss Institute Faculty Startup Fund to P.Y.
Footnotes
Supplementary Information is linked to the online version of the paper at xxx
Author Contributions H.Q. designed and conducted the experiments, performed data analysis, and wrote the paper; M. G. designed and performed experiments. Y.D., H.B., C.G. assisted in designing and performing experiments. P.Y. and A.K. conceived, designed and supervised the study, interpreted the data and prepared the manuscript.
Additional Information Reprints and permissions information is available at xxx.
The authors declare competing financial interests in the form of a pending provisional patent (U.S. Provisional Application No.: 61/727,344, Nucleic acid hydrogel self-assembly).
References
- 1.Whitesides GM, Grzybowski B. Self-assembly at all scales. Science. 2002;295:2418–2421. doi: 10.1126/science.1070821. [DOI] [PubMed] [Google Scholar]
- 2.Breen TL, Tien J, Oliver SR, Hadzic T, Whitesides GM. Design and self-assembly of open, regular, 3D mesostructures. Science. 1999;284:948–951. doi: 10.1126/science.284.5416.948. [DOI] [PubMed] [Google Scholar]
- 3.Boncheva M, et al. Magnetic self-assembly of three-dimensional surfaces from planar sheets. Proc Natl Acad Sci U S A. 2005;102:3924–3929. doi: 10.1073/pnas.0500807102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark TD, Ferrigno R, Tien J, Paul KE, Whitesides GM. Template-directed self-assembly of 10-microm-sized hexagonal plates. J Am Chem Soc. 2002;124:5419–5426. doi: 10.1021/ja020056o. [DOI] [PubMed] [Google Scholar]
- 5.Du Y, Lo E, Ali S, Khademhosseini A. Directed assembly of cell-laden microgels for fabrication of 3D tissue constructs. Proc Natl Acad Sci U S A. 2008;105:9522–9527. doi: 10.1073/pnas.0801866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowden N, Terfort A, Carbeck J, Whitesides GM. Self-Assembly of Mesoscale Objects into Ordered Two-Dimensional Arrays. Science. 1997;276:233–235. doi: 10.1126/science.276.5310.233. [DOI] [PubMed] [Google Scholar]
- 7.Harada A, Kobayashi R, Takashima Y, Hashidzume A, Yamaguchi H. Macroscopic self-assembly through molecular recognition. Nat Chem. 3:34–37. doi: 10.1038/nchem.893. [DOI] [PubMed] [Google Scholar]
- 8.Seeman NC. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 9.Lin C, Liu Y, Rinker S, Yan H. DNA tile based self-assembly: building complex nanoarchitectures. Chemphyschem: a European journal of chemical physics and physical chemistry. 2006;7:1641–1647. doi: 10.1002/cphc.200600260. [DOI] [PubMed] [Google Scholar]
- 10.Aldaye FA, Palmer AL, Sleiman HF. Assembling materials with DNA as the guide. Science. 2008;321:1795–1799. doi: 10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]
- 11.Torring T, Voigt NV, Nangreave J, Yan H, Gothelf KV. DNA origami: a quantum leap for self-assembly of complex structures. Chemical Society reviews. 2011;40:5636–5646. doi: 10.1039/c1cs15057j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen JH, Seeman NC. Synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 13.Fu TJ, Seeman NC. DNA double-crossover molecules. Biochemistry. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 14.Winfree E, Liu F, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 15.Chworos A, et al. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 16.Rothemund PW, Papadakis N, Winfree E. Algorithmic self-assembly of DNA Sierpinski triangles. PLoS biology. 2004;2:e424. doi: 10.1371/journal.pbio.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shih WM, Quispe JD, Joyce GF. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 18.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 19.He Y, et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 20.Yin P, et al. Programming DNA tube circumferences. Science. 2008;321:824–826. doi: 10.1126/science.1157312. [DOI] [PubMed] [Google Scholar]
- 21.Andersen ES, et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 22.Dietz H, Douglas SM, Shih WM. Folding DNA into Twisted and Curved Nanoscale Shapes. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Douglas SM, et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng J, et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature. 2009;461:74–77. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han D, et al. DNA origami with complex curvatures in three-dimensional space. Science. 2011;332:342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 26.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 27.Wei B, Dai M, Yin P. Complex shapes self-assembled from single-stranded DNA tiles. Nature. 2012;485:623–626. doi: 10.1038/nature11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ke Y, Ong LL, Shih WM, Yin P. Three-dimensional structures self-assembled from DNA bricks. Science. 2012;338:1177–1183. doi: 10.1126/science.1227268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han D, et al. DNA gridiron nanostructures based on four-arm junctions. Science. 2013;339:1412–1415. doi: 10.1126/science.1232252. [DOI] [PubMed] [Google Scholar]
