Abstract
Pumilio/fem-3 mRNA binding factor (FBF) proteins are characterized by a sequence-specific RNA-binding domain. This unique single-stranded RNA recognition module, whose sequence specificity can be reprogrammed, has been fused with functional modules to engineer protein factors with various functions. Here we summarize the advancement in developing RNA regulatory tools and opportunities for the future.
Keywords: PUF protein, RNA, RNA-binding protein, splicing, protein engineering, X-ray crystallography, translation, RNA localization, RNA regulation, protein-RNA interaction
Introduction
Isaac Newton based his first Rule of Reasoning in Philosophy on the teleological principle of purposeful design and assertion that “Nature is pleased with simplicity.” Such simplicity is often illustrated in Nature’s elegant solution to control gene expression with a modular molecule. In many key gene expression steps, such as DNA transcription or RNA splicing, the regulatory proteins usually comprise a nucleic acid binding module to recognize target and a functional module to activate or inhibit the biochemical process. This simple design principle has also been adopted by humans to engineer protein factors to manipulate DNA/RNA. For specific recognition of RNA, Nature evolved an RNA-binding module, the PUF (Pumilio/FBF) scaffold, whose specificity, itself simple and modular, is determined by combination of functional base recognition repeats. This unique RNA-binding mode allows one to reprogram the specificity of the PUF scaffold with a small number of mutations, thus generating novel engineered protein factors to modulate RNA metabolism. In this review, we present the design principles, current progress, and future outlook of engineering protein factors with a PUF domain as the RNA-binding scaffold. A broader review of many different RNA-binding modules and their utility for engineering specificity accompanies our more focused report [1].
Drosophila melanogaster Pumilio and Caenorhabiditis elegans FBF are the founding members of the PUF family of RNA regulatory proteins. Pumilio is encoded by a maternal gene required for abdominal development in Drosophila [2]. FBF was selected by yeast 3-hybrid as a protein that binds to an RNA sequence in the 3′ untranslated region (UTR) of fem-3 mRNA [3, 4], regulating the switch from spermatogenesis to oogenesis in C. elegans hermaphrodites. Molecular cloning revealed that Pumilio and FBF proteins share a conserved RNA-binding domain with eight ~36 amino acid sequence repeats, which was named a Pumilio Homology Domain or PUF domain [4–7]. The known target RNAs suggested that both proteins recognize a sequence in the 3′ UTR containing a conserved UGUR sequence (where R is a purine) [4, 6, 8–11]. Pumilio and FBF regulate stability or translation of target mRNAs by recruiting effector protein complexes to their target RNAs. The PUF proteins themselves seem to lack additional functional modules, and instead protein-protein interactions, often with the RNA-binding domain, assemble different complexes on the target RNAs [12–16]. The formation of complexes may also fine tune RNA specificity [17]. PUF proteins utilize these activities to regulate stem cell maintenance, cell proliferation and differentiation, and stress response (reviewed in [18, 19]).
RNA recognition code for native PUF proteins
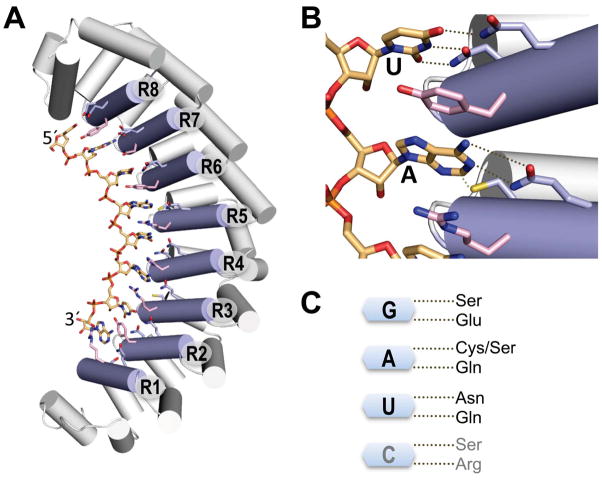
The first crystal structures of PUF proteins revealed how the repeated sequences form a series of α-helical repeats that assemble an arc of RNA-recognition helices (Fig. 1A) [20, 21]. A structure of human Pumilio1 in complex with an RNA ligand showed how each of the eight RNA-recognition helices recognizes one base using amino acid side chains at specific positions in the second of three α-helices in each PUF repeat (Fig. 1B) [22]. Two of these side chains interact specifically with an edge of the base while a third side chain forms a stacking interaction with the base. The second helix in each PUF repeat includes a 5-residue sequence, designated here as 12xx5, where the side chain at the 2nd position stacks with the recognized base and the side chains at the 1st and 5th positions contact the edge of the RNA base [22, 23].
Fig. 1.
PUF domain RNA interaction scaffold. A. Ribbon diagram of a crystal structure of human Pumilio 1 RNA-binding domain in complex with RNA ligand (5′-UGUACAUA). RNA interaction helices are shown as light blue cylinders and labeled R1–R8. Edge-interacting side chains from each repeat are colored light blue, stacking side chains are colored pink, and RNA is colored peach. For stick representations, nitrogen atoms are blue, oxygen atoms are red, and sulfur atoms are yellow. B. Interaction of PUF repeats with uracil or adenine bases. Hydrogen bonds or van der Waals contacts are indicated by dotted lines. C. PUF RNA interaction code. Edge-interacting side chains that specify base recognition are shown. G, A, and U were derived from natural PUF proteins while C recognition was selected by screening.
Mutagenesis confirmed that Nature had provided a remarkably simple code for base recognition: glutamate and serine at the 1st and 5th positions recognize guanine; glutamine and cysteine/serine recognize adenine; and glutamine and asparagine recognize uracil (Fig. 1C) [22, 24]. No code for cytosine was apparent from Nature, but one of the repeats (repeat 4) can accept any base. In vitro binding affinity is tight due to the many stacking interactions between protein side chains and RNA bases. Wildtype human Pumilio1 binds to cognate RNA with a Kd of 0.5 nM, while designed Pumilio1 mutants bind to their cognate RNAs with Kds ranging from as tight as ~4 pM to 18 nM. The effect on binding of a single non-cognate repeat is largest when a base other than G is presented opposite a G-specific repeat (30–150-fold weaker) and smallest when a G is presented opposite an A-specific repeat (1.5–3-fold weaker) [24]. Other non-cognate mismatches reduce binding 10–20 fold [24, 25], indicating the importance of most repeats for specificity. With this RNA recognition code, in principle, the RNA-binding specificity of human Pumilio1 can be manipulated by site-directed mutagenesis to recognize various RNA targets containing guanine, adenine and uracil.
The identification of this recognition code and the ability to modify specificity of a PUF repeat has facilitated the identification of the RNA recognition properties of other PUF proteins. PUF proteins comprise a relatively small family of RNA-binding proteins with few members in any organism. Humans and other mammals have two while Drosophila have one. S. cerevisiae with six and C. elegans with nine are larger families. This is in contrast to RNA recognition motif (RRM) proteins or KH (hnRNP K homology) domains, where hundreds of proteins are found in an organism. Most PUF proteins are predicted to have eight PUF repeats like human Pumilio1, and the base recognition side chains are highly conserved. However, the RNA target sequences of these proteins are considerably more diverse than would be expected from the strong conservation. The core target sequences begin with a 5′ UGUR, but may contain from 8–10 bases with differing 3′ sequences.
To reconcile this inconsistency, the Wickens lab at University of Wisconsin used the design-ability of PUF repeat base recognition to determine the mode of recognition for additional C. elegans PUF proteins [26, 27]. Beginning with FBF, the Wickens lab used the yeast three-hybrid assay to measure RNA binding of cleverly designed PUF proteins. They had determined that FBF with 8 PUF repeats bound to a core RNA sequence with 9 bases [28], but was the ‘extra’ base positioned within the sequence or at the 3′ end? To answer this question, they created designed FBF proteins where a specific repeat was mutated to recognize a different base, and they determined the sequence specificity of the designed proteins [27]. The change in sequence specificity vs. wildtype FBF established the register of PUF repeats and RNA bases. They concluded that FBF targets possess an ‘extra’ base at the 6th of 9 bases that is not recognized by the protein, and crystal structures have confirmed it [29, 30].
Additional studies of the RNA target specificities of different PUF proteins have revealed how Nature expands recognition properties using an assortment of variations on the theme of the 1 PUF repeat:1 RNA base recognition observed for Pumilio 1 [26–28, 30–35]. Crystal structures of PUF proteins with RNA ligands demonstrate that extra or other non-specifically recognized bases within natural PUF target sequences may stack directly with adjacent bases and/or be flipped away from the PUF protein recognition surface [23, 29, 30, 36, 37]. A specialized binding pocket in a subset of PUF proteins recognizes a cytosine base upstream of the core sequence [30, 38], and forming a complex with a partner protein can modify the recognition motif [17]. The combinations of such variations produce the unique specificity of natural target recognition by PUF proteins.
The accumulated body of evidence from genome-wide RNA target identification, X-ray crystal structures, in vitro RNA-binding assays, and systematic mutagenesis studies indicate that PUF protein repeats toward the N- and C-termini recognize conserved RNA sequence elements, while plasticity in RNA recognition is focused mainly at the central repeats 4 and 5. The conserved 5′ UGUR sequence in PUF protein target sequences is recognized by repeats 5–8 (Fig. 1A). A downstream AU sequence is also conserved and recognized by repeats 2 and 3 in many PUF proteins. Valley, et al. examined systematically the effect of alanine mutation of either edge-interacting or base-stacking side chains on RNA-binding by C. elegans FBF and yeast Puf3p and Puf4p [34]. They found that repeats 4 and 5 can often tolerate alanine substitutions and proposed a “two-handed model” to describe PUF-RNA interactions, where the N- and C-terminal PUF repeats “grasp” the conserved 3′ and 5′ sequences of RNA binding sites, respectively. Protein-specific variations in RNA recognition are accommodated between them.
In the case of yeast Puf4p, plasticity in recognition allows binding to target RNAs with two different recognition patterns. In many target sites, the 7th of 9 bases is flipped away from the RNA-binding surface (7-flipped), driven by recognition of an adenosine at position 6 by repeat 3 (5′-UGUAUA-n7-UA, where n7 is any base at position 7). In a second set of target sites, the recognition pattern produces flipping of the 6th base (6-flipped) with an adenosine at position 7 recognized by repeat 3 (5′-UGUAU-n6-AUA). The sequence of these 6-flipped sites also matches the target sites of another yeast PUF protein, Mpt5p. Thus, Puf4p and Mpt5p may dually regulate target mRNAs with six-flipped sites, while seven-flipped sites are not recognized by Mpt5p and regulated only by Puf4p.
Stacking interactions in PUF-RNA recognition contribute substantially to the binding energy, but the role of these interactions in binding specificity of PUF proteins is less well understood. RNA sequence specificity can be changed by site-directed mutagenesis of only the two edge-interacting side chains [24]. However, it appears that the identity of the third amino acid side chain that stacks with the bases has been optimized for the natural specificities of PUF proteins [39]. Substitution of the stacking side chains in C. elegans FBF broadens the range of RNA bases that can be tolerated at a particular position. The simple approach of designing PUF specificity by altering edge-interacting side chains is typically successful, but a better understanding of how stacking side chains contribute to specificity may enhance target selectivity of designed proteins.
Expanding the modular binding code for cytosine
With the natural modularity and recognition code of human Pumilio 1, its PUF domain can be engineered to target an eight-nucleotide RNA sequence containing adenine, uracil and guanine [22, 24]. By fusing the PUF domain with a functional domain, these ‘designer’ PUFs have been applied to track RNA localization in cells [40, 41], regulate alternative splicing [25, 42], and modulate translational regulation [43]. Opportunities remain to develop novel tools for biomedical research with potential therapeutic applications (see below).
A limitation to the application of designer PUFs had been that no recognition code for cytosine specificity was found in Nature. However, two groups recently succeeded in identifying combinations of amino acid side chains that specify cytosine recognition by a PUF repeat, expanding the PUF recognition code and permitting the creation of PUFs directed to any 8-base RNA sequence [25, 44]. Both groups used yeast three-hybrid screening of a PUF domain library with random sequences at the 1st and 5th positions of the RNA interaction motif in repeat 6 to select proteins that bound a cytosine at the corresponding 3rd base of the RNA target. Dong, et al. found that a combination of serine at the 1st position and arginine at the 5th position specify cytosine [25]. A crystal structure showed hydrogen bonding between the side chain of arginine and the O2 and N3 positions of the cytosine. The serine residue positions the arginine, but does not contact the RNA. Using a similar strategy, Filipovska, et al. selected five PUF variants that specify cytosine binding [44]. Each variant pairs an arginine at the 5th position with a small side chain at the 1st position. The ‘C code’ could be transferred to different PUF repeats, and tyrosine, histidine, or arginine side chains at the 2nd position of the RNA interaction sequence are capable of forming stacking interactions for cytosine recognition [25]. The identification of the C code makes it now possible to design PUF domains that bind any 8-base RNA target sequence and increases the potential for use of engineered PUF proteins in research and therapeutics.
Engineered factors with PUF as RNA-binding scaffold
The unique RNA-binding mode of the PUF domain makes it possible to engineer artificial proteins to modulate RNA metabolism. Following the composition of natural proteins, these artificial proteins employ a modular design by fusing a functional module (protein domains with desired activities) to a target recognition module (modified PUF domains with re-designed RNA binding specificity). Compared with other domains that specifically bind single-stranded RNA, such as RRM, Zn finger and pentatricopeptide repeats (PPR), the PUF domain has the advantage of having a fully determined modular binding code. The RRM and Zn finger domains only recognize short RNA motifs (3-4 nt), and their binding specificity cannot be reprogrammed. However, it is conceivable that several RRMs or Zn fingers can be tandemly linked to recognize a longer RNA. Similar to the PUF domain, the PPR domain can recognize RNA in a modular fashion and several researchers have begun to make progress in solving its modular binding code [45]. Once the binding code is completed, the PPR may be used similarly to PUF as a modular RNA binding scaffold in engineered factors. The main disadvantage of PUF is that the native protein contains only eight repeats that recognize 8-nt target, however it is possible to expand the length of PUF recognition sites (described below). Varying the choice of functional modules, an assortment of PUF factors has been engineered since 2007 and more factors with novel activities are expected to be generated in the future.
Engineered PUF proteins as specific RNA probes
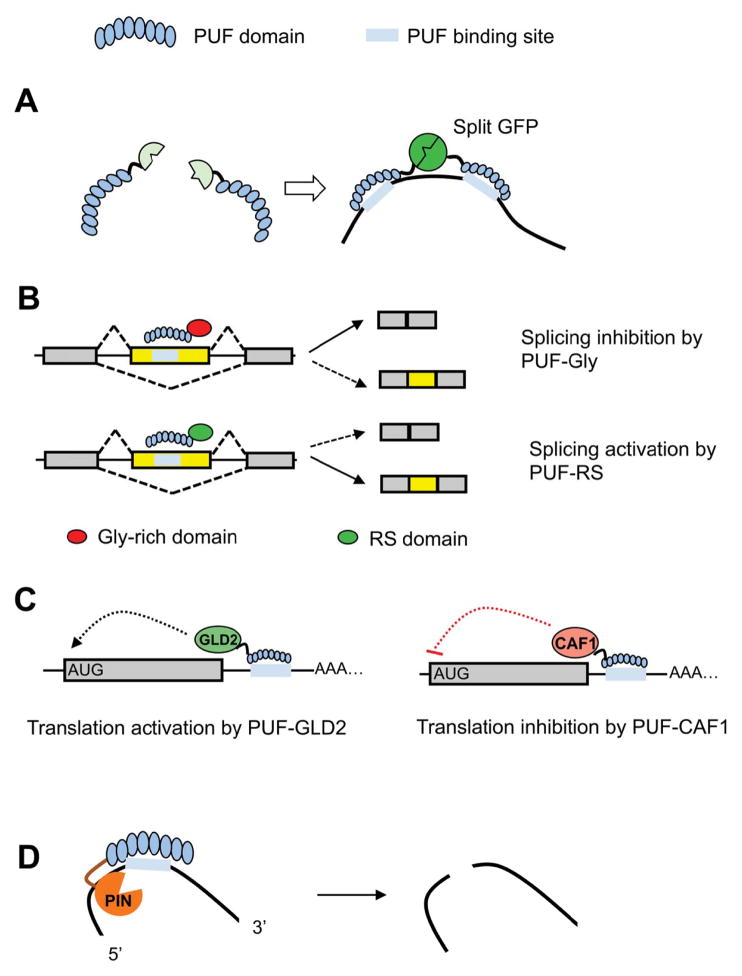
The first application of artificial PUF proteins was as a fluorescent marker to visualize RNAs in live cells [40]. A highly effective approach to visualize RNAs in live cells is to use a fusion protein of green fluorescent protein (GFP) fused to bacteriophage coat protein MS2 that recognizes reporter RNAs containing multiple copies (e.g. 24 copies) of MS2 hairpin sequences [46, 47](reviewed in [48]). However, the GFP-MS2 fusion protein requires artificial RNA targets with MS2 hairpins. To visualize endogenous RNA targets, Ozawa, et al. designed a split GFP system that comprises two fusion proteins, each containing a PUF domain and either an N-terminal or C-terminal fragment of GFP (Fig. 2A). When the PUF domains recognize the adjacent 8-nt sequences in the same RNA, the two fusion proteins are brought together so that the split GFP fragments assemble and fluoresce. The specificity of this system is increased by requiring binding to two 8-nt sites separated by a small spacer. This design has been used to visualize mitochondrial RNA in cultured mammalian cells [40], cytosolic β-actin mRNA [49], and a modified version using intact GFP fused with two PUFs was also used to detect cytosolic β-actin mRNA [50]. A similar approach was also developed by Tilsner, et al, who combined PUF domains with split mCitrine and used it to reveal plant viral RNA localization [41]. Another variation of this method used PUF domains fused with split luciferase to detect ssRNA in vitro [51], however this approach has not yet been tested in live cells.
Fig. 2.
Engineered PUF factors. A. Fluorescent probe for in vivo RNA labeling. Combination of split GFP (or other fluorescent protein) with PUF scaffold generated an RNA probe to visualize RNA in live cells. B. Engineered splicing factors. Combination of a PUF scaffold with an RS domain or a Gly-rich domain generated splicing factors that activate or inhibit splicing of alternative exon. C. Modulation of translation. Fusion of GLD2 or CAF1 with PUF domain produced novel factors that can activate or inhibit mRNA translation. D. Artificial site-specific RNA endonuclease. Combination of PUF domain with a non-specific RNA endonuclease (PIN domain) can produce a new class of enzymes that specifically recognize and cleave RNA.
The use of split fluorescent protein reduces the noise level of RNA detection, because the fluorescent protein is assembled by the RNA target. However, because of the limited efficiency in the co-folding of two GFP fragments and only one GFP per RNA, this system produces lower fluorescent signal compared to the GFP-MS2. On the other hand, the PUF-split GFP can detect a single endogenous RNA target, whereas the GFP-MS2 requires a reporter RNA with multiple MS2 target sites to produce a higher signal over the diffuse background noise of unbound fluorescent protein. For some special cases, it should be possible to use the GFP-PUF fusion protein directly. Like the GFP-MS2 fusion protein, the GFP-PUF will produce a weak fluorescence background. However, if the target contains multiple copies of the recognition site, the GFP-PUF may form bright foci on the target RNA, like the GFP-MS2 protein forms a bright spot on reporter RNAs with consecutive MS2 hairpins [47]. Several trinucleotide expansions have been found to cause neurodegenerative diseases (such as CAG repeats in Huntington disease and CUG repeats in myotonic dystrophy, reviewed in [52]). These pathogenic RNA repeats usually contain hundreds of copies of the trinucleotide, thus a GFP-PUF recognizing these repeated sequences may provide a new way to visualize the RNA and study the dynamics of such pathogenic RNA in live cells.
Engineered PUF proteins as regulatory factors for gene expression
Mammalian gene expression is controlled by multiple regulatory steps during mRNA processing by protein factors with modular configuration: separable RNA recognition and functional domains. A common method to study the function of RNA regulatory proteins is to tether the protein or a fragment to a specific RNA target and analyze the consequences. Using this engineering principle, artificial regulatory factors can be derived by combining a PUF domain with different effector domains. For example, Wang, et al. developed engineered splicing factors (ESFs) by fusing a PUF domain with Arg/Ser-rich (RS) domains of SRSF1 or the glycine-rich (Gly) domain of hnRNP A1 (Fig. 2B) [42]. The resulting ESFs can function either as splicing activators (PUF-RS) or inhibitors (PUF-Gly), and can specifically control different types of alternative splicing. Specifically, PUF-RS promotes exon inclusion when binding to an alternative exon, but inhibits splicing when binding downstream of an alternative exon. PUF-Gly, on the other hand, inhibits exon inclusion when binding to the target RNA either inside or downstream of the alternative exon [42, 53]. When binding to the region between two alternative 5′ or 3′ splice sites, PUF-RS promotes use of the proximal splice site to favor the longer splicing isoform, whereas PUF-Gly inhibits proximal site usage to induce the shorter isoform [42].
Using this general approach, a PUF-Gly ESF was generated to shift splicing of the Bcl-x pre-mRNA from the anti-apoptotic long isoform (Bcl-xL) to the pro-apoptotic short isoform (Bcl-xS). The altered splice isoform distribution was sufficient to sensitize several cancer cell lines to multiple anti-cancer drugs [42]. With the identification of the C code, both PUF-RS and PUF-Gly ESFs were engineered to shift splicing of VEGF-A pre-mRNA from the angiogenic to anti-angiogenic isoform [25].
Another elegant use of the modular configuration for RNA regulation was developed by Cooke, et al. to alter translation [43]. They attached a PUF domain to either a translational activator, GLD2, or a translational repressor, CAF1 (Fig. 2C). The resulting PUF-GLD2 engineered proteins specifically recognized their RNA target and activated translation and induced polyadenylation in Xenopus oocytes, whereas the PUF-CAF1 fusion protein repressed translation and directed deadenylation. The coordinate regulation of translation and/or stability of mRNA by PUF proteins and miRNAs [54–57] suggests that engineered PUF proteins may be designed to use as antagonizers or enhancers of miRNA regulation.
Engineered PUF proteins as novel RNA endonucleases
Specific cleavage of RNAs is critical for in vitro manipulation of RNA and for in vivo gene silencing. However, a simple enzyme that cleaves RNA in a sequence-specific manner has not been found in Nature despite extensive investigations. Most RNA endonucleases either have limited sequence specificity (e.g., RNase A or RNase T1 [58–60]) or recognize their targets by specific structures (e.g., tRNA splicing endonuclease [61]) or through guide RNA that pairs with target (e.g., Argonaute proteins [62]). By fusing a PUF domain with a non-specific endoribonuclease domain (PIN domain of Smg6p), Choudhury, et al. engineered artificial site-specific RNA endonucleases (ASREs) that specifically recognize RNA substrates and efficiently cleave near the binding sites both in vitro and in cultured cells (Fig. 2D) [63]. The digested products have 5′-phosphate and 3′-hydroxyl groups, making it possible to religate. Two ASREs were designed to silence specifically the expression of a bacterial gene or human mitochondrial mRNAs that contain one or two binding sites of designer PUFs. Since PUF domains recognize their targets through an 8-nt sequence, comparable length to the seed match of siRNA, engineered ASREs may serve as an RNA silencing tool complementary to RNAi, which will be effective in organisms or cellular compartments where RNAi machinery is not present.
Future applications of PUF-based designer factors
Mammalian gene expression contains multiple RNA processing stages including RNA splicing, polyadenylation, editing, translocation, translation and degradation, which are under tight control by a variety of RNA-binding proteins. The reprogrammable RNA-binding scaffold of the PUF domain makes it possible, and tempting, to create artificial factors that can specifically modulate target RNAs at each processing stage. A benefit of manipulating gene expression at the RNA level rather than the DNA level is that the effect is often non-permanent and reversible. The factors can be engineered by fusing designer PUF domains with a protein factor (or its functional domain) with known regulatory activity on RNA metabolism. In theory, any protein that has demonstrated activity in a tethering experiment can be fused with PUF domains to produce novel artificial factors that target endogenous RNA targets. Minimal functional domains can be determined by fusing fragments of known regulatory factors to PUF domains and testing activity. Alternatively, new functional modules can be identified by constructing a cDNA library of PUF domain fusions and screening for RNA regulation. A previous review has proposed several ideas for potential applications of engineered RNA-binding proteins [64], some of which have been achieved successfully using the PUF scaffold (Fig. 2). Here we propose other potential applications using the engineered PUF factors in humans, and similar applications can be adapted to other organisms.
Engineering RNA editing enzymes
RNA editing is commonly found in many eukaryotes to generate specific sequence substitutions and changes in gene expression levels, resulting in increase of RNA and protein diversity. Adenosine-to-inosine (A-to-I) editing represents the most important class of RNA editing in human and affects function of many genes, especially in the central nervous system (recently reviewed in [65]). The native substrate for A-to-I editing is a double-stranded RNA (dsRNA) region, recognized by the adenosine deaminase acting on RNA (ADAR). Canonical ADAR proteins have a conserved catalytic deaminase domain and one or more dsRNA-binding domains (dsRBDs), however there is no report on whether these domains function in a modular fashion [65]. Thus we propose replacing the dsRBDs with a PUF domain and testing whether the resulting fusion factors have deaminase activities against single-stranded RNA (Fig. 3A). With optimization, the engineered enzyme may specifically recognize an RNA target by sequence and deaminate a nearby adenosine.
Fig. 3.
Future engineered factors with PUF scaffold as RNA recognition module. A. Novel enzyme for single-stranded RNA editing. Sequence specificity may be determined by a PUF binding site rather than double-stranded RNA. B. Next generation engineered splicing factors. With recently identified functional domains that control splicing in diverse fashion, new splicing factors may be generated to fine tune alternative splicing. C. Engineered PUF factors to manipulate alternative polyadenylation. By combining a PUF domain with regulatory proteins/domains for alternative polyadenylation sites, artificial factors may be constructed to inhibit or activate certain polyadenylation sites and thus change the 3′ UTR of mRNA. D. Engineered RNA transporters. New proteins can be engineered by combining a PUF domain with protein translocation signals to transport RNA into different cellular compartments. E. Engineered PUF factors to control RNA degradation. The PUF domain can be linked with additional domains to specifically recruit RNA stabilization or destabilization proteins, thus controlling RNA half-life. F. Engineered PUF factors to control transcription elongation. By specifically recognizing the nascent RNA transcript, new PUF factors can be engineered to induce transcription pausing or release paused RNA polymerases.
Next generation engineered splicing factors
Most human genes undergo alternative splicing to produce multiple isoforms with distinct activities. This process is tightly regulated, and the mis-regulation of splicing is a common cause of human diseases [66]. The specific manipulation of alternative splicing will provide a useful way to fine tune gene function. Many known splicing factors have modular activities in which they recognize a short RNA element in the target with an RNA-binding module (e.g., RRM or KH domain) and alter splicing with a functional module. The best-known functional domains for splicing modulation are the RS domains and Gly-rich domains, which were used to develop the first generation of engineered splicing factors [25, 42]. However, other domains in splicing regulatory factors may have distinct activities in regulating splicing. For example, we found that the C-terminal domain of several SR proteins can activate or inhibit splicing when binding to different pre-mRNA regions [67], the alanine-rich motif of RBM4 can inhibit splicing, and the proline-rich motif of DAZAP1 can enhance splicing [68]. These new functional domains can be used to derive additional types of artificial splicing factors to fine tune alterative splicing (Fig. 3B).
Engineered factors to control alternative polyadenylation
Many human genes contain multiple polyadenylation sites. The use of different polyadenylation sites creates mRNA isoforms with different 3′ UTRs that regulate the translation efficiency and stability of mRNA. Like alternative splicing, control of alternative polyadenylation can have multifaceted effects on biological processes, and shortening of the 3′ UTR through proximal polyadenylation site usage is closely regulated in cell proliferation and differentiation [69–71]. Thus reprogramming the 3′ UTR region by modulating polyadenylation site selection may provide another way to manipulate gene expression. Several recent reports suggested that alternative polyadenylation is regulated by trans-acting factors that recognize cis-elements near polyadenylation sites [72–74]. Some of these factors can control both splicing and poly-adenylation (eg. U1A, PTB) [72, 74]. Therefore we expect engineered PUF fusion proteins to be similarly successful in modulating the use of alternative polyadenylation sites (Fig. 3C).
Engineered factors to control RNA localization
The transport of RNA to correct intracellular locations is an essential step for the expression of both coding and non-coding messages, as the function of RNA is highly localized. While there are many known RNA elements that govern their transport and final localization, other recognition elements are unknown and the detailed mechanisms are unclear. On the other hand, protein localization signals are better understood and well characterized signal peptides or fragments have been established that are responsible for protein localization in nuclear or cytoplasmic compartments or for protein secretion. Therefore, by combining these established protein localization signals with PUF domains, we may create engineered factors to specifically transport RNA (Fig. 3D). For example, a PUF domain fused with a nuclear localization sequence (NLS) may be created to block nuclear-to-cytoplasmic RNA transport. Such engineered proteins can be designed to recognize the loop-region of a specific pre-miRNA, thus retaining the pre-miRNA in the nucleus and preventing its processing to mature form in the cytoplasm, thereby inhibiting the miRNA function.
Engineered PUF factors to affect RNA stability
Turnover of mRNA is continuous in the cytoplasm and often regulated by protein factors that recognize the 3′ UTR region. A well-studied case is directed by AU-rich elements in the 3′ UTR of short-lived mRNA, which destabilize the messages (reviewed in [75]). Several factors positively or negatively regulate this process, and PUF fusion proteins with fragments of these factors may function as engineered factors to modulate RNA stability (Fig. 3E).
Engineered PUF transcriptional regulators
RNA transcription can be controlled at the initiation-to-elongation switch of RNA polymerase II through a transcriptional pausing step after the initiation and synthesis of a small RNA fragment (reviewed recently in [76]). Such pausing is released to continue transcription, which is often controlled by transcription elongation factors. Using PUF engineered factors that recognize the nascent transcripts, it may be possible to design factors that specifically control transcription at the initiation-to-elongation switch by promoting or releasing the paused RNA polymerase II (Fig. 3F). For example, the release of paused RNA polymerase II can be mediated by CDK9 and Cyclin T1, which form a tightly complex to promote transcription elongation [77]. It may be possible to recruit CDK9 through a PUF-CDK9 fusion protein to the nascent transcripts, which in turn releases paused polymerase II and activates transcription elongation.
Additional challenges for engineered PUF factors
Expanding binding specificity
Most naturally occurring PUF proteins have eight RNA-binding repeats, which recognize 8 bases in target mRNA. Some PUF domains can accommodate one or two extra bases, however, the specificity of the extra bases cannot be reprogrammed. Thus the sequence specificity of engineered PUF factors is largely determined by an 8-nt PUF binding site, which is comparable to the specificity of miRNA that recognize targets by 7-nt seed match. However, like miRNA, designer PUF factors may have off-target effects, since on average any given 8-nt sequence will occur by chance in sequences with 48 bases or 64 kb in length.
Nevertheless it is possible to increase the specificity of engineered PUF factors. One obvious approach is to use two PUF domains in combination, which in theory provides a recognition site of 16 nts. This design was used first for the visualization of RNA in vivo with two fusion proteins of PUF domains and split GFP fragments [40] and later was used by creating a fusion protein containing two PUF domains separated by an intact GFP protein [50]. Specificity for a 16-nt sequence should ensure the recognition of a unique target in the human transcriptome. However, engineered factors with tandem PUF domains may recognize an 8-nt ‘half-site’ with sufficient affinity for functional activity. An alternative approach is to construct an expanded PUF domain by inserting more PUF repeats into a native PUF domain. For example, a PUF domain with 16 repeats was demonstrated to bind cognate RNA target in a yeast-three-hybrid assay [44], and it should be possible to construct PUF domains with different numbers of repeats using similar design. We have generated expanded PUF domains with nine, ten or twelve PUF repeats and found that these novel PUF scaffolds can indeed recognize cognate targets (Zhang and Wang, unpublished results).
While most applications will benefit from a PUF domain with increased specificity, in certain cases the engineered PUF factors may need to recognize multiple diverse targets with a degenerate binding code (i.e. a decreased specificity). This requires generating PUF domains that recognize shorter target sequences. Two strategies may solve this problem. One is to identify amino acid side chain combinations in PUF repeats that can tolerate multiple or all bases to produce a binding code for “N”. This degenerate code can be used in combination with the specific base recognition code to make new PUF domains that specifically recognize sequences shorter than 8-nt while maintaining binding affinity. We suggest it may be possible to screen for a degenerate code of PUF-RNA interaction. Another approach is to construct a PUF scaffold with fewer than 8 PUF repeats. The minimum number of repeats in a stable PUF scaffold is unknown, but we speculate that a minimal number may be required for correct folding. Using a PUF scaffold with fewer repeats also decreases the size of the engineered protein, which may be beneficial for in vivo applications and simplify the design.
Minimizing off-target effects
The control of off-target effects is an important concern for any method that specifically targets endogenous gene expression, especially when the engineered PUF factors are used in vivo. In addition to using a PUF scaffold with higher specificity (e.g., a scaffold containing additional repeats), careful experimental design should minimize off-target effects.
The first opportunity to minimize off-target effects is in the design stage by identifying a target sequence with fewer off-target hits in unrelated RNAs. Since the sequence of the human genome is not random, some sequence patterns are more common than others. When selecting PUF recognition sites in RNA targets, it will be helpful to search the transcriptome and choose relatively uncommon sites. While off-target effects are impossible to eliminate, such a search will estimate the frequency of possible off-target sites. Another concern in selecting target sequences is evaluating how RNA structure may affect PUF binding. While the PUF scaffold can recognize an RNA target sequence in a double-stranded region [44, 54] (Wang, unpublished result), the occurrence of extensive hairpin structures in the binding site greatly decreases binding affinity (Wang, unpublished result). Therefore, we should select target sites in less structured regions, whereas possible off-target sites in more structured regions may not pose a problem.
Another method to reduce off-target effects is to control the expression of the engineered PUF factors by expressing them at the same time and place as their targets. For example, by designing an ASRE with mitochondrial targeting signals to silence mitochondrial RNA, we produced a protein with low off-target effects against nuclear or cytoplasmic RNA, as the ASRE is undetectable outside mitochondria [63]. A wide array of gene expression tools have been developed over the years to fine tune expression of exogenous proteins (such as cell type specific vectors). They should be helpful tools to fine tune the expression pattern of the engineered PUF factors.
In vivo delivery of engineered PUF factors
Since engineered PUF factors are typical proteins, they should be deliverable to live animals using established gene therapy vectors. For application in cultured cells or animal models, the lentiviral system is a good choice for ease of use and robust expression in most cell types. For therapeutic applications in humans, adeno-associated virus (AAV) is a favorable choice in gene therapy, because it lacks known pathogenicity, can infect non-dividing cells, and has many serotypes that allow specific gene delivery into different tissues. The AAV can stably integrate into the host cell genome at a specific site (designated AAVS1) in human chromosome 19 [78, 79], however the non-integrating AAV vectors are used for all gene therapy applications. The drawback of AAV is its small package capacity, but the current engineered PUF factors are small enough to be packed by AAV.
Several non-viral methods may be used to deliver DNAs or proteins into animals, and some of these methods are under testing for use in humans. These methods include different nano-particles and cell penetration peptides, and more in depth information on in vivo delivery methods can be found in related reviews [80–82]. These methods can be applied, at least in principle, for in vivo delivery of the engineered PUF factors. Since the application of engineered PUF factors is still in early stages, we expect the technology for protein delivery to advance ahead of the in vivo application of these factors in live animals or humans.
Final comments
Thus far engineered factors combining a PUF scaffold with a functional module have proven to be versatile tools for manipulating and understanding RNA metabolism. Recent advances allowing design of the PUF scaffold to recognize any RNA sequence expand the potential to apply this technology to regulate RNA regulatory processes. Starting with the gift of Nature’s design, continued adherence to the rule of simplicity may be prudent in moving this new field forward.
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (TMTH).
Abbreviations
- FBF
fem-3 mRNA binding factor
- PUF
Pumilio/FBF
- RRM
RNA recognition motif
- KH
hnRNP K homology
- PPR
pentatricopeptide repeat
- GFP
green fluorescent protein
- ESF
engineered splicing factor
- RS
arginine/serine
- ASRE
artificial site-specific RNA endonuclease
- ADAR
adenosine deaminase acting on RNA
- dsRBD
double-stranded RNA-binding domain
- AAV
adeno-associated virus
References
- 1.Chen Y, Varani G. Engineering RNA-binding proteins for biology. FEBS J. 2013 doi: 10.1111/febs.12375. in this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehmann R, Nusslein-Volhard C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development. 1991;112:679–691. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- 3.Ahringer J, Kimble J. Control of the sperm-oocyte switch in Caenorhabditis elegans hermaphrodites by the fem-3 3′ untranslated region. Nature. 1991;349:346–348. doi: 10.1038/349346a0. [DOI] [PubMed] [Google Scholar]
- 4.Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- 5.Macdonald PM. The Drosophila pumilio gene: an unusually long transcription unit and an unusual protein. Development. 1992;114:221–232. doi: 10.1242/dev.114.1.221. [DOI] [PubMed] [Google Scholar]
- 6.Zamore PD, Williamson JR, Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 7.Barker DD, Wang C, Moore J, Dickinson LK, Lehmann R. Pumilio is essential for function but not for distribution of the Drosophila abdominal determinant Nanos. Genes Dev. 1992;6:2312–2326. doi: 10.1101/gad.6.12a.2312. [DOI] [PubMed] [Google Scholar]
- 8.Zamore PD, Bartel DP, Lehmann R, Williamson JR. The PUMILIO-RNA interaction: a single RNA-binding domain monomer recognizes a bipartite target sequence. Biochemistry. 1999;38:596–604. doi: 10.1021/bi982264s. [DOI] [PubMed] [Google Scholar]
- 9.Murata Y, Wharton RP. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell. 1995;80:747–756. doi: 10.1016/0092-8674(95)90353-4. [DOI] [PubMed] [Google Scholar]
- 10.Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wharton RP, Sonoda J, Lee T, Patterson M, Murata Y. The Pumilio RNA-binding domain is also a translational regulator. Mol Cell. 1998;1:863–872. doi: 10.1016/s1097-2765(00)80085-4. [DOI] [PubMed] [Google Scholar]
- 12.Van Etten J, Schagat TL, Hrit J, Weidmann CA, Brumbaugh J, Coon JJ, Goldstrohm AC. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J Biol Chem. 2012;287:36370–36383. doi: 10.1074/jbc.M112.373522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blewett NH, Goldstrohm AC. A eukaryotic translation initiation factor 4E-binding protein promotes mRNA decapping and is required for PUF repression. Mol Cell Biol. 2012;32:4181–4194. doi: 10.1128/MCB.00483-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weidmann CA, Goldstrohm AC. Drosophila Pumilio protein contains multiple autonomous repression domains that regulate mRNAs independently of Nanos and brain tumor. Mol Cell Biol. 2012;32:527–540. doi: 10.1128/MCB.06052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstrohm AC, Seay DJ, Hook BA, Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J Biol Chem. 2007;282:109–114. doi: 10.1074/jbc.M609413200. [DOI] [PubMed] [Google Scholar]
- 16.Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 17.Campbell ZT, Bhimsaria D, Valley CT, Rodriguez-Martinez JA, Menichelli E, Williamson JR, Ansari AZ, Wickens M. Cooperativity in RNA-protein interactions: global analysis of RNA binding specificity. Cell Rep. 2012;1:570–581. doi: 10.1016/j.celrep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller MA, Olivas WM. Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip Rev RNA. 2011;2:471–492. doi: 10.1002/wrna.69. [DOI] [PubMed] [Google Scholar]
- 19.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [pii] [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Zamore PD, Hall TMT. Crystal structure of a Pumilio homology domain. Mol Cell. 2001;7:855–865. doi: 10.1016/s1097-2765(01)00229-5. [pii] [DOI] [PubMed] [Google Scholar]
- 21.Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–289. doi: 10.1016/s0092-8674(01)00318-x. [pii] [DOI] [PubMed] [Google Scholar]
- 22.Wang X, McLachlan J, Zamore PD, Hall TMT. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [pii] [DOI] [PubMed] [Google Scholar]
- 23.Lu G, Hall TMT. Alternate modes of cognate RNA recognition by human PUMILIO proteins. Structure. 2011;19:361–367. doi: 10.1016/j.str.2010.12.019. S0969-2126(11)00029-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheong CG, Hall TMT. Engineering RNA sequence specificity of Pumilio repeats. Proc Natl Acad Sci U S A. 2006;103:13635–13639. doi: 10.1073/pnas.0606294103. 0606294103 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong S, Wang Y, Cassidy-Amstutz C, Lu G, Bigler R, Jezyk MR, Li C, Hall TMT, Wang Z. Specific and modular binding code for cytosine recognition in Pumilio/FBF (PUF) RNA-binding domains. J Biol Chem. 2011;286:26732–26742. doi: 10.1074/jbc.M111.244889. M111.244889 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koh YY, Opperman L, Stumpf C, Mandan A, Keles S, Wickens M. A single C. elegans PUF protein binds RNA in multiple modes. RNA. 2009;15:1090–1099. doi: 10.1261/rna.1545309. rna.1545309 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Opperman L, Hook B, DeFino M, Bernstein DS, Wickens M. A single spacer nucleotide determines the specificities of two mRNA regulatory proteins. Nat Struct Mol Biol. 2005;12:945–951. doi: 10.1038/nsmb1010. [DOI] [PubMed] [Google Scholar]
- 28.Bernstein D, Hook B, Hajarnavis A, Opperman L, Wickens M. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA. 2005;11:447–458. doi: 10.1261/rna.7255805. 11/4/447 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Opperman L, Wickens M, Hall TMT. Structural basis for specific recognition of multiple mRNA targets by a PUF regulatory protein. Proc Natl Acad Sci U S A. 2009;106:20186–20191. doi: 10.1073/pnas.0812076106. 0812076106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu C, Kershner A, Wang Y, Holley CP, Wilinski D, Keles S, Kimble J, Wickens M, Hall TMT. Divergence of Pumilio/fem-3 mRNA binding factor (PUF) protein specificity through variations in an RNA-binding pocket. J Biol Chem. 2012;287:6949–6957. doi: 10.1074/jbc.M111.326264. M111.326264 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kershner AM, Kimble J. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc Natl Acad Sci U S A. 2010;107:3936–3941. doi: 10.1073/pnas.1000495107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stumpf CR, Kimble J, Wickens M. A Caenorhabditis elegans PUF protein family with distinct RNA binding specificity. RNA. 2008;14:1550–1557. doi: 10.1261/rna.1095908. rna.1095908 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valley CT, Porter DF, Qiu C, Campbell ZT, Hall TMT, Wickens M. Patterns and plasticity in RNA-protein interactions enable recruitment of multiple proteins through a single site. Proc Natl Acad Sci U S A. 2012;109:6054–6059. doi: 10.1073/pnas.1200521109. 1200521109 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yosefzon Y, Koh YY, Chritton JJ, Lande A, Leibovich L, Barziv L, Petzold C, Yakhini Z, Mandel-Gutfreund Y, Wickens M, et al. Divergent RNA binding specificity of yeast Puf2p. RNA. 2011;17:1479–1488. doi: 10.1261/rna.2700311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta YK, Nair DT, Wharton RP, Aggarwal AK. Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity. Structure. 2008;16:549–557. doi: 10.1016/j.str.2008.01.006. S0969-2126(08)00053-1 [pii] [DOI] [PubMed] [Google Scholar]
- 37.Miller MT, Higgin JJ, Hall TMT. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat Struct Mol Biol. 2008;15:397–402. doi: 10.1038/nsmb.1390. nsmb.1390 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu D, Stumpf CR, Krahn JM, Wickens M, Hall TMT. A 5′ cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs. Proc Natl Acad Sci U S A. 2009;106:20192–20197. doi: 10.1073/pnas.0812079106. 0812079106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koh YY, Wang Y, Qiu C, Opperman L, Gross L, Hall TMT, Wickens M. Stacking interactions in PUF-RNA complexes. RNA. 2011;17:718–727. doi: 10.1261/rna.2540311. rna.2540311 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozawa T, Natori Y, Sato M, Umezawa Y. Imaging dynamics of endogenous mitochondrial RNA in single living cells. Nat Methods. 2007;4:413–419. doi: 10.1038/nmeth1030. nmeth1030 [pii] [DOI] [PubMed] [Google Scholar]
- 41.Tilsner J, Linnik O, Christensen NM, Bell K, Roberts IM, Lacomme C, Oparka KJ. Live-cell imaging of viral RNA genomes using a Pumilio-based reporter. Plant J. 2009;57:758–770. doi: 10.1111/j.1365-313X.2008.03720.x. TPJ3720 [pii] [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Cheong CG, Hall TMT, Wang Z. Engineering splicing factors with designed specificities. Nat Methods. 2009;6:825–830. doi: 10.1038/nmeth.1379. nmeth.1379 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooke A, Prigge A, Opperman L, Wickens M. Targeted translational regulation using the PUF protein family scaffold. Proc Natl Acad Sci U S A. 2011;108:15870–15875. doi: 10.1073/pnas.1105151108. 1105151108 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filipovska A, Razif MF, Nygard KK, Rackham O. A universal code for RNA recognition by PUF proteins. Nat Chem Biol. 2011;7:425–427. doi: 10.1038/nchembio.577. nchembio.577 [pii] [DOI] [PubMed] [Google Scholar]
- 45.Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, Small I. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012;8:e1002910. doi: 10.1371/journal.pgen.1002910. PGENETICS-D-12-01004 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods. 2011;8:165–170. doi: 10.1038/nmeth.1551. nmeth.1551 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park HY, Trcek T, Wells AL, Chao JA, Singer RH. An unbiased analysis method to quantify mRNA localization reveals its correlation with cell motility. Cell Rep. 2012;1:179–184. doi: 10.1016/j.celrep.2011.12.009. S2211-1247(11)00019-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Querido E, Chartrand P. Using fluorescent proteins to study mRNA trafficking in living cells. Methods Cell Biol. 2008;85:273–292. doi: 10.1016/S0091-679X(08)85012-1. S0091-679X(08)85012-1 [pii] [DOI] [PubMed] [Google Scholar]
- 49.Yamada T, Yoshimura H, Inaguma A, Ozawa T. Visualization of nonengineered single mRNAs in living cells using genetically encoded fluorescent probes. Anal Chem. 2011;83:5708–5714. doi: 10.1021/ac2009405. [DOI] [PubMed] [Google Scholar]
- 50.Yoshimura H, Inaguma A, Yamada T, Ozawa T. Fluorescent probes for imaging endogenous beta-actin mRNA in living cells using fluorescent protein-tagged pumilio. ACS Chem Biol. 2012;7:999–1005. doi: 10.1021/cb200474a. [DOI] [PubMed] [Google Scholar]
- 51.Furman JL, Badran AH, Ajulo O, Porter JR, Stains CI, Segal DJ, Ghosh I. Toward a general approach for RNA-templated hierarchical assembly of split-proteins. J Am Chem Soc. 2010;132:11692–11701. doi: 10.1021/ja104395b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galka-Marciniak P, Urbanek MO, Krzyzosiak WJ. Triplet repeats in transcripts: structural insights into RNA toxicity. Biol Chem. 2012;393:1299–1315. doi: 10.1515/hsz-2012-0218. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Xiao X, Zhang J, Choudhury R, Robertson A, Li K, Ma M, Burge CB, Wang Z. A complex network of factors with overlapping affinities represses splicing through intronic elements. Nat Struct Mol Biol. 2013;20:36–45. doi: 10.1038/nsmb.2459. nsmb.2459 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12:1014–1020. doi: 10.1038/ncb2105. ncb2105 [pii] [DOI] [PubMed] [Google Scholar]
- 55.Leibovich L, Mandel-Gutfreund Y, Yakhini Z. A structural-based statistical approach suggests a cooperative activity of PUM1 and miR-410 in human 3′-untranslated regions. Silence. 2010;1:17. doi: 10.1186/1758-907X-1-17. 1758-907X-1-17 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miles WO, Tschop K, Herr A, Ji JY, Dyson NJ. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev. 2012;26:356–368. doi: 10.1101/gad.182568.111. 26/4/356 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nolde MJ, Saka N, Reinert KL, Slack FJ. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3′UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev Biol. 2007;305:551–563. doi: 10.1016/j.ydbio.2007.02.040. S0012-1606(07)00171-6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marshall GR, Feng JA, Kuster DJ. Back to the future: ribonuclease A. Biopolymers. 2008;90:259–277. doi: 10.1002/bip.20845. [DOI] [PubMed] [Google Scholar]
- 59.Yoshida H. The ribonuclease T1 family. Meth Enzymol. 2001;341:28–41. doi: 10.1016/s0076-6879(01)41143-8. [DOI] [PubMed] [Google Scholar]
- 60.Beintema JJ, Kleineidam RG. The ribonuclease A superfamily: general discussion. Cell Mol Life Sci. 1998;54:825–832. doi: 10.1007/s000180050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Calvin K, Li H. RNA-splicing endonuclease structure and function. Cell Mol Life Sci. 2008;65:1176–1185. doi: 10.1007/s00018-008-7393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parker JS. How to slice: snapshots of Argonaute in action. Silence. 2010;1:3. doi: 10.1186/1758-907X-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choudhury R, Tsai YS, Dominguez D, Wang Y, Wang Z. Engineering RNA endonucleases with customized sequence specificities. Nat Commun. 2012;3:1147. doi: 10.1038/ncomms2154. ncomms2154 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mackay JP, Font J, Segal DJ. The prospects for designer single-stranded RNA-binding proteins. Nat Struct Mol Biol. 2011;18:256–261. doi: 10.1038/nsmb.2005. nsmb.2005 [pii] [DOI] [PubMed] [Google Scholar]
- 65.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. S1471-4914(12)00101-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Y, Xiao X, Zhang J, Choudhury R, Robertson A, Li K, Ma M, Burge CB, Wang Z. A complex network of factors with overlapping affinities represses splicing through intronic elements. Nat Struct Mol Biol. 2012 doi: 10.1038/nsmb.2459. nsmb.2459 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y, Ma M, Xiao X, Wang Z. Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules. Nat Struct Mol Biol. 2012;19:1044–1052. doi: 10.1038/nsmb.2377. nsmb.2377 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ji Z, Tian B. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One. 2009;4:e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. 320/5883/1643 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thomsen S, Azzam G, Kaschula R, Williams LS, Alonso CR. Developmental RNA processing of 3′UTRs in Hox mRNAs as a context-dependent mechanism modulating visibility to microRNAs. Development. 2010;137:2951–2960. doi: 10.1242/dev.047324. dev.047324 [pii] [DOI] [PubMed] [Google Scholar]
- 72.Hall-Pogar T, Liang S, Hague LK, Lutz CS. Specific trans-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3′-UTR. RNA. 2007;13:1103–1115. doi: 10.1261/rna.577707. rna.577707 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Newnham CM, Hall-Pogar T, Liang S, Wu J, Tian B, Hu J, Lutz CS. Alternative polyadenylation of MeCP2: Influence of cis-acting elements and trans-acting factors. RNA Biol. 2010;7:361–372. doi: 10.4161/rna.7.3.11564. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peterson ML, Bingham GL, Cowan C. Multiple features contribute to the use of the immunoglobulin M secretion-specific poly(A) signal but are not required for developmental regulation. Mol Cell Biol. 2006;26:6762–6771. doi: 10.1128/MCB.00889-06. 26/18/6762 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.von Roretz C, Di Marco S, Mazroui R, Gallouzi IE. Turnover of AU-rich-containing mRNAs during stress: a matter of survival. Wiley Interdiscip Rev RNA. 2011;2:336–347. doi: 10.1002/wrna.55. [DOI] [PubMed] [Google Scholar]
- 76.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Larochelle S, Amat R, Glover-Cutter K, Sanso M, Zhang C, Allen JJ, Shokat KM, Bentley DL, Fisher RP. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat Struct Mol Biol. 2012;19:1108–1115. doi: 10.1038/nsmb.2399. nsmb.2399 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kotin RM, Siniscalco M, Samulski RJ, Zhu XD, Hunter L, Laughlin CA, McLaughlin S, Muzyczka N, Rocchi M, Berns KI. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci U S A. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Surosky RT, Urabe M, Godwin SG, McQuiston SA, Kurtzman GJ, Ozawa K, Natsoulis G. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J Virol. 1997;71:7951–7959. doi: 10.1128/jvi.71.10.7951-7959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Y, Wang J, Satterle A, Wu Q, Liu F. Gene transfer to skeletal muscle by site-specific delivery of electroporation and ultrasound. Biochem Biophys Res Commun. 2012;424:203–207. doi: 10.1016/j.bbrc.2012.06.090. [DOI] [PubMed] [Google Scholar]
- 81.Elzoghby AO, Samy WM, Elgindy NA. Protein-based nanocarriers as promising drug and gene delivery systems. J Control Release. 2012;161:38–49. doi: 10.1016/j.jconrel.2012.04.036. [DOI] [PubMed] [Google Scholar]
- 82.Koren E, Torchilin VP. Cell-penetrating peptides: breaking through to the other side. Trends Mol Med. 2012;18:385–393. doi: 10.1016/j.molmed.