Abstract
Claudins are tight junction membrane proteins that are expressed in epithelia and endothelia and form paracellular barriers and pores that determine tight junction permeability. This review summarizes our current knowledge of this large protein family and discusses recent advances in our understanding of their structure and physiological functions.
I. INTRODUCTION
Claudins are integral membrane proteins found in tight junctions of all epithelia and endothelia. Claudins were first identified in 1998 in a purified junctional fraction from chicken liver by Furuse in the lab of the late Shoichiro Tsukita (95). They appear to be major structural components of the tight junction. Overexpression of claudins in fibroblasts, which normally lack tight junctions, is sufficient to reconstitute tight junction-like networks of strands (98, 198). It is also now clear that claudins constitute both paracellular barriers and pores and thereby play a key role in determining the permeability properties of epithelial and endothelial cells. The major evidence for this comes from a multitude of studies showing that overexpression, knockdown or knockout of claudins, and both naturally occurring and experimentally introduced mutations of claudins, consistently cause changes in paracellular permeability. Furthermore, the effect of mutations that alter charged residues in the extracellular domains of claudins, or that introduce cysteines accessible to covalent modification by extracellular probes, indicate that the extracellular domains of claudins must line the paracellular pore. In this article, we review the biology of claudins and discuss what is known about how they affect tight junction permeability and their role in human diseases, focusing primarily on epithelial tissues. The reader is referred to two recent reviews for a detailed discussion of the regulation of junctional permeability in endothelia (28, 377).
II. TIGHT JUNCTIONS AND THEIR ROLE IN PARACELLULAR PERMEABILITY
A. Tight Junctions Form the Seal Between Epithelial Cells
Epithelia are sheets of cells that line body cavities and external surfaces in multicellular organisms. A key function of epithelia is to act as a physical and chemical barrier. For example, the epidermis acts as the skin barrier to the exterior, the intestinal epithelium acts as a barrier to bacterial toxins in the gut lumen, and the urinary bladder has to act as a barrier to water and electrolytes in urine. At the same time, epithelia also allow selective transport of solutes and water between compartments. Transepithelial transport may occur via two routes: transcellular, meaning transport occurs through the cell, crossing the apical and basolateral plasma membranes; and paracellular, which refers to transport in between cells. Frömter and Diamond (88) were the first to demonstrate that the paracellular permeability pathway corresponds to the intercellular space. They used conductance scanning to show that the transepithelial conductance in Necturus gall bladder, a leaky epithelial tissue, was maximum at the junction between neighboring cells.
Epithelial cells are attached to each other at their lateral membranes by a complex of intercellular junctions (84). The most apical of the intercellular junctions is the tight junction, or zona occludens. Anatomically the tight junction appears as a series of appositions or “kisses” between the exocytoplasmic leaflets of the lateral membranes of adjacent cells. These appositions extend in a beltlike network that surrounds each cell and attaches it to the neighboring cell to form a continuous seal. It is now well established that the tight junction is the major determinant of paracellular permeability. Electron-dense molecules to which the paracellular pathway is impermeable, such as hemoglobin, colloidal lanthanum, and ruthenium red, have been shown to freely diffuse along the intercellular space but stop at the level of the tight junction, indicating that this is the site of the permeability barrier (84, 233, 397).
In addition to acting as a selective barrier to permeability between aqueous compartments, the epithelial tight junction has been proposed to have one other function, which is to act as a fence within the cell membrane, i.e., to mechanically restrict diffusion of proteins and lipids within the plane of the lipid bilayer (69). Such a fence would ensure that the lipid and protein components of the plasma membrane remain separated in distinct apical and basolateral domains at the tight junction, a prerequisite for directional transepithelial transport. Several observations indicate that the membrane-spanning proteins of the tight junction act as a diffusion barrier within the membrane and that fence and gate function are simultaneouly affected (see, e.g., Refs. 137, 376). Yet, other manipulations can dissociate the barrier and fence functions of the tight junction. Thus transient ATP depletion (24, 227) and depolymerization of the cortical actin network (341) disrupt barrier function while preserving fence function. This suggests that the fence function is mechanistically independent of barrier function. There is now increasing experimental evidence that tight junctions and claudins are not required for fence function and hence for upholding cell polarization (23, 137, 158, 227, 250, 361).
B. Tight Junctions Are Constituted of a Complex of Proteins
Early models of the tight junction posited that it was composed purely of lipid organized into inverted cylindrical micelles, that constituted the tight junction strands (171, 290). There now exist three lines of evidence that refute this. First, tight junction strands were found to be resistant to deoxycholate, an ionic detergent that should solubilize lipids (333). Second, the lipid model predicts that the outer leaflets of the bilayers of adjacent cells are continuous, yet no transfer of glycolipid or a fluorescent probe could ever be demonstrated (267, 375, 376). Finally, biochemical characterization of the tight junction has now revealed that it is composed of a complex of multiple proteins that include transmembrane proteins, cytoplasmic plaque proteins, signaling proteins, and adapters that link it to the actin cytoskeleton.
The transmembrane proteins are of particular interest. Because they are the only components of the junction that have intramembranous and extracellular portions, it is likely that they mediate all three major functions of the tight junction: barrier, pore, and fence. The transmembrane proteins of tight junctions fall into three groups: the single transmembrane domain proteins, including JAM (junctional adhesion molecule), Crb3 (Crumbs protein homolog 3), and CAR (coxsackievirus and adenovirus receptor); the triple transmembrane domain protein, Bves (blood vessel epicardial substance); and the four-transmembrane domain proteins of the claudin and TAMP (tight junction-associated MARVEL proteins) families, which include occludin, tricellulin, and MarvelD3. Of these, the overwhelming weight of evidence suggests that claudins are the major determinants of paracellular permeability.
III. CLAUDIN GENE FAMILY
A. Evolution of Tight Junctions
Although the necessity to seal body compartments exists in all animals, tight junctions seem to be a relatively new evolutionary achievement. Their presence in all vertebrates and in the invertebrate tunicates (109, 202) appeared to indicate that they are common to all chordates; however, they are not found in the cephalochordate Branchiostoma (203, 393, 394). Lower invertebrates possess various types of septate junctions, a type of cell-cell contact that, in cross-sections, has a ladderlike appearance, with the bars of the ladder spanning an intercellular gap of ∼15–18 nm (109). Vertebrates possess septatelike junctions in the axo-glial paranodal junctions (141; for review, see Ref. 293). Whereas invertebrate septate junctions and vertebrate septatelike paranodal junctions are seen as having “evolved from a common ancestral precursor” (141), the relationship between septate junctions and tight junctions is yet unclear. Hemichordates and echinoderms possess two distinct types of septate junctions. One type, called anastomosing septate junction, forms a meshlike network between two adjacent cells (110), the morphology of which resembles vertebrate tight junctions (109). This type of septate junctions appears to be also present in cephalochordates (393). These findings were taken as an indication that septate junctions may be the fore-runners of chordate tight junctions (109). However, there are reports of truly tight junction-like structures in arthropods, especially in the arachnid hemolymph-CNS barrier (201, 204). Lane and Chandler (201) therefore see tight and septate junctions as independent structures, both being already present in invertebrates, whereas Green and Bergquist (109) consider arthropod tight junction-like structures as independent, parallel developments to chordate tight junctions (109).
B. Phylogeny of Claudins
The discovery of the first claudin as a strand-forming tight junction protein was published by Shoichiro Tsukita and co-workers in 1998 (95). The number of proteins recognized as members of the claudin family has been growing continually since then. As suggested by the morphological studies, claudins have been reported or at least predicted for all vertebrates and for tunicates, but, according to the NCBI database, do not appear to be present in cephalochordates and in lower invertebrates. Whereas for most species only a few claudins are listed, indicating that these are chance observations, more systematic investigations exist especially in fish (e.g., Takifugu rubriens, Danio rerio, Salmo salar), birds (Gallus gallus), and mammals (e.g., Mus musculus, Rattus norvegicus, Bos taurus, Homo sapiens).
Fish appear to possess an exceptionally wide range of different claudins, e.g., 56 different claudins have been reported for the puffer fish Takifugu rubripes (218). This high number appears to be due to gene duplication processes that are independent from the proposed whole genome duplication in teleost fish (165, 218). This may be due to the requirements specific to fish, such as osmoregulation and adaptation to different water salinities. Fish that travel from fresh to sea water or back have to undergo dramatic changes in the tight junction composition especially of their kidney and gills, but also of their intestine and skin to adapt to the reversal of osmotic and ionic gradients (25, 46, 74, 80, 173, 354).
Similarly, terrestrial amphibians have to adapt to different osmotic conditions when they develop from water-dwelling tadpoles to land-dwelling adults. However, the effects of this transition on tight junction structure and composition, e.g., in tadpole gills, adult lungs, or amphibian skin with its remarkably high transport activity, do not appear to have been investigated yet. Indeed, most amphibian claudins reported in the NCBI database to date are from Xenopus laevis and Xenopus tropicalis, which are purely aquatic animals.
There are thought to be 27 mammalian claudin genes, including three distant members that were recently discovered (246, 407), although disagreement exists as to whether they should all be classified as claudins (224). Not all of these genes are found in all mammalian species. Claudin-13, for example, is present in rodents but does not exist in humans so that there is now evidence for 26 human claudins, the physiological role of which will be described in more detail in section VI. As indicated in TABLE 1, the nomenclature of claudins 21 and 25–27 is still a matter of debate, and the present review will follow the nomenclature suggested by Mineta et al. (246).
Table 1.
Characteristics of human claudin transcripts and protein products
| Claudins and Their Isoforms | Characteristics of Transcripts Variants | Characteristics of Resulting Isoforms | PDZ-Binding Motif | Number of Amino Acids (N-T1-E1-T2-I-T3-E2-T4-C) | Molecular Mass, kDa | mRNA ID |
|---|---|---|---|---|---|---|
| 1 | 4 exons | Yes | 211 | 22.8 | NM_021101.4 | |
| 4 exons in CDS | (7-21-53-21-13-21-27-21-27) | |||||
| 2 | 2 exons | Variants 1–3 encode the same protein | Yes | 230 | 24.4 | NM_020384.3 |
| 1 exon in CDS | (7-21-53-21-14-21-25-21-47) | NM_001171092.1 | ||||
| 3 variants using alternate 5′ noncoding exons | NM_001171095.1 | |||||
| 3 | 1 exon | Yes | 220 (8-21-51-21-14-21-23-21-40) | 23.3 | NM_001306.3 | |
| 4 | 1 exon | Yes | 209 (7-21-53-21-15-21-22-21-28) | 22.1 | NM_001305.3 | |
| 5 | Variant 1: 1 exon | Variants 1 and 2 encode the same protein | Yes | 303 (92-21-53-21-20-21-16-21-38) | 31.6 | NM_001130861.1 |
| Variant 2: 2 exons | Long and short version due to two start codons within the intracellular NH2 terminus | NM_003277.3 | ||||
| Both: 1 exon in CDS | ||||||
| Variant 2 lacks segment in 5′ UTR | 218 (7-21-53-21-20-21-16-21-38) | 23.2 | ||||
| 6 | 2 exons | Yes | 220 (7-21-53-21-14-21-23-21-39) | 23.3 | NM_021195.4 | |
| 1 exon in CDS | ||||||
| 7 (isoform 1) | Variant 1: 4 exons | Variants 1 and 2 encode the same isoform | Yes | 211 (7-21-53-21-15-21-22-21-30) | 22.4 | NM_001307.5 |
| Variant 2: 5 exons | NM_001185022.1 | |||||
| 4 exons in CDS | ||||||
| Two alternate 5′ UTR sequences. Variants 1 and 2 | ||||||
| 7 (isoform 2) | 3 exons | Shorter and distinct COOH terminus | No* | 145 (topology not available) | 15.2 | NM_001185023.1 |
| 3 exons in CDS | ||||||
| Lacks exon 3 in the 3′ CDS | ||||||
| 8 | 1 exon | Yes | 225 (7-21-53-21-15-21-28-21-38) | 24.8 | NM_199328.2 | |
| 9 | 1 exon | Yes | 217 (7-21-53-21-14-21-22-21-37) | 22.9 | NM_020982.3 | |
| 10a | 5 exons | Yes | 226 (0-21-57-21-14-21-24-21-47) | 24.3 | NM_182848.3 | |
| 5 exons in CDS | ||||||
| 10a_i1 | 5 exons | Lacks an internal segment near the NH2 terminus (within ECL1), compared with isoform 10a | Yes | 207 (0-21-38-21-14-21-24-21-47) | 22.2 | NM_001160100.1 |
| 5 exons in CDS | ||||||
| Variant a_v1 uses an alternate in-frame splice site in the 5′ coding region, compared with variant a. | ||||||
| 10b | 5 exons | Longer and distinct NH2 terminus, compared with isoform 10a | Yes | 228 (0-21-59-21-14-21-24-21-47) | 24.5 | NM_006984.4 |
| 5 exons in CDS | ||||||
| differs in the 5′ UTR and 5' coding region, uses alternate promoter and exon 1, compared with variant a. | ||||||
| 11 (isoform 1) | 3 exons | Yes | 207 (1-21-60-21-19-21-14-21-29) | 22 | NM_005602.5 | |
| 3 exon in CDS | ||||||
| 11 (isoform 2) | 2 exons | shorter NH2 terminus, lacking aa 1-84 of variant 1 | Yes | 123 (topology not available) | 12.9 | NM_001185056.1 |
| 2 exon in CDS | ||||||
| Alternate 5′ exon, resulting in downstream in-frame AUG start codon | ||||||
| 12 | Variant 1: 5 exons | Variants 1-3 encode the same protein | No* | 244 (10-21-56-21-27-21-18-21-49) | 27.1 | NM_001185072.2 |
| Variants 2 and 3: 3 exons | NM_001185073.2 | |||||
| 1 exon in CDS | NM_012129.4 | |||||
| Variants 2 and 3 use alternate splice site and/or lack an exon in 5′ UTR | ||||||
| 14 | Variants 1-4: 3 exons | Variants 1-5 encode the same protein | Yes | 239 (7-21-53-21-13-21-26-21-56) | 25.7 | NM_144492.2 |
| Variant 5: 2 exons | NM_012130.2 | |||||
| 1 exon in CDS | NM_001146077.1 | |||||
| 5 variants differing in 5′ UTR | NM_001146078.1 | |||||
| Alternative names: | NM_001146079.1 | |||||
| variant 1 =α, 2 = ε, 3 =δ, 4 = γ, 5 =β | ||||||
| 15 | Variant 1: 6 exons | Variants 1 and 2 encode the same protein | Yes | 228 (3-21-55-21-19-21-17-21-50) | 24.4 | NM_001185080.1 |
| Variant 2: 5 exons | NM_014343.2 | |||||
| 5 exons in CDS | ||||||
| Variant 2 has shorter and alternate 5′ UTR | ||||||
| 16 | 5 exons | Long and short version due to two start codons within the intracellular NH2 terminus | Yes | 305 (73-21-56-21-14-21-33-21-45) | 33.8 | NM_006580.3 |
| 5 exons in CDS | 235 (3-21-56-21-14-21-33-21-45) | 26.1 | ||||
| Two start codons | ||||||
| 16 | 2 exons | Lacks an internal segment, shorter and distinct COOH terminus | No* | 119 (topology not available) | 13.5 | DQ305102 |
| 2 exons in CDS | ||||||
| Lacks exons 2 to 4, frame shift results in earlier stop codon | ||||||
| 17 | 1 exon | Yes* | 224 (7-21-53-21-22-21-19-21-39) | 24.6 | NM_012131.2 | |
| 18A1 (also: 18-1) | 5 exons | Yes | 261 (6-21-53-21-21-21-31-21-66) | 27.9 | NM_016369.3 | |
| 5 exons in CDS | ||||||
| 18A2 (also: 18-2) | 5 exons | Same size but different NH2 terminus, as compared with isoform A1 | Yes | 261 (6-21-53-21-21-21-31-21-66) | 27.7 | NM_001002026.2 |
| 5 exons in CDS | ||||||
| alternate 5′ exon | ||||||
| 19a | 5 exons | No* | 224 (7-21-53-21-15-21-22-21-43) | 23.2 | NM_148960.2 | |
| 5 exons in CDS | ||||||
| 19b | 4 exons | Shorter and distinct COOH terminus compared with isoform a | Yes | 211 (7-21-53-21-15-21-22-21-30) | 22.1 | NM_001123395.1 |
| 4 exons in CDS | ||||||
| Additional segment in coding region compared with variant 1. | ||||||
| 19c | 3 exons | Shorter and distinct COOH terminus compared with isoform a | Yes* | 218 (topology not available) | 22.7 | NM_001185117.1 |
| 3 exons in CDS | ||||||
| Lacks exon in CDS, which results in frame-shift, and contains additional segment in the 3′ region compared with variant 1. | ||||||
| 20 | 2 exons | Yes* | 219 (7-21-53-21-16-21-21-21-38) | 23.5 | NM_001001346.3 | |
| 1 exon in CDS | ||||||
| 21 | 1 exon in CDS | No* | 229 (10-21-50-21-22-21-19-21-44) | 25.4 | NM_001101389.1 | |
| 22 | 1 exon | Yes | 220 (10-20-51-21-15-21-26-21-35) | 24.5 | NM_001111319.1 | |
| 1 exon in CDS | ||||||
| 23 | 1 exon | Yes | 292 (3-21-57-21-8-21-29-21-111) | 31.9 | NM_194284.2 | |
| 1 exon in CDS | ||||||
| 24 | 1 exon in CDS | (Uniprot: 205 aa, COOH terminus only 23 aa) | No* | 220 (10-21-50-21-15-21-23-21-38) | 24.4 | NM_001185149.1 |
| 25 | 6 exons | Long and short version due to two start codons within the intracellular NH2 terminus | No* | 276 (27-21-115-21-13-21-20-21-17) | 31.1 | NM_001040182.1 |
| 6 exon in CDS | 253 (4-21-115-21-13-21-20-21-17) | 28.6 | ||||
| 26 | 1 exon in CDS | No* | 223 (6-21-78-21-7-21-34-21-14) | 24.2 | NM_001146336.1 | |
| 27 | 1 exon in CDS | No* | 208 (topology not available) | 21.6 |
Presence of a PDZ-binding motif determined using the model suggested by Stiffler et al. (334); presence of all other motifs determined by Stiffler et al. (334). Number of amino acids is according to the uniprot database (http://www.uniprot.org/): N, intracellular NH2 terminus; T1 to T4, transmembrane regions 1 to 4; E1, E2, first and second extracellular loop, respectively; I, intracellular loop; C, intracellular COOH terminus. Molecular mass was calculated from amino acid sequence (http://www.bioinformatics.org/sms/prot_mw.html). Nomenclature follows the proposal of Mineta et al. (246) and Lal-Nag and Morin (200), taking sequence NP_001094859 as claudin-21 (NCBI: putative claudin-25), sequence NP_001035272 as claudin-25 (NCBI: claudin domain-containing protein 1, CLDND1), sequence NP_001139808 as claudin-26 (NCBI: transmembrane protein 114, TMEM114). As described by Mineta et al. (246), since August 2010 the sequence for claudin-27 (XP_946151) is no longer available through the NCBI database.
C. Structure of Claudin Genes and Their Encoded Proteins
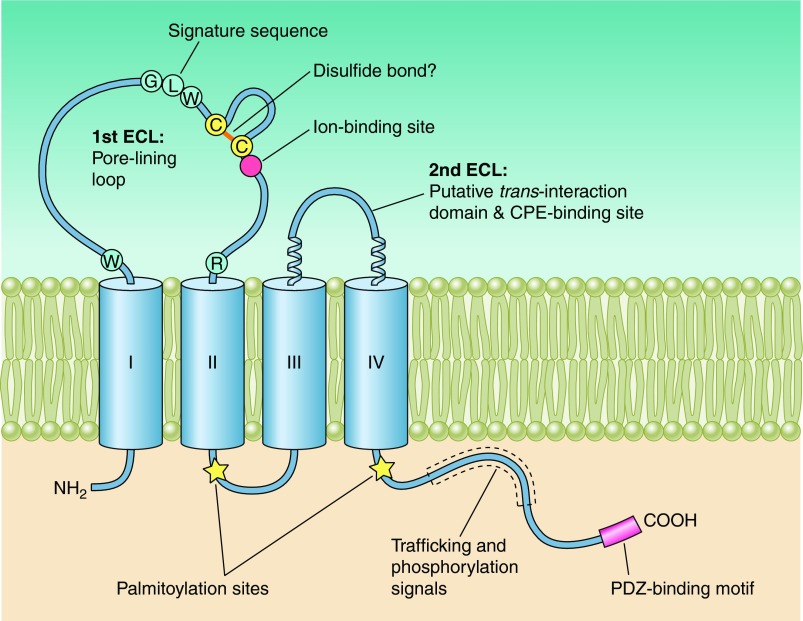
Human claudins possess between 207 and 305 amino acids and have calculated molecular masses of 21–34 kDa (see TABLE 1). Hydropathy plots indicate four transmembrane helices (TM1–4) and the general structure of all claudins consists of an intracellular NH2 terminus that, with the possible exception of claudin-5, -16, and -25, is very short, together with a longer intracellular COOH terminus, two extracellular loops (ECL1, which is larger, and a smaller ECL2), and one short intracellular loop. Claudin-5, -16, and -25 appear to be unusual, due to their long NH2 terminus and, consequently, high molecular mass. However, human mRNAs encoding these three claudins possess two start codons that potentially give rise to a short and long protein version each. The differences lie exclusively within the intracellular NH2 termini of these proteins (claudin-5, 7 vs. 92 amino acids; claudin-16, 3 vs. 73 amino acids; claudin-25, 4 vs. 27 amino acids), and it is still a matter of debate which version is the physiologically relevant form. Further typical features of the claudin family include a signature sequence within ECL1 (see below and sect. VII) and a COOH-terminal PDZ-binding motif, through which the majority of human claudins (334), with the probable exception of claudin-12, -19a, -21, and -24 to -27, (120), are able to interact with PDZ domains of tight-junction associated scaffolding/adapter proteins. PDZ stands for PSD95 (postsynaptic density protein), Dlg1 (Drosophila disc large tumor suppressor), and ZO-1 (zonula occludens-1 protein), the first three proteins, in which these domains had been discovered. Tight junction-associated PDZ domain proteins include ZO-1, -2, and -3, MUPP1, and MAGI-1 to -3. As will be discussed in section VIII, these adaptor proteins also directly or indirectly bind actin and thus anchor the tight junction within the cytoskeleton.
mRNAs encoding mammalian claudins are built from between 1 and 6 exons. The coding sequences (CDS) of most mammalian claudin mRNAs are encoded by only one exon. However, the CDS encoding claudins 1, 7, 10, 11, 15, 16, 18, 19, and 25 are composed of several exons. For some of these mRNAs, splice variants have been reported that result in different protein isoforms, exhibiting distinct expression pattens and/or function. In general, four types of splice variants can be distinguished.
1. Splicing of first coding exon
The genes CLDN10, CLDN11, and CLDN18 possess alternative first exons. In both CLDN10 and CLDN18, these give rise to two full length isoforms that differ in their intracellular NH2 terminus, TM1 and most of ECL1. Whereas ECL1 of claudin-18–1 and claudin-18–2 are very similar (87% identity), ECL1 of claudin-10a and claudin-10b differ greatly (∼32% identity). For both claudins, the respective isoforms have distinct tissue distribution (see sect. VI). In CLDN11, use of the alternative exon 1 results in a downstream in-frame start codon. The resulting isoform 2 is a truncated isoform 1 that lacks the first 84 amino acids, corresponding to the intracellular NH2 terminus, TM1, and the complete ECL1.
2. Splicing of last coding exon
In contrast, claudin-7 and claudin-19 isoforms differ in their COOH-terminal parts. Claudin-19a and -19b are very similar as they share the first 209 amino acids. Claudin-19a then continues with a further 15 amino acids, whereas claudin-19b possesses another 2 amino acids that complete the sequence to a full PDZ binding motif. Thus the only difference between these two isoforms is the length of the intracellular COOH terminus including the absence (19a) or presence (19b) of the PDZ-binding motif. Claudin-19c shares the first 130 amino acids with isoforms a and b. Consequently, it differs in its sequence downstream of the intracellular loop. This sequence is of similar length as in isoform a and b, but does not possess a PDZ binding motif at its COOH terminus. Similarly, claudin-7 isoform 1 and 2 share the first 130 amino acids. Isoform 1 then continues for another 81 amino acids, completing TM3, ECL2, TM4, and the intracellular COOH terminus. In contrast, isoform 2 continues only for a further 15 amino acids and does not posses a COOH-terminal PDZ binding motif (425).
3. Splicing of internal exons
A further group of splice variants arises from skipped exons or from deletions due to the use of alternative splice sites. One example is CLDN10a variant 1 that lacks 57 bp at the border between exon 1a and 2. The resulting isoform, claudin-10a_i1, lacks 19 amino acids within ECL1, including one of the conserved cysteines (122). However, this cysteine may be functionally replaced by a downstream cysteine within ECL1. As described in section VI, this isoform functionally differs from isoform 10a. In mouse kidney, evidence for three further splice variants (10a_v2, 10a_v3, 10b_v) was found (122). They result from variants 10a, 10_v1, and 10b, respectively, by an in-frame skipping of exon 4. This exon encodes part of ECL2, TM3, and the first eight amino acids of the intracellular COOH terminus. For claudin-18, two further murine isoforms (A1.2, NP 001181851; A2.2, NP 001181852) are listed in the NCBI database. They are truncated versions of isoforms A1.1 and A2.1, respectively, lacking exon 5 and therefore 56 COOH-terminal amino acids due to a 12-bp insertion derived from the 5′ end of intron 4 (269). The very short COOH terminus does not possess a PDZ binding motif, and the physiological relevance of these truncated isoforms is unknown. In human salivary glands, Kriegs et al. (194) report the presence of a claudin-16 splice variant that consists of exons one and five of CLDN16. Skipping exons 2–4 causes a frame shift so that the resulting isoform consists only of the intracellular NH2 terminus and part of ECL1 plus 11 further amino acids that are not contained by the full-length claudin-16.
4. Splicing of the 5′ untranslated region
Finally, splice variants within the 5′ untranslated region have been reported for claudin-2, -5, -7, -12, -14, and -15; however, the resulting proteins are unaffected. The physiological roles of these splice variants are unknown.
D. Claudins as Members of the PMP-22/EMP/MP20/Claudin Superfamily
Claudins belong to the greater PMP-22/EMP/MP20/Claudin (pfam00822) family which itself is part of the cl15676 superfamily. PMP-22 (peripheral myelin protein 22) is a protein necessary for peripheral nerve myelin formation and is found to be highly expressed in Schwann cells. Defects in PMP-22 cause peripheral neuropathies. Overexpression of PMP-22 in MDCK cells causes an increase in transepithelial resistance (307), whereas PMP-22 knock-out causes a loss of the focal distribution of juxta-paranodal K+ channels and of the adhesion junction protein E-cadherin (266), indicating a role for PMP-22 in both gate and fence function. Not much is known about the function of EMP-1 to -3 (epithelial membrane proteins 1 to 3). EMPs are reported to be involved in cell growth, cell cycle, and cell differentiation and to play a role in tumorigenesis (for review, see Ref. 168). MP20 (lens fiber membrane intrinsic protein, also called MP17, MP18, or MP19) is one of the most abundant proteins within the lens fiber membrane and missense mutations in its gene, LIM2, cause autosomal recessive congenital cataract (294). During lens differentiation, membrane insertion of MP20 correlates with the formation of an extracellular diffusion barrier (114). Using single particle electron microscopy, Gonen et al. (102) found that in the absence of divalent cations, MP20 formed tetramers, whereas in the presence of divalent cations these tetramers associated into higher-order species. Three-dimensional reconstructions of MP20 particles resulted in four distinct types, and it is intriguing to speculate that claudins might associate in a similar way. Further members of the pfam00822 family are the gamma subunits of voltage-dependent calcium channels, CACNG1 to -8. A subset of the CACNGs, γ2 (stargazin), γ3, γ4, and γ8, are subsumed as TARPs (transmembrane AMPA receptor regulatory proteins), as they affect the electrical properties of AMPA receptors, as well as their trafficking and mediate their anchoring by interacting with the synaptic scaffolding protein, PSD-95, through their COOH-terminal PDZ-binding motif (for review, see Ref. 311).
All of the pfam00822 proteins are characterized by four transmembrane regions. In contrast to claudins and CACNGs, the COOH termini of PMP-22, EMPs, and MP20 are short and do not appear to posses PDZ binding motifs. Furthermore, ECL1 of PMP-22 and EMP-1 contains one and two consensus sequences for N-linked glycosylation, respectively, and glycosylation has been experimentally verified (for review, see Ref. 168). Unifying features of the family are the presence of a PMP22/claudin-domain with the consensus pattern [LIVMF]-[LIVMFC]-[LIVMF](2)-[SA]-[TL]-x(2)-[DNKS]-x-W-x(9,13)-[LIV]-W-x(2)-[CG] (signature 1) and [RQ]-[AVS]-x-[MC]-[IV]-l-[SA]-x-[LI]-x(4)-[GSA]-[LIVMF]-[LIVMFS]-[LIVMF] (signature 2, prosite PDOC00939). Signature 1 in part overlaps with the consensus sequence of the claudin family, [GN]-l-W-x(2)-C-x(7,9)-[STDENQH]-C (prosite PS01346, bold marks overlap, see FIGURE 1). Members of the pfam00822 family have also been found in lower invertebrates. In Drosophila, the three pfam00822 family members megatrachea, sinuous, and kune-kune are directly involved in septate junction organization and epithelial barrier function (29, 264, 409). The function of Caenorhabditis elegans PMP22/claudin-domain proteins in cell adhesion and polarization has recently been reviewed by Simske and Hardin (325). In yeast, there are reports for several “claudin-like” proteins that are, however, not classified as pfam00822 family members by the NCBI database. These proteins are involved in membrane organization [Sur7p (8, 71, 415); Dni1p (55)] and processes leading to membrane fusion [Dni1p (55)] and Ca2+ influx during mating [Fig1p, (3, 82)].
Figure 1.
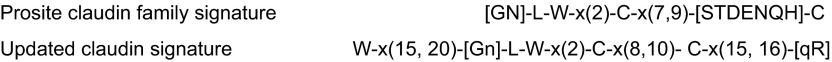
Claudin family signature sequence in the predicted 1st extracellular loop. Top: Prosite pattern (accession no. PS01346), which is 89% specific for claudin family members (323). Ambiguities are indicated by listing acceptable amino acids at a given position between square parentheses, “x” is used for a position where any amino acid is accepted, with the range in the number of repetitions indicated in round parentheses. Bottom: updated version to include two other highly conserved residues located upstream and downstream of the classic signature. In addition to standard Prosite syntax, we use uppercase letters to indicate the more frequent residue at a position of ambiguity and lowercase to indicate the rarer one.
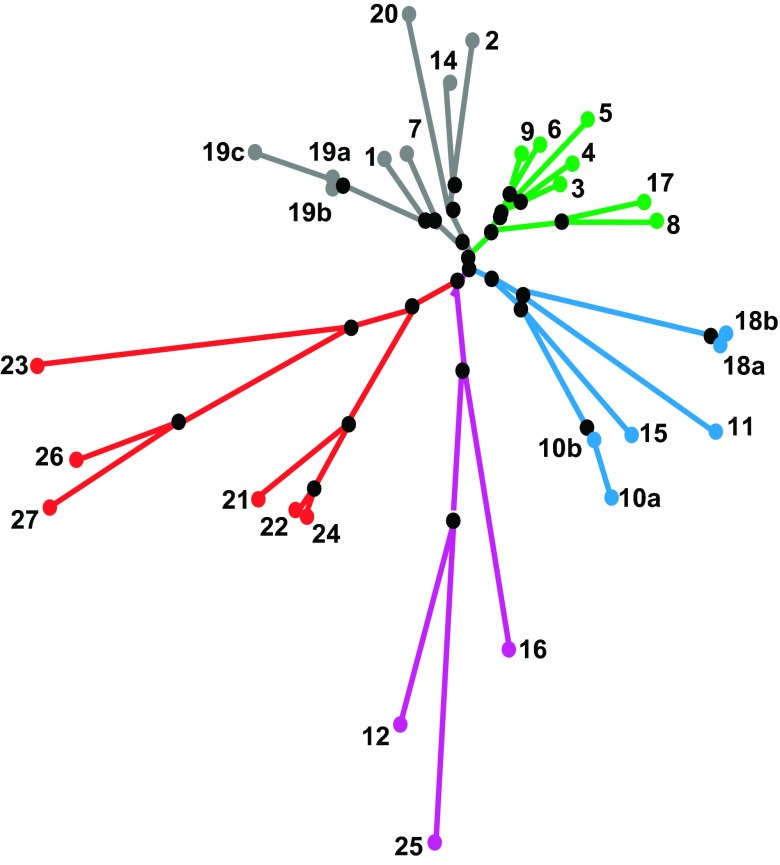
A great variety of different phylogenetic trees has been proposed for pfam00822 family members (e.g., Refs. 51, 164, 224, 264, 311, 325, 342, 366, 385, 409) and specifically for claudins (138, 148, 190, 193, 200, 218, 246, 350), based on sequence similarity either within one species or from various species. Günzel and Fromm (120) recently compared different claudin classification concepts and suggested a phylogenetic tree that sorts human claudins into eight subgroups, which form four major clusters: cluster I (subgroups A/B) claudin-3, -4, -5, -6, -9/claudin-8, -17; cluster II (subgroups D/E) claudin-1, -7, -19/claudin-2, -14, 20; cluster III (subgroups F) claudin-10, -11, -15, -18; and cluster IV (subgroups C/G/H) claudin-21, -22, -24/claudin-12, -16, -25/claudin-23, -26, -27 (FIGURE 2).
Figure 2.
Phylogenetic tree of the 26 human claudin genes. [From Günzel and Fromm (120). Copyright 2012, with permission from John Wiley & Sons, Inc.]
IV. EXPRESSION OF CLAUDINS
A. Epithelial Tissues
Claudins are expressed in all known epithelial tissues. Furthermore, in all epithelia there appear to be multiple different claudins expressed simultaneously.
1. Kidney
TABLE 2 summarizes the distribution of claudins along the mammalian nephron. In the adult glomerulus, visceral epithelial cells (podocytes) form a specialized intercellular junction, the slit diaphragm. True tight junctions do form between immature podocytes in the fetal glomerulus and are of uncertain functional significance (149, 300). These disappear during development, but can reappear during nephrotic states, coincident with effacement of the foot processes and obliteration of the slit pore (44, 295, 309). Tight junction proteins such as occludin, JAM-A, cingulin, and ZO-1 are known to be expressed at the base of the slit diaphragm (94). Claudin-5 is the major claudin expressed in the glomerulus, as assessed by quantitative real-time PCR, and is found throughout the plasma membrane of podocytes (189). Claudin-6 has also been detected in podocytes, where it is found at the basal membrane, and at the base of the slit diaphragm (423). In experimental nephrosis induced by puromycin, claudin-6 is upregulated and concentrated at the apparent tight junctions between podocytes; claudin-5 is also recruited to the same location, although its overall expression level is not upregulated. The parietal epithelium of Bowman's capsule, which acts as a barrier to macromolecules (277), consists of squamous cells whose tight junctions express claudin-1 (186, 278).
Table 2.
Claudin expression along the nephron
| Nephron Segment | Claudins | Reference Nos. |
|---|---|---|
| Glomerulus | 5, 6 (podocyte); 1 (parietal epithelium) | 186, 189, 423 |
| Proximal tubule | 2, 10a, 17 (adult); 6, 9 (neonate) | 1, 79, 122, 186, 373 |
| Thin descending limb, upper segment | 2 | 79, 186 |
| Thin descending limb, lower segment | 7, 8 | 211 |
| Thin ascending limb | 3, 4, 16, 19 | 14, 186 |
| Thick ascending limb | 3, 10a, 10b, 16, 18, 19 | 14, 122, 144, 186, 191, 324 |
| Macula densa | 10 | 373 |
| Distal tubule, connecting tubule, collecting duct | 7, 8, 10 | 7, 211, 373 |
| Collecting duct | 3, 4, 7, 8, 10a (cortical CD) or 10b (medullary CD), 14, 18 | 30, 122, 144, 186, 211, 373 |
CD, collecting duct.
The primary role of the proximal tubule is bulk reclamation of solutes that evade the glomerular filter. Na+, K+, Ca2+, Mg2+ and Cl− are thought to be reabsorbed, at least in part, paracellularly. The claudins that have been found to be expressed in the proximal tubule are claudin-2 (79, 183, 186), claudin-10a (122, 183, 186, 261, 373), and claudin-17 (196). Antibodies to claudin-11 were initially reported to stain proximal tubules from mice (186) and humans (183), but the data in mice are now in doubt because the antibody used was later found to cross-react with claudin-10 (261). The neonatal proximal tubule, which has a lower chloride permeability than the adult (297), also expresses claudins 6 and 9 (1).
The thick ascending limb of Henle is a quantitatively important site for paracellular reabsorption of the divalent cations Ca2+ and Mg2+ (123). In addition, some of the Na+ reabsorbed in the medullary part of this nephron segment is also thought to occur paracellularly, thus enhancing the metabolic efficiency of transcellular Na+ reabsorption. The thick ascending limb is known to express claudins 10, 14, 16, 18, and 19 (14, 103, 144, 186, 191, 324, 373), and possibly 3 (144, 186). Claudin-16 (also known as paracellin) and claudin-19 are of particular interest because mutations in these genes cause familial hypercalciuric hypomagnesemia with nephrocalcinosis (FHHNC), an autosomal recessive disorder characterized by renal Ca2+ and Mg2+ wasting (191, 324) (see below). The aldosterone-sensitive distal nephron, which encompasses the distal convoluted tubule, connecting segment and collecting duct, is the last section of the nephron and is responsible for fine-tuning the urinary composition. It expresses claudins 3, 4, 7, 8, 14, and 18 (30, 169, 245, 365, 418). It also expresses claudin-10 (373), predominantly 10a in the cortical collecting duct and 10b in the outer and inner medullary collecting duct (122).
The urinary bladder epithelium is probably the tightest mammalian epithelium (transepithelial resistance 75,000 to >100,000 Ω·cm2; Ref. 209, for review see Ref. 208). mRNA of five claudins was detected in mouse bladder tissue (claudin-2, -4, -8, -12, and -13), but only three of these claudins, claudin-4, -8, and -12, were localized within the tight junctions of uroepithelial cells (2).
2. Gastrointestinal tract
In the rat stomach, the expression of claudins 2–5 has been examined (298). Claudin-3 is most strongly expressed in the surface epithelial cells of the stomach, predominantly along the basolateral membrane, while claudin-4 is expressed mainly at the tight junction in proximal gastric glands, and claudin-5 is uniformly expressed from the base of the glands to the surface and, like claudin-3, is located on the basolateral membrane. Claudin-2 was not detected in the stomach. In addition, claudins 12 (138) and 23 (178) as well as claudin-18, splice variant 2 (269), are also known to be highly expressed in human stomach.
Expression of claudins 1–19 has been examined throughout the rat and mouse intestine (91, 139, 298) and of claudins 20–24 in the mouse upper small intestine (344). All claudins were detectable by RT-PCR except 6, 16, 19, 22, and 24. Claudins 2, 3, 7, and 15 are the most highly expressed. Some claudins exhibit a striking regional distribution. Claudin-8 expression increases progressively from small intestine to colon, while claudin-15 decreases along the same axis. Expression of claudins 2, 5, 7, and 10 peaks in the ileocecal region, claudin-13 was only detectable in the colon, and claudin-18 was only in the duodenum and jejunum. Several claudins displayed selective expression along the crypt-villus axis. Claudin-2 is confined to the deep crypts (91, 230, 298). Claudin-10 and -15 are expressed throughout the epithelium in small intestine but are localized to crypts in the colon (91, 139). By immunofluorescence detection of the subcellular distribution, claudins 2, 8, 10, 12, 15, and 18 were found to be restricted to the tight junction while claudins 1, 3, 4, 5, and 7 were frequently found along the basolateral membranes (91, 139, 298). Western blots of human sigmoid colon biopsies have confirmed the expression of claudins 1–5, 7, and 8 (42, 420).
Developmental changes in claudin expression during neonatal life have been observed in the mouse jejunum (139). Claudin-19 was detected predominantly in the first 2 wk after birth and was no longer expressed after 28 days. Claudin-2 was expressed throughout the epithelium at high levels at birth, but decreased markedly in expression and became progressively restricted to the crypts over 90 days. In contrast, claudins 3, 4, 7, and 15 increased progressively in expression during early life, and claudin-15 was found to migrate from the crypts to the surface epithelium over this period. Expression of selected claudins in embryonic development has been examined in the chick intestine (284). Claudins 3, 5, and 16 were found to exhibit peak expression at 20 days prehatch and then decline rapidly after birth. Interestingly claudin-16 protein in this study was detected in goblet cells.
In the liver, claudin-1 is expressed at the tight junctions of hepatocytes and also along the lateral membrane of bile duct cholangiocytes (97, 125). Claudin-2 is expressed in hepatocyte tight junctions with a progressive increase from periportal to pericentral hepatocytes, claudin-3 is expressed uniformly in hepatocyte and cholangiocyte tight junctions, claudin-5 is expressed in endothelial cells of the portal veins and hepatic arteries, and claudin-4 was not detected (298). Claudin-7 is expressed in the basolateral membrane of cholangiocytes but not in hepatocytes (167). There is also evidence for liver expression of claudins 6 (424), 8 (253), 9 (424), and 14 (398). In the gall bladder epithelium, claudins 1–4 and 10 are expressed strongly and claudins 7 and 8 weakly (206, 265).
3. Respiratory tract
In the mammalian proximal airways, claudins 1, 3, 4, 5, 7, 10, and 18 (splice variant 1) have been shown to be expressed in bronchi and bronchioles (58, 170, 251, 269). Claudin-2 was not detected in one study (58) but in two others was found in cytoplasmic granules of airway epithelial cells (170, 251). Claudin-7 was found to be expressed basolaterally, as in other epithelia, and was barely, if at all, detected in the tight junction (33, 170). Claudin-10 expression appeared to be confined to Clara cells (421).
In the distal lung, claudins 3, 4, 5, 7, 8, 15, and 18 have been shown to be expressed (170, 253, 269, 303, 382). Claudins 3, 4, and 7 are predominantly expressed in alveolar type II cells (170, 382), while claudin-5 is expressed in most alveolar epithelial cells (382). Freshly isolated alveolar type II cells express claudins 3 and 5 most abundantly (47) as well as claudin-18 (405), but after primary culture for 1 wk and transdifferentiation to alveolar type I cells, claudins 4 and 7 are markedly upregulated (47). Claudin-1 could also be detected in the lungs by RT-PCR (47) and immunostaining (382) but only at very low levels.
4. Other epithelia
TABLE 3 summarizes the claudins that have been shown to be expressed in other epithelial tissues in mammalian species.
Table 3.
Claudin expression in gastrointestinal and respiratory tract and other mammalian epithelial tissues
| Tissue | Claudin | Reference Nos. |
|---|---|---|
| Stomach | 3, 4, 5, 12, 18, 23 | See text |
| Intestine | 1–5, 7, 8, 10, 12, 13 (rodent), 15, 18-2, 20, 21, 23 | See text |
| Liver | 1–3, 5–9, 14 | See text |
| Gall bladder | 1–4,10 > 7, 8 | See text |
| Respiratory tract | 1, 3–5, 7, 10, 18-1 (proximal); 3–5, 7, 8, 15, 18-1 (distal) | See text |
| Epidermis | 1, 4, 7 > 3, 5, 8, 11, 12, 17 | 38, 160 |
| Eye | 1, 4, 7 (cornea & conjunctiva); 10 (conjunctiva) | 414 |
| Salivary gland | 10 > 1, 2, 3, 4, 7, 8, 12 | 112, 229, 288 |
| Mammary gland | 1–5, 7, 8, 15, 16 | 33, 34, 166, 231 |
| Taste bud | 4, 6, 7, 8 | 244 |
| Exocrine pancreas | 1–5, 7 | 61, 298 |
| Retinal pigment epithelium | 19 > 3, 10 (human); 1 > 3 (rodent) | 286, 410 |
| Choroid plexus | 1, 2, 5, 11 | 217, 254, 402 |
| Cochlea | 1, 2, 3, 8, 9, 10, 12, 14, 18 (Organ of Corti & striae vascularis marginal cells); 11 (striae vascularis basal cells) | 185 |
| Ovary | 1, 5 | 427 |
| Prostate | 1, 3, 4, 5, 7, 8, 10 | 314 |
| Epididymis | 4, 7 > 2, 5, 10 | 72 |
| Seminiferous tubule | 3, 5, 11 | 254, 257 |
| Urinary bladder | 4, 8, 12 | 2 |
B. Endothelia
Vascular endothelial cells also have tight junctions and express multiple claudins. Claudin-5 is by far the predominant claudin (255, 270), but claudins 1 (212), 3 (401), and 12 (270, 279) have also been reported. In a purified preparation of brain capillary endothelial cells, claudins 10 and 22 were also reported to be expressed at significant levels (280). In brain capillary endothelial cells, claudin-5 mRNA levels are almost 600-fold higher than claudin-3 (280). In general, the number of claudin isoforms that have so far been identified in endothelia is far less than those in epithelia, suggesting that there are probably going to turn out to be a number of epithelium-specific isoforms, including most of the pore-forming claudins.
C. Other Tissues
In addition to epithelia and endothelia, claudins are also found in a variety of other cell types. Claudin-11 and -19 are expressed in interlamellar strands of myelin sheaths in the central and peripheral nervous system, respectively, where they serve to insulate myelinated nerves and facilitate nerve conduction (108, 249, 254). Claudin-4 is expressed in pancreatic islet cells (298), which have tight junction-like strands on their cell surface (282). In mouse, claudin-13 is expressed in hematopoietic tissues, including the bone marrow, thymus, and spleen (350). Claudins have also been described in lymphocytes and monocytes (225), dendritic cells (197, 428), thymocytes (181), osteoblasts and osteoclasts (216, 404), astrocytes, and even neurons (73, 304) under certain situations.
D. Subcellular Localization
In almost all cells, multiple claudin isoforms are expressed simultaneously at the tight junction, and distributed among all the faces that form cell-cell contacts. Interestingly, though, Dieter cells (DC) and outer hair cells (OHC) in the organ of Corti form a hybrid tight junction/adherens junction in which claudin-14, claudin-6/9 partition into distinct subdomains of the junctional complex (273). Furthermore, claudin-14 is restricted to heterotypic DC-OHC contacts and is absent from homotypic DC-DC contacts. This hierarchy of claudin localization seems to be unique to this junction.
As mentioned above, claudins are also frequently found outside the tight junction, usually at the lateral membrane, with claudin-7 being a particularly striking example. Non-tight junction claudin localization has three general functions: 1) formation of unconventional adhesive cell contacts. For example, claudin-1 mediates contacts between dendritic cells and the epidermis (197), and claudin-2 mediates contacts between metastatic breast cancer cells and hepatocytes (339). 2) Interaction with other cell surface receptors such as other cell adhesion molecules (199), tetraspanins (131, 192, 199, 355), integrins (70, 355), and even the T cell receptor (181). 3) Intracellular signaling: for example, claudin-18 in osteoclasts inhibits ZO-2 signaling and thereby inhibits osteoclast differentiation and bone resorption (216). There are also reports of nuclear localization of claudins (85), suggesting the intriguing possibility that, like other junctional proteins such as ZO1/ZONAB, ZO2, and beta catenin, they may be more directly involved in regulating gene expression, perhaps in the context of cancer development. However, their actual role, if any, in the nucleus is still unknown.
V. ROLE OF CLAUDINS IN PARACELLULAR BARRIER AND PORE FORMATION
A. Evidence That Tight Junctions and Claudins Regulate Paracellular Permeability
Evidence for the role of tight junctions as intercellular barriers was historically based on observations from two different fields: electron microscopy and electrophysiology. Conventional transmission electron microscopic images of sections through epithelial cell layers showed regions within the lateral space, close to the apical pole of the cells, where the membranes of neighboring cells appeared to fuse in a series of “kissing points” (54, 84; for review, see Refs. 68, 252, 357). Freeze-fracture electron microscopy revealed a complex network of tight junction strands; after the discovery of tight junction proteins, immuno-gold staining proved that it is these proteins that constitute tight junction strands (90, 95, 418). Knowledge of the molecular identity of tight junction proteins also opened ample possibilities to apply further optical methods such as immunofluorescence staining or direct fusion with fluorescent proteins combined with confocal laser scanning microscopy and, recently, spectral position determination microscopy (179). Transfection of nonpolarized cells (fibroblasts, HEK293 cells) with tight junction proteins demonstrated that claudins are able to spontaneously form tight junction-like strands in contacts of these cells, whereas other tight junction proteins, such as occludin, are not (95, 99, 291).
Electrophysiological studies on epithelial layers demonstrated that formation of tight junctions is accompanied by an increase in transepithelial resistance. Direct correlation of transepithelial resistance with the number of tight junction strands observed in freeze-fracture images indicated that there is a logarithmic relationship, rather than the linear relationship expected from simply adding unit resistances for each strand in series (53, 54, 150, 223). Claude (53) interpreted this relationship in terms of an open probability of tight junction strands which she estimated to be 0.4.
Several studies, however, found similar numbers of tight junction strands in preparations that greatly differed in transepithelial resistance (96, 104, 332). Furthermore, diffusion potential measurements across epithelia demonstrated that the high paracellular conductance of some low-resistance epithelia was charge-selective (cation preferring: Refs. 87, 113, 213; anion-preferring: Ref. 327). In addition, flux studies with charged and uncharged solutes of different sizes showed that the paracellular pathway had two different size limits (135, 187). Interestingly, the radius of the small pore was very similar in Caco-2 and T84 cells (4.3 Å; Ref. 387), even though transepithelial resistance of T84 cell layers was almost a factor of 10 larger than that of Caco-2 cells. These observations indicate that a low resistance does not necessarily mean an “imperfect” TJ but that conductance for certain ions may be a built-in property of TJ strands.
In 1977, Diamond (69) coined the expression “gate” to refer to the ability of tight junctions to specifically regulate paracellular transport. The expression gate includes two aspect of paracellular barrier function: 1) the ability of tight junctions to restrict paracellular diffusion processes, and 2) the aspect of paracellular channel function, i.e., the ability to selectively allow paracellular transport of certain ion species. The concept of claudins forming paracellular pores was put forward almost immediately after their discovery (99, 324, 403). Shortly afterwards, claudin-2 was demonstrated to act as a paracellular cation channel (10, 96). The ability of certain claudins to form either paracellular barriers or act as paracellular ion channels will be further discussed in the next sections.
B. Barrier- Versus Pore-Forming Claudins
When considering barrier or channel properties of a certain claudin, it has to be kept in mind that all claudins that insert into the cell membrane and interact with claudins from neighboring cells contribute towards a barrier, if compared with the situation of cells without any tight junctions. In that respect, all claudins are barrier-forming proteins. However, if there is already a preexisting TJ, then barrier-forming claudins should cause a further increase in transepithelial resistance, whereas pore-forming claudins should cause a decrease when inserted into this TJ. Conversely, knock-down of a barrier-forming claudin should cause a decrease in transepithelial resistance, knock-down of a pore-forming claudin an increase. Specific pore formation through claudins has to be further distinguished from the formation of an nonspecific paracellular leak. Thus a pore should specifically increase paracellular permeability either for molecules of a certain size or of a certain charge (or both) but leave the epithelial barrier function against macromolecules intact.
So far, only few claudins unequivocally qualify as pore-forming claudins: claudin-2, -10b, and -15 as cation pores and claudin-10a and -17 as anion pores (10, 122, 159, 196, 343, 364, 373, 416). Others have been reported to form pores only when specifically interacting with another claudin. Thus a combination of claudin-16/-19 has been reported to act as cation pore and claudin-4/-8 as anion pore (145, 146). TABLE 4 summarizes our current knowledge of the permeability properties of claudin family members.
Table 4.
Putative ion permeability characteristics of claudin isoforms
| Claudin | Reference Nos. |
|---|---|
| Cation-selective claudins | |
| Predominantly pore-forming (↑permeability to cations) | |
| Claudin-2 | 10, 96, 416 |
| Claudin-10b | 373 |
| Claudin-15a | 57, 159, 343, 364 |
| (Claudin-16)b | 119, 143, 152, 180 |
| Predominantly barrier-forming (↓permeability to anions) | |
| Claudin-7 | 6, 7 |
| Claudin-19c | 145 |
| Anion-selective claudins | |
| Predominantly pore-forming (↑permeability to anions) | |
| Claudin-7 | 142 |
| Claudin-10a | 373 |
| Claudin-17 | 196 |
| Predominantly barrier-forming (↓permeability to cations) | |
| Claudin-1 | 160, 238 |
| Claudin-3 | 245 |
| Claudin-4d | 142, 146, 365 |
| Claudin-5 | 395 |
| Claudin-6 | 317 |
| Claudin-8d | 16, 418 |
| Claudin-9 | 317 |
| Claudin-11d | 364 |
| Claudin-14 | 30 |
| Claudin-18-2 | 169 |
Based on in vitro overexpression or knockdown studies in cultured cell lines. We assume that permeability and selectivity are properties intrinsic to individual claudin isoforms and ignore the possible confounding effect of heteromeric interactions between isoforms. Pore-forming claudins refer to those that predominantly decrease transepithelial resistance (TER) or increase solute permeability, while barrier-forming claudins refer to those that predominantly increase TER or decrease solute permeability. The distinction is somewhat arbitrary since most claudins probably have some finite permeability to most solutes, and the observable phenotype is highly dependent on the properties of the host cell line (16, 364).
Acts as a Cl− barrier in MDCK II cells but as a Na channel in LLC-PK1, MDCK I cells and in mouse intestine.
Conflicting data as discussed in detail in section VI.
Conflicting data also exist suggesting claudin-19 can act as a Na+ barrier (14).
Acts as a Na+ barrier in MDCK II cells but as a Cl− pore in LLC-PK1 and/or collecting duct cells.
C. Charge Selectivity of Claudins
Claudins, whether acting predominantly as barriers or pores, are capable of altering the charge selectivity of the paracellular conductance. This selectivity is thought to be conferred by charged sites within the paracellular pathway formed by the claudin. Electrophysiological measurements, such as diffusion potential measurements (“dilution” and “bi-ionic” potentials, Ref. 27) have been the principal approach to investigate charge selectivity properties. The theoretical basis for ion channel selectivity was first developed by Eisenman (77) to describe the equilibrium ion selectivity of glass electrodes. It was soon found that these concepts could also be applied to the interaction of permeating ions with electrostatic sites in biological membranes. In Eisenman's theory, ion selectivity is dependent on the thermodynamic equilibrium between ions that are hydrated in free solution and dehydrated ions that can enter the pore and be stabilized by interaction with electrostatic binding site(s). He deduced that at electrostatic binding sites with low electrical field strengths, the equilibrium is dominated by dehydration energies and hence favors large, weakly hydrated ions, while high field strength binding sites favor smaller dehydrated ions because of their higher binding energies. Thus permeability sequences for monovalent cations parallel their hydrated ion radius for weakly interacting pores (permeability PLi < PNa < PK < PRb < PCs, Eisenman sequence I), whereas they parallel their dehydrated ion radius for strongly interacting pores (PCs < PRb < PK < PNa < PLi, Eisenman sequence XI).
Eisenman sequences were determined in the 1970s in leaky epithelia such as rabbit gallbladder (27, 406) and cultured kidney tubule cell lines (45), and it was already concluded that there must be a charge-selective paracellular permeability with strong electrostatic interaction sites. Similar results were found by Tang and Goodenough in their reinvestigation of paracellular permeability properties (346). Claudins are now known to be largely responsible for these properties. Amasheh et al. (10) overexpressed claudin-2 in MDCK C7 cells, which normally exhibit a PNa/PCl ∼1, and observed that PNa/PCl greatly increased to values >5. The increase in permeability was accompanied by a drop in transepithelial resistance (10, 96), indicating that the presence of junctional claudin-2 caused the formation of cation-selective channels sufficient to transform a “tight” tight junction into a leaky one. Yu et al. (416) determined the claudin-2-induced permeability sequence in MDCK I cells as PK > PRb > PNa > PLi >> PCs, resembling Eisenman sequences V–VIII. This supported the idea that the cation selectivity of claudin-2 was conferred by a moderately strong negatively charged binding site. Transfection of MDCK C7 cells with claudin-10b resulted in an increase in Eisenman sequence from IV in vector transfected control cells (PK > PRb > PCs > PNa > PLi) to X (PNa > PLi > PK > PRb > PCs), consistent with the presence of a strong interaction site in that claudin. In contrast, the anion channel claudin-17 showed less interaction between channel and permeating ion than the vector transfected MDCK C7 control cells, as it changed the permeability sequence observed under control conditions (PF > PCl > PBr > PI) to PCl > PBr > PJ > PI (196).
There is now strong evidence to suggest that the charge selectivity of claudin-based ion channels depends mainly on certain charged amino acids within the first extracellular loop (see sect. VII).
D. Water Selectivity of Claudins
To date, claudin-2 is the only claudin that has been demonstrated to enhance paracellular water permeability upon overexpression in MDCK C7 cells (306). Neither overexpression of claudin-10b nor of claudin-17 altered water permeability of MDCK C7 cell layers (196, 306). Interestingly, not only were osmotic gradients able to drive water across claudin-2 transfected cell layers, but also NaCl gradients that were osmotically compensated by the addition of mannitol. Conversely, water movement elicited by osmotic gradients was accompanied by Na+ flux (306), suggesting that water and Na+ were transported through the same pore. That claudin-2 is permeable to water as well as Na+ is not that surprising given that its pore radius is 3.25–4 Å (see below, Refs. 370, 416) and the diameter of a water molecule is 2.8 Å. What is unclear is whether this property plays any important physiological role or whether it is simply an unavoidable leak that is a consequence of the size of the cation pore.
E. Size Selectivity of Claudins
Most studies of tight junction permeability have found the presence of two populations of pores, corresponding to a small pore or restrictive pathway, that is permeable to small ions and neutral solutes, and a much larger pore or nonrestrictive pathway (sometimes referred to as the “leak” pathway) that is permeable to macromolecules. Tight junctional pore size was first investigated in Caco-2 cells and two types of pores were found (4.3–4.6 Å and 14.6 Å in radius, Refs. 187, 387). In a different approach, Guo et al. (124) modeled permeabilities for rat proximal tubule epithelium. They suggested a large slitlike pore type with a height of 19.6 nm, possibly resulting from TJ strand breaks, and a small circular pore type with a pore radius of 6.68 Å. They explained the larger dimensions by the fact that they took hydrodynamic resistance due to the pore walls into consideration in their calculations, whereas such interactions were neglected in other approaches. In accordance with Guo et al. (124), Saitoh et al. (312) estimated the TJ pore radius of Caco-2 cells to be 7 Å.
Watson et al. (387) used polyethylene glycol (PEG) profiling, i.e., simultaneous flux measurement of 24 PEGs of different size, to investigate alterations in paracellular pore size induced by EGTA and sodium caprate in Caco-2 and T84 cell layers. They found that EGTA increased pore size, whereas sodium caprate increased pore number but did not affect pore size. In a subsequent study Watson et al. (386) investigated effects of IFN-γ on T84 barrier function. Again, they found evidence for two different pore populations (restrictive pathway, pore radius ∼4.5 Å; nonrestrictive pathway, pore radius >23 Å). IFN-γ increased the abundance of the larger pore population but had no effects on the pores with smaller diameters (386). Van Itallie et al. (370) used a similar profiling technique to investigate pore sizes in five different epithelial cell lines and in porcine ileum and estimated an apparent pore size of ∼4 Å radius for the smallest pore population in all preparations. All preparations had a further population with a pore radius >>8 Å. However, in ileum and Caco-2 cells there was evidence for a third population with a radius of ∼6.5 Å.
Claudins seem to be mainly responsible for the small pore (∼4 Å) pathway. Overexpression of claudin-2 in MDCK II as well as MDCK C7 cells (cells with and without endogenous claudin-2 expression, respectively) increased the abundance of this pore population (370). However, knock-down of claudin-2 in MDCK II cells did not alter pore abundance, although it effectively increased transepithelial resistance. Neither was pore abundance affected by the overexpression of claudin-4, -14, or -18, conditions under which transpithelial resistance was increased. Finally, mannitol permeability was found to be unaffected by claudin-2 overexpression, indicating that mannitol, with a radius of 4.2 Å is not able to pass through claudin-2-based pores (370). The lack of correlation between transepithelial resistance and PEG or mannitol permeability indicates that there may be several populations of small pores in parallel and that some pore types effectively exclude uncharged molecules.
In an alternative approach, Yu et al. (416) estimated the pore radius of claudin-2-based pores to be ∼3.25 Å in their narrowest point. In contrast, claudin-17-based pore diameters have been estimated from permeabilities for different organic anions to be ∼5 Å (196). Comparison of pyruvate permeabilities of claudin-17- and claudin-10a-transfected cell layers suggest that claudin-10a-based pores are considerably narrower. Pyruvate dimensions have been reported to be 4.09 × 5.73 × 6.82 Å [effective radius = 0.5 × (4.09 × 5.73 × 6.82)1/3 Å = 2.71 Å; Ref. 310]. Whereas claudin-17 greatly increased both pyruvate and NO3− permeability (196), claudin-10a increased NO3− permeability but decreased pyruvate permeability (122), when overexpressed in MDCK C7 cells.
F. Role of Background in Phenotype of Claudin
In many cases it is impossible to allot a claudin to the group of either pore- or barrier-forming TJ proteins, because effects of overexpression or knock-down on transepithelial resistance differ in different preparations. Obviously, in these cases, properties depend on the TJ protein background within the chosen cell systems. Several mechanisms may underlie such behavior.
1. Masking
Insertion of a pore-forming claudin into an originally leaky TJ, or of a barrier-forming claudin into an originally tight TJ may prevent detection of specific properties. Thus the cation pore-forming properties of claudin-10b were obvious in MDCK C7 cells which form very tight TJs, whereas they could not be detected in MDCK II cells which exhibit high endogenous expression of the cation pore-forming claudin-2 (122, 373). In contrast, anion pore formation of claudin-10a was obvious in the cation-preferring MDCK II cells but only just detectable in MDCK C7 cells (122, 373).
2. Displacement
Overexpression of one claudin may cause displacement of an endogenous claudin from the TJ. If the displaced claudin has a strong effect on ion permeability, the observed changes may be (erroneously) attributed to the overexpressed claudin. Displacement was, for example, responsible for at least part of the tightening effects of claudin-8 which was observed upon overexpression in MDCK II cells (15). In a subsequent study it was demonstrated that claudin-8 had displaced claudin-2 from the TJ and that the removal of this pore-forming claudin had caused at least part of the observed tightening (16).
3. Specific interaction
Recently, evidence has been presented that some claudins specifically need another claudin to translocate to and insert into the TJ and/or to be fully functional within the TJ. Two such pairs have been described: claudin-16/-19 (144, 145), which together form a cation pore, and claudin-4/-8 (146), which form an anion pore. Studies in cell culture as well as in a knock-down mouse model suggested that claudin-16 is only inserted into the TJ, if claudin-19 is also present and vice versa (144, 145). As described below, however, these findings are controversial. For two other pairs of potentially interacting claudins, claudin-10a/-10b and claudin-10a/-10a_i1, interaction is not necessary for correct insertion into the TJ but solely modulates existing pore properties (122). Thus claudin-10a and -10b, when interacting through their extracellular loops in coculture experiments, still form cation channels, but these channels fail to remove the hydration shells of the permeating ions, resulting in Eisenman sequence I for monovalent cations. Claudin-10a_i1 slightly alters the anion pore properties of claudin-10a, preventing the restriction of the pore for organic anions (122).
G. Gating of Claudin Pores
A fundamental property of transmembrane channels at the molecular level is gating of the pore. This constitutes a mechanism by which channel permeability can be regulated by voltage, extracellular ligands, and intracellular or intramembrane signals. Whether paracellular claudin pores also exhibit gating is unknown. Claude found a logarithmic relationship between the number of TJ strands and the transepithelial resistance (TER). She inferred that there might be individual channels in each strand that gate, with each have a finite probability of being open (53). However, subsequent studies have not found a consistent relationship between the number of strands and TER. With the discovery of claudins, the idea that the permeability properties of the individual claudins determined TER became increasingly accepted. However, the question of whether claudins exhibit gating remained unanswered. The field has been hampered by the lack of a method to assess claudin permeability at the single molecule level. Weber et al. recently presented preliminary data showing that they can patch clamp the apical membrane in the region of the TJ and identify stochastic conductance increases consistent with gating of individual claudin channels (391). This promises to be a powerful method to study regulation of claudin permeability by modulation of its gate.
H. Role of Claudins in Nonselective Leak
Two major hypotheses have been put forward to explain paracellular permeability or leak of macromolecules. The first hypothesis is that TJ strands occasionally exhibit breaks across which macromolecules are able to diffuse from TJ mesh to TJ mesh and then reseal, without the necessity to permanently open up a large gap across the entire TJ system (12, 96). Indeed, such dynamic breaking and reannealing has been observed in tight junction-like strands that form in fibroblasts transfected with claudin-1, but whether this happens physiologically in native epithelia is unknown (318). A corollary of this is that claudins which increase strand dynamics and favor the appearance of gaps could perhaps increase permeability to macromolecules. Findings that suggest that this hypothesis holds true may come from electron microscopic studies employing lanthanum as an electron-dense paracellular marker. Images from several publications show that lanthanum is able to pass several tight junctional “kissing points” before it is stopped, often only at the apical-most TJ strand when added basolaterally (86, 101, 263). One uncertainty in the interpretation of these data is that the possibility that La3+ might pass through the specific claudin pore pathway is not excluded. Its unhydrated radius is 1.15 Å and thus small enough to pass these channels, and even its hydrated radius is close to the estimates for claudin-based pores (4.52 Å, Ref. 268). Interestingly, the fact that La3+ usually does stop at the apical-most strand may indicate that composition of strands differs within the basal to apical span of a TJ, but to date there are no experiments that prove such differences between the strands of one TJ.
An alternative hypothesis is that the leak occurs at the tricellular junction. This is based on the observation that moderate overexpression of tricellulin specifically reduces paracellular permeability to macromolecules, but not to small inorganic ions. Under these conditions, tricellulin displays a strictly tricellular distribution. The lack of effect on ion permeability can be explained by the scarcity of tricellular junctions compared with the abundant claudin-based bicellular pores that dominate ion permeability. Electron microscopy supports this hypothesis: at points where three cells meet, the TJ extends further towards the basolateral side of the cell layer and forms a tube with a length of ∼1 μm and a diameter of ∼10 nm (330). It is not yet clear whether claudins contribute towards the structure of the tricellular TJ.
In summary, it is clear that loss of TJ integrity increases the passage of macromolecules so that conditions that cause a withdrawal of claudins from the TJ, such as exposure to EGTA, increase nonselective leak (370, 387). However, whether any claudins play a specific role in paracellular permeability to macromolecules remains yet unsolved.
VI. PHYSIOLOGICAL AND IN VIVO ROLE OF INDIVIDUAL CLAUDINS
A. Claudin-1
Claudin-1 is expressed ubiquitously in most tissues of the body (95). In in vitro overexpression studies, it acts predominantly as a “barrier claudin” to increase TER (115, 160, 238), suggesting that it may play a general role in epithelial barrier function. Claudin-1 is expressed in the epidermis and clearly plays an important role in the skin barrier (38, 97). Knockout of claudin-1 in mice leads to loss of the tight junctional barrier to water and macromolecules at the stratum granulosum of the epidermis. As a consequence, these mice die of dehydration in the neonatal period (97). Downregulation of claudin-1 is also found in other skin diseases associated with dry skin, including atopic dermatitis and psoriasis (63, 388). Claudin-1 is also expressed in cholangiocytes and hepatocytes and acts as a barrier in the hepatic biliary ducts (115), presumably against bile salt leakage. Consistent with this, loss of function mutations in claudin-1 cause a human disease, neonatal sclerosing cholangitis associated with ichthyosis (125).
B. Claudin-2
Claudin-2 is highly expressed in leaky epithelial tissues, including the proximal tubule of the kidney (79, 186) and the intestinal crypts (298). Its in vitro properties are among the most well studied of all the claudins. When overexpressed in epithelial cell lines, it acts as a high-conductance, cation-permeable paracellular pore (10, 96, 364, 416). The pore has a radius of 3–4 Å (370, 416), is also permeable to water (306), and is partially permeable to and inhibited by calcium (416, 417).
In the kidney, global knockout of claudin-2 in mice leads to a reduction in the Na+ conductance of proximal tubule S2 segments and reduced net reabsorption of Na+, Cl−, and water (261). Under normal circumstances with free access to water and food, the claudin-2 null mice displayed normal appearance, activity, growth, and normal serum and urine biochemistry. However, fractional excretions of Na+ and Cl− were increased when they were challenged with a saline load. These observations suggest that claudin-2 acts as the paracellular cation pore for passive reabsorption of Na+ in the late proximal tubule, which secondarily drives Cl− and water reaborption in this segment. This seems to be necessary for maximal NaCl excretory capacity. Interestingly, claudin-2 null mice also had hypercalciuria, which suggests that claudin-2 also plays a role in reabsorbing Ca2+ paracellularly in the proximal tubule. They also had both higher urine volume and lower urine osmolality than normal littermates, which may reflect loss of water permeability.
The small intestine of claudin-2 knockout mice exhibits reduced transepithelial conductance and Na+ permeability that is more striking in infants, in whom claudin-2 is also expressed in the villus, than in adults, in whom claudin-2 is confined exclusively to the crypts (343). In adult claudin-2 knockout mice there is no overt intestinal phenotype, suggesting either that claudin-2 plays a minimal role under baseline conditions (e.g., in the absence of secretagogues) or that in its absence, other intestinal claudins (see claudin-15 below) can substitute for it.
C. Claudin-3
Claudin-3 is expressed in a wide variety of epithelia, such as respiratory, urinary and gastrointestinal tract, and salivary and mammary glands. It is also a constituent of the blood-brain and blood-testis barrier. A great wealth of studies indicate that claudin-3 acts as a barrier-forming TJ protein. Furuse et al. (96) overexpressed claudin-3 in very high resistance MDCK I cells and found no change in transepithelial resistance. Resistances in untransfected cell layers had already been extremely high (>10,000 Ohm·cm2) so that a further increase might not have been possible. However, claudin-3 overexpression did not induce any decrease in transepithelial resistance (as did claudin-2 within the same study) and due to the very high resistance of these cells, any conductance added to the paracellular pathway should have caused a breakdown of such a barrier. Thus pore formation through claudin-3 could be ruled out. Coyne et al. (58) stably expressed claudin-3 in NIH/3T3 fibroblasts and observed a reduction in paracellular permeability for extremely large molecules (2,000 kDa dextran) and the development of a small resistance across the transfected fibroblast cell layer. Milatz et al. (245) overexpressed claudin-3 in low-resistance MDCK II cells. They found an increase in transepithelial resistance that, by use of two-path impedance spectroscopy, could be solely attributed to a 9- to 15-fold increase in paracellular resistance. Conversely, siRNA-induced, as well as oxidative-stress induced decrease of claudin-3 expression in MKN28 gastric epithelial cells (132), ochratoxin A (a fungal molds toxin) induced loss of claudin-3 from tight junctions of Caco-2 cells layers (239) as well as a TRPV4 activation-induced downregulation of claudin-3 in HC11 mammary cells (301) caused a decrease in barrier function. In Sertoli cells of mouse testis, claudin-3 is transiently expressed during puberty, when the blood-testis barrier develops as judged by decreasing biotin permeability. In mice with a conditional androgen receptor knock-down, Sertoli cells lack claudin-3 expression, and biotin permeability remains high (241). Claudin-3 is also a constituent of the blood-brain barrier, and it is selectively lost from endothelial tight junction under conditions that cause a loss of blood-brain barrier function, such as autoimmune encephalomyelitis and in glioblastoma tumors (401).
In contrast, there are recent reports from alveolar epithelial cells, in which claudin-3 appears to be greatly upregulated under conditions of decreased barrier function (47, 247). When overexpressed in alveolar epithelial cells, transepithelial resistance was found to decrease and fluxes of paracellular marker substances (600 Da calcein; 10 kDa dextran) increased (247). However, during these experiments, short-circuit current (Isc) was also increased so that at least part of the observed effects are probably due to transcellular effects.
D. Claudin-4
Claudin-4 is most strongly expressed in the kidney, primarily in the collecting ducts, and in the lung. In vitro, depending on the cell type investigated, it acts either as a general barrier (243), as a Na+ barrier without affecting Cl− permeability (364, 365) or, when interacting with claudin-8, as a Cl−/anion pore (146). In the kidney, claudin-4 has been postulated to constitute the passive Cl− permeability in the collecting duct and thereby facilitate NaCl reabsorption driven by electrogenic Na+ transport through the epithelial Na+ channel, ENaC (146). In the lung, it has been suggested that claudin-4 maintains a tight alveolar Na+ barrier and so prevents leakage of fluid and electrolytes into the alveolar space. Consistent with this, treatment of mice with a COOH-terminal peptide of Clostridium perfringens enterotoxin (CPE) that removes claudin-4 (but also claudin-3) from the tight junction inhibited air space fluid clearance and exacerbated ventilator-induced pulmonary edema (405). Furthermore, in ex vivo perfused lung specimens from human donors, claudin-4 expression was positively correlated with alveolar fluid clearance (303). In the colon, there is indirect evidence that claudin-4 acts to tighten the paracellular pathway, as claudin-4 is downregulated under various conditions that cause increased permeability (for review, see Ref. 120).
E. Claudin-5
Claudin-5 is one of only three claudins that have a long NH2 terminus. Interestingly, Wang et al. (382) observed two anti-claudin-5 antibody-reactive bands in their Western blots, and only the lower band protein was upregulated in the presence of methanandamide. It is intriguing to speculate that the two bands may reflect the long and short version of claudin-5 generated by the two start codons within the CLDN5 gene. Claudin-5 is the predominant claudin expressed in endothelial tight junctions (255). Endothelial tight junctions appear to be particularly important in vascular endothelium of the blood-brain barrier, where they have been shown to be tight and impermeable to macromolecules (299), unlike the looser cell junctions found in endothelia of other tissues (175). Claudin-5 plays an important role in the blood-brain barrier to small molecules. Knockout of claudin-5 in mice allowed rapid extravasation of tracers with molecular mass below ∼800 Da into brain and spinal cord parenchyma, while in control mice these tracers were restricted to the intravascular compartment (270). Claudin-5 null mice died within 10 h of birth, but the underlying cause was unclear. Claudin-5 is also expressed in some epithelial tissues, most notably in alveolar epithelium of the lung (253, 382), but its physiological role there is unknown.
F. Claudin-6
Claudin-6 is one of the earliest molecules expressed during epithelial differentiation, and its expression is largely confined to embryonic and fetal life (1, 133, 358). In vitro, it appears to act as a Na+ and Cl− barrier when transfected into MDCK II cells (317). Tight junctions containing claudins 4 and 6 are expressed in the trophectoderm, which covers the surface of the blastocyst, and form a permeability seal. This is thought to be important for vectorial transport of ions and water to generate the blastocoel cavity (32). Consistent with this, culture of mouse embryos with CPE, which inhibited claudin-4 and -6 expression, induced breakdown of the trophectoderm barrier to FITC-dextran and inhibition of normal blastocyst formation (256). However, claudin-6 knockout does not affect mouse development, viability, and behavior (13) so that the CPE effect may be primarily due to the removal of claudin-4. On the other hand, when mice were made transgenic to induce unregulated overexpression of claudin-6 in the epidermis, they exhibited loss of the normal skin barrier to macromolecules with consequent transepidermal water loss and neonatal death (360). This indicates not only that the presence of claudin-6 is important in normal epithelial barrier development, but that the precise balance in the amount of its expression is also important.
G. Claudin-7
Claudin-7 is expressed in a variety of epithelial tissues, with the lung and the aldosterone-sensitive distal nephron of the kidney having the highest levels (7, 211, 253). In vitro studies of its function have come to conflicting conclusions. Alexandre et al. (7) found that overexpression of claudin-7 in LLC-PK cells caused a decrease in the permeability to Cl− and a simultaneous slight increase in the permeability to Na+, suggesting that it is an cation-selective claudin. On the other hand, Hou et al. (142) found that knockdown of claudin-7 in MDCK and LLC-PK cells increased Na+ permeability or decreased Cl− permeability, respectively, suggesting the opposite conclusion, that claudin-7 is anion selective. In the light of the recent discovery, that claudin-17 acts as a strong anion channel and is highly expressed in both MDCK II and LLC-PK1 cells, the discrepancies between the two studies may be secondary to potential changes in claudin-17 (196). Mice with complete knockout of claudin-7 have urinary Na+, Cl−, and K+ wasting, dehydration, and growth retardation, and findings consistent with secondary hyperaldosteronism, and die within 12 days of birth (349). Since the distal nephron, and specifically the connecting tubule and collecting duct, mediates electrogenic Na+ reabsorption, accompanied by passive paracellular Cl− transport, these findings appear to favor the hypothesis that claudin-7 acts as a paracellular Cl− pore (316). The other relatively unique feature of claudin-7 is that it is also strongly expressed at the basolateral membrane in all tissues that have been examined (58, 72, 91, 139, 211), and is known to form a complex with the epithelial cell adhesion molecule EpCAM (271). This could potentially play a role in regulating cell-cell adhesion, cell motility, and tumor progression.
H. Claudin-8
Claudin-8 is expressed in a variety of epithelial tissues, with the lung and the tight junctions of the aldosterone-sensitive distal nephron of the kidney having the highest levels (186, 211, 253). It is proposed to be an anion-selective claudin that acts either as a Na+ barrier (418) or Cl− pore depending on the cell type (146). Together with claudin-4 and -12, it forms the impermeable tight junctions of the urinary bladder (2). In collecting duct cells, claudin-8 recruits claudin-4 to form a complex at the tight junction that mediates Cl− transport (146). In vivo, this would be predicted to facilitate distal nephron NaCl reabsorption. In the colon, aldosterone-induced ENaC activation causes an upregulation of claudin-8 which seals the paracellular pathway, as indicated by the specific increase in paracellular resistance (11). Similar to the situation in the distal nephron, this sealing would prevent absorbed Na+ from leaking back into the gut lumen and thus enhance net Na+ absorption. Interestingly, upregulation of claudin-8 by aldosterone is an indirect effect that requires Na+ influx into the cells, as it can be prevented by the ENaC inhibitor, amiloride (11).
I. Claudin-9
Claudin-9 is selectively expressed in tight junctions of the cochlea (185, 262) where it separates the high K+ endolymph from the low K+ perilymph, with the latter bathing the outer sensory hair cells. In vitro overexpression of claudin-9 led to augmented Na+ and K+ barrier function (262, 317), indicating that its physiological role is to protect the outer hair cells from exposure to high K+ concentrations. Consistent with this, a random ethylnitrosourea-induced missense mutation in claudin-9 in mice led to deafness associated with high perilymph K+ concentrations and widespread loss of outer hair cells (262).
J. Claudin-10
Claudin-10 is expressed in many tissues including renal tubules (186), the vasa recta of the kidney medulla, intestine (161), salivary gland (133), and epididymis (117). Claudin-10a appears to be restricted to the kidney (122, 373) so other tissues presumably express claudin-10b. The physiological function of the two major claudin-10 isoforms was initially investigated by Van Itallie et al. (373) who found that claudin-10a acts as an anion pore. Claudin-10b had no strong ion selectivity in LLC-PK1 and MDCK II cells but was found to act as a strong cation-permeating claudin in MDCK C7 cells (122) and MDCK I cells (159). Both isoforms differ from other known channel-forming claudins, in that they are not only charge selective but also strictly size selective. Thus claudin-10b greatly increased permeability for Li+, Na+, K+, but only marginally for Cs+ (122) and not at all for a 4 kDa dextran (159) nor for water (306).
K. Claudin-11
Claudin-11 is expressed by oligodendrocytes and localized in parallel arrays of autotypic tight junction strands within the myelin sheaths of neurons (254). It is also expressed at junctions between Sertoli cells, which constitute the blood-testis barrier, and basal cells of the striae vascularis. Mice with knockout of claudin-11 have slowed nerve conduction and consequent hindlimb weakness (108). It has been suggested that claudin-11 plays occluding and resistive roles by sealing the edges of the membrane along the length of the myelin sheath and at paranodes, limiting diffusion between the interstitium and intramyelinic space. By diminishing the paracellular current pathway between membrane layers in the myelin sheath, claudin-11-based junctions decrease the capacitive charge and hence the time taken to fully charge, which favors the rapid regeneration and propagation of action potentials between nodes of Ranvier (66, 107). Male claudin-11 knockout mice are also sterile, and have loss of normal differentiation of spermatocytes (108). The blood-testis barrier has been postulated to function as a shield for differentiating spermatocytes against immune surveillance, and also to exclude blood-derived components from the adluminal space that might otherwise disrupt spermatogenesis. However, since the claudin-11 knockout mice had no autoimmune disorder, it is likely that the function of claudin-11 is primarily to act as a barrier to blood-derived small molecules. Furthermore, claudin-11 is essential to hearing as claudin-11 knockout mice displayed inner ear deafness due to a disappearance of tight junctions from the basal cells and the resulting loss of endocochlear potential (106, 185).
L. Claudin-12
Claudin-12 is an unusual member of the claudin family, as it is one of the few claudins that does not possess a PDZ binding motif. In all phylogenetic trees it appears to be only distantly related to all other claudins. Claudin-12 is expressed in tight junctions throughout the intestinal epithelia. In vitro, it increases calcium permeability and is upregulated by vitamin D (92), suggesting a role in vitamin D-dependent paracellular absorption of calcium, but its role in the whole animal has yet to be investigated.
M. Claudin-13
As already mentioned, claudin-13 is found in rodents but not in primates. In neonatal mice, claudin-13 is expressed in kidney (1), whereas in adult mice it is expressed in colon surface epithelium and in the urinary bladder (91). In the latter, however, claudin-13 is not localized within the TJ (2). Interestingly, as mentioned above, claudin-13 is also expressed in hematopoietic tissues, where it appears to be part of a stress-induced erythropoiesis pathway (350).
N. Claudin-14
Claudin-14 is expressed in the cochlea, where it is located at the tight junctions between inner and outer hair cells and supporting cells (30). The tight junctions in these cells separate the K-rich endolymph apically from basolateral compartments that contain K-poor fluid, including the cortilymph confined to the space of Nuel that bathes the basolateral side of outer hair cells. Mutations in claudin-14 cause nonsyndromic deafness in humans (398). Likewise, in mice knockout of the claudin-14 gene causes deafness that is associated with degeneration of the outer hair cells (30). In vitro, claudin-14 acts as a strong cation barrier (30). Thus it is likely that claudin-14 functions in the cochlea as a paracellular barrier to K. Loss of claudin-14 would be predicted to expose the basolateral surface of outer hair cells to abnormally elevated K+ concentrations, which may be cause cell death.
Claudin-14 is also highly expressed in the kidney (398). Although there have been suggestions that it is localized to the collecting ducts or proximal tubules (30, 78), the most extensive analysis so far has found it predominantly in the thick ascending limb (103). In this tubule segment, it interacts directly with claudin-16 and indirectly with claudin-19 (see below) to reduce their paracellular Na+ permeability and, in doing so, is thought to reduce the driving force for divalent cation reabsorption. Consistent with this, claudin-14 knockout mice were found to be hypercalciuric and hypermagnesiuric. Furthermore, high-Ca2+ diet and high extracellular Ca2+ increased claudin-14 protein levels by downregulating two microRNAs, miR-9 and miR-374, that normally suppress claudin-14 expression (103). Thus claudin-14 acts physiologically as a negative regulator of renal Ca2+ reabsorption. Carriers of common sequence variants in claudin-14 have an increased risk for kidney stones as well as reduced bone mineral density (351).
O. Claudin-15
Claudin-15 is highly expressed throughout the intestine and is found at both villus and crypt cell junctions in adults. In vitro it can behave predominantly as a Na+ channel or a Cl− barrier, depending on the cellular context (57, 159, 364). Knockout of claudin-15 in mice causes congenital enlargement of the small intestine (344). Interestingly, in zebrafish, claudin-15-mediated paracellular Na+ secretion by intestinal epithelial cells has also been proposed to play a role in development of a normal intestinal lumen (26). Zebrafish with a loss-of-function mutation in the transcription factor tcf2 have marked downregulation of claudin-15 and develop multiple intestinal lumens. However, the exact mechanism for development of megaintestine in claudin-15-knockout mice is unclear.
In adulthood, claudin-15 knockout mice also have lower small intestinal lumen Na+ concentrations and higher K+ concentrations, and impaired Na-dependent glucose absorption (343). This suggests that claudin-15 normally plays an important role in paracellular Na+ secretion, presumably driven by the transepithelial electrical potential from transcellular Cl− secretion, and in paracellular K+ absorption, presumably driven by the K+ concentration gradient established by fluid reabsorption. It was suggested that the impaired glucose reabsorption in the claudin-15 knockout mice may be due to the limiting concentration of luminal Na+ (343).
P. Claudin-16
Claudin-16 was originally named paracellin-1 (324) and described as a protein the defect of which causes familial hypomagnesemia, hypercalciuria, and nephrocalcinosis (FHHNC) in affected patients. It has since been found to be expressed in a variety of tissues including kidney (324), salivary gland (194), and mammary gland (231). Due to two potential in-frame start codons, claudin-16 is one of three claudins that, in many species (including primates, elephant, panda, dog, guinea pig), may exist in a long and a short variant. In humans, it is a matter of debate whether the long version exists under physiological conditions. Some mammals, such as mouse, rat, or pig, only possess the short version while others (e.g., naked mole rat, horse) appear to possess only a long version. Hou et al. (143) reported that upon transfection into the porcine cell line, LLC-PK1, only the short version correctly inserted into the tight junctions, whereas Günzel et al. (119), upon transfection in the canine cell line MDCK-C7, found no difference between the localization and function of the short and the long version.
Most research on claudin-16 deals with its role in renal reabsorption of divalent cations because its mutations underlie FHHNC. To date, on the order of 50 different claudin-16 mutations causing FHHNC have been reported (for review, see Ref. 121). Along the nephron, claudin-16 expression has been demonstrated in the thick ascending limb of Henle's loop (TAL), but there are also reports for additional expression in the distal convoluted tubule (DCT) and cortical collecting duct (324, 392). Both TAL and DCT are important for Ca2+ and Mg2+ reabsorption [for review, see de Rouffignac and Quamme (64)]. The TAL is responsible for mass transport, and ∼70% of all filtered Mg2+ and 20% of filtered Ca2+ is reabsorbed here, probably along the paracellular pathway. The driving force for this is the lumen-positive electrical potential, which is generated by two distinct mechanisms: 1) reabsorption of Na+, K+, and Cl− through the apical electroneutral Na-K-2Cl transporter together with recycling of K+ back into the lumen through ROMK K+ channels; and 2) backleak of reabsorbed Na+ into the lumen through the paracellular pathway, creating a diffusion potential. The DCT is responsible for the “fine tuning” of Ca2+ and Mg2+ homeostasis, and transport is active and occurs along the transcellular route.
The physiological role of claudin-16 in the renal tubule is still controversial. The initial assumption that claudin-16 acts as a major paracellular channel for divalent cations has not been unequivocally verified experimentally (119, 143). Some studies reported that claudin-16 causes a minor increase in Mg2+(180) or Ca2+(152) transport when overexpressed in MDCK cells (for review, see Ref. 121). Results by Hou et al. (143) from in vitro overexpression as well as from a knock-down mouse model (147) suggested that claudin-16 may primarily increase Na+ permeability. The claudin-16 knock-down mice had renal Ca2+ and Mg2+ wasting, phenocopying FHHNC. Since paracellular Na+ permeability of the TAL is required for backleak of reabsorbed Na+ and hence generation of the lumen-positive potential, it was proposed that claudin-16 might affect paracellular Ca2+ and Mg2+ transport by altering the driving force in the TAL. However, Kausalya et al. (180) did not find any effect on Na+ permeability in claudin-16-transfected cells. Furthermore, a claudin-16 knockout mouse model was recently reported that also exhibits renal divalent cation wasting, yet the Na+ permeability of the TAL was unaffected (399). Interestingly, for both mouse models (147, 399), the experiments on isolated TAL tubules were carried out by the same laboratory. It therefore seems unlikely that the effect on Na+ permeability observed by Hou et al. (147) is the main reason for the Mg2+ wasting observed in both mouse models. Further hypotheses as to how claudin-16 may modulate Mg2+ transport include interactions with Cl− channels as well as with the calcium sensing receptor (119, 155) but, clearly, more research is necessary to solve the role of claudin-16 in the transport of divalent cations.
Finally, claudin-16 deletions are observed in Japanese Black Cattle that exhibit renal tubular dysplasia causing growth retardation and finally renal failure (275, although one report suggests that claudin-16 may not necessarily be the responsible gene, Ref. 336). Most interestingly, although affected animals exhibit complete absence of claudin-16 from the thick ascending limb of Henle (281), they do not display altered serum or urine Mg2+ and Ca2+ levels (275).
Q. Claudin-17
Claudin-17 appears to be predominantly present in the kidney, but also, to some extent, in the brain (196). In the kidney, expression is highest in the most proximal segments of the nephron, less in the loop of Henle, sparse and irregular in the distal tubule, and absent in the collecting duct. Expression was also found in low-resistance renal cell lines such as LLC-PK1, MDCK C11, and MDCK II but not in high-resistance renal or colonic cell lines (196). Overexpression in high-resistance MDCK C7 cells as well as knock-down in LLC-PK1 cells indicated that claudin-17 acts as an anion channel. Charge selectivity is higher than in all other paracellular anion channel candidates known to date, but size selective is low: claudin-17 is highly permeable to all halide anions as well as HCO3−, NO3−, and small organic anions such as pyruvate. Size limit is on the order of ∼300 Da, as fluorescein permeability is only marginally increased in claudin-17 transfected cells (196).
R. Claudin-18
Claudin-18 is a major constituent of tight junctions in lung and stomach epithelia. In the lung, the splice variant 18–1 is predominantly expressed, while in stomach epithelium, the major variant is claudin-18–2 (269). Overexpression of claudin-18–2 in MDCK II cells induces a selective sealing of the tight junction against Na+ and especially against H+ (169). Claudin-18–2 is strongly expressed in the specialized columnar epithelium of Barrett's esophagus, suggesting that it may be a specific adaptation to protect the epithelium against low pH (169). This is further supported by the recent finding that claudin-18–2 knockout mice develop atrophic gastritis within 3 days after birth, which is due to the paracellular H+ leak observed in their stomach epithelia (134). Interestingly, claudin-18–2 is also expressed in bone cells, and knockout of the claudin-18 gene in mice causes osteoporosis due to increased bone resorption by osteoclasts (216).
S. Claudin-19
Claudin-19 is expressed in the peripheral nervous system, kidney, and retinal pigment epithelium (191, 207, 221, 249, 286). Overexpression and knock-down studies on claudin-19 in various cell types (MDCK II, LLCPK1, RPE, Refs. 14, 145, 286, 383) demonstrated that the presence of claudin-19 correlates with and increases paracellular barrier function. However, depending on the cell type, this effect may be due to a tightening effect against cations (14) or against anions (145).
In peripheral neurons, claudin-19 is located at the interlamellar junctions of myelin sheaths. Claudin-19 knockout caused peripheral neuropathy in mice that was attributed to insufficient electrical sealing of Schwann cells (249).
In the kidney, claudin-19 is expressed mainly in the TAL and, perhaps to a lesser extent, in the DCT (14, 191). Mutations in claudin-19 are responsible for a form of FHHNC that is associated with severe ocular defects (191). Claudin-19 knock-down in mice was found to cause greatly increased excretion of K+, Mg2+, and Ca2+, causing decreased plasma Mg2+ levels, without significant changes in plasma K+ and Ca2+ levels (144). In the kidney as well as in an in vitro system, Hou et al. (144, 145) demonstrated that claudin-16 and -19 need to interact to be correctly inserted into the TJ and to be fully functional, whereas in human RPE claudin-19 is expressed and functional in the absence of claudin-16 (287). In their model, claudin-16 and -19 have additive effects to increase the relative permeability of the TAL to Na+ versus Cl−, and thus are both required to generate the electrical driving force for divalent cation reabsorption in this tubule segment.
T. Claudin-20 to -27
Not much is known about tissue distribution or physiological function of claudins 20–27. As summarized in TABLE 1, some of these claudins have unusual structural features, such as an unusually long NH2 terminus (claudin-25), COOH terminus (claudin-23), or ECL1 (claudin-25, -26); unusually short COOH terminus (claudin-25, -26); or lack of a PDZ-binding motif (claudin-21, -24, -25, -26, -27).
In chick retinal pigment epithelium, claudin-20 was found to be upregulated during embryonic development. Claudin-20, -22, and -23 mRNA have been found to be expressed in mouse brain capillary endothelial cells and especially the levels of claudin-22 mRNA suggest a role for this claudin in the formation of the blood-brain barrier (280). Claudin-20 and -23 mRNA was detected in mouse small intestine (344). Furthermore, claudin-23 expression has been reported for skin, placenta, stomach, and germinal center B cells. It appears to be dysregulated in several diseases such as gastric and colonic cancer (178) as well as atopic dermatitis (63).
Claudin-21 and claudins 24–27 are expressed in the intestine, but also in the stomach (claudin-21, -24, -25), liver (claudin-21, -25, -27), kidney (claudin-21, -24, -25), heart (claudin-24, -25), and brain (claudin-25, -26). During mouse development between day E.7 and E.17, claudin-21 and -24 show increasing expression levels, whereas expression levels of claudins-26 and -27 decrease and claudin-25 is highly expressed throughout this period (246). Maher et al. (224) report TMEM114 (claudin-26) and TMEM235 (which is a member of the pfam00822 family but not identical to any of the claudins reported by Ref. 246) to be expressed in brain and eye. TMEM114 appears to play an essential role in eye development, as it is expressed in the developing eye of human, mouse, and frog and Tmem114 knock-down in frog causes developmental ocular disorder. When transfected into MDCK I cells, Mineta et al. (246) found claudin-25 (CLDND1) and -26 (TMEM114) to colocalize with ZO-1 despite their predicted lack of direct interaction, whereas claudin-24 and -27 remained in intracellular compartments. In contrast, Maher et al. (224) found TMEM114 (claudin-26) and TMEM235 within the membrane of MDCK II cells, but not within the tight junction. Maher et al. (224) further demonstrate that TMEM114 and -235 are glycosylated and conclude from their phylogenetic considerations that TMEM114 and -235 should rather be placed within the CACNG than within the claudin family.
U. Teleological Purpose of Paracellular Transport
As outlined above, we are beginning to understand the role of individual claudins in determining the magnitude and selectivity of paracellular permeability in a variety of tissues. However, a more fundamental question is: Why is paracellular transport even necessary at all? In the case of epithelial tissues that expend considerable effort on vectorial solute and water transport, one reason may be that paracellular transport increases the efficiency of transepithelial transport. For example, in the kidney, energy and O2 consumption are largely dictated by the rate of tubular Na reabsorption. Transcellular transport of Na is dependent on the potential energy generated by the Na+-K+-ATPase, which uses one molecule of ATP to drive the transport of three Na+. Mitochondria generate six molecules of ATP for each molecule of O2 consumed. Thus the expected suprabasal renal oxygen consumption (Qo2), as a function of Na reabsorption (TNa) is 1:18. In fact, measured values of suprabasal Qo2/TNa are in the range of 1:25 to 1:29 (182, 188, 352, 356), indicating that the kidney is much more oxygen efficient than expected. One proposed explanation for this is that the PT can leverage the excess free energy in solute gradients established by active transcellular transport to drive passive paracellular reabsorption of Na, Cl, and other solutes (65, 235), presumably mediated by claudin proteins.
VII. STRUCTURE-FUNCTION STUDIES OF CLAUDINS
The tertiary and quaternary structures of claudins have not yet been directly solved. However, there is substantial indirect evidence from biochemical, mutagenesis, binding, and modeling studies to make some inferences about their structure and function (FIGURE 3).
Figure 3.
Model of claudin protein showing predicted topology and secondary structure as well as putative functional domains. Roman numerals indicate the predicted α-helical transmembrane domains.
A. First Extracellular Loop (ECL1)
The predicted ECL1 in claudins lies between the first and second transmembrane helices and is a long loop of 42–56 residues. This loop contains the highly conserved signature motif of claudins (FIGURE 1). This suggests that the ECL1 likely has a well-defined structure and serves an important conserved function of claudins.
1. ECL1 forms the lining of the paracellular pore
Chimera studies in which the extracellular loops of claudin-2 and claudin-4 were swapped in various combinations showed that the ECL1 confers the charge selectivity and magnitude of small ion permeability of the paracellular pathway (56). In claudin-4 and -15, specific charged residues, primarily in the second half of the ECL1, could be identified as being responsible for determining charge selectivity. Charge-reversing mutations at these sites reversed the charge selectivity of the paracellular pathway (57). Together, these findings strongly suggest that the ECL1 forms the lining of the paracellular pore for small ions.
2. ECL1 contains a specific ion-binding site
By studying multiple claudins with different charge selectivities, as well as charge reversal mutations, Van Itallie et al. (364) found a strong correlation between the net charge on the second half of ECL1 and paracellular charge selectivity. This concept is also supported by the difference in ion selectivity of the two major splice variants of claudin-10: 10a which is anion selective and 10b which is cation selective. These isoforms differ in their NH2-terminal region, encompassing the first transmembrane domain and most of ECL1 (122, 373). In humans, ECL1 of claudin-10a contains one negatively charged and seven positively charged amino acids, ECL1 of claudin-10b five negatively and four positively charged amino acids, resulting in a preponderance of positive charge within ECL1 of the anion-preferring claudin-10a, but of negative charge for the cation-preferring claudin-10b. A mutational approach, exchanging individual positively charged amino acids of claudin-10a by negatively charged amino acids demonstrated that certain positions within ECL1 (mouse sequence: R33, R62) are important for charge selectivity of the resulting channel. A simple explanation might be that this entire region of the loop determines the general electrostatic environment within the paracellular pore. However, there are a number of reasons to believe such a model may be oversimplified.
First, most of the early mutagenesis studies used charge-reversing mutations that not only abolish a charged side chain, but introduce a new, oppositely charged moiety (57). This complicates interpretation of the experiment as the mutants not only inform on the function of the native residue but could also manifest artifactual effects from the newly introduced charge. Second, these early studies did not distinguish between surface charge effects and electrostatic effects within the pore itself. Third, subsequent studies that have surveyed the role of charged residues in this region have generally indicated that some, but not all, of them contribute to paracellular ion selectivity (143, 416). As a consequence, we now favor a model in which the ECL1 forms a well-defined structure in which a subset of the residues face into the pore lumen; among these pore-lining residues are specific charged amino acid(s) that act as binding sites for permeating ions.
The role of charged amino acids in the ECL1 in determining paracellular charge selectivity has been most carefully examined by Yu et al. for claudin-2 (416). By using an inducible expression system in high-resistance MDCK I cells, the permeability through claudin-2 pores could be estimated. The relative permeability for Na+ over Cl− (PNa/PCl) for claudin-2 was found to be 7.5. With the use of charge-neutralizing mutations of all three acidic amino-acids in the ECL1, it was shown that only D65 played an important role in ion permeation and that the carboxylate side chain on this residue conferred about two-thirds of the cation selectivity. Several lines of evidence prove that this is a bona fide ion binding site: 1) mutation of D65 to asparagine (D65N) increased the activation energy for Na+ permeation. 2) The selectivity pattern of claudin-2 for alkali metal cations exhibited a high-order Eisenman sequence that was decreased in the D65N mutant. 3) The permeability of claudin-2 for the divalent cation Ca2+ was disproportionately decreased by D65N, relative to monovalent cations. 4) Titration by extracellular acidification abrogated Na+ permeability of wild-type claudin-2 but not the D65N mutant. 5) The cation permeability conferred by D65 was independent of ionic strength, excluding a surface charge effect. This site, located immediately after the second conserved extracellular cysteine (C64), appears to be generally important for determining charge selectivity in claudins. Mutations of the charged residues at this site also alter charge selectivity of claudin-4, -10a, -15, and -17 (57, 146, 196, 373) (FIGURE 4, TABLE 5).
Figure 4.
Alignment of the predicted first extracellular loop of human claudins. Residues within ECL1 were delineated using TMHMM (195). Sequence alignment was then performed with MUSCLE (76) and visualized in Jalview. Claudins 25–27 have lower homology and are not included in the alignment. Residues are color coded according to the following scheme: red, acidic; dark blue, basic; light blue, aromatic; green, polar; yellow, cysteine; purple, proline; cream, hydrophobic. Top: residue numbering for claudin-1 (identical to claudins 2, 4–9, 14, 17, 19, and 20). Bottom: consensus sequence of the claudin signature pattern.
Table 5.
Role and localization of residues in ECL1 as determined by mutagenesis studies
| Classification | Claudin-1* (59) | Claudin-2+ (17, 416) | Claudin-4 (57, 146) | Claudin-5 (395) | Claudin-7 (6) | Claudin-10a (373) | Claudin-15 (57) | Claudin-16 (143) | Claudin-17 (196) |
|---|---|---|---|---|---|---|---|---|---|
| F | R31A no effect on HCV entry | R31T no effect | |||||||
| E | Y35A no effect on HCV entry | Y35C located at vestibule | R32D reduce Cl− selectivity | ||||||
| C? | D38A abolish HCV entry | D38R reduce Cl− barrier | D104S reduce Na+ selectivity | ||||||
| F | D105S reduce Na+ selectivity slightly | ||||||||
| F | E108T no effect | ||||||||
| F | E48Q no effect | E46K no effect | R114T no effect | ||||||
| B | G49A abolish HCV entry | ||||||||
| B | L50A abolish HCV entry | ||||||||
| B | W51A abolish HCV entry | ||||||||
| F | S53A no effect on HCV entry | E53Q no effect | E53K reduce Cl− barrier slightly | E119T reduce Na+ selectivity slightly | |||||
| B | C54A abolish HCV entry | C54S loss of Na+ barrier | |||||||
| E | H57C located at vestibule | D55R confer Cl− selectivity | |||||||
| B | C64A abolish HCV entry | C64S loss of Na+ and mannitol barrier | |||||||
| C | D65N abolish Na+ binding site | K65D, K65T confer Na+ selectivity | R59D reduce Cl− selectivity | E64K confer Cl− selectivity | D132S no effect | K65E and K65A abolish Cl− selectivity | |||
| D | I66C exposed to pore lumen | E133T no effect | |||||||
| F | D68N no effect | D135S no effect | |||||||
| F | D76N no effect | D76N no effect | |||||||
| A | R81T weak ER expression | R149L/T weak ER expression |
Classification refers to tentative assignment of role at each position: A, conserved residue essential for expression, trafficking, and presumably for global protein folding; B, conserved residue essential for claudin function at the plasma membrane; C, charged residue that acts as intrapore binding site; D, other pore-lining residue; E, residue at vestibule which may confer surface charge effects; F, residue that appears nonessential.
Claudin function assessed by ability to mediate cellular entry of hepatitis C virus (HCV).
Residue locations inferred based on accessibility to and block by methanethiosulfonate reagents. Reference numbers are given in parentheses.
3. Mapping of ECL1 residues by cysteine mutagenesis
The location of several residues in claudin-2 has been investigated by cysteine-scanning mutagenesis (17). I66 is located within the narrow part of the pore with its side chain facing the lumen, presumably just adjacent to D65. D65, in addition to being within the pore, was shown to lie close to the intermolecular interface between adjacent subunits. Y35 and H57 are exposed outside the pore, presumably in the vestibule. Interestingly, charged residues in other claudins located at the homologous positions appear to have weak effects on charge selectivity (TABLE 5), suggesting that they may exert surface charge effects at the pore vestibule (111).
4. Conserved residues in the signature sequence play important structural roles
The ECL1 contains a highly conserved claudin signature motif. Central to this is a triplet of amino acids, G-L-W. Mutation of any of these residues individually in claudin-1, which is a receptor for hepatitis C virus (HCV), abolishes HCV binding (59). Interestingly, the mutants still traffic normally to the plasma membrane and have preserved cis-interactions but may have impaired ability to form trans-interactions. Mutation of all three residues in claudin-2 simultaneously to alanine (A3) abolishes its ability to reach the plasma membrane (371). Interestingly though, the A3 mutant still has the ability to form dimers (presumably in cis) that are detectable by native PAGE, just like wild-type claudin-2. This suggests that the G-L-W motif may play an important role common to all claudins in stabilizing the correctly folded structure of the ECL1 and hence stabilizing trans-interactions.
Another notable feature of the claudin signature is a pair of conserved extracellular cysteines. Claudins 1–24 have two cysteines in ECL1 that start shortly after the G-L-W motif and are spaced 9–11 residues apart (FIGURES 1 AND 4). Claudins 25–27 also have two cysteines in ECL1, but the second cysteine is in an aberrant position, located 5, 6, or 25 residues downstream from the first one. In claudin-1, mutation of either conserved cysteine abolishes HCV binding, mimicking the effect of mutating the G-L-W motif (59). In claudin-5, mutation of either cysteine individually, or both together, abolished the ability of the claudin to form a barrier to small ions, while preserving its ability to traffic to the tight junction (395). This suggests that these cysteines also play an important role in stabilizing ECL1 in its correct fold. Furthermore, the fact that the cysteines are always conserved as a pair, that mutation of either one has the same functional effect, and that mutation of both together has no additive effect over mutation of either one individually, all suggest that they are probably engaged in an intramolecular disulfide bond with each other.
Finally, at the very end of ECL1, just before the predicted second transmembrane helix, is a highly conserved arginine residue found in 23 of 27 claudins. Mutation of this residue in claudin-4 and -16 led to reduction in expression level and retention in the endoplasmic reticulum. The most likely reason is that this residue is critical for global folding of claudin proteins.
B. Second Extracellular Loop (ECL2)
1. Role of ECL2 in trans-interactions
The amino acid sequence of ECL2 is quite highly conserved (FIGURE 5). A previous chimera study had already showed that ECL2 is not involved in determining paracellular permeability (56). When the ECL2 of claudin-5 was fused to maltose-binding protein and expressed in bacteria, it was found to dimerize (36). This suggests that ECL2 mediates claudin-claudin interactions.
Figure 5.
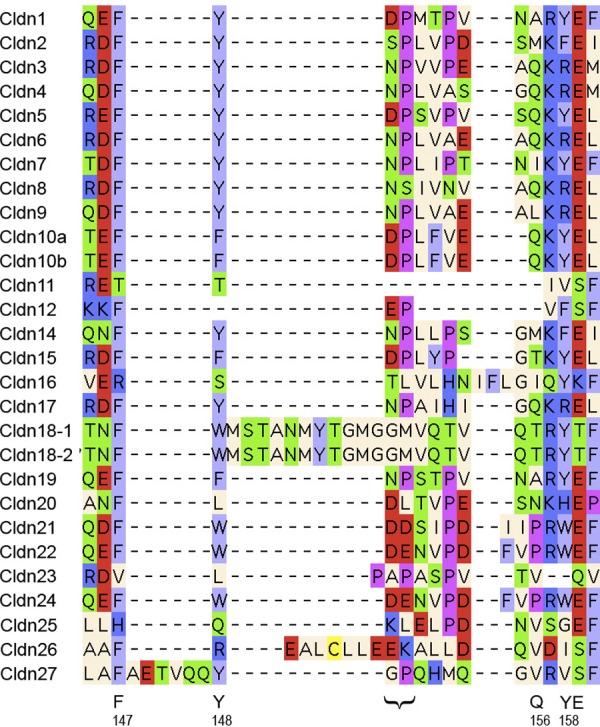
Alignment of the predicted second extracellular loop of human claudins. See legend to FIG. 4 for methods and color code. Bottom: letters indicate residues proposed to participate in trans-interactions, numbered according to the claudin-5 sequence. The bracket indicates the N-P motif proposed to bind to CPE.
Piontek et al. (292) have done the most extensive analysis of the ECL2. They mutated each residue in ECL2 of claudin-5 and transfected it into HEK cells. Several mutations disrupted the normal enrichment of claudin-5 at cell contacts with other claudin-5-expressing cells, leading to a homogeneous distribution throughout the plasma membrane. Importantly, these mutations did not affect the proximity of claudin-5 molecules in cis, as measured by FRET. It was therefore concluded that the residues at these positions are probably important for trans-interaction (FIGURE 5).
The ECL2 structure was then modeled based on homology to an 18-residue fold within 2BDV (hypothetical protein BB2244, from Bordetella bronchiseptica). In this model, ECL2 forms a helix-turn-helix, where the predicted helices are extramembranous extensions of transmembrane domains 3 and 4, and the central turn region is anchored by two proline residues. Three of the residues proposed to participate in trans-interactions, F147, Y148, and Y158, form an aromatic core on one face of the loop that could potentially undergo hydrophobic interactions with the aromatic core from another ECL2 to form a claudin dimer. However, the role of the two hydrophilic residues proposed to participate in trans-interactions, Q156 and E159, which are located on the opposite side to the aromatic core, is unclear.
The functional importance of trans-interaction was demonstrated by expressing the Y148A, Y158A, and E159Q mutants in MDCK II cells (289). In contrast to wild-type claudin-5, none of the mutants increased TER. Furthermore, all mutants increased the paracellular leak to 10 kDa FITC-dextran, suggesting that mutant claudin proteins that are unable to interact in trans somehow disrupt the paracellular barrier to macromolecules.
In contrast to these functional studies in claudin-5, Lim et al. (215) studied trans-interactions in claudin-2 by an entirely in vitro technique and found that it was mediated by ECL1 and not ECL2. They bound purified claudin-2 protein (or an extracellular loop polypeptide) to the cantilever tip of an atomic force microscope and also to the surface of glass cover slips. The force of adhesion between single molecules of claudin-2 was then measured by force spectroscopy. By this technique, they found ECL1-ECL1 trans-interactions, but no such interaction between ECL2-ECL2 or ECL1-ECL2.
2. ECL2 contains the binding site for Clostridium perfringens enterotoxin
Claudin-3 and -4 were identified early on as receptors for Clostridium perfringens enterotoxin (CPE), a causative agent for food poisoning in humans (176, 177). The COOH-terminal half of CPE binds claudins (129, 329), while the NH2-terminal half forms pores in the plasma membrane and induces cell death. Claudin-6, -7, -8, and -14 are also able to bind CPE, whereas claudin-1, -2, and -5 do not. With the use of chimeras of claudin-3 and claudin-1, it was shown that the ECL2 was responsible for binding CPE (93). Using peptide mapping, Winkler et al. (400) narrowed down the binding region to a five-residue peptide in the predicted turn region of ECL2 (N-P-L-V-P in claudin-3). Within this region, the N-P motif (FIGURE 5) was shown to be completely conserved in all CPE-binding claudins, whereas the N was replaced by D or S in non-CPE-binding claudins. This residue was shown to be critical since replacing N at this position could restore CPE binding to nonbinding claudins (302).
The structure of CPE has now been solved by X-ray crystallography (39, 184, 367). The COOH-terminal domain of CPE contains two hydrophobic cavities formed by triplets of tyrosine and leucine residues, respectively. With the use of mutagenesis and molecular modeling, it has been proposed that L150/151 of claudin-3/-4 binds to the “triple tyrosine pit” while P152/A153 binds to the “triple leucine pit” (380).
C. Cytoplasmic Tail
The COOH-terminal tail of claudins is long, highly divergent in sequence between different claudins, and is predicted to be intracellular and largely disordered.
1. Role in trafficking to tight junction
Truncation of the cytoplasmic tail in claudin-1, -5, -6, and -16 impairs trafficking to the tight junction leading to intracellular retention, predominantly in the endoplasmic reticulum (ER) (20, 260, 308) and subsequent degradation via the proteasome (260). Overexpression of the truncated mutant of claudin-6 was shown to induce the unfolded protein response (20). This could indicate that the cytoplasmic tail is in some way important for protein folding, but could simply be an artifact of protein overexpression. An alternative explanation for ER retention of the truncation mutants is that the cytoplasmic tail normally binds to other molecules that facilitate exit of claudin proteins from the ER. In claudin-1 and -5, serial truncations indicated that removal of just the PDZ-binding domain did not affect normal trafficking and that truncation of most of the cytoplasmic tail (24 amino acids in claudin-1, 34 in claudin-5) was needed to see an effect on trafficking, suggesting that the 15–20 juxtamembrane cytosolic residues were most critical. Importantly, none of these truncations removed the cysteines located just after the fourth transmembrane domain that are palmitoylated. Thus the cytoplasmic tail, independent of PDZ binding or palmitoylation, seems to play an important role common to many or possibly all claudins in exit from the ER and trafficking to the tight junction.
2. Role in protein degradation
Van Itallie et al. (368) noted that claudin-2 has a threefold longer protein half-life than claudin-4. Using domain-swapping chimeras they showed that the cytoplasmic tail (but not the PDZ domain) was able to specify the protein half-life (368). However, the mechanism for this is currently unknown.
3. PDZ-binding domain
The end of the COOH terminus contains a hydrophobic dipeptide motif in all the claudins except claudin-12, -22, -25, and -27. This motif (Y-V in most claudins) binds to PDZ domains on several tight junction scaffolding proteins, namely, ZO1, ZO2, and ZO3 (163) and MUPP1 (127), although the functional importance of this interaction remains uncertain (see sect. VIII for further discussion).
4. Palmitoylation and phosphorylation
The region of the cytoplasmic tail just after the fourth transmembrane domain, as well as the region of the intracellular loop just after the second transmembrane domain, contain conserved pairs of cysteines that, in claudin-14, have been shown to be palmitoylated. Inhibition of palmitoylation impaired the ability of claudin-14 to localize correctly at the tight junction, suggesting an important role of palmitoylation in protein trafficking (369). The cytoplasmic tails of most claudins also contain a large number of predicted phosphorylation sites, which are not highly conserved but are mostly unique to each claudin. The role of phosphorylation in modulating function of specific claudins is discussed in detail in section VIII.
D. Stoichiometry of Claudins
When claudins are transfected into fibroblasts, which lack tight junction proteins, they polymerize into tight junction-like strands at cell-cell contacts and adhere to adjacent cells (198). Thus the numerical stoichiometry of claudin polymers could in theory be very large. It is likely, though, that within these polymers there exist discrete, lower order complexes consisting of claudin oligomers.
By freeze-fracture electron microscopy, both the native tight junction (330) and tight junction-like strands reconstituted by claudin transfection into fibroblasts (99) appear as strands of particles ∼10 nm in diameter and spaced 18 nm apart, center to center. These particles are presumed to represent claudin proteins, in which case their large size suggests that they represent multimeric complexes. In comparison, connexin subunits, which like claudins also have four transmembrane domains, assemble into hexamers that are ∼8–9 nm in diameter (PDB ID: 2ZW3) and also appear as ∼10 nm particles by electron microscopy (331). The nicotinic acetylcholine receptor (PDB ID: 2BG9) is formed from a pentamer of four transmembrane domain subunits and spans a molecular diameter of 7–8 nm. It is therefore likely that claudins also assemble into oligomers.
Mitic et al. (248) found that claudin-4 that was expressed in Sf9 cells and solubilized with perfluoro-octanoic acid formed hexamers. Van Itallie et al. (371) showed that in native polyacrylamide gels, claudin-2 solubilized in dodecylmaltoside formed stable dimers. These dimers appeared to be due to cis-interaction between two subunits and the site of interaction was most likely the second transmembrane domain. In addition, though, higher molecular weight complexes could be detected, both in cell lines and native liver tissue, containing multiple claudins and occludin. Angelow and Yu (18) found specific evidence that the paracellular pore is surrounded by multiple claudins subunits, based on the results of cysteine scanning mutagenesis experiments performed in claudin-2. Mutation of an intrapore residue, D65, to cysteine led to formation of dimers by disulfide bonding, suggesting that this residue is located close to the intermolecular interface. Furthermore, by comparing the stoichiometry of binding of thiol-reactive methanethiosulfonate molecules to a cysteine located within the narrow part of the pore versus a cysteine located at the vestibule, they deduced that the claudin subunit stoichiometry of the pore must be quite large.
In summary, claudins engage in both cis- and trans-interactions both with other claudins and also potentially with occludin or other tight junction membrane proteins, and by these means assemble into complexes that vary in their size and stability. These range from quite stable cis-dimers to less stable, higher order oligomers that surround the paracellular pore, to very long strandlike polymers.
VIII. REGULATION OF CLAUDINS
Regulation of claudins and thus of TJ properties occurs on various levels, such as regulation of transcription, posttranslational modification, interaction with cytoplasmic scaffolding proteins, and interactions with claudins within the same membrane (cis-interaction) as well as with claudins of neighboring cells (trans-interaction). Together, these processes determine tight junction assembly, remodeling/modulation, and degradation.
A. Transcriptional Regulation
Major regulators of claudin expression comprise the TNF-α/NF-κB and TGF-β-Smad/Snail pathways, but PPARγ, SP1, HNF-1α, HNF-4α, CDX1, CDX2, GATA-4, and Grhl2 have also been reported to be involved.
1. TNF-α/NF-κB
Intestinal epithelia from patients suffering from inflammatory bowel diseases display characteristic changes in their claudin expression pattern: a downregulation of the “tightening” claudins 1, 3, 4, 5, 7, and/or 8 together with an upregulation of the cation pore- and water channel-forming claudin-2 (for review, see Ref. 120). Similarly, Epple et al. (81) observed downregulation of claudin-1, upregulation of claudin-2, and barrier impairment in duodenal biopsies from HIV-infected, untreated patients. Under experimental conditions, similar changes were elicited by an application of cytokines that are elevated under these conditions, including TNF-α, IFN-γ, and IL-13 (81, 120). Amasheh et al. (9) were able to demonstrate that TNF-α-induced downregulation of claudin-1 and upregulation of claudin-2 in the intestinal cell line HT-29/B6 was mediated by NF-κB and that these changes were antagonized by the herbal compound berberine. Likewise, TNF-α acts through NF-κB to impair the blood-brain barrier (BBB) by a downregulation of claudin-5 (22, 40, 41, 335, 384). Inhibition of NF-κB by FoxO4 was found to protect mice against colonic injury and inflammation (426). Transgenic mice with epithelial-specific overexpression of an NF-κB super repressor (IκBα with mutated ubiquitination sites) were protected against T cell-induced diarrhea through protection against opening of the paracellular pathway (347). Furthermore, Tanshinone IIA, a substance that downregulates NF-κB expression, improves BBB tightness, and protects rat brains against focal ischemia (384). In contrast to effects in intestinal epithelia and BBB, upregulation of NF-κB was found to strengthen tight junction barrier properties in nasal epithelia (upregluation of claudin-4, Ref. 234) and in activated dendritic cells during their penetration of epithelia in Th2-type inflammatory responses (upregulation of claudin-7, Ref. 174). FIGURE 6 summarizes the effects of TNF-α and NF-κB on claudin expression and epithelial permeability.
Figure 6.
Summary of NF-κB-mediated TNF-α and PPAR-γ effects on claudin expression and epithelial permeability. In most epithelia, increases in NF-κB cause an increase in epithelial permeability, due to decreased expression in tightening tight junction proteins and/or increased expression in pore-forming claudin-2 (red arrows). Direct or indirect inhibition of NF-κB causes opposite effects (green arrows). In a few, exceptional cases, NF-κB was observed to upregulate tightening claudins, causing decreases in epithelial permeability (blue arrows). Figure is based on References 9, 22, 40, 41, 81, 120, 174, 210, 234, 237, 274, 335, 347, 378, 379, 384, 426. See text for detailed description.
2. PPARγ
In urothelial cells and nasal epithelia, PPARγ upregulates claudin-1, -3, and -4, but downregulates claudin-2 (274, 378, 379) (FIGURE 6). The action of PPARγ is PKC dependent (274). Varley et al. (378) further report that the increase in claudin-4 is due to inhibition of proteasomal degradation and that, as a consequence, claudin-5 is stabilized within the tight junction through specific interaction with claudin-4. HIV-induced downregulation of PPARγ in lung tissue causes an upregulation of NF-κB and a downregulation of claudin-5 and thus compromises barrier integrity of pulmonary capillaries, and these effects can be reversed by PPARγ agonists (210) (see FIGURE 6). Similarly, PPARα appears to be barrier protective, and a knock-down of PPARα causes an upregulation of claudin-2 and enhanced effects of DSS-induced colitis in a PPARα knock-down mouse model (237).
3. TGF-β- Smad/snail
Epithelial-to-mesenchymal transition (EMT) is of central importance for cell migratory behavior and thus for invasiveness and metastasis in cancer. Several pathways intertwine to regulate EMT, and many of the transcription factors involved have been reported to directly affect expression of claudins. Although many details are not yet fully resolved and at present even appear to be contradicting, a general pattern emerges and is depicted in FIGURE 7. TGF-β signaling pathways play a crucial role in EMT but also in vascular development and in maintaining intestinal barrier function (67, 126, 136, 283, 353). The latter function may be directly mediated by nutrients, as, e.g., milk and dairy products contain high amounts of TGF-β (136). In endothelial cells, TGF-β activates ALK5 which acts through SMAD2/3 and downregulates claudin-5 expression (283), whereas inhibition of ALK5 using SB431542 was observed to increase claudin-3 and -5 expression (305). SMAD2 and/or 3 may further interact with SMAD4, and an inverse relationship between SMAD2/3/4 and claudin expression has been observed in various further systems. For example, in colonic cancer loss of SMAD4 causes upregulation of claudin-1, a condition which correlates with tumour growth, metastasis, and poor outcome (322). SMAD signaling is interrupted by the inhibitory SMAD7 and SMAD7 overexpression in colon adenocarcinoma cells elicited increased expression of claudin-1, -4, and -7 (126). The mode of action of SMAD in claudin repression was investigated in Sertoli cells. Here, claudin-11 promoter activation occurs through a GATA, NF-YA, and CREB complex that is able to bind to the GATA/NF-Y motif of the claudin-11 promoter. Binding, and therefore promoter activation, was inhibited by SMAD3 and SMAD4, a process that involves histone deacetylation at the site of the claudin-11 promoter region (220). In breast cancer cells, SMAD3 and -4 form a complex with SNAIL1 and repress claudin-3 expression, probably through binding to SMAD binding elements that were found adjacent to SNAIL binding E-box sites on the claudin-3 promoter (381). Additionally, promoter hypermethylation has been shown to play an important role in the downregulation of claudin-4 during overactivation of TGF-β/SMAD signaling in breast cancer cells (285). It is noteworthy that in contrast to colonic cancer, metastasis and poor outcome in breast cancer are correlated with a downregulation of claudins (381). In accordance with these findings, invasive breast tumor cells express low levels of claudin-1 and high levels of SNAIL and SLUG, both of which are able to bind to E-boxes of the claudin-1 promoter and thereby to repress claudin-1 expression (232).
Figure 7.
Summary of TGF-β-dependent and -independent SMAD/SNAIL/SLUG-signalling on claudin expression. Upregulation of SMAD2, SMAD3, SMAD4, SNAIL1 and/or SLUG by TGFβ (upper part), Wnt, β-catenin or HGF (lower part) causes a general downregulation of claudin expression, irrespective of claudin function (tightening or pore-forming, red arrows). Effects are transmitted by different effects on claudin promoters. Inhibition of SMAD2, SMAD3, and/or SMAD4 upregulation (e.g., by SMAD7), or interference with promoter binding, causes claudin upregulation (green arrows). TGF-β may also differentially affect certain claudins in a SMAD-independent way via the MEKK/p38/MAP kinase pathway (blue arrows). Based on References 31, 67, 116, 126, 136, 157, 220, 232, 240, 276, 283, 285, 305, 322, 353, 362, 381, 422. See text for detailed description.
SMAD4 may also act independently of TGF-β through interaction with the Wnt/β-catenin pathway, with β-catenin directly affecting claudin-1 expression in colonic cells (322, 353). Conversely, TGF-β was shown to act not only through SMAD but also independently of SMAD through MEKK/p38/MAP kinase. This caused upregulation of claudin-4 in colonic cells (136) but downregulation of claudin-11 in Sertoli cells (219). TGF-β can also directly affect SNAIL1 through MEK and PI3 kinase (240). Similarly, SNAIL1 is regulated by HGF (through MAP kinase/Egr1, Ref. 116) or by RON (422). Increased expression of SNAIL1 downregulates the expression of various claudins [claudin-1, (31, 232, 240, 276, 362, 422); claudin-2, (240); claudin-3, (116, 157); claudin-4, (67, 157); claudin-7, (157, 362)] either by direct binding to the respective promoters (157, 232) or indirectly (276).
4. CDX1 and CDX2
CDX1 and CDX2 are able to activate the claudin-2 promoter (83, 100, 228, 313, 315, 338, 412). In addition, CDX2 upregulates claudin-3 and -4 (319). HNF-1α and GATA-4 are able to augment the action of CDX2, but not of CDX1 (83, 313). HNF-1α appears to play a role in tissue-specific expression of claudin-2 as it was necessary for claudin-2 expression in ileum and liver, but not in kidney (313). Both CDX1 and CDX2 effects underlie further modulation through various regulatory pathways (Wnt, LEF-1/β-catenin, MAP-kinase, p38 MAP-kinase, PI3K, MEK; Refs. 228, 338, 412), e.g., via cytokines such as IL-1β or IL-6.
5. SP1
Promoters of claudin-1, -4 and -19 were found to contain SP1 binding sites (75, 140, 221), and SP1 is necessary for the expression of these claudins. Regulation of claudin-4 expression by SP1 is modulated through epigenetic modifications near the SP1 binding site of the claudin-4 promoter. High claudin-4 expression correlates with low methylation levels but high histone H3 acetylation, and vice versa(140). In MDCK cells, upregulation of SP1 and consequently of claudin-4 was induced by EGF through activation of the MEK/ERK pathway (151). Conversely, knock-down of claudin-2 in A549 cells caused a decrease in SP1 but not in c-Fos, c-Jun, NF-kB p65, claudin-1, occludin, or E-cadherin. The decrease in SP1 caused a reduced promoter activity of MMP9 and was associated with a reduced migratory behavior of the cells (156).
6. Grhl2
During development, claudin-4 expression levels in mouse embryos (and claudin-b levels of otic epithelial cells in zebra fish) are determined by Grhl2 (128, 396). Mutation of Grhl2 causes defects in the apical junctional complex of gut endoderm, surface ectoderm, and otic epithelium, and these defects coincide with the occurrence of neural tube defects (396). In breast tumor cells with low levels of claudin expression, Grhl2 expression is low and overexpression of Grhl2 in such cells induces MET (52).
7. Further regulatory pathways
These include the action of HNF-4α, which upregulates the expression of claudin-6 and -7 in mouse F9 cells and induces translocation of these claudins to the tight junction, together with the cell polarity proteins PAR-6, aPKC, CRB3 and Pals1, but not PAR-3 or PATJ (48, 320). HNF-4α-induced formation of tight junctions was demonstrated to underlie establishment of a diffusion barrier between the apical and basolateral membrane compartment, i.e., fence function (48). HNF-4α expression is thus essential for the formation of epithelial polarity (320). It further inhibits cell proliferation which is one of the prerequisites for the development of stable epithelial cell layers (49). Claudin-5 expression was recently described to be modulated by members of the ETS transcription factor family (ETV5, Ref. 258; ERG, Ref. 419). Expression of the lung specific isoform of claudin-18 is regulated through T/EBP/NKX2.1 via two specific binding sites within the promoter of this claudin-18 isoform (269). Ouabain at levels found endogenously in humans (10 nM) has been found to upregulate protein expression of claudin-1, -2, and -4 and enhance their junctional localization, coincident with a decrease in paracellular permeability. This seems to be mediated by c-Src and ERK1/2 but exactly how is unclear (205).
8. From pathways to networks
Although presented here as separate pathways, it has to be emphasized that all these pathways intertwine and should rather be regarded as a complex network with several metastable states and multiple possibilities to move from one state to another. The emerging connections between claudin dysregulation and the development of cancer or the probability of metastasis call for a more thorough understanding of the underlying regulatory network, e.g., by using systems biologic modeling (see, e.g., review in Ref. 326).
B. Posttranslational Modification
Reports on posttranslational modifications of claudins comprise phosphorylation, palmitoylation, and ubiquitinylation. In addition, O-linked N-acetylglucosamine modification (O-glycosylation) sites, N-linked glycosylation sites, and further phosphorylation sites are predicted from computational studies. The effects of such modifications on tight junction assembly, maintenance, and degradation have recently been reviewed by Chiba et al. (50), González-Mariscal et al. (105), and Blasig et al. (35).
1. Phosphorylation
González-Mariscal et al. (105) carried out a computer-aided search on possible claudin phosphorylation sites for various kinases (PKC, PKA, MAP kinase, WNK, MLCK, c-Src, RhoK, and the Eph receptor family) using several phosphorylation site detection tools. They found between 0 (claudin-20) and 10 (claudin-2) such phosphorylation sites on claudins 1 to 22. Some of these sites have already been described to be of physiological importance in independent experimental studies: claudin-1 is a target of MAP kinase, aPKC and PKA, and dephosphorylation is achieved by PP2A. MAP kinase induced phosphorylation at position T203, as well as aPKC and PKA induced phosphorylation promotes claudin-1 insertion into the tight junction (85, 89, 272). Conversely, removal of claudin-1 from the tight junction was observed upon PP2A-induced dephosphorylation (272) and mutation of PKA phosphorylation sites to mimic the nonphosphorylated form (85). In claudin-2, S208 was predicted to be a cPKC/nPKC/p38/Erk1/2/JNK target (105). In a recent study on MDCK II cells, claudin-2 phosphorylation in this position was verified and found to depend on as well as to promote localization of this claudin within the plasma membrane (374). Dephosphorylation could be induced by increases in intracellular cAMP levels (through application of dibutyryl cAMP, forskolin, or prostaglandin E2) (374). Claudin-4 was phosphorylated through PKC (60) or aPKC (at S194 and S195 in human and mouse claudin-4, respectively; Ref. 19), and the latter process proved to be essential for tight junction formation. In contrast, PKA-dependent phophorylation of claudin-3 or mutation of T192 to mimic phosphorylation (60) as well as EphA2 phosphorylation of claudin-4 (position T208, Ref. 345) induced the removal of these claudins from the TJ and resulted in TJ breakdown. Effects of PKA-dependent phosphorylation in endothelial cells are yet contradicting. Both PKA and RhoK are able to phosphorylate claudin-5 at position T207 (162, 328, 411). Although this phosphorylation caused translocation of claudin-5 into the tight junction, the process was accompanied by functional tight junction breakdown (e.g., decrease in transendothelial resistance and increased permeability to mannitol, Refs. 328, 411). S217 of claudin-16 is phosphorylated by PKA, and this was observed to cause insertion of claudin-16 into the tight junction. Conversely, dephosphorylation of S217 led to claudin-16 translocation to lysosomes (154). PKA-dependent claudin-16 abundance within the tight junction was regulated by changes in cAMP through the basolateral Ca2+-sensing receptor and thus directly by extracellular Ca2+ concentrations (155). Claudin-16 localization is further regulated by a Mg2+-dependent phosphorylation at positions T225 and T233 through the MEK/ERK pathway (153). Finally, WNK4-induced phosphorylation of claudin-1 to -4 (413) as well as claudin-7 (at position S207, Ref. 348) caused an increased paracellular Cl− permeability of the affected cells layers (see also Ref. 172).
2. Palmitoylation
As already mentioned, tight junctional localization of some claudins can also be promoted by palmitoylation. Thus, in claudin-14, two palmitoylation sites were identified within the cytoplasmic loop and two further sites within the cytoplasmic COOH terminus, close to the fourth transmembrane region (369). Palmitoylation also occurs in claudin-1, -2, and -4, but not, e.g., in occludin (222, 369), and palmitoylation sites are predicted for claudin-3 (43).
3. O-glycosylation
O-glycosylation has been postulated for the cytoplasmic COOH termini of claudin-1, -3, and -4 (4, 43) using computational approaches. Some of the predicted O-glycosylation sites are so-called Yin Yang sites [e.g., T191, S192, S205, S206 and possibly also T190 and T195 of claudin-1, (4, 43); S199, S203, T204 and possibly T192 of claudin-3 and T189, S194 and probably also S198 and S206 of claudin-4 (43)], that allow phosphorylation as well as O-glycosylation and that potentially could act as switches to regulate claudin localization.
4. N-linked glycosylation
N-linked glycosylation sites are predicted in the first extracellular loops of many members of the PMP-22/EMP/MP20/claudin family, including claudin-1 and -12 (296). Furthermore, claudin-26 (TMEM114) was found to be N-glycosylated at position N54 and N88 (224). N-glycosylation is required for correct membrane localization of the PMP-22/EMP/MP20/claudin family member Cacng2 (stargazin) (296) and of claudin-26 (224). However, there is no experimental evidence that any other claudins are glycosylated so the relevance for this is unclear. Indeed, N-glycosylation of claudin-26 was taken as an indication that this protein does not belong to the claudin family but should rather be placed among the CACNG (calcium channel gamma subunit) family (224).
5. Ubiquitylation and SUMOylation
Ubiquitylation of claudins is still a matter of debate. According to the PhosphoSitePlus database (http://www.phosphosite.org/proteinSearchSubmitAction.do), claudin-1, -3, -4, -5, -6, -7, and -11 contain ubiquitylation sites. However, Asaka et al. (21) failed to detect claudin-1 ubiquitylation despite the fact that proteasome-dependent claudin-1 localization at the plasma membrane was observed. In contrast, Takahashi et al. (340) demonstrated that LNX1p80 induced polyubiquitylation of claudin-1 and found indications for ubiquitylation-dependent claudin endocytosis and lysosomal degradation. In accordance with these findings, LNX1p80 overexpression in MDCK cells caused a reduction in the number of tight junction strands and in the amount of detergent-insoluble claudins (340). Claudin-5 is also polyubiquitylated, on lysine 199, but in this case it targets the protein for proteasomal degradation (226).
SUMOylation by SUMO-1 (small ubiquitin-like modifier 1) has recently been demonstrated for lysine 218 of claudin-2 (372). Moreover, overexpression of SUMO-1 in MDCK cells caused a reduction in claudin-2 expression. Confocal microscopy revealed that the reduction primarily concerned the amount of claudin-2 localized in the lateral membrane rather than in the tight junction. However, it is yet unknown which signals affect SUMO-1 expression in MDCK cells under physiological conditions. Furthermore, SUMO-1 may also have indirect effects as, in parallel with claudin-2, claudin-4 was also downregulated during SUMO-1 overexpression, yet claudin-4 does not possess a SUMOylation consensus sequence (372).
C. Interaction With Scaffolding Proteins
The majority of claudins possess PDZ binding motifs at their COOH terminus with which they are able to bind to various tight junction-associated proteins. These scaffolding/adapter proteins form the link between the transmembrane tight junction proteins and the cytoskeleton. They comprise ZO-1, -2, and -3; MAGI-1, -2, and -3; MUPP-1; Par3 and Par6; PALS1; and PATJ (for review, see Refs. 5, 118, 130, 236, 242). The presence or absence of PDZ binding motifs in claudins 1–11, 13–16, 18, 19 (isoform b), 22, and 23 was studied in detail by Stiffler et al. (334). These authors detected interactions with PDZ domain 1 of ZO-1 (all claudins investigated except claudin-13, -22, -23), PDZ domain 1 of ZO-2 (claudins 1 to 4, 7 to 10, 14, 15, 18, and 19b), PDZ domain 1 of ZO-3 (claudin-8), MAGI-1 and -3 (claudin-23), MAGI-2 (claudin-16 and -23), and MUPP-1 (claudin-3, -8, and -14) but not any interactions with Par3 and Par6. Stiffler et al. (334) used their binding data to develop a PDZ prediction model. This model was used by Günzel and Fromm (120) as well as in the present review to predict the presence or absence of PDZ binding motifs of further claudins and claudin isoforms (see TABLE 1). The absence of a PDZ binding motif is predicted for claudin-12, -19a, -21, and -24 to -27 as well as for the truncated isoforms of claudin-7 and -16, whereas binding is predicted for PDZ domain 1 of ZO-1 (claudin-17, -19c, -20), PDZ domain 1 of ZO-2 (claudin-17, -20), and PDZ domain 10 of MUPP-1 (claudin-20).
The physiological importance of an interaction via the PDZ binding motif can be inferred from patients carrying a mutation within the PDZ binding motif of claudin-16. These patients suffer from a mild, self-limiting childhood form of FHHNC (259) as the claudin is able to reach the cell surface but is rapidly internalized and transported to lysosomes (180).
D. Claudin-Claudin Interactions
Several types of interactions between the claudins within one tight junction can be envisaged: homo- and heteromeric interactions that may occur in cis or in trans. Similarly, claudins may interact with other transmembrane tight junction proteins, such as members of the TAMP family. Considering the number of different proteins even within one single tight junction, this generates a multitude of possible combinations, all of which may have specific consequences for the functionality of the resulting tight junction. Only a few interactions have yet been investigated, and knowledge is still quite sketchy (for reviews, see Refs. 120, 321), especially as techniques employed to investigate interactions greatly differ and do not necessarily generate comparable results. For example, there is evidence that claudin-1 is able to trans-interact with claudin-1 and -3 (99, 291) but not with claudin-2 or claudin-4 (62, 99), but the ability of claudin-1 and -5 to trans-interact is a matter of debate (58, 62, 291). Claudin-5/claudin-5 trans-interactions have been demonstrated to originate from hydrophobic interactions between amino acids of the second extracellular loops and as the involved amino acids are conserved in many claudins, these findings may be of general validity (292). However, investigations employing single-molecule force spectroscopy to study claudin-2/claudin-2 trans-interactions indicated that overall adhesion strength is to a large extent due to electrostatic interactions (214). Cis-interactions in claudin-2 homodimerization occur through the second transmembrane region (371). In contrast, claudin-4 appears to be unable to form homodimers (371). Interaction between claudin-16/-19 and between claudin-4/-8 have been reported to produce paracellular cation and anion permeabilities, respectively (145, 146); however, the mode of these interactions is yet unsolved. Many open questions regarding claudin interactions will probably only be solved once detailed structural information on claudins and claudin multimers becomes available.
IX. CLAUDINS AND HUMAN DISEASE
As described in section VI, claudin mutations are known to cause four Mendelian inherited disorders, neonatal sclerosing cholangitis with ichthyosis (claudin 1 mutations, OMIM 607626), autosomal recessive, nonsyndromic deafness (claudin-14, OMIM 614035), familial hypomagnesemic hypercalciuria with nephrocalcinosis (FHHNC, due to claudin-16 mutations, OMIM 248250), and FHHNC with ocular involvement (claudin-19, OMIM 248190). In addition, polymorphisms in claudin genes have been found to be associated with polygenic diseases, including claudin-1 with atopic dermatitis (63), claudin-5 with schizophrenia (337, 408), and claudin-14 in kidney stone disease (351).
Acquired abnormalities of claudin expression or localization are likely to play important roles in clinical disease. For example, claudin-2 is consistently upregulated in human inflammatory bowel disease (389, 420), as well as in animal models (390), and likely contributes significantly to the increased intestinal ion permeability in this disease, which causes leak flux diarrhea.
Aberrant expression of claudins is also common in epithelial cancers. Different cancer sites and histological subtypes seem to affect different claudins. In general though, the most common claudins to be affected are claudin-3 and -4, which are usually upregulated in cancers, and claudin-1 and -7 which can be both up- and downregulated. In vitro studies suggest that dysregulation of claudin expression may play a pathogenic role in tumorigenesis, but the mechanisms seem to vary between different cancers and claudin isoforms and are mostly poorly understood. The interested reader is referred to two recent reviews for a detailed discussion (359, 363).
Finally, claudins expressed on hepatocytes have a role as co-receptors for infection by the hepatitis C virus (37).
X. SUMMARY
In summary, 14 years after the initial discovery of claudins we now know that there are ∼27 mammalian genes. We know that they form the paracellular barrier and pore at the tight junction of endothelial cells. We know a great deal about the distribution of claudins in different tissues, and for some, we now understand their role in normal physiological function and disease processes. At the molecular level we have a rudimentary understanding of how they interact to form the paracellular barrier and understand some differences in the functions of the protein domains.
There remain, though, many exciting challenges in this field. We still do not know the tertiary and quaternary structure of claudins and how exactly they interact to form tight junction strands, so high-resolution structural information is needed. We have limited understanding of how different claudins interact with each other or with occludin and tricellulin, and the functional importance of this. There is no readily accessible technique to observe molecular level permeation events through claudin pores. We know very little about additional roles for claudins other than determining paracellular permeability, which might include potential interactions with other intracellular, intramembrane, or extracellular molecules and signaling functions. We have scant knowledge of how claudins are regulated and in particular the role of the cytoplasmic tail and of posttranslational modifications. We have barely begun the task of dissecting out the physiological role of each claudin in each tissue where it is expressed, and wider generation and dissemination of both global and tissue-specific claudin knockout mouse models is needed. Finally, it is likely that many more roles will be identified for claudins in the pathogenesis of inherited, particularly polygenic disease, and in acquired human diseases.
GRANTS
This work was supported by National Institutes of Health Grants DK062283 and U01GM094627 (to A. Yu) and Deutsche Forschungsgemeinschaft Research Unit FOR 721/2 (to D. Günzel).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
ACKNOWLEDGMENTS
Address for reprint requests and other correspondence: A. S. L. Yu, Kidney Institute, University of Kansas Medical Center, 3901 Rainbow Blvd., 6018 WHE, Kansas City, KS 66160-3018 (e-mail: ayu@kumc.edu).
REFERENCES
- 1. Abuazza G, Becker A, Williams SS, Chakravarty S, Truong HT, Lin F, Baum M. Claudins 6, 9, 13 are developmentally expressed renal tight junction proteins. Am J Physiol Renal Physiol 291: F1132–F1141, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Acharya P, Beckel J, Ruiz WG, Wang E, Rojas R, Birder L, Apodaca G. Distribution of the tight junction proteins ZO-1, occludin, claudin-4, -8, and -12 in bladder epithelium. Am J Physiol Renal Physiol 287: F305–F318, 2004 [DOI] [PubMed] [Google Scholar]
- 3. Aguilar PS, Engel A, Walter P. The plasma membrane proteins Prm1 and Fig. 1 ascertain fidelity of membrane fusion during yeast mating. Mol Biol Cell 18: 547–556, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ahmad W, Shabbiri K, Ijaz B, Asad S, Sarwar MT, Gull S, Kausar H, Fouzia K, Shahid I, Hassan S. Claudin-1 required for HCV virus entry has high potential for phosphorylation and O-glycosylation. Virol J 8: 229, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aijaz S, Balda MS, Matter K. Tight junctions: molecular architecture and function. Int Rev Cytol 248: 261–298, 2006 [DOI] [PubMed] [Google Scholar]
- 6. Alexandre MD, Jeansonne BG, Renegar RH, Tatum R, Chen YH. The first extracellular domain of claudin-7 affects paracellular Cl- permeability. Biochem Biophys Res Commun 357: 87–91, 2007 [DOI] [PubMed] [Google Scholar]
- 7. Alexandre MD, Lu Q, Chen YH. Overexpression of claudin-7 decreases the paracellular Cl− conductance and increases the paracellular Na+ conductance in LLC-PK1 cells. J Cell Sci 118: 2683–2693, 2005 [DOI] [PubMed] [Google Scholar]
- 8. Alvarez FJ, Douglas LM, Rosebrock A, Konopka JB. The Sur7 protein regulates plasma membrane organization and prevents intracellular cell wall growth in Candida albicans. Mol Biol Cell 19: 5214–5225, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Amasheh M, Fromm A, Krug SM, Amasheh S, Andres S, Zeitz M, Fromm M, Schulzke JD. TNFalpha-induced and berberine-antagonized tight junction barrier impairment via tyrosine kinase, Akt and NFkappaB signaling. J Cell Sci 123: 4145–4155, 2010 [DOI] [PubMed] [Google Scholar]
- 10. Amasheh S, Meiri N, Gitter A.H, Schoneberg T, Mankertz J, Schulzke J.D, Fromm M. Claudin-2 expression induces cation-selective channels in tight junctions of epithelial cells. J Cell Sci 115: 4969–4976, 2002 [DOI] [PubMed] [Google Scholar]
- 11. Amasheh S, Milatz S, Krug SM, Bergs M, Amasheh M, Schulzke JD, Fromm M. Na+ absorption defends from paracellular back-leakage by claudin-8 upregulation. Biochem Biophys Res Commun 378: 45–50, 2009 [DOI] [PubMed] [Google Scholar]
- 12. Anderson JM, Van Itallie CM, Fanning AS. Setting up a selective barrier at the apical junction complex. Curr Opin Cell Biol 16: 140–145, 2004 [DOI] [PubMed] [Google Scholar]
- 13. Anderson WJ, Zhou Q, Alcalde V, Kaneko OF, Blank LJ, Sherwood RI, Guseh JS, Rajagopal J, Melton DA. Genetic targeting of the endoderm with claudin-6CreER. Dev Dyn 237: 504–512, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Angelow S, El-Husseini R, Kanzawa SA, Yu AS. Renal localization and function of the tight junction protein, claudin-19. Am J Physiol Renal Physiol 293: F166–F177, 2007 [DOI] [PubMed] [Google Scholar]
- 15. Angelow S, Kim KJ, Yu AS. Claudin-8 modulates paracellular permeability to acidic and basic ions in MDCK II cells. J Physiol 571: 15–26, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Angelow S, Schneeberger EE, Yu AS. Claudin-8 expression in renal epithelial cells augments the paracellular barrier by replacing endogenous claudin-2. J Membr Biol 215: 147–159, 2007 [DOI] [PubMed] [Google Scholar]
- 17. Angelow S, Yu AS. Cysteine mutagenesis to study the structure of claudin-2 paracellular pores. Ann NY Acad Sci 1165: 143–147, 2009 [DOI] [PubMed] [Google Scholar]
- 18. Angelow S, Yu AS. Structure-function studies of claudin extracellular domains by cysteine-scanning mutagenesis. J Biol Chem 284: 29205–29217, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aono S, Hirai Y. Phosphorylation of claudin-4 is required for tight junction formation in a human keratinocyte cell line. Exp Cell Res 314: 3326–3339, 2008 [DOI] [PubMed] [Google Scholar]
- 20. Arabzadeh A, Troy TC, Turksen K. Role of the Cldn6 cytoplasmic tail domain in membrane targeting and epidermal differentiation in vivo. Mol Cell Biol 26: 5876–5887, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Asaka M, Hirase T, Hashimoto-Komatsu A, Node K. Rab5a-mediated localization of claudin-1 is regulated by proteasomes in endothelial cells. Am J Physiol Cell Physiol 300: C87–C96, 2011 [DOI] [PubMed] [Google Scholar]
- 22. Aslam M, Ahmad N, Srivastava R, Hemmer B. TNF-alpha induced NFkappaB signaling and p65 (RelA) overexpression repress Cldn5 promoter in mouse brain endothelial cells. Cytokine 57: 269–275, 2012 [DOI] [PubMed] [Google Scholar]
- 23. Baas AF, Kuipers J, van der Wel NN, Batlle E, Koerten HK, Peters PJ, Clevers HC. Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell 116: 457–466, 2004 [DOI] [PubMed] [Google Scholar]
- 24. Bacallao R, Garfinkel A, Monke S, Zampighi G, Mandel LJ. ATP depletion: a novel method to study junctional properties in epithelial tissues. I. Rearrangement of the actin cytoskeleton. J Cell Sci 107: 3301–3313, 1994 [DOI] [PubMed] [Google Scholar]
- 25. Bagherie-Lachidan M, Wright SI, Kelly SP. Claudin-3 tight junction proteins in Tetraodon nigroviridis: cloning, tissue-specific expression, and a role in hydromineral balance. Am J Physiol Regul Integr Comp Physiol 294: R1638–R1647, 2008 [DOI] [PubMed] [Google Scholar]
- 26. Bagnat M, Cheung ID, Mostov KE, Stainier DY. Genetic control of single lumen formation in the zebrafish gut. Nat Cell Biol 9: 954–960, 2007 [DOI] [PubMed] [Google Scholar]
- 27. Barry PH, Diamond JM, Wright EM. The mechanism of cation permeation in the rabbit gallbladder: dilution potentials and bi-ionic potentials. J Membr Biol 4: 358–394, 1971 [DOI] [PubMed] [Google Scholar]
- 28. Bazzoni G. Endothelial tight junctions: permeable barriers of the vessel wall. Thromb Haemost 95: 36–42, 2006 [PubMed] [Google Scholar]
- 29. Behr M, Riedel D, Schuh R. The claudin-like megatrachea is essential in septate junctions for the epithelial barrier function in Drosophila. Dev Cell 5: 611–620, 2003 [DOI] [PubMed] [Google Scholar]
- 30. Ben-Yosef T, Belyantseva IA, Saunders TL, Hughes ED, Kawamoto K, Van Itallie CM, Beyer LA, Halsey K, Gardner DJ, Wilcox ER, Rasmussen J, Anderson JM, Dolan DF, Forge A, Raphael Y, Camper SA, Friedman TB. Claudin 14 knockout mice, a model for autosomal recessive deafness DFNB29, are deaf due to cochlear hair cell degeneration. Hum Mol Genet 12: 2049–2061, 2003 [DOI] [PubMed] [Google Scholar]
- 31. Bezdekova M, Brychtova S, Sedlakova E, Langova K, Brychta T, Belej K. Analysis of snail-1, e-cadherin and claudin-1 expression in colorectal adenomas and carcinomas. Int J Mol Sci 13: 1632–1643, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Biggers JD, Bell JE, Benos DJ. Mammalian blastocyst: transport functions in a developing epithelium. Am J Physiol Cell Physiol 255: C419–C432, 1988 [DOI] [PubMed] [Google Scholar]
- 33. Blackman B, Russell T, Nordeen SK, Medina D, Neville MC. Claudin 7 expression and localization in the normal murine mammary gland and murine mammary tumors. Breast Cancer Res 7: R248–255, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blanchard AA, Watson PH, Shiu RP, Leygue E, Nistor A, Wong P, Myal Y. Differential expression of claudin 1, 3, and 4 during normal mammary gland development in the mouse. DNA Cell Biol 25: 79–86, 2006 [DOI] [PubMed] [Google Scholar]
- 35. Blasig IE, Bellmann C, Cording J, Del Vecchio G, Zwanziger D, Huber O, Haseloff RF. Occludin protein family: oxidative stress and reducing conditions. Antioxid Redox Signal 15: 1195–1219, 2011 [DOI] [PubMed] [Google Scholar]
- 36. Blasig IE, Winkler L, Lassowski B, Mueller S.L, Zuleger N, Krause E, Krause G, Gast K, Kolbe M, Piontek J. On the self-association potential of transmembrane tight junction proteins. Cell Mol Life Sci 63: 505–514, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bonander N, Jamshad M, Hu K, Farquhar MJ, Stamataki Z, Balfe P, McKeating JA, Bill RM. Structural characterization of CD81-Claudin-1 hepatitis C virus receptor complexes. Biochem Soc Trans 39: 537–540, 2011 [DOI] [PubMed] [Google Scholar]
- 38. Brandner JM, Kief S, Grund C, Rendl M, Houdek P, Kuhn C, Tschachler E, Franke WW, Moll I. Organization and formation of the tight junction system in human epidermis and cultured keratinocytes. Eur J Cell Biol 81: 253–263, 2002 [DOI] [PubMed] [Google Scholar]
- 39. Briggs DC, Naylor CE, Smedley JG, 3rd, Lukoyanova N, Robertson S, Moss DS, McClane BA, Basak AK. Structure of the food-poisoning Clostridium perfringens enterotoxin reveals similarity to the aerolysin-like pore-forming toxins. J Mol Biol 413: 138–149, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brown RC, Mark KS, Egleton RD, Huber JD, Burroughs AR, Davis TP. Protection against hypoxia-induced increase in blood-brain barrier permeability: role of tight junction proteins and NFkappaB. J Cell Sci 116: 693–700, 2003 [DOI] [PubMed] [Google Scholar]
- 41. Burek M, Forster CY. Cloning and characterization of the murine claudin-5 promoter. Mol Cell Endocrinol 298: 19–24, 2009 [DOI] [PubMed] [Google Scholar]
- 42. Burgel N, Bojarski C, Mankertz J, Zeitz M, Fromm M, Schulzke JD. Mechanisms of diarrhea in collagenous colitis. Gastroenterology 123: 433–443, 2002 [DOI] [PubMed] [Google Scholar]
- 43. Butt AM, Khan IB, Hussain M, Idress M, Lu J, Tong Y. Role of post translational modifications and novel crosstalk between phosphorylation and O-beta-GlcNAc modifications in human claudin-1, -3 and -4. Mol Biol Rep 39: 1359–1369, 2011 [DOI] [PubMed] [Google Scholar]
- 44. Caulfield JP, Reid JJ, Farquhar MG. Alterations of the glomerular epithelium in acute aminonucleoside nephrosis. Evidence for formation of occluding junctions and epithelial cell detachment. Lab Invest 34: 43–59, 1976 [PubMed] [Google Scholar]
- 45. Cereijido M, Robbins ES, Dolan WJ, Rotunno CA, Sabatini DD. Polarized monolayers formed by epithelial cells on a permeable and translucent support. J Cell Biol 77: 853–880, 1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chasiotis H, Kolosov D, Kelly SP. Permeability properties of the teleost gill epithelium under ion-poor conditions. Am J Physiol Regul Integr Comp Physiol 302: R727–R739, 2012 [DOI] [PubMed] [Google Scholar]
- 47. Chen SP, Zhou B, Willis BC, Sandoval AJ, Liebler JM, Kim KJ, Ann DK, Crandall ED, Borok Z. Effects of transdifferentiation and EGF on claudin isoform expression in alveolar epithelial cells. J Appl Physiol 98: 322–328, 2005 [DOI] [PubMed] [Google Scholar]
- 48. Chiba H, Gotoh T, Kojima T, Satohisa S, Kikuchi K, Osanai M, Sawada N. Hepatocyte nuclear factor (HNF)-4alpha triggers formation of functional tight junctions and establishment of polarized epithelial morphology in F9 embryonal carcinoma cells. Exp Cell Res 286: 288–297, 2003 [DOI] [PubMed] [Google Scholar]
- 49. Chiba H, Itoh T, Satohisa S, Sakai N, Noguchi H, Osanai M, Kojima T, Sawada N. Activation of p21CIP1/WAF1 gene expression and inhibition of cell proliferation by overexpression of hepatocyte nuclear factor-4alpha. Exp Cell Res 302: 11–21, 2005 [DOI] [PubMed] [Google Scholar]
- 50. Chiba H, Osanai M, Murata M, Kojima T, Sawada N. Transmembrane proteins of tight junctions. Biochim Biophys Acta 1778: 588–600, 2008 [DOI] [PubMed] [Google Scholar]
- 51. Christophe-Hobertus C, Szpirer C, Guyon R, Christophe D. Identification of the gene encoding brain cell membrane protein 1 (BCMP1), a putative four-transmembrane protein distantly related to the peripheral myelin protein 22/epithelial membrane proteins and the claudins. BMC Genomics 2: 3, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cieply B, Riley PT, Pifer PM, Widmeyer J, Addison JB, Ivanov AV, Denvir J, Frisch SM. Suppression of the epithelial-mesenchymal transition by grainyhead-like-2. Cancer Res 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Claude P. Morphological factors influencing transepithelial permeability: a model for the resistance of the zonula occludens. J Membr Biol 39: 219–232, 1978 [DOI] [PubMed] [Google Scholar]
- 54. Claude P, Goodenough DA. Fracture faces of zonulae occludentes from “tight” and “leaky” epithelia. J Cell Biol 58: 390–400, 1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Clemente-Ramos JA, Martin-Garcia R, Sharifmoghadam MR, Konomi M, Osumi M, Valdivieso MH. The tetraspan protein Dni1p is required for correct membrane organization and cell wall remodelling during mating in Schizosaccharomyces pombe. Mol Microbiol 73: 695–709, 2009 [DOI] [PubMed] [Google Scholar]
- 56. Colegio OR, Van Itallie C, Rahner C, Anderson JM. Claudin extracellular domains determine paracellular charge selectivity and resistance but not tight junction fibril architecture. Am J Physiol Cell Physiol 284: C1346–C1354, 2003 [DOI] [PubMed] [Google Scholar]
- 57. Colegio OR, Van Itallie CM, McCrea HJ, Rahner C, Anderson JM. Claudins create charge-selective channels in the paracellular pathway between epithelial cells. Am J Physiol Cell Physiol 283: C142–C147, 2002 [DOI] [PubMed] [Google Scholar]
- 58. Coyne CB, Gambling TM, Boucher RC, Carson JL, Johnson LG. Role of claudin interactions in airway tight junctional permeability. Am J Physiol Lung Cell Mol Physiol 285: L1166–L1178, 2003 [DOI] [PubMed] [Google Scholar]
- 59. Cukierman L, Meertens L, Bertaux C, Kajumo F, Dragic T. Residues in a highly conserved claudin-1 motif are required for hepatitis C virus entry and mediate the formation of cell-cell contacts. J Virol 83: 5477–5484, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. D'Souza T, Agarwal R, Morin PJ. Phosphorylation of claudin-3 at threonine 192 by cAMP-dependent protein kinase regulates tight junction barrier function in ovarian cancer cells. J Biol Chem 280: 26233–26240, 2005 [DOI] [PubMed] [Google Scholar]
- 61. D'Souza T, Sherman-Baust CA, Poosala S, Mullin JM, Morin PJ. Age-related changes of claudin expression in mouse liver, kidney, and pancreas. J Gerontol A Biol Sci Med Sci 64: 1146–1153, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Daugherty BL, Ward C, Smith T, Ritzenthaler JD, Koval M. Regulation of heterotypic claudin compatibility. J Biol Chem 282: 30005–30013, 2007 [DOI] [PubMed] [Google Scholar]
- 63. De Benedetto A, Rafaels NM, McGirt LY, Ivanov AI, Georas SN, Cheadle C, Berger AE, Zhang K, Vidyasagar S, Yoshida T, Boguniewicz M, Hata T, Schneider LC, Hanifin JM, Gallo RL, Novak N, Weidinger S, Beaty TH, Leung DY, Barnes KC, Beck LA. Tight junction defects in patients with atopic dermatitis. J Allergy Clin Immunol 127: 773–786, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. De Rouffignac C, Quamme G. Renal magnesium handling and its hormonal control. Physiol Rev 74: 305–322, 1994 [DOI] [PubMed] [Google Scholar]
- 65. Deng A, Miracle CM, Lortie M, Satriano J, Gabbai FB, Munger KA, Thomson SC, Blantz RC. Kidney oxygen consumption, carbonic anhydrase, and proton secretion. Am J Physiol Renal Physiol 290: F1009–F1015, 2006 [DOI] [PubMed] [Google Scholar]
- 66. Devaux J, Gow A. Tight junctions potentiate the insulative properties of small CNS myelinated axons. J Cell Biol 183: 909–921, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dhasarathy A, Phadke D, Mav D, Shah RR, Wade PA. The transcription factors Snail and Slug activate the transforming growth factor-beta signaling pathway in breast cancer. PLoS One 6: e26514, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Diamond JM. Tight and leaky junctions of epithelia: a perspective on kisses in the dark. Federation Proc 33: 2220–2224, 1974 [PubMed] [Google Scholar]
- 69. Diamond JM. Twenty-first Bowditch lecture. The epithelial junction: bridge, gate, and fence. Physiologist 20: 10–18, 1977 [PubMed] [Google Scholar]
- 70. Ding L, Lu Z, Foreman O, Tatum R, Lu Q, Renegar R, Cao J, Chen YH. Inflammation and disruption of the mucosal architecture in claudin-7-deficient mice. Gastroenterology 142: 305–315, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Douglas LM, Wang HX, Keppler-Ross S, Dean N, Konopka JB. Sur7 promotes plasma membrane organization and is needed for resistance to stressful conditions and to the invasive growth and virulence of Candida albicans. MBio 3: 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dube E, Chan PT, Hermo L, Cyr DG. Gene expression profiling and its relevance to the blood-epididymal barrier in the human epididymis. Biol Reprod 76: 1034–1044, 2007 [DOI] [PubMed] [Google Scholar]
- 73. Duffy HS, John GR, Lee SC, Brosnan CF, Spray DC. Reciprocal regulation of the junctional proteins claudin-1 and connexin43 by interleukin-1beta in primary human fetal astrocytes. J Neurosci 20: RC114, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Duffy N.M, Bui P, Bagherie-Lachidan M, Kelly SP. Epithelial remodeling and claudin mRNA abundance in the gill and kidney of puffer fish (Tetraodon biocellatus) acclimated to altered environmental ion levels. J Comp Physiol B 181: 219–238, 2011 [DOI] [PubMed] [Google Scholar]
- 75. Dufresne J, Cyr DG. Activation of an SP binding site is crucial for the expression of claudin 1 in rat epididymal principal cells. Biol Reprod 76: 825–832, 2007 [DOI] [PubMed] [Google Scholar]
- 76. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Eisenman G. Cation selective glass electrodes and their mode of operation. Biophys J 2: 259–323, 1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Elkouby-Naor L, Abassi Z, Lagziel A, Gow A, Ben-Yosef T. Double gene deletion reveals lack of cooperation between claudin 11 and claudin 14 tight junction proteins. Cell Tissue Res 333: 427–438, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Enck AH, Berger UV, Yu AS. Claudin-2 is selectively expressed in proximal nephron in mouse kidney. Am J Physiol Renal Physiol 281: F966–F974, 2001 [DOI] [PubMed] [Google Scholar]
- 80. Engelund MB, Yu AS, Li J, Madsen SS, Faergeman NJ, Tipsmark CK. Functional characterization and localization of a gill-specific claudin isoform in Atlantic salmon. Am J Physiol Regul Integr Comp Physiol 302: R300–R311, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Epple HJ, Schneider T, Troeger H, Kunkel D, Allers K, Moos V, Amasheh M, Loddenkemper C, Fromm M, Zeitz M, Schulzke J.D. Impairment of the intestinal barrier is evident in untreated but absent in suppressively treated HIV-infected patients. Gut 58: 220–227, 2009 [DOI] [PubMed] [Google Scholar]
- 82. Erdman S, Lin L, Malczynski M, Snyder M. Pheromone-regulated genes required for yeast mating differentiation. J Cell Biol 140: 461–483, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Escaffit F, Boudreau F, Beaulieu JF. Differential expression of claudin-2 along the human intestine: implication of GATA-4 in the maintenance of claudin-2 in differentiating cells. J Cell Physiol 203: 15–26, 2005 [DOI] [PubMed] [Google Scholar]
- 84. Farquhar MG, Palade GE. Junctional complexes in various epithelia. J Cell Biol 17: 375–412, 1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. French AD, Fiori JL, Camilli TC, Leotlela PD, O'Connell MP, Frank BP, Subaran S, Indig FE, Taub DD, Weeraratna AT. PKC and PKA phosphorylation affect the subcellular localization of claudin-1 in melanoma cells. Int J Med Sci 6: 93–101, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Friend DS, Gilula NB. Variations in tight and gap junctions in mammalian tissues. J Cell Biol 53: 758–776, 1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Frizzell RA, Schultz SG. Ionic conductances of extracellular shunt pathway in rabbit ileum. Influence of shunt on transmural sodium transport and electrical potential differences. J Gen Physiol 59: 318–346, 1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Frömter E, Diamond J. Route of passive ion permeation in epithelia. Nat New Biol 235: 9–13, 1972 [DOI] [PubMed] [Google Scholar]
- 89. Fujibe M, Chiba H, Kojima T, Soma T, Wada T, Yamashita T, Sawada N. Thr203 of claudin-1, a putative phosphorylation site for MAP kinase, is required to promote the barrier function of tight junctions. Exp Cell Res 295: 36–47, 2004 [DOI] [PubMed] [Google Scholar]
- 90. Fujimoto K. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. Application to the immunogold labeling of intercellular junctional complexes. J Cell Sci 108: 3443–3449, 1995 [DOI] [PubMed] [Google Scholar]
- 91. Fujita H, Chiba H, Yokozaki H, Sakai N, Sugimoto K, Wada T, Kojima T, Yamashita T, Sawada N. Differential expression and subcellular localization of claudin-7, -8, -12, -13, and -15 along the mouse intestine. J Histochem Cytochem 54: 933–944, 2006 [DOI] [PubMed] [Google Scholar]
- 92. Fujita H, Sugimoto K, Inatomi S, Maeda T, Osanai M, Uchiyama Y, Yamamoto Y, Wada T, Kojima T, Yokozaki H, Yamashita T, Kato S, Sawada N, Chiba H. Tight junction proteins claudin-2 and -12 are critical for vitamin D-dependent Ca2+ absorption between enterocytes. Mol Biol Cell 19: 1912–1921, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Fujita K, Katahira J, Horiguchi Y, Sonoda N, Furuse M, Tsukita S. Clostridium perfringens enterotoxin binds to the second extracellular loop of claudin-3, a tight junction integral membrane protein. FEBS Lett 476: 258–261, 2000 [DOI] [PubMed] [Google Scholar]
- 94. Fukasawa H, Bornheimer S, Kudlicka K, Farquhar MG. Slit diaphragms contain tight junction proteins. J Am Soc Nephrol 20: 1491–1503, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol 141: 1539–1550, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Furuse M, Furuse K, Sasaki H, Tsukita S. Conversion of zonulae occludentes from tight to leaky strand type by introducing claudin-2 into madin-darby canine kidney I cells. J Cell Biol 153: 263–272, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Furuse M, Hata M, Furuse K, Yoshida Y, Haratake A, Sugitani Y, Noda T, Kubo A, Tsukita S. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: a lesson from claudin-1-deficient mice. J Cell Biol 156: 1099–1111, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Furuse M, Sasaki H, Fujimoto K, Tsukita S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J Cell Biol 143: 391–401, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Furuse M, Sasaki H, Tsukita S. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J Cell Biol 147: 891–903, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gendron F.P, Mongrain S, Laprise P, McMahon S, Dubois C.M, Blais M, Asselin C, Rivard N. The CDX2 transcription factor regulates furin expression during intestinal epithelial cell differentiation. Am J Physiol Gastrointest Liver Physiol 290: G310–G318, 2006 [DOI] [PubMed] [Google Scholar]
- 101. Giacomelli F, Wiener J. Specialized junction in the distal convoluted tubule of rat kidney. Anat Rec 185: 197–207, 1976 [DOI] [PubMed] [Google Scholar]
- 102. Gonen T, Hite R.K, Cheng Y, Petre B.M, Kistler J, Walz T. Polymorphic assemblies and crystalline arrays of lens tetraspanin MP20. J Mol Biol 376: 380–392, 2008 [DOI] [PubMed] [Google Scholar]
- 103. Gong Y, Renigunta V, Himmerkus N, Zhang J, Renigunta A, Bleich M, Hou J. Claudin-14 regulates renal Ca2+ transport in response to CaSR signalling via a novel microRNA pathway. EMBO J 31: 1999–2012, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. González-Mariscal L, Chávez de Ramirez B, Lázaro AMC. Establishment of tight junctions between cells from different animal species and different sealing capacities J Membr Biol 107: 43–56, 1989 [DOI] [PubMed] [Google Scholar]
- 105. González-Mariscal L, Garay E, Quirós M. Regulation of claudins by posttranslational modifications and cell-signaling cascades. Curr Topics Membr 65: 113–150, 2010 [Google Scholar]
- 106. Gow A, Davies C, Southwood CM, Frolenkov G, Chrustowski M, Ng L, Yamauchi D, Marcus DC, Kachar B. Deafness in Claudin 11-null mice reveals the critical contribution of basal cell tight junctions to stria vascularis function. J Neuroscie 24: 7051–7062, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Gow A, Devaux J. A model of tight junction function in central nervous system myelinated axons. Neuron Glia Biol 4: 307–317, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Gow A, Southwood CM, Li JS, Pariali M, Riordan GP, Brodie SE, Danias J, Bronstein JM, Kachar B, Lazzarini RA. CNS myelin and sertoli cell tight junction strands are absent in Osp/claudin-11 null mice. Cell 99: 649–659, 1999 [DOI] [PubMed] [Google Scholar]
- 109. Green CR, Bergquist PR. Phylogenetic-relationships within the invertebrata in relation to the structure of septate junctions and the development of occluding junctional types. J Cell Sci 53: 279–305, 1982 [Google Scholar]
- 110. Green CR, Bergquist PR, Bullivant S. An anastomosing septate junction in endothelial cells of the phylum echinodermata. J Ultrastruct Res 68: 72–80, 1979 [DOI] [PubMed] [Google Scholar]
- 111. Green WN, Andersen OS. Surface charges and ion channel function. Annu Rev Physiol 53: 341–359, 1991 [DOI] [PubMed] [Google Scholar]
- 112. Greenwell-Wild T, Moutsopoulos NM, Gliozzi M, Kapsogeorgou E, Rangel Z, Munson PJ, Moutsopoulos HM, Wahl SM. Chitinases in the salivary glands and circulation of patients with Sjogren's syndrome: macrophage harbingers of disease severity. Arthritis Rheum 63: 3103–3115, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Greger R. Cation selectivity of the isolated perfused cortical thick ascending limb of Henle's loop of rabbit kidney. Pflügers Arch 390: 30–37, 1981 [DOI] [PubMed] [Google Scholar]
- 114. Grey AC, Jacobs MD, Gonen T, Kistler J, Donaldson PJ. Insertion of MP20 into lens fibre cell plasma membranes correlates with the formation of an extracellular diffusion barrier. Exp Eye Res 77: 567–574, 2003 [DOI] [PubMed] [Google Scholar]
- 115. Grosse B, Cassio D, Yousef N, Bernardo C, Jacquemin E, Gonzales E. Claudin-1 involved in neonatal ichthyosis-sclerosing cholangitis syndrome, regulates hepatic paracellular permeability. Hepatology 2011 [DOI] [PubMed] [Google Scholar]
- 116. Grotegut S, von Schweinitz D, Christofori G, Lehembre F. Hepatocyte growth factor induces cell scattering through MAPK/Egr-1-mediated upregulation of Snail. EMBO J 25: 3534–3545, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Guan X, Inai T, Shibata Y. Segment-specific expression of tight junction proteins, claudin-2 and -10, in the rat epididymal epithelium. Arch Histol Cytol 68: 213–225, 2005 [DOI] [PubMed] [Google Scholar]
- 118. Guillemot L, Paschoud S, Pulimeno P, Foglia A, Citi S. The cytoplasmic plaque of tight junctions: a scaffolding and signalling center. Biochim Biophys Acta 1778: 601–613, 2008 [DOI] [PubMed] [Google Scholar]
- 119. Günzel D, Amasheh S, Pfaffenbach S, Richter JF, Kausalya PJ, Hunziker W, Fromm M. Claudin-16 affects transcellular Cl− secretion in MDCK cells. J Physiol 587: 3777–3793, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Günzel D, Fromm M. Claudins and other tight junction proteins. Comp Physiol 2: 1819–1852, 2012 [DOI] [PubMed] [Google Scholar]
- 121. Günzel D, Haisch L, Pfaffenbach S, Krug S.M, Milatz S, Amasheh S, Hunziker W, Müller D. Claudin function in the thick ascending limb of Henle's loop. Ann NY Acad Sci 1165: 152–162, 2009 [DOI] [PubMed] [Google Scholar]
- 122. Günzel D, Stuiver M, Kausalya PJ, Haisch L, Krug SM, Rosenthal R, Meij IC, Hunziker W, Fromm M, Muller D. Claudin-10 exists in six alternatively spliced isoforms that exhibit distinct localization and function. J Cell Sci 122: 1507–1517, 2009 [DOI] [PubMed] [Google Scholar]
- 123. Günzel D, Yu AS. Function and regulation of claudins in the thick ascending limb of Henle. Pflügers Arch 458: 77–88, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Guo P, Weinstein AM, Weinbaum S. A dual-pathway ultrastructural model for the tight junction of rat proximal tubule epithelium. Am J Physiol Renal Physiol 285: F241–F257, 2003 [DOI] [PubMed] [Google Scholar]
- 125. Hadj-Rabia S, Baala L, Vabres P, Hamel-Teillac D, Jacquemin E, Fabre M, Lyonnet S, De Prost Y, Munnich A, Hadchouel M, Smahi A. Claudin-1 gene mutations in neonatal sclerosing cholangitis associated with ichthyosis: a tight junction disease. Gastroenterology 127: 1386–1390, 2004 [DOI] [PubMed] [Google Scholar]
- 126. Halder SK, Rachakonda G, Deane NG, Datta PK. Smad7 induces hepatic metastasis in colorectal cancer. Br J Cancer 99: 957–965, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Hamazaki Y, Itoh M, Sasaki H, Furuse M, Tsukita S. Multi-PDZ domain protein 1 (MUPP1) is concentrated at tight junctions through its possible interaction with claudin-1 and junctional adhesion molecule. J Biol Chem 277: 455–461, 2002 [DOI] [PubMed] [Google Scholar]
- 128. Han Y, Mu Y, Li X, Xu P, Tong J, Liu Z, Ma T, Zeng G, Yang S, Du J, Meng A. Grhl2 deficiency impairs otic development and hearing ability in a zebrafish model of the progressive dominant hearing loss DFNA28. Hum Mol Genet 20: 3213–3226, 2011 [DOI] [PubMed] [Google Scholar]
- 129. Hanna PC, Mietzner TA, Schoolnik GK, McClane BA. Localization of the receptor-binding region of Clostridium perfringens enterotoxin utilizing cloned toxin fragments and synthetic peptides. The 30 C-terminal amino acids define a functional binding region. J Biol Chem 266: 11037–11043, 1991 [PubMed] [Google Scholar]
- 130. Harder JL, Margolis B. SnapShot: tight and adherens junction signaling. Cell 133: 1118, 2008 [DOI] [PubMed] [Google Scholar]
- 131. Harris HJ, Davis C, Mullins JG, Hu K, Goodall M, Farquhar MJ, Mee CJ, McCaffrey K, Young S, Drummer H, Balfe P, McKeating JA. Claudin association with CD81 defines hepatitis C virus entry. J Biol Chem 285: 21092–21102, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Hashimoto K, Oshima T, Tomita T, Kim Y, Matsumoto T, Joh T, Miwa H. Oxidative stress induces gastric epithelial permeability through claudin-3. Biochem Biophys Res Commun 376: 154–157, 2008 [DOI] [PubMed] [Google Scholar]
- 133. Hashizume A, Ueno T, Furuse M, Tsukita S, Nakanishi Y, Hieda Y. Expression patterns of claudin family of tight junction membrane proteins in developing mouse submandibular gland. Dev Dyn 231: 425–431, 2004 [DOI] [PubMed] [Google Scholar]
- 134. Hayashi D, Tamura A, Tanaka H, Yamazaki Y, Watanabe S, Suzuki K, Suzuki K, Sentani K, Yasui W, Rakugi H, Isaka Y, Tsukita S. Deficiency of claudin-18 causes paracellular H+ leakage, up-regulation of interleukin-1beta, and atrophic gastritis in mice. Gastroenterology 142: 292–304, 2012 [DOI] [PubMed] [Google Scholar]
- 135. He YL, Murby S, Warhurst G, Gifford L, Walker D, Ayrton J, Eastmond R, Rowland M. Species differences in size discrimination in the paracellular pathway reflected by oral bioavailability of poly(ethylene glycol) and D-peptides. J Pharm Sci 87: 626–633, 1998 [DOI] [PubMed] [Google Scholar]
- 136. Hering NA, Andres S, Fromm A, van Tol EA, Amasheh M, Mankertz J, Fromm M, Schulzke JD. Transforming growth factor-beta, a whey protein component, strengthens the intestinal barrier by upregulating claudin-4 in HT-29/B6 cells. J Nutr 141: 783–789, 2011 [DOI] [PubMed] [Google Scholar]
- 137. Hernandez S, Chavez Munguia B, Gonzalez-Mariscal L. ZO-2 silencing in epithelial cells perturbs the gate and fence function of tight junctions and leads to an atypical monolayer architecture. Exp Cell Res 313: 1533–1547, 2007 [DOI] [PubMed] [Google Scholar]
- 138. Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer 6: 186, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Holmes JL, Van Itallie CM, Rasmussen JE, Anderson JM. Claudin profiling in the mouse during postnatal intestinal development and along the gastrointestinal tract reveals complex expression patterns. Gene Expr Patterns 6: 581–588, 2006 [DOI] [PubMed] [Google Scholar]
- 140. Honda H, Pazin MJ, Ji H, Wernyj RP, Morin PJ. Crucial roles of Sp1 and epigenetic modifications in the regulation of the CLDN4 promoter in ovarian cancer cells. J Biol Chem 281: 21433–21444, 2006 [DOI] [PubMed] [Google Scholar]
- 141. Hortsch M, Margolis B. Septate and paranodal junctions: kissing cousins. Trends Cell Biol 13: 557–561, 2003 [DOI] [PubMed] [Google Scholar]
- 142. Hou J, Gomes AS, Paul DL, Goodenough DA. Study of claudin function by RNA interference. J Biol Chem 281: 36117–36123, 2006 [DOI] [PubMed] [Google Scholar]
- 143. Hou J, Paul DL, Goodenough DA. Paracellin-1 and the modulation of ion selectivity of tight junctions. J Cell Sci 118: 5109–5118, 2005 [DOI] [PubMed] [Google Scholar]
- 144. Hou J, Renigunta A, Gomes AS, Hou M, Paul DL, Waldegger S, Goodenough DA. Claudin-16 and claudin-19 interaction is required for their assembly into tight junctions and for renal reabsorption of magnesium. Proc Natl Acad Sci USA 106: 15350–15355, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Hou J, Renigunta A, Konrad M, Gomes AS, Schneeberger EE, Paul DL, Waldegger S, Goodenough DA. Claudin-16 and claudin-19 interact and form a cation-selective tight junction complex. J Clin Invest 118: 619–628, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Hou J, Renigunta A, Yang J, Waldegger S. Claudin-4 forms paracellular chloride channel in the kidney and requires claudin-8 for tight junction localization. Proc Natl Acad Sci USA 107: 18010–18015, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Hou J, Shan Q, Wang T, Gomes AS, Yan Q, Paul DL, Bleich M, Goodenough DA. Transgenic RNAi depletion of claudin-16 and the renal handling of magnesium. J Biol Chem 282: 17114–17122, 2007 [DOI] [PubMed] [Google Scholar]
- 148. Hua VB, Chang AB, Tchieu JH, Kumar NM, Nielsen PA, Saier MH., Jr Sequence and phylogenetic analyses of 4 TMS junctional proteins of animals: connexins, innexins, claudins and occludins. J Membr Biol 194: 59–76, 2003 [DOI] [PubMed] [Google Scholar]
- 149. Humbert F, Montesano R, Perrelet A, Orci L. Junctions in developing human and rat kidney: a freeze-fracture study. J Ultrastruct Res 56: 202–214, 1976 [DOI] [PubMed] [Google Scholar]
- 150. Hyun CS, Chen CW, Shinowara NL, Palaia T, Fallick FS, Martello LA, Mueenuddin M, Donovan VM, Teichberg S. Morphological factors influencing transepithelial conductance in a rabbit model of ileitis. Gastroenterology 109: 13–23, 1995 [DOI] [PubMed] [Google Scholar]
- 151. Ikari A, Atomi K, Takiguchi A, Yamazaki Y, Miwa M, Sugatani J. Epidermal growth factor increases claudin-4 expression mediated by Sp1 elevation in MDCK cells. Biochem Biophys Res Commun 384: 306–310, 2009 [DOI] [PubMed] [Google Scholar]
- 152. Ikari A, Hirai N, Shiroma M, Harada H, Sakai H, Hayashi H, Suzuki Y, Degawa M, Takagi K. Association of paracellin-1 with ZO-1 augments the reabsorption of divalent cations in renal epithelial cells. J Biol Chem 279: 54826–54832, 2004 [DOI] [PubMed] [Google Scholar]
- 153. Ikari A, Kinjo K, Atomi K, Sasaki Y, Yamazaki Y, Sugatani J. Extracellular Mg2+ regulates the tight junctional localization of claudin-16 mediated by ERK-dependent phosphorylation. Biochim Biophys Acta 1798: 415–421, 2010 [DOI] [PubMed] [Google Scholar]
- 154. Ikari A, Matsumoto S, Harada H, Takagi K, Hayashi H, Suzuki Y, Degawa M, Miwa M. Phosphorylation of paracellin-1 at Ser217 by protein kinase A is essential for localization in tight junctions. J Cell Sci 119: 1781–1789, 2006 [DOI] [PubMed] [Google Scholar]
- 155. Ikari A, Okude C, Sawada H, Sasaki Y, Yamazaki Y, Sugatani J, Degawa M, Miwa M. Activation of a polyvalent cation-sensing receptor decreases magnesium transport via claudin-16. Biochim Biophys Acta 1778: 283–290, 2008 [DOI] [PubMed] [Google Scholar]
- 156. Ikari A, Sato T, Takiguchi A, Atomi K, Yamazaki Y, Sugatani J. Claudin-2 knockdown decreases matrix metalloproteinase-9 activity and cell migration via suppression of nuclear Sp1 in A549 cells. Life Sci 88: 628–633, 2011 [DOI] [PubMed] [Google Scholar]
- 157. Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci 116: 1959–1967, 2003 [DOI] [PubMed] [Google Scholar]
- 158. Ikenouchi J, Suzuki M, Umeda K, Ikeda K, Taguchi R, Kobayashi T, Sato SB, Stolz DB, Umeda M. Lipid polarity is maintained in absence of tight junctions. J Biol Chem 287: 9525–9533, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Inai T, Kamimura T, Hirose E, Iida H, Shibata Y. The protoplasmic or exoplasmic face association of tight junction particles cannot predict paracellular permeability or heterotypic claudin compatibility. Eur J Cell Biol 89: 547–556, 2010 [DOI] [PubMed] [Google Scholar]
- 160. Inai T, Kobayashi J, Shibata Y. Claudin-1 contributes to the epithelial barrier function in MDCK cells. Eur J Cell Biol 78: 849–855, 1999 [DOI] [PubMed] [Google Scholar]
- 161. Inai T, Sengoku A, Guan X, Hirose E, Iida H, Shibata Y. Heterogeneity in expression and subcellular localization of tight junction proteins, claudin-10 and -15, examined by RT-PCR and immunofluorescence microscopy. Arch Histol Cytol 68: 349–360, 2005 [DOI] [PubMed] [Google Scholar]
- 162. Ishizaki T, Chiba H, Kojima T, Fujibe M, Soma T, Miyajima H, Nagasawa K, Wada I, Sawada N. Cyclic AMP induces phosphorylation of claudin-5 immunoprecipitates and expression of claudin-5 gene in blood-brain-barrier endothelial cells via protein kinase A-dependent and -independent pathways. Exp Cell Res 290: 275–288, 2003 [DOI] [PubMed] [Google Scholar]
- 163. Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, ZO-3, and with the COOH termini of claudins. J Cell Biol 147: 1351–1363, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Itou J, Suyama M, Imamura Y, Deguchi T, Fujimori K, Yuba S, Kawarabayasi Y, Kawasaki T. Functional and comparative genomics analyses of pmp22 in medaka fish. BMC Neurosci 10: 60, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, Nicaud S, Jaffe D, Fisher S, Lutfalla G, Dossat C, Segurens B, Dasilva C, Salanoubat M, Levy M, Boudet N, Castellano S, Anthouard V, Jubin C, Castelli V, Katinka M, Vacherie B, Biemont C, Skalli Z, Cattolico L, Poulain J, De Berardinis V, Cruaud C, Duprat S, Brottier P, Coutanceau JP, Gouzy J, Parra G, Lardier G, Chapple C, McKernan KJ, McEwan P, Bosak S, Kellis M, Volff JN, Guigo R, Zody MC, Mesirov J, Lindblad-Toh K, Birren B, Nusbaum C, Kahn D, Robinson-Rechavi M, Laudet V, Schachter V, Quetier F, Saurin W, Scarpelli C, Wincker P, Lander ES, Weissenbach J, Roest Crollius H. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431: 946–957, 2004 [DOI] [PubMed] [Google Scholar]
- 166. Jakab C, Halasz J, Szasz A.M, Kiss A, Schaff Z, Rusvai M, Galfi P, Kulka J. Expression of claudin-1, -2, -3, -4, -5 and -7 proteins in benign and malignant canine mammary gland epithelial tumours. J Comp Pathol 139: 238–245, 2008 [DOI] [PubMed] [Google Scholar]
- 167. Jakab C, Kiss A, Schaff Z, Szabo Z, Rusvai M, Galfi P, Szabara A, Sterczer A, Kulka J. Claudin-7 protein differentiates canine cholangiocarcinoma from hepatocellular carcinoma. Histol Histopathol 25: 857–864, 2010 [DOI] [PubMed] [Google Scholar]
- 168. Jetten AM, Suter U. The peripheral myelin protein 22 and epithelial membrane protein family. Prog Nucleic Acid Res Mol Biol 64: 97–129, 2000 [DOI] [PubMed] [Google Scholar]
- 169. Jovov B, Van Itallie CM, Shaheen NJ, Carson JL, Gambling TM, Anderson JM, Orlando RC. Claudin-18: a dominant tight junction protein in Barrett's esophagus and likely contributor to its acid resistance. Am J Physiol Gastrointest Liver Physiol 293: G1106–G1113, 2007 [DOI] [PubMed] [Google Scholar]
- 170. Kaarteenaho-Wiik R, Soini Y. Claudin-1, -2, -3, -4, -5, and -7 in usual interstitial pneumonia and sarcoidosis. J Histochem Cytochem 57: 187–195, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Kachar B, Reese TS. Evidence for the lipidic nature of tight junction strands. Nature 296: 464–466, 1982 [DOI] [PubMed] [Google Scholar]
- 172. Kahle KT, Macgregor GG, Wilson FH, Van Hoek AN, Brown D, Ardito T, Kashgarian M, Giebisch G, Hebert SC, Boulpaep EL, Lifton RP. Paracellular Cl− permeability is regulated by WNK4 kinase: insight into normal physiology and hypertension. Proc Natl Acad Sci USA 101: 14877–14882, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Kalujnaia S, McWilliam IS, Zaguinaiko VA, Feilen AL, Nicholson J, Hazon N, Cutler CP, Cramb G. Transcriptomic approach to the study of osmoregulation in the European eel Anguilla anguilla. Physiol Genomics 31: 385–401, 2007 [DOI] [PubMed] [Google Scholar]
- 174. Kamekura R, Kojima T, Takashima A, Koizumi J, Ogasawara N, Go M, Takano K, Murata M, Tanaka S, Ichimiya S, Himi T, Sawada N. Thymic stromal lymphopoietin induces tight junction protein claudin-7 via NF-kappaB in dendritic cells. Histochem Cell Biol 133: 339–348, 2010 [DOI] [PubMed] [Google Scholar]
- 175. Karnovsky MJ. The ultrastructural basis of capillary permeability studied with peroxidase as a tracer. J Cell Biol 35: 213–236, 1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Katahira J, Inoue N, Horiguchi Y, Matsuda M, Sugimoto N. Molecular cloning and functional characterization of the receptor for Clostridium perfringens enterotoxin. J Cell Biol 136: 1239–1247, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Katahira J, Sugiyama H, Inoue N, Horiguchi Y, Matsuda M, Sugimoto N. Clostridium perfringens enterotoxin utilizes two structurally related membrane proteins as functional receptors in vivo. J Biol Chem 272: 26652–26658, 1997 [DOI] [PubMed] [Google Scholar]
- 178. Katoh M. CLDN23 gene, frequently down-regulated in intestinal-type gastric cancer, is a novel member of CLAUDIN gene family. Int J Mol Med 11: 683–689, 2003 [PubMed] [Google Scholar]
- 179. Kaufmann R, Piontek J, Grull F, Kirchgessner M, Rossa J, Wolburg H, Blasig IE, Cremer C. Visualization and quantitative analysis of reconstituted tight junctions using localization microscopy. PLoS One 7: e31128, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Kausalya P.J, Amasheh S, Günzel D, Wurps H, Muller D, Fromm M, Hunziker W. Disease-associated mutations affect intracellular traffic and paracellular Mg2+ transport function of Claudin-16. J Clin Invest 116: 878–891, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Kawai Y, Hamazaki Y, Fujita H, Fujita A, Sato T, Furuse M, Fujimoto T, Jetten A.M, Agata Y, Minato N. Claudin-4 induction by E-protein activity in later stages of CD4/8 double-positive thymocytes to increase positive selection efficiency. Proc Natl Acad Sci USA 108: 4075–4080, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Kiil F, Aukland K, Refsum HE. Renal sodium transport and oxygen consumption. Am J Physiol 201: 511–516, 1961 [DOI] [PubMed] [Google Scholar]
- 183. Kirk A, Campbell S, Bass P, Mason J, Collins J. Differential expression of claudin tight junction proteins in the human cortical nephron. Nephrol Dial Transplant 25: 2107–2119, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Kitadokoro K, Nishimura K, Kamitani S, Fukui-Miyazaki A, Toshima H, Abe H, Kamata Y, Sugita-Konishi Y, Yamamoto S, Karatani H, Horiguchi Y. Crystal structure of Clostridium perfringens enterotoxin displays features of beta-pore-forming toxins. J Biol Chem 286: 19549–19555, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Kitajiri SI, Furuse M, Morita K, Saishin-Kiuchi Y, Kido H, Ito J, Tsukita S. Expression patterns of claudins, tight junction adhesion molecules, in the inner ear. Hear Res 187: 25–34, 2004 [DOI] [PubMed] [Google Scholar]
- 186. Kiuchi-Saishin Y, Gotoh S, Furuse M, Takasuga A, Tano Y, Tsukita S. Differential expression patterns of claudins, tight junction membrane proteins, in mouse nephron segments. J Am Soc Nephrol 13: 875–886, 2002 [DOI] [PubMed] [Google Scholar]
- 187. Knipp GT, Ho NF, Barsuhn CL, Borchardt RT. Paracellular diffusion in Caco-2 cell monolayers: effect of perturbation on the transport of hydrophilic compounds that vary in charge and size. J Pharm Sci 86: 1105–1110, 1997 [DOI] [PubMed] [Google Scholar]
- 188. Knox FG, Fleming JS, Rennie DW. Effects of osmotic diuresis on sodium reabsorption and oxygen consumption of kidney. Am J Physiol 210: 751–759, 1966 [DOI] [PubMed] [Google Scholar]
- 189. Koda R, Zhao L, Yaoita E, Yoshida Y, Tsukita S, Tamura A, Nameta M, Zhang Y, Fujinaka H, Magdeldin S, Xu B, Narita I, Yamamoto T. Novel expression of claudin-5 in glomerular podocytes. Cell Tissue Res 343: 637–648, 2011 [DOI] [PubMed] [Google Scholar]
- 190. Kollmar R, Nakamura SK, Kappler JA, Hudspeth AJ. Expression and phylogeny of claudins in vertebrate primordia. Proc Natl Acad Sci USA 98: 10196–10201, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Konrad M, Schaller A, Seelow D, Pandey A.V, Waldegger S, Lesslauer A, Vitzthum H, Suzuki Y, Luk J.M, Becker C, Schlingmann K.P, Schmid M, Rodriguez-Soriano J, Ariceta G, Cano F, Enriquez R, Juppner H, Bakkaloglu S.A, Hediger M.A, Gallati S, Neuhauss S.C, Nurnberg P, Weber S. Mutations in the tight-junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. Am J Hum Genet 79: 949–957, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Kovalenko OV, Yang XH, Hemler ME. A novel cysteine cross-linking method reveals a direct association between claudin-1 and tetraspanin CD9. Mol Cell Proteomics 6: 1855–1867, 2007 [DOI] [PubMed] [Google Scholar]
- 193. Krause G, Winkler L, Mueller SL, Haseloff RF, Piontek J, Blasig IE. Structure and function of claudins. Biochim Biophys Acta 1778: 631–645, 2008 [DOI] [PubMed] [Google Scholar]
- 194. Kriegs JO, Homann V, Kinne-Saffran E, Kinne RK. Identification and subcellular localization of paracellin-1 (claudin-16) in human salivary glands. Histochem Cell Biol 128: 45–53, 2007 [DOI] [PubMed] [Google Scholar]
- 195. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580, 2001 [DOI] [PubMed] [Google Scholar]
- 196. Krug SM, Günzel D, Conrad MP, Rosenthal R, Fromm A, Amasheh S, Schulzke JD, Fromm M. Claudin-17 forms tight junction channels with distinct anion selectivity. Cell Mol Life Sci 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Kubo A, Nagao K, Yokouchi M, Sasaki H, Amagai M. External antigen uptake by Langerhans cells with reorganization of epidermal tight junction barriers. J Exp Med 206: 2937–2946, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Kubota K, Furuse M, Sasaki H, Sonoda N, Fujita K, Nagafuchi A, Tsukita S. Ca2+-independent cell-adhesion activity of claudins, a family of integral membrane proteins localized at tight junctions. Curr Biol 9: 1035–1038, 1999 [DOI] [PubMed] [Google Scholar]
- 199. Kuhn S, Koch M, Nubel T, Ladwein M, Antolovic D, Klingbeil P, Hildebrand D, Moldenhauer G, Langbein L, Franke WW, Weitz J, Zoller M. A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol Cancer Res 5: 553–567, 2007 [DOI] [PubMed] [Google Scholar]
- 200. Lal-Nag M, Morin PJ. The claudins. Genome Biol 10: 235, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Lane NJ, Chandler HJ. Definitive evidence for the existence of tight junctions in invertebrates. J Cell Biol 86: 765–774, 1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Lane NJ, Dallai R, Burighel P, Martinucci GB. Tight and gap junctions in the intestinal tract of tunicates (Urochordata): a freeze-fracture study. J Cell Sci 84: 1–17, 1986 [DOI] [PubMed] [Google Scholar]
- 203. Lane NJ, Dallai R, Martinucci GB, Burighel P. Cell junctions in amphioxus (Cephalochordata): a thin section and freeze-fracture study. Tissue Cell 19: 399–411, 1987 [DOI] [PubMed] [Google Scholar]
- 204. Lane NJ, Skaer HI. Intercellular junctions in insect tissues. Adv Insect Physiol 15: 35–213, 1980 [Google Scholar]
- 205. Larre I, Lazaro A, Contreras R.G, Balda M.S, Matter K, Flores-Maldonado C, Ponce A, Flores-Benitez D, Rincon-Heredia R, Padilla-Benavides T, Castillo A, Shoshani L, Cereijido M. Ouabain modulates epithelial cell tight junction. Proc Natl Acad Sci USA 107: 11387–11392, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Laurila JJ, Karttunen T, Koivukangas V, Laurila PA, Syrjala H, Saarnio J, Soini Y, Ala-Kokko TI. Tight junction proteins in gallbladder epithelium: different expression in acute acalculous and calculous cholecystitis. J Histochem Cytochem 55: 567–573, 2007 [DOI] [PubMed] [Google Scholar]
- 207. Lee NP, Tong MK, Leung PP, Chan VW, Leung S, Tam PC, Chan KW, Lee KF, Yeung WS, Luk JM. Kidney claudin-19: localization in distal tubules and collecting ducts and dysregulation in polycystic renal disease. FEBS Lett 580: 923–931, 2006 [DOI] [PubMed] [Google Scholar]
- 208. Lewis SA. Everything you wanted to know about the bladder epithelium but were afraid to ask. Am J Physiol Renal Physiol 278: F867–F874, 2000 [DOI] [PubMed] [Google Scholar]
- 209. Lewis SA, Diamond JM. Na+ transport by rabbit urinary bladder, a tight epithelium. J Membr Biol 28: 1–40, 1976 [DOI] [PubMed] [Google Scholar]
- 210. Li H, Singh S, Gorantla S, Potula R, Persidsky Y, Poluektova L, Kanmogne GD. Dysregulation of claudin-5 in HIV-induced interstitial pneumonitis and lung vascular injury: protective role of PPAR-gamma. Am J Respir Crit Care Med 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Li WY, Huey CL, Yu AS. Expression of claudin-7 and -8 along the mouse nephron. Am J Physiol Renal Physiol 286: F1063–F1071, 2004 [DOI] [PubMed] [Google Scholar]
- 212. Liebner S, Fischmann A, Rascher G, Duffner F, Grote EH, Kalbacher H, Wolburg H. Claudin-1 and claudin-5 expression and tight junction morphology are altered in blood vessels of human glioblastoma multiforme. Acta Neuropathol 100: 323–331, 2000 [DOI] [PubMed] [Google Scholar]
- 213. Lim JJ, Liebovitch LS, Fischbarg J. Ionic selectivity of the paracellular shunt path across rabbit corneal endothelium. J Membr Biol 73: 95–102, 1983 [DOI] [PubMed] [Google Scholar]
- 214. Lim TS, Vedula SR, Hui S, Kausalya PJ, Hunziker W, Lim CT. Probing effects of pH change on dynamic response of Claudin-2 mediated adhesion using single molecule force spectroscopy. Exp Cell Res 314: 2643–2651, 2008 [DOI] [PubMed] [Google Scholar]
- 215. Lim TS, Vedula SR, Hunziker W, Lim CT. Kinetics of adhesion mediated by extracellular loops of claudin-2 as revealed by single-molecule force spectroscopy. J Mol Biol 381: 681–691, 2008 [DOI] [PubMed] [Google Scholar]
- 216. Linares GR, Brommage R, Powell DR, Xing W, Chen ST, Alshbool FZ, Lau KW, Wergedal JE, Mohan S. Claudin 18 is a novel negative regulator of bone resorption and osteoclast differentiation. J Bone Miner Res 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Lippoldt A, Liebner S, Andbjer B, Kalbacher H, Wolburg H, Haller H, Fuxe K. Organization of choroid plexus epithelial and endothelial cell tight junctions and regulation of claudin-1, -2 and -5 expression by protein kinase C. Neuroreport 11: 1427–1431, 2000 [DOI] [PubMed] [Google Scholar]
- 218. Loh YH, Christoffels A, Brenner S, Hunziker W, Venkatesh B. Extensive expansion of the claudin gene family in the teleost fish, Fugu rubripes. Genome Res 14: 1248–1257, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Lui WY, Lee WM, Cheng CY. Transforming growth factor beta3 regulates the dynamics of Sertoli cell tight junctions via the p38 mitogen-activated protein kinase pathway. Biol Reprod 68: 1597–1612, 2003 [DOI] [PubMed] [Google Scholar]
- 220. Lui WY, Wong EW, Guan Y, Lee WM. Dual transcriptional control of claudin-11 via an overlapping GATA/NF-Y motif: positive regulation through the interaction of GATA, NF-YA, CREB and negative regulation through the interaction of Smad, HDAC1, and mSin3A. J Cell Physiol 211: 638–648, 2007 [DOI] [PubMed] [Google Scholar]
- 221. Luk JM, Tong MK, Mok BW, Tam PC, Yeung WS, Lee KF. Sp1 site is crucial for the mouse claudin-19 gene expression in the kidney cells. FEBS Lett 578: 251–256, 2004 [DOI] [PubMed] [Google Scholar]
- 222. Lynch RD, Francis SA, McCarthy KM, Casas E, Thiele C, Schneeberger EE. Cholesterol depletion alters detergent-specific solubility profiles of selected tight junction proteins and the phosphorylation of occludin. Exp Cell Res 313: 2597–2610, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Madara JL, Dharmsathaphorn K. Occluding junction structure-function relationships in a cultured epithelial monolayer. J Cell Biol 101: 2124–2133, 1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Maher GJ, Hilton EN, Urquhart JE, Davidson AE, Spencer HL, Black GC, Manson FD. The cataract-associated protein TMEM114, TMEM235, are glycosylated transmembrane proteins that are distinct from claudin family members. FEBS Lett 585: 2187–2192, 2011 [DOI] [PubMed] [Google Scholar]
- 225. Mandel I, Paperna T, Glass-Marmor L, Volkowich A, Badarny S, Schwartz I, Vardi P, Koren I, Miller A. Tight junction proteins expression and modulation in immune cells and multiple sclerosis. J Cell Mol Med 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Mandel I, Paperna T, Volkowich A, Merhav M, Glass-Marmor L, Miller A. The ubiquitin-proteasome pathway regulates claudin 5 degradation. J Cell Biochem 2012 [DOI] [PubMed] [Google Scholar]
- 227. Mandel LJ, Bacallao R, Zampighi G. Uncoupling of the molecular “fence” and paracellular “gate” functions in epithelial tight junctions. Nature 361: 552–555, 1993 [DOI] [PubMed] [Google Scholar]
- 228. Mankertz J, Hillenbrand B, Tavalali S, Huber O, Fromm M, Schulzke JD. Functional crosstalk between Wnt signaling and Cdx-related transcriptional activation in the regulation of the claudin-2 promoter activity. Biochem Biophys Res Commun 314: 1001–1007, 2004 [DOI] [PubMed] [Google Scholar]
- 229. Maria OM, Kim JW, Gerstenhaber JA, Baum BJ, Tran SD. Distribution of tight junction proteins in adult human salivary glands. J Histochem Cytochem 56: 1093–1098, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Mariadason JM, Nicholas C, L'Italien KE, Zhuang M, Smartt HJ, Heerdt BG, Yang W, Corner GA, Wilson AJ, Klampfer L, Arango D, Augenlicht LH. Gene expression profiling of intestinal epithelial cell maturation along the crypt-villus axis. Gastroenterology 128: 1081–1088, 2005 [DOI] [PubMed] [Google Scholar]
- 231. Markov AG, Kruglova NM, Fomina YA, Fromm M, Amasheh S. Altered expression of tight junction proteins in mammary epithelium after discontinued suckling in mice. Pflügers Arch 2011 [DOI] [PubMed] [Google Scholar]
- 232. Martinez-Estrada O.M, Culleres A, Soriano F.X, Peinado H, Bolos V, Martinez F.O, Reina M, Cano A, Fabre M, Vilaro S. The transcription factors Slug and Snail act as repressors of Claudin-1 expression in epithelial cells. Biochem J 394: 449–457, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Martinez-Palomo A, Erlij D, Bracho H. Localization of permeability barriers in the frog skin epithelium. J Cell Biol 50: 277–287, 1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Masaki T, Kojima T, Okabayashi T, Ogasawara N, Ohkuni T, Obata K, Takasawa A, Murata M, Tanaka S, Hirakawa S, Fuchimoto J, Ninomiya T, Fujii N, Tsutsumi H, Himi T, Sawada N. A nuclear factor-kappaB signaling pathway via protein kinase C delta regulates replication of respiratory syncytial virus in polarized normal human nasal epithelial cells. Mol Biol Cell 22: 2144–2156, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Mathisen O, Monclair T, Kiil F. Oxygen requirement of bicarbonate-dependent sodium reabsorption in the dog kidney. Am J Physiol Renal Fluid Electrolyte Physiol 238: F175–F180, 1980 [DOI] [PubMed] [Google Scholar]
- 236. Matter K, Aijaz S, Tsapara A, Balda MS. Mammalian tight junctions in the regulation of epithelial differentiation and proliferation. Curr Opin Cell Biol 17: 453–458, 2005 [DOI] [PubMed] [Google Scholar]
- 237. Mazzon E, Cuzzocrea S. Absence of functional peroxisome proliferator-activated receptor-alpha enhanced ileum permeability during experimental colitis. Shock 28: 192–201, 2007 [DOI] [PubMed] [Google Scholar]
- 238. McCarthy KM, Francis SA, McCormack JM, Lai J, Rogers RA, Skare IB, Lynch RD, Schneeberger EE. Inducible expression of claudin-1-myc but not occludin-VSV-G results in aberrant tight junction strand formation in MDCK cells. J Cell Sci 113: 3387–3398, 2000 [DOI] [PubMed] [Google Scholar]
- 239. McLaughlin J, Padfield PJ, Burt JP, O'Neill CA. Ochratoxin A increases permeability through tight junctions by removal of specific claudin isoforms. Am J Physiol Cell Physiol 287: C1412–C1417, 2004 [DOI] [PubMed] [Google Scholar]
- 240. Medici D, Hay ED, Goodenough DA. Cooperation between snail and LEF-1 transcription factors is essential for TGF-beta1-induced epithelial-mesenchymal transition. Mol Biol Cell 17: 1871–1879, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Meng J, Holdcraft RW, Shima JE, Griswold MD, Braun RE. Androgens regulate the permeability of the blood-testis barrier. Proc Natl Acad Sci USA 102: 16696–16700, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Michel D, Arsanto JP, Massey-Harroche D, Beclin C, Wijnholds J, Le Bivic A. PATJ connects and stabilizes apical and lateral components of tight junctions in human intestinal cells. J Cell Sci 118: 4049–4057, 2005 [DOI] [PubMed] [Google Scholar]
- 243. Michikawa H, Fujita-Yoshigaki J, Sugiya H. Enhancement of barrier function by overexpression of claudin-4 in tight junctions of submandibular gland cells. Cell Tissue Res 334: 255–264, 2008 [DOI] [PubMed] [Google Scholar]
- 244. Michlig S, Damak S, Le Coutre J. Claudin-based permeability barriers in taste buds. J Comp Neurol 502: 1003–1011, 2007 [DOI] [PubMed] [Google Scholar]
- 245. Milatz S, Krug SM, Rosenthal R, Günzel D, Muller D, Schulzke JD, Amasheh S, Fromm M. Claudin-3 acts as a sealing component of the tight junction for ions of either charge and uncharged solutes. Biochim Biophys Acta 2010 [DOI] [PubMed] [Google Scholar]
- 246. Mineta K, Yamamoto Y, Yamazaki Y, Tanaka H, Tada Y, Saito K, Tamura A, Igarashi M, Endo T, Takeuchi K, Tsukita S. Predicted expansion of the claudin multigene family. FEBS Lett 585: 606–612, 2011 [DOI] [PubMed] [Google Scholar]
- 247. Mitchell LA, Overgaard CE, Ward C, Margulies SS, Koval M. Differential effects of claudin-3 and claudin-4 on alveolar epithelial barrier function. Am J Physiol Lung Cell Mol Physiol 301: L40–L49, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Mitic LL, Unger VM, Anderson JM. Expression, solubilization, and biochemical characterization of the tight junction transmembrane protein claudin-4. Protein Sci 12: 218–227, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Miyamoto T, Morita K, Takemoto D, Takeuchi K, Kitano Y, Miyakawa T, Nakayama K, Okamura Y, Sasaki H, Miyachi Y, Furuse M, Tsukita S. Tight junctions in Schwann cells of peripheral myelinated axons: a lesson from claudin-19-deficient mice. J Cell Biol 169: 527–538, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Mo D, Potter BA, Bertrand CA, Hildebrand JD, Bruns JR, Weisz OA. Nucleofection disrupts tight junction fence function to alter membrane polarity of renal epithelial cells. Am J Physiol Renal Physiol 299: F1178–F1184, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Moldvay J, Jackel M, Paska C, Soltesz I, Schaff Z, Kiss A. Distinct claudin expression profile in histologic subtypes of lung cancer. Lung Cancer 57: 159–167, 2007 [DOI] [PubMed] [Google Scholar]
- 252. Moreno JH, Diamond JM. Cation permeation mechanisms and cation selectivity in “tight junctions” of gallbladder epithelium. Membranes 3: 383–497, 1975 [PubMed] [Google Scholar]
- 253. Morita K, Furuse M, Fujimoto K, Tsukita S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci USA 96: 511–516, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Morita K, Sasaki H, Fujimoto K, Furuse M, Tsukita S. Claudin-11/OSP-based tight junctions of myelin sheaths in brain and Sertoli cells in testis. J Cell Biol 145: 579–588, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Morita K, Sasaki H, Furuse M, Tsukita S. Endothelial claudin: claudin-5/TMVCF constitutes tight junction strands in endothelial cells. J Cell Biol 147: 185–194, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Moriwaki K, Tsukita S, Furuse M. Tight junctions containing claudin 4 and 6 are essential for blastocyst formation in preimplantation mouse embryos. Dev Biol 312: 509–522, 2007 [DOI] [PubMed] [Google Scholar]
- 257. Morrow CM, Mruk D, Cheng CY, Hess RA. Claudin and occludin expression and function in the seminiferous epithelium. Philos Trans R Soc Lond B Biol Sci 365: 1679–1696, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Morrow CM, Tyagi G, Simon L, Carnes K, Murphy KM, Cooke PS, Hofmann MC, Hess RA. Claudin 5 expression in mouse seminiferous epithelium is dependent upon the transcription factor ets variant 5 and contributes to blood-testis barrier function. Biol Reprod 81: 871–879, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Muller D, Kausalya PJ, Claverie-Martin F, Meij IC, Eggert P, Garcia-Nieto V, Hunziker W. A novel claudin 16 mutation associated with childhood hypercalciuria abolishes binding to ZO-1 and results in lysosomal mistargeting. Am J Hum Genet 73: 1293–1301, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Muller D, Kausalya PJ, Meij IC, Hunziker W. Familial hypomagnesemia with hypercalciuria and nephrocalcinosis: blocking endocytosis restores surface expression of a novel Claudin-16 mutant that lacks the entire C-terminal cytosolic tail. Hum Mol Genet 15: 1049–1058, 2006 [DOI] [PubMed] [Google Scholar]
- 261. Muto S, Hata M, Taniguchi J, Tsuruoka S, Moriwaki K, Saitou M, Furuse K, Sasaki H, Fujimura A, Imai M, Kusano E, Tsukita S, Furuse M. Claudin-2-deficient mice are defective in the leaky and cation-selective paracellular permeability properties of renal proximal tubules. Proc Natl Acad Sci USA 107: 8011–8016, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Nakano Y, Kim SH, Kim HM, Sanneman JD, Zhang Y, Smith RJ, Marcus DC, Wangemann P, Nessler RA, Banfi B. A claudin-9-based ion permeability barrier is essential for hearing. PLoS Genet 5: e1000610, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Neaves WB. Permeability of Sertoli cell tight junctions to lanthanum after ligation of ductus deferens and ductuli efferentes. J Cell Biol 59: 559–572, 1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Nelson KS, Furuse M, Beitel GJ. The Drosophila Claudin Kune-kune is required for septate junction organization and tracheal tube size control. Genetics 185: 831–839, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Nemeth Z, Szasz AM, Tatrai P, Nemeth J, Gyorffy H, Somoracz A, Szijarto A, Kupcsulik P, Kiss A, Schaff Z. Claudin-1, -2, -3, -4, -7, -8, -10 protein expression in biliary tract cancers. J Histochem Cytochem 57: 113–121, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Neuberg DH, Sancho S, Suter U. Altered molecular architecture of peripheral nerves in mice lacking the peripheral myelin protein 22 or connexin32. J Neurosci Res 58: 612–623, 1999 [DOI] [PubMed] [Google Scholar]
- 267. Nichols GE, Borgman CA, Young WW., Jr On tight junction structure: Forssman glycolipid does not flow between MDCK cells in an intact epithelial monolayer. Biochem Biophys Res Commun 138: 1163–1169, 1986 [DOI] [PubMed] [Google Scholar]
- 268. Nightingale ER. Phenomenological theory of ion solvation. Effective radii of hydrated ions. J Phys Chem 63: 1381–1387, 1959 [Google Scholar]
- 269. Niimi T, Nagashima K, Ward JM, Minoo P, Zimonjic DB, Popescu NC, Kimura S. Claudin-18, a novel downstream target gene for the T/EBP/NKX2.1 homeodomain transcription factor, encodes lung- and stomach-specific isoforms through alternative splicing. Mol Cell Biol 21: 7380–7390, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Nitta T, Hata M, Gotoh S, Seo Y, Sasaki H, Hashimoto N, Furuse M, Tsukita S. Size-selective loosening of the blood-brain barrier in claudin-5-deficient mice. J Cell Biol 161: 653–660, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Nubel T, Preobraschenski J, Tuncay H, Weiss T, Kuhn S, Ladwein M, Langbein L, Zoller M. Claudin-7 regulates EpCAM-mediated functions in tumor progression. Mol Cancer Res 7: 285–299, 2009 [DOI] [PubMed] [Google Scholar]
- 272. Nunbhakdi-Craig V, Machleidt T, Ogris E, Bellotto D, White CL, 3rd, Sontag E. Protein phosphatase 2A associates with and regulates atypical PKC and the epithelial tight junction complex. J Cell Biol 158: 967–978, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Nunes FD, Lopez LN, Lin HW, Davies C, Azevedo RB, Gow A, Kachar B. Distinct subdomain organization and molecular composition of a tight junction with adherens junction features. J Cell Sci 119: 4819–4827, 2006 [DOI] [PubMed] [Google Scholar]
- 274. Ogasawara N, Kojima T, Go M, Ohkuni T, Koizumi J, Kamekura R, Masaki T, Murata M, Tanaka S, Fuchimoto J, Himi T, Sawada N. PPARgamma agonists upregulate the barrier function of tight junctions via a PKC pathway in human nasal epithelial cells. Pharmacol Res 61: 489–498, 2010 [DOI] [PubMed] [Google Scholar]
- 275. Ohba Y, Kitagawa H, Kitoh K, Sasaki Y, Takami M, Shinkai Y, Kunieda T. A deletion of the paracellin-1 gene is responsible for renal tubular dysplasia in cattle. Genomics 68: 229–236, 2000 [DOI] [PubMed] [Google Scholar]
- 276. Ohkubo T, Ozawa M. The transcription factor Snail downregulates the tight junction components independently of E-cadherin downregulation. J Cell Sci 117: 1675–1685, 2004 [DOI] [PubMed] [Google Scholar]
- 277. Ohse T, Chang AM, Pippin JW, Jarad G, Hudkins KL, Alpers CE, Miner JH, Shankland SJ. A new function for parietal epithelial cells: a second glomerular barrier. Am J Physiol Renal Physiol 297: F1566–F1574, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Ohse T, Pippin JW, Vaughan MR, Brinkkoetter PT, Krofft RD, Shankland SJ. Establishment of conditionally immortalized mouse glomerular parietal epithelial cells in culture. J Am Soc Nephrol 19: 1879–1890, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Ohtsuki S, Sato S, Yamaguchi H, Kamoi M, Asashima T, Terasaki T. Exogenous expression of claudin-5 induces barrier properties in cultured rat brain capillary endothelial cells. J Cell Physiol 210: 81–86, 2007 [DOI] [PubMed] [Google Scholar]
- 280. Ohtsuki S, Yamaguchi H, Katsukura Y, Asashima T, Terasaki T. mRNA expression levels of tight junction protein genes in mouse brain capillary endothelial cells highly purified by magnetic cell sorting. J Neurochem 104: 147–154, 2008 [DOI] [PubMed] [Google Scholar]
- 281. Okada K, Ishikawa N, Fujimori K, Goryo M, Ikeda M, Sasaki J, Watanabe D, Takasuga A, Hirano T, Sugimoto Y. Abnormal development of nephrons in claudin-16-defective Japanese black cattle. J Vet Med Sci 67: 171–178, 2005 [DOI] [PubMed] [Google Scholar]
- 282. Orci L, Amherdt M, Henquin JC, Lambert AE, Unger RH, Renold AE. Pronase effect on pancreatic beta cell secretion and morphology. Science 180: 647–649, 1973 [DOI] [PubMed] [Google Scholar]
- 283. Ota T, Fujii M, Sugizaki T, Ishii M, Miyazawa K, Aburatani H, Miyazono K. Targets of transcriptional regulation by two distinct type I receptors for transforming growth factor-beta in human umbilical vein endothelial cells. J Cell Physiol 193: 299–318, 2002 [DOI] [PubMed] [Google Scholar]
- 284. Ozden O, Black BL, Ashwell CM, Tipsmark CK, Borski RJ, Grubb BJ. Developmental profile of claudin-3, -5, and -16 proteins in the epithelium of chick intestine. Anat Rec 293: 1175–1183, 2010 [DOI] [PubMed] [Google Scholar]
- 285. Papageorgis P, Lambert AW, Ozturk S, Gao F, Pan H, Manne U, Alekseyev YO, Thiagalingam A, Abdolmaleky HM, Lenburg M, Thiagalingam S. Smad signaling is required to maintain epigenetic silencing during breast cancer progression. Cancer Res 70: 968–978, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Peng S, Adelman RA, Rizzolo LJ. Minimal effects of VEGF and anti-VEGF drugs on the permeability or selectivity of RPE tight junctions. Invest Ophthalmol Vis Sci 51: 3216–3225, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Peng S, Rao VS, Adelman RA, Rizzolo LJ. Claudin-19 and the barrier properties of the human retinal pigment epithelium. Invest Ophthalmol Vis Sci 52: 1392–1403, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Peppi M, Ghabriel MN. Tissue-specific expression of the tight junction proteins claudins and occludin in the rat salivary glands. J Anat 205: 257–266, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Piehl C, Piontek J, Cording J, Wolburg H, Blasig IE. Participation of the second extracellular loop of claudin-5 in paracellular tightening against ions, small and large molecules. Cell Mol Life Sci 67: 2131–2140, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Pinto da Silva P, Kachar B. On tight-junction structure. Cell 28: 441–450, 1982 [DOI] [PubMed] [Google Scholar]
- 291. Piontek J, Fritzsche S, Cording J, Richter S, Hartwig J, Walter M, Yu D, Turner JR, Gehring C, Rahn HP, Wolburg H, Blasig IE. Elucidating the principles of the molecular organization of heteropolymeric tight junction strands. Cell Mol Life Sci 68: 3903–3918, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Piontek J, Winkler L, Wolburg H, Muller SL, Zuleger N, Piehl C, Wiesner B, Krause G, Blasig IE. Formation of tight junction: determinants of homophilic interaction between classic claudins. FASEB J 22: 146–158, 2008 [DOI] [PubMed] [Google Scholar]
- 293. Poliak S, Peles E. The local differentiation of myelinated axons at nodes of Ranvier. Nat Rev Neurosci 4: 968–980, 2003 [DOI] [PubMed] [Google Scholar]
- 294. Ponnam S.P, Ramesha K, Tejwani S, Matalia J, Kannabiran C. A missense mutation in LIM2 causes autosomal recessive congenital cataract. Mol Vis 14: 1204–1208, 2008 [PMC free article] [PubMed] [Google Scholar]
- 295. Pricam C, Humbert F, Perrelet A, Amherdt M, Orci L. Intercellular junctions in podocytes of the nephrotic glomerulus as seen with freeze-fracture. Lab Invest 33: 209–218, 1975 [PubMed] [Google Scholar]
- 296. Price M.G, Davis C.F, Deng F, Burgess D.L. The alpha-amino-3-hydroxyl-5-methyl-4-isoxazolepropionate receptor trafficking regulator “stargazin” is related to the claudin family of proteins by its ability to mediate cell-cell adhesion. J Biol Chem 280: 19711–19720, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Quigley R, Baum M. Developmental changes in rabbit proximal straight tubule paracellular permeability. Am J Physiol Renal Physiol 283: F525–F531, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Rahner C, Mitic LL, Anderson JM. Heterogeneity in expression and subcellular localization of claudins 2, 3, 4, and 5 in the rat liver, pancreas, and gut. Gastroenterology 120: 411–422, 2001 [DOI] [PubMed] [Google Scholar]
- 299. Reese TS, Karnovsky MJ. Fine structural localization of a blood-brain barrier to exogenous peroxidase. J Cell Biol 34: 207–217, 1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Reeves W, Caulfield JP, Farquhar MG. Differentiation of epithelial foot processes and filtration slits: sequential appearance of occluding junctions, epithelial polyanion, and slit membranes in developing glomeruli. Lab Invest 39: 90–100, 1978 [PubMed] [Google Scholar]
- 301. Reiter B, Kraft R, Günzel D, Zeissig S, Schulzke JD, Fromm M, Harteneck C. TRPV4-mediated regulation of epithelial permeability. FASEB J 20: 1802–1812, 2006 [DOI] [PubMed] [Google Scholar]
- 302. Robertson SL, Smedley JG, 3rd, McClane BA. Identification of a claudin-4 residue important for mediating the host cell binding and action of Clostridium perfringens enterotoxin. Infection Immunity 78: 505–517, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Rokkam D, Lafemina MJ, Lee JW, Matthay MA, Frank JA. Claudin-4 levels are associated with intact alveolar fluid clearance in human lungs. Am J Pathol 179: 1081–1087, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Romanitan MO, Popescu BO, Spulber S, Bajenaru O, Popescu LM, Winblad B, Bogdanovic N. Altered expression of claudin family proteins in Alzheimer's disease and vascular dementia brains. J Cell Mol Med 14: 1088–1100, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Ronaldson PT, Demarco KM, Sanchez-Covarrubias L, Solinsky CM, Davis TP. Transforming growth factor-beta signaling alters substrate permeability and tight junction protein expression at the blood-brain barrier during inflammatory pain. J Cereb Blood Flow Metab 29: 1084–1098, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Rosenthal R, Milatz S, Krug SM, Oelrich B, Schulzke JD, Amasheh S, Günzel D, Fromm M. Claudin-2, a component of the tight junction, forms a paracellular water channel. J Cell Sci 123: 1913–1921, 2010 [DOI] [PubMed] [Google Scholar]
- 307. Roux KJ, Amici SA, Fletcher BS, Notterpek L. Modulation of epithelial morphology, monolayer permeability, and cell migration by growth arrest specific 3/peripheral myelin protein 22. Mol Biol Cell 16: 1142–1151, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Ruffer C, Gerke V. The C-terminal cytoplasmic tail of claudins 1 and 5 but not its PDZ-binding motif is required for apical localization at epithelial and endothelial tight junctions. Eur J Cell Biol 83: 135–144, 2004 [DOI] [PubMed] [Google Scholar]
- 309. Ryan GB, Leventhal M, Karnovsky MJ. A freeze-fracture study of the junctions between glomerular epithelial cells in aminonucleoside nephrosis. Lab Invest 32: 397–403, 1975 [PubMed] [Google Scholar]
- 310. Sabirov RZ, Okada Y. Wide nanoscopic pore of maxi-anion channel suits its function as an ATP-conductive pathway. Biophys J 87: 1672–1685, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Sager C, Tapken D, Kott S, Hollmann M. Functional modulation of AMPA receptors by transmembrane AMPA receptor regulatory proteins. Neuroscience 158: 45–54, 2009 [DOI] [PubMed] [Google Scholar]
- 312. Saitoh R, Sugano K, Takata N, Tachibana T, Higashida A, Nabuchi Y, Aso Y. Correction of permeability with pore radius of tight junctions in Caco-2 monolayers improves the prediction of the dose fraction of hydrophilic drugs absorbed by humans. Pharm Res 21: 749–755, 2004 [DOI] [PubMed] [Google Scholar]
- 313. Sakaguchi T, Gu X, Golden HM, Suh E, Rhoads DB, Reinecker HC. Cloning of the human claudin-2 5′-flanking region revealed a TATA-less promoter with conserved binding sites in mouse and human for caudal-related homeodomain proteins and hepatocyte nuclear factor-1alpha. J Biol Chem 277: 21361–21370, 2002 [DOI] [PubMed] [Google Scholar]
- 314. Sakai N, Chiba H, Fujita H, Akashi Y, Osanai M, Kojima T, Sawada N. Expression patterns of claudin family of tight-junction proteins in the mouse prostate. Histochem Cell Biol 127: 457–462, 2007 [DOI] [PubMed] [Google Scholar]
- 315. Sakamoto H, Mutoh H, Sugano K. Expression of Claudin-2 in intestinal metaplastic mucosa of Cdx2-transgenic mouse stomach. Scand J Gastroenterol 45: 1273–1280, 2010 [DOI] [PubMed] [Google Scholar]
- 316. Sansom SC, Weinman EJ, O'Neil RG. Microelectrode assessment of chloride-conductive properties of cortical collecting duct. Am J Physiol Renal Fluid Electrolyte Physiol 247: F291–F302, 1984 [DOI] [PubMed] [Google Scholar]
- 317. Sas D, Hu M, Moe OW, Baum M. Effect of claudins 6 and 9 on paracellular permeability in MDCK II cells. Am J Physiol Regul Integr Comp Physiol 295: R1713–R1719, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Sasaki H, Matsui C, Furuse K, Mimori-Kiyosue Y, Furuse M, Tsukita S. Dynamic behavior of paired claudin strands within apposing plasma membranes. Proc Natl Acad Sci USA 100: 3971–3976, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Satake S, Semba S, Matsuda Y, Usami Y, Chiba H, Sawada N, Kasuga M, Yokozaki H. Cdx2 transcription factor regulates claudin-3 and claudin-4 expression during intestinal differentiation of gastric carcinoma. Pathol Int 58: 156–163, 2008 [DOI] [PubMed] [Google Scholar]
- 320. Satohisa S, Chiba H, Osanai M, Ohno S, Kojima T, Saito T, Sawada N. Behavior of tight-junction, adherens-junction and cell polarity proteins during HNF-4alpha-induced epithelial polarization. Exp Cell Res 310: 66–78, 2005 [DOI] [PubMed] [Google Scholar]
- 321. Schulzke JD, Günzel D, John LJ, Fromm M. Perspectives in tight junction research. Ann NY Acad Sci. In press [DOI] [PubMed] [Google Scholar]
- 322. Shiou SR, Singh AB, Moorthy K, Datta PK, Washington MK, Beauchamp RD, Dhawan P. Smad4 regulates claudin-1 expression in a transforming growth factor-beta-independent manner in colon cancer cells. Cancer Res 67: 1571–1579, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res 38: D161–166, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Simon DB, Lu Y, Choate KA, Velazquez H, Al-Sabban E, Praga M, Casari G, Bettinelli A, Colussi G, Rodriguez-Soriano J, McCredie D, Milford D, Sanjad S, Lifton RP. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science 285: 103–106, 1999 [DOI] [PubMed] [Google Scholar]
- 325. Simske JS, Hardin J. Claudin family proteins in Caenorhabditis elegans. Methods Mol Biol 762: 147–169, 2011 [DOI] [PubMed] [Google Scholar]
- 326. Singh AB, Sharma A, Dhawan P. Claudin family of proteins and cancer: an overview. J Oncol 2010: 541957, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Slegers JF, Moons WM, Idzerda PP, Stadhouders AM. The contribution of a chloride shunt to the transmucosal potential of the rabbit submaxillary duct. J Membr Biol 25: 213–236, 1975 [DOI] [PubMed] [Google Scholar]
- 328. Soma T, Chiba H, Kato-Mori Y, Wada T, Yamashita T, Kojima T, Sawada N. Thr(207) of claudin-5 is involved in size-selective loosening of the endothelial barrier by cyclic AMP. Exp Cell Res 300: 202–212, 2004 [DOI] [PubMed] [Google Scholar]
- 329. Sonoda N, Furuse M, Sasaki H, Yonemura S, Katahira J, Horiguchi Y, Tsukita S. Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: evidence for direct involvement of claudins in tight junction barrier. J Cell Biol 147: 195–204, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Staehelin LA. Further observations on the fine structure of freeze-cleaved tight junctions. J Cell Sci 13: 763–786, 1973 [DOI] [PubMed] [Google Scholar]
- 331. Staehelin LA. Three types of gap junctions interconnecting intestinal epithelial cells visualized by freeze-etching. Proc Natl Acad Sci USA 69: 1318–1321, 1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332. Stevenson BR, Anderson JM, Goodenough DA, Mooseker MS. Tight junction structure and ZO-1 content are identical in two strains of Madin-Darby canine kidney cells which differ in transepithelial resistance. J Cell Biol 107: 2401–2408, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. Stevenson BR, Goodenough DA. Zonulae occludentes in junctional complex-enriched fractions from mouse liver: preliminary morphological and biochemical characterization. J Cell Biol 98: 1209–1221, 1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334. Stiffler MA, Chen JR, Grantcharova VP, Lei Y, Fuchs D, Allen JE, Zaslavskaia LA, MacBeath G. PDZ domain binding selectivity is optimized across the mouse proteome. Science 317: 364–369, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335. Stone KP, Kastin AJ, Pan W. NFkB is an unexpected major mediator of interleukin-15 signaling in cerebral endothelia. Cell Physiol Biochem 28: 115–124, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336. Sugiyama A, Ozaki K, Miyazaki, Tanabe Y, Takeuchi T, Narama I. Renal dysplasia unrelated to claudin-16 deficiency in Japanese Black cattle. J Comp Pathol 137: 71–77, 2007 [DOI] [PubMed] [Google Scholar]
- 337. Sun ZY, Wei J, Xie L, Shen Y, Liu SZ, Ju GZ, Shi JP, Yu YQ, Zhang X, Xu Q, Hemmings GP. The CLDN5 locus may be involved in the vulnerability to schizophrenia. Eur Psychiatry 19: 354–357, 2004 [DOI] [PubMed] [Google Scholar]
- 338. Suzuki T, Yoshinaga N, Tanabe S. Interleukin-6 (IL-6) regulates claudin-2 expression and tight junction permeability in intestinal epithelium. J Biol Chem 286: 31263–31271, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339. Tabaries S, Dupuy F, Dong Z, Monast A, Annis MG, Spicer J, Ferri LE, Omeroglu A, Basik M, Amir E, Clemons M, Siegel PM. Claudin-2 promotes breast cancer liver metastasis by facilitating tumor cell interactions with hepatocytes. Mol Cell Biol 32: 2979–2991, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340. Takahashi S, Iwamoto N, Sasaki H, Ohashi M, Oda Y, Tsukita S, Furuse M. The E3 ubiquitin ligase LNX1p80 promotes the removal of claudins from tight junctions in MDCK cells. J Cell Sci 122: 985–994, 2009 [DOI] [PubMed] [Google Scholar]
- 341. Takakuwa R, Kokai Y, Kojima T, Akatsuka T, Tobioka H, Sawada N, Mori M. Uncoupling of gate and fence functions of MDCK cells by the actin-depolymerizing reagent mycalolide B. Exp Cell Res 257: 238–244, 2000 [DOI] [PubMed] [Google Scholar]
- 342. Takeda Y, Notsu T, Kitamura K, Uyemura K. Functional analysis for peripheral myelin protein PASII/PMP22: is it a member of claudin superfamily? Neurochem Res 26: 599–607, 2001 [DOI] [PubMed] [Google Scholar]
- 343. Tamura A, Hayashi H, Imasato M, Yamazaki Y, Hagiwara A, Wada M, Noda T, Watanabe M, Suzuki Y, Tsukita S. Loss of claudin-15, but not claudin-2, causes Na+ deficiency and glucose malabsorption in mouse small intestine. Gastroenterology 140: 913–923, 2011 [DOI] [PubMed] [Google Scholar]
- 344. Tamura A, Kitano Y, Hata M, Katsuno T, Moriwaki K, Sasaki H, Hayashi H, Suzuki Y, Noda T, Furuse M, Tsukita S. Megaintestine in claudin-15-deficient mice. Gastroenterology 134: 523–534, 2008 [DOI] [PubMed] [Google Scholar]
- 345. Tanaka M, Kamata R, Sakai R. EphA2 phosphorylates the cytoplasmic tail of Claudin-4 and mediates paracellular permeability. J Biol Chem 280: 42375–42382, 2005 [DOI] [PubMed] [Google Scholar]
- 346. Tang VW, Goodenough DA. Paracellular ion channel at the tight junction. Biophys J 84: 1660–1673, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347. Tang Y, Clayburgh DR, Mittal N, Goretsky T, Dirisina R, Zhang Z, Kron M, Ivancic D, Katzman RB, Grimm G, Lee G, Fryer J, Nusrat A, Turner JR, Barrett TA. Epithelial NF-kappaB enhances transmucosal fluid movement by altering tight junction protein composition after T cell activation. Am J Pathol 176: 158–167, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348. Tatum R, Zhang Y, Lu Q, Kim K, Jeansonne BG, Chen YH. WNK4 phosphorylates ser(206) of claudin-7 and promotes paracellular Cl− permeability. FEBS Lett 581: 3887–3891, 2007 [DOI] [PubMed] [Google Scholar]
- 349. Tatum R, Zhang Y, Salleng K, Lu Z, Lin JJ, Lu Q, Jeansonne BG, Ding L, Chen YH. Renal salt wasting and chronic dehydration in claudin-7-deficient mice. Am J Physiol Renal Physiol 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350. Thompson PD, Tipney H, Brass A, Noyes H, Kemp S, Naessens J, Tassabehji M. Claudin 13, a member of the claudin family regulated in mouse stress induced erythropoiesis. PLoS One 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351. Thorleifsson G, Holm H, Edvardsson V, Walters GB, Styrkarsdottir U, Gudbjartsson DF, Sulem P, Halldorsson BV, de Vegt F, d'Ancona FC, den Heijer M, Franzson L, Christiansen C, Alexandersen P, Rafnar T, Kristjansson K, Sigurdsson G, Kiemeney LA, Bodvarsson M, Indridason OS, Palsson R, Kong A, Thorsteinsdottir U, Stefansson K. Sequence variants in the CLDN14 gene associate with kidney stones and bone mineral density. Nat Genet 41: 926–930, 2009 [DOI] [PubMed] [Google Scholar]
- 352. Thurau K. Renal Na-reabsorption and O2-uptake in dogs during hypoxia and hydrochlorothiazide infusion. Proc Soc Exp Biol Med 106: 714–717, 1961 [DOI] [PubMed] [Google Scholar]
- 353. Tian X, Du H, Fu X, Li K, Li A, Zhang Y. Smad4 restoration leads to a suppression of Wnt/beta-catenin signaling activity and migration capacity in human colon carcinoma cells. Biochem Biophys Res Commun 380: 478–483, 2009 [DOI] [PubMed] [Google Scholar]
- 354. Tipsmark CK, Baltzegar DA, Ozden O, Grubb BJ, Borski RJ. Salinity regulates claudin mRNA and protein expression in the teleost gill. Am J Physiol Regul Integr Comp Physiol 294: R1004–R1014, 2008 [DOI] [PubMed] [Google Scholar]
- 355. Tiwari-Woodruff SK, Kaplan R, Kornblum HI, Bronstein JM. Developmental expression of OAP-1/Tspan-3, a member of the tetraspanin superfamily. J Neurosci Res 77: 166–173, 2004 [DOI] [PubMed] [Google Scholar]
- 356. Torelli G, Milla E, Faelli A, Costantini S. Energy requirement for sodium reabsorption in the in vivo rabbit kidney. Am J Physiol 211: 576–580, 1966 [DOI] [PubMed] [Google Scholar]
- 357. Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol 2: 285–293, 2001 [DOI] [PubMed] [Google Scholar]
- 358. Turksen K, Troy TC. Claudin-6: a novel tight junction molecule is developmentally regulated in mouse embryonic epithelium. Dev Dynamics 222: 292–300, 2001 [DOI] [PubMed] [Google Scholar]
- 359. Turksen K, Troy TC. Junctions gone bad: claudins and loss of the barrier in cancer. Biochim Biophys Acta 1816: 73–79, 2011 [DOI] [PubMed] [Google Scholar]
- 360. Turksen K, Troy TC. Permeability barrier dysfunction in transgenic mice overexpressing claudin 6. Development 129: 1775–1784, 2002 [DOI] [PubMed] [Google Scholar]
- 361. Umeda K, Ikenouchi J, Katahira-Tayama S, Furuse K, Sasaki H, Nakayama M, Matsui T, Tsukita S, Furuse M. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell 126: 741–754, 2006 [DOI] [PubMed] [Google Scholar]
- 362. Usami Y, Satake S, Nakayama F, Matsumoto M, Ohnuma K, Komori T, Semba S, Ito A, Yokozaki H. Snail-associated epithelial-mesenchymal transition promotes oesophageal squamous cell carcinoma motility and progression. J Pathol 215: 330–339, 2008 [DOI] [PubMed] [Google Scholar]
- 363. Valle BL, Morin PJ. Claudins in cancer biology. In: Claudins , edited by Yu AS. Philadelphia, PA: Elsevier, 2010, p. 293–333 [Google Scholar]
- 364. Van Itallie C, Fanning AS, Anderson JM. Reversal of charge selectivity in cation or anion selective epithelial lines by expression of different claudins. Am J Physiol Renal Physiol 286: F1078–F1084, 2003 [DOI] [PubMed] [Google Scholar]
- 365. Van Itallie C, Rahner C, Anderson JM. Regulated expression of claudin-4 decreases paracellular conductance through a selective decrease in sodium permeability. J Clin Invest 107: 1319–1327, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366. Van Itallie CM, Anderson JM. The molecular physiology of tight junction pores. Physiology 19: 331–338, 2004 [DOI] [PubMed] [Google Scholar]
- 367. Van Itallie CM, Betts L, Smedley JG, 3rd, McClane BA, Anderson JM. Structure of the claudin-binding domain of Clostridium perfringens enterotoxin. J Biol Chem 283: 268–274, 2008 [DOI] [PubMed] [Google Scholar]
- 368. Van Itallie CM, Colegio OR, Anderson JM. The cytoplasmic tails of claudins can influence tight junction barrier properties through effects on protein stability. J Membr Biol 199: 29–38, 2004 [DOI] [PubMed] [Google Scholar]
- 369. Van Itallie CM, Gambling TM, Carson JL, Anderson JM. Palmitoylation of claudins is required for efficient tight-junction localization. J Cell Sci 118: 1427–1436, 2005 [DOI] [PubMed] [Google Scholar]
- 370. Van Itallie CM, Holmes J, Bridges A, Gookin JL, Coccaro MR, Proctor W, Colegio OR, Anderson JM. The density of small tight junction pores varies among cell types and is increased by expression of claudin-2. J Cell Sci 121: 298–305, 2008 [DOI] [PubMed] [Google Scholar]
- 371. Van Itallie CM, Mitic LL, Anderson JM. Claudin-2 forms homodimers and is a component of a high molecular weight protein complex. J Biol Chem 286: 3442–3450, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372. Van Itallie CM, Mitic LL, Anderson JM. SUMOylation of claudin-2. Ann NY Acad Sci 1258: 60–64, 2012 [DOI] [PubMed] [Google Scholar]
- 373. Van Itallie CM, Rogan S, Yu AS, Seminario-Vidal L, Holmes J, Anderson JM. Two splice variants of claudin-10 in the kidney create paracellular pores with different ion selectivities. Am J Physiol Renal Physiol 291: F1288–F1299, 2006 [DOI] [PubMed] [Google Scholar]
- 374. Van Itallie CM, Tietgens AJ, Logrande K, Aponte A, Gucek M, Anderson JM. Phosphorylation of claudin-2 on serine 208 promotes membrane retention and reduces trafficking to lysosomes. J Cell Sci 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375. Van Meer G, Gumbiner B, Simons K. The tight junction does not allow lipid molecules to diffuse from one epithelial cell to the next. Nature 322: 639–641, 1986 [DOI] [PubMed] [Google Scholar]
- 376. Van Meer G, Simons K. The function of tight junctions in maintaining differences in lipid composition between the apical and the basolateral cell surface domains of MDCK cells. EMBO J 5: 1455–1464, 1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377. Vandenbroucke E, Mehta D, Minshall R, Malik AB. Regulation of endothelial junctional permeability. Ann NY Acad Sci 1123: 134–145, 2008 [DOI] [PubMed] [Google Scholar]
- 378. Varley CL, Garthwaite MA, Cross W, Hinley J, Trejdosiewicz LK, Southgate J. PPARgamma-regulated tight junction development during human urothelial cytodifferentiation. J Cell Physiol 208: 407–417, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379. Varley CL, Southgate J. Effects of PPAR agonists on proliferation and differentiation in human urothelium. Exp Toxicol Pathol 60: 435–441, 2008 [DOI] [PubMed] [Google Scholar]
- 380. Veshnyakova A, Piontek J, Protze J, Waziri N, Heise I, Krause G. Mechanism of Clostridium perfringens enterotoxin interaction with claudin-3/-4 suggests structural modifications of the toxin to target specific claudins. J Biol Chem 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381. Vincent T, Neve EP, Johnson JR, Kukalev A, Rojo F, Albanell J, Pietras K, Virtanen I, Philipson L, Leopold PL, Crystal RG, de Herreros AG, Moustakas A, Pettersson RF, Fuxe J. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat Cell Biol 11: 943–950, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382. Wang F, Daugherty B, Keise LL, Wei Z, Foley JP, Savani RC, Koval M. Heterogeneity of claudin expression by alveolar epithelial cells. Am J Respir Cell Mol Biol 29: 62–70, 2003 [DOI] [PubMed] [Google Scholar]
- 383. Wang FE, Zhang C, Maminishkis A, Dong L, Zhi C, Li R, Zhao J, Majerciak V, Gaur AB, Chen S, Miller SS. MicroRNA-204/211 alters epithelial physiology. FASEB J 24: 1552–1571, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384. Wang L, Zhang X, Liu L, Cui L, Yang R, Li M, Du W. Tanshinone II A down-regulates HMGB1, RAGE, TLR4, and NF-kappaB expression, ameliorates BBB permeability and endothelial cell function, protects rat brains against focal ischemia. Brain Res 1321: 143–151, 2010 [DOI] [PubMed] [Google Scholar]
- 385. Wang MH, Luo GM, Li F, Wu SL, Jiang JX, Huang CQ. Isolation and characterization of a novel four-transmembrane protein PMP22CD specifically expressed in the testis. Int J Mol Sci 7: 425–437, 2006 [Google Scholar]
- 386. Watson CJ, Hoare CJ, Garrod DR, Carlson GL, Warhurst G. Interferon-gamma selectively increases epithelial permeability to large molecules by activating different populations of paracellular pores. J Cell Sci 118: 5221–5230, 2005 [DOI] [PubMed] [Google Scholar]
- 387. Watson CJ, Rowland M, Warhurst G. Functional modeling of tight junctions in intestinal cell monolayers using polyethylene glycol oligomers. Am J Physiol Cell Physiol 281: C388–C397, 2001 [DOI] [PubMed] [Google Scholar]
- 388. Watson RE, Poddar R, Walker JM, McGuill I, Hoare LM, Griffiths CE, O'Neill CA. Altered claudin expression is a feature of chronic plaque psoriasis. J Pathol 212: 450–458, 2007 [DOI] [PubMed] [Google Scholar]
- 389. Weber CR, Nalle SC, Tretiakova M, Rubin DT, Turner JR. Claudin-1 and claudin-2 expression is elevated in inflammatory bowel disease and may contribute to early neoplastic transformation. Lab Invest 88: 1110–1120, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390. Weber CR, Raleigh DR, Su L, Shen L, Sullivan EA, Wang Y, Turner JR. Epithelial myosin light chain kinase activation induces mucosal interleukin-13 expression to alter tight junction ion selectivity. J Biol Chem 285: 12037–12046, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391. Weber CR, Shen L, Wang Y, Wang Y, Nelson DJ, Turner JR. Identification of discrete single tight junction opening/closing events with ion channel-like properties FASEB J 26: 1107, 2012 [Google Scholar]
- 392. Weber S, Schlingmann KP, Peters M, Nejsum LN, Nielsen S, Engel H, Grzeschik KH, Seyberth HW, Grone HJ, Nusing R, Konrad M. Primary gene structure and expression studies of rodent paracellin-1. J Am Soc Nephrol 12: 2664–2672, 2001 [DOI] [PubMed] [Google Scholar]
- 393. Welsch U. Freeze fracture-observations and TEM-observations on the plasma-membrane and associated structures of the epidermis of Branchiostoma lanceolatum (Cephalochordata). Zoomorphology 103: 209–218, 1983 [Google Scholar]
- 394. Welsch U. Septate junctions with paired septa in the cephalochordate Branchiostoma lanceolatum. Cell Tissue Res 229: 175–181, 1983 [DOI] [PubMed] [Google Scholar]
- 395. Wen H, Watry DD, Marcondes MC, Fox HS. Selective decrease in paracellular conductance of tight junctions: role of the first extracellular domain of claudin-5. Mol Cell Biol 24: 8408–8417, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396. Werth M, Walentin K, Aue A, Schonheit J, Wuebken A, Pode-Shakked N, Vilianovitch L, Erdmann B, Dekel B, Bader M, Barasch J, Rosenbauer F, Luft FC, Schmidt-Ott KM. The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development 137: 3835–3845, 2010 [DOI] [PubMed] [Google Scholar]
- 397. Whittembury G, Rawlins FA. Evidence of a paracellular pathway for ion flow in the kidney proximal tubule. Electromicroscopic demonstration of lanthanum precipitate in the tight junction. Pflügers Arch 330: 302–309, 1971 [DOI] [PubMed] [Google Scholar]
- 398. Wilcox ER, Burton QL, Naz S, Riazuddin S, Smith TN, Ploplis B, Belyantseva I, Ben-Yosef T, Liburd NA, Morell RJ, Kachar B, Wu DK, Griffith AJ, Friedman TB. Mutations in the gene encoding tight junction claudin-14 cause autosomal recessive deafness DFNB29. Cell 104: 165–172, 2001 [DOI] [PubMed] [Google Scholar]
- 399. Will C, Breiderhoff T, Thumfart J, Stuiver M, Kopplin K, Sommer K, Günzel D, Querfeld U, Meij IC, Shan Q, Bleich M, Willnow TE, Müller D. Targeted deletion of murine Cldn16 identifies extra- and intrarenal compensatory mechanisms of Ca2+ and Mg2+ wasting. Am J Physiol Renal Physiol 2010 [DOI] [PubMed] [Google Scholar]
- 400. Winkler L, Gehring C, Wenzel A, Muller SL, Piehl C, Krause G, Blasig IE, Piontek J. Molecular determinants of the interaction between Clostridium perfringens enterotoxin fragments and claudin-3. J Biol Chem 284: 18863–18872, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401. Wolburg H, Wolburg-Buchholz K, Kraus J, Rascher-Eggstein G, Liebner S, Hamm S, Duffner F, Grote EH, Risau W, Engelhardt B. Localization of claudin-3 in tight junctions of the blood-brain barrier is selectively lost during experimental autoimmune encephalomyelitis and human glioblastoma multiforme. Acta Neuropathol 105: 586–592, 2003 [DOI] [PubMed] [Google Scholar]
- 402. Wolburg H, Wolburg-Buchholz K, Liebner S, Engelhardt B. Claudin-1, claudin-2 and claudin-11 are present in tight junctions of choroid plexus epithelium of the mouse. Neurosci Lett 307: 77–80, 2001 [DOI] [PubMed] [Google Scholar]
- 403. Wong V, Goodenough DA. Paracellular channels! Science 285: 62, 1999 [DOI] [PubMed] [Google Scholar]
- 404. Wongdee K, Pandaranandaka J, Teerapornpuntakit J, Tudpor K, Thongbunchoo J, Thongon N, Jantarajit W, Krishnamra N, Charoenphandhu N. Osteoblasts express claudins and tight junction-associated proteins. Histochem Cell Biol 130: 79–90, 2008 [DOI] [PubMed] [Google Scholar]
- 405. Wray C, Mao Y, Pan J, Chandrasena A, Piasta F, Frank JA. Claudin-4 augments alveolar epithelial barrier function and is induced in acute lung injury. Am J Physiol Lung Cell Mol Physiol 297: L219–L227, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406. Wright EM, Barry PH, Diamond JM. The mechanism of cation permeation in the rabbit gallbladder: conductances, the current-voltage relation, the concentration dependence of anion-cation discrimination, and the calcium competition effect. J Membr Biol 4: 331–357, 1971 [DOI] [PubMed] [Google Scholar]
- 407. Wu J, Helftenbein G, Koslowski M, Sahin U, Tureci O. Identification of new claudin family members by a novel PSI-BLAST based approach with enhanced specificity. Proteins 65: 808–815, 2006 [DOI] [PubMed] [Google Scholar]
- 408. Wu N, Zhang X, Jin S, Liu S, Ju G, Wang Z, Liu L, Ye L, Wei J. A weak association of the CLDN5 locus with schizophrenia in Chinese case-control samples. Psychiatry Res 178: 223, 2010 [DOI] [PubMed] [Google Scholar]
- 409. Wu VM, Schulte J, Hirschi A, Tepass U, Beitel GJ. Sinuous is a Drosophila claudin required for septate junction organization and epithelial tube size control. J Cell Biol 164: 313–323, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410. Xu H, Dawson R, Crane IJ, Liversidge J. Leukocyte diapedesis in vivo induces transient loss of tight junction protein at the blood-retina barrier. Invest Ophthalmol Vis Sci 46: 2487–2494, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411. Yamamoto M, Ramirez SH, Sato S, Kiyota T, Cerny RL, Kaibuchi K, Persidsky Y, Ikezu T. Phosphorylation of claudin-5 and occludin by rho kinase in brain endothelial cells. Am J Pathol 172: 521–533, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412. Yamamoto T, Kojima T, Murata M, Takano K, Go M, Chiba H, Sawada N. IL-1beta regulates expression of Cx32, occludin, and claudin-2 of rat hepatocytes via distinct signal transduction pathways. Exp Cell Res 299: 427–441, 2004 [DOI] [PubMed] [Google Scholar]
- 413. Yamauchi K, Rai T, Kobayashi K, Sohara E, Suzuki T, Itoh T, Suda S, Hayama A, Sasaki S, Uchida S. Disease-causing mutant WNK4 increases paracellular chloride permeability and phosphorylates claudins. Proc Natl Acad Sci USA 101: 4690–4694, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414. Yoshida Y, Ban Y, Kinoshita S. Tight junction transmembrane protein claudin subtype expression and distribution in human corneal and conjunctival epithelium. Invest Ophthalmol Vis Sci 50: 2103–2108, 2009 [DOI] [PubMed] [Google Scholar]
- 415. Young ME, Karpova TS, Brugger B, Moschenross DM, Wang GK, Schneiter R, Wieland FT, Cooper JA. The Sur7p family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, is involved in sporulation. Mol Cell Biol 22: 927–934, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416. Yu AS, Cheng MH, Angelow S, Günzel D, Kanzawa SA, Schneeberger EE, Fromm M, Coalson RD. Molecular basis for cation selectivity in claudin-2-based paracellular pores: identification of an electrostatic interaction site. J Gen Physiol 133: 111–127, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417. Yu AS, Cheng MH, Coalson RD. Calcium inhibits paracellular sodium conductance through claudin-2 by competitive binding. J Biol Chem 285: 37060–37069, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418. Yu AS, Enck AH, Lencer WI, Schneeberger EE. Claudin-8 expression in MDCK cells augments the paracellular barrier to cation permeation. J Biol Chem 278: 17350–17359, 2003 [DOI] [PubMed] [Google Scholar]
- 419. Yuan L, Le Bras A, Sacharidou A, Itagaki K, Zhan Y, Kondo M, Carman CV, Davis G.E, Aird WC, Oettgen P. ETS-related gene (ERG) controls endothelial cell permeability via transcriptional regulation of the claudin 5 (CLDN5) gene. J Biol Chem 287: 6582–6591, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420. Zeissig S, Burgel N, Günzel D, Richter J, Mankertz J, Wahnschaffe U, Kroesen AJ, Zeitz M, Fromm M, Schulzke JD. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn's disease. Gut 56: 61–72, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421. Zemke AC, Snyder JC, Brockway BL, Drake JA, Reynolds SD, Kaminski N, Stripp BR. Molecular staging of epithelial maturation using secretory cell-specific genes as markers. Am J Respir Cell Mol Biol 40: 340–348, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422. Zhang K, Yao HP, Wang MH. Activation of RON differentially regulates claudin expression and localization: role of claudin-1 in RON-mediated epithelial cell motility. Carcinogenesis 29: 552–559, 2008 [DOI] [PubMed] [Google Scholar]
- 423. Zhao L, Yaoita E, Nameta M, Zhang Y, Cuellar LM, Fujinaka H, Xu B, Yoshida Y, Hatakeyama K, Yamamoto T. Claudin-6 localized in tight junctions of rat podocytes. Am J Physiol Regul Integr Comp Physiol 2008 [DOI] [PubMed] [Google Scholar]
- 424. Zheng A, Yuan F, Li Y, Zhu F, Hou P, Li J, Song X, Ding M, Deng H. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J Virol 81: 12465–12471, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425. Zheng JY, Yu D, Foroohar M, Ko E, Chan J, Kim N, Chiu R, Pang S. Regulation of the expression of the prostate-specific antigen by claudin-7. J Membr Biol 194: 187–197, 2003 [DOI] [PubMed] [Google Scholar]
- 426. Zhou W, Cao Q, Peng Y, Zhang QJ, Castrillon DH, DePinho RA, Liu ZP. FoxO4 inhibits NF-kappaB and protects mice against colonic injury and inflammation. Gastroenterology 137: 1403–1414, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427. Zhu Y, Brannstrom M, Janson PO, Sundfeldt K. Differences in expression patterns of the tight junction proteins,claudin 1, 3, 4 and 5, in human ovarian surface epithelium compared with epithelia in inclusion cysts and epithelial ovarian tumours. Int J Cancer 118: 1884–1891, 2006 [DOI] [PubMed] [Google Scholar]
- 428. Zimmerli SC, Hauser C. Langerhans cells and lymph node dendritic cells express the tight junction component claudin-1. J Invest Dermatol 127: 2381–2390, 2007 [DOI] [PubMed] [Google Scholar]