- 30.Yurke B, Turberfield AJ, Mills AP, Jr, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 31.Yan H, Zhang X, Shen Z, Seeman NC. A robust DNA mechanical device controlled by hybridization topology. Nature. 2002;415:62–65. doi: 10.1038/415062a. [DOI] [PubMed] [Google Scholar]
- 32.Ding B, Seeman NC. Operation of a DNA robot arm inserted into a 2D DNA crystalline substrate. Science. 2006;314:1583–1585. doi: 10.1126/science.1131372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-free nucleic acid logic circuits. Science. 2006;314:1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 34.Zhang DY, Turberfield AJ, Yurke B, Winfree E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science. 2007;318:1121–1125. doi: 10.1126/science.1148532. [DOI] [PubMed] [Google Scholar]
- 35.Yin P, Choi HM, Calvert CR, Pierce NA. Programming biomolecular self-assembly pathways. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 36.Omabegho T, Sha R, Seeman NC. A bipedal DNA Brownian motor with coordinated legs. Science. 2009;324:67–71. doi: 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lund K, et al. Molecular robots guided by prescriptive landscapes. Nature. 2010;465:206–210. doi: 10.1038/nature09012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu H, Chao J, Xiao SJ, Seeman NC. A proximity-based programmable DNA nanoscale assembly line. Nature. 2010;465:202–205. doi: 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian L, Winfree E, Bruck J. Neural network computation with DNA strand displacement cascades. Nature. 2011;475:368–372. doi: 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]
- 40.Qian L, Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 41.Douglas SM, Bachelet I, Church GM. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 42.Um SH, et al. Enzyme-catalysed assembly of DNA hydrogel. Nat Mater. 2006;5:797–801. doi: 10.1038/nmat1741. [DOI] [PubMed] [Google Scholar]
- 43.Park N, et al. High-yield cell-free protein production from P-gel. Nature protocols. 2009;4:1759–1770. doi: 10.1038/nprot.2009.174. [DOI] [PubMed] [Google Scholar]
- 44.Park N, Um SH, Funabashi H, Xu J, Luo D. A cell-free protein-producing gel. Nat Mater. 2009;8:432–437. doi: 10.1038/nmat2419. [DOI] [PubMed] [Google Scholar]
- 45.Lee JB, et al. A mechanical metamaterial made from a DNA hydrogel. Nat Nanotechnol. 2012;7:816–820. doi: 10.1038/nnano.2012.211. [DOI] [PubMed] [Google Scholar]
- 46.Lin C, et al. Submicrometre geometrically encoded fluorescent barcodes self-assembled from DNA. Nat Chem. 2012;4:832–839. doi: 10.1038/nchem.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 48.Nykypanchuk D, Maye MM, van der Lelie D, Gang O. DNA-guided crystallization of colloidal nanoparticles. Nature. 2008;451:549–552. doi: 10.1038/nature06560. [DOI] [PubMed] [Google Scholar]
- 49.Sharma J, et al. Control of Self-Assembly of DNA Tubules Through Integration of Gold Nanoparticles. Science. 2009;323:112–116. doi: 10.1126/science.1165831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hung AM, et al. Large-area spatially ordered arrays of gold nanoparticles directed by lithographically confined DNA origami. Nat Nanotechnol. 2010;5:121–126. doi: 10.1038/nnano.2009.450. [DOI] [PubMed] [Google Scholar]
- 51.Macfarlane RJ, et al. Nanoparticle superlattice engineering with DNA. Science. 2011;334:204–208. doi: 10.1126/science.1210493. [DOI] [PubMed] [Google Scholar]
- 52.Maune HT, et al. Self-assembly of carbon nanotubes into two-dimensional geometries using DNA origami templates. Nature Nanotechnology. 2010;5:61–66. doi: 10.1038/nnano.2009.311. [DOI] [PubMed] [Google Scholar]
- 53.Beales PA, Nam J, Vanderlick TK. Specific adhesion between DNA-functionalized “Janus’ vesicles: size-limited clusters. Soft Matter. 2011;7:1747–1755. [Google Scholar]
- 54.Gartner ZJ, Bertozzi CR. Programmed assembly of 3-dimensional microtissues with defined cellular connectivity. Proc Natl Acad Sci U S A. 2009;106:4606–4610. doi: 10.1073/pnas.0900717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li CY, Wood DK, Hsu CM, Bhatia SN. DNA-templated assembly of droplet-derived PEG microtissues. Lab on a chip. 2011;11:2967–2975. doi: 10.1039/c1lc20318e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schlingman DJ, Mack AH, Mochrie SG, Regan L. A new method for the covalent attachment of DNA to a surface for single-molecule studies. Colloids and surfaces B, Biointerfaces. 2011;83:91–95. doi: 10.1016/j.colsurfb.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schopf E, Chen Y. Attomole DNA detection assay via rolling circle amplification and single molecule detection. Anal Biochem. 2010;397:115–117. doi: 10.1016/j.ab.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 58.Zhang DY. Towards Domain-Based Sequence Design for DNA Strand Displacement Reactions. Lecture Notes in Computer Science. 2011;6518:162–175. [Google Scholar]
- 59.Dirks RM, Bios JS, Schaeffer JM, Winfree E, Pierce NA. Thermodynamic analysis of interacting nucleic acid strands. SIAM Rev. 2007;49:65–88. [Google Scholar]
- 60.Qi H, et al. Patterned differentiation of individual embryoid bodies in spatially organized 3D hybrid microgels. Adv Mater. 2010;22:5276–5281. doi: 10.1002/adma.201002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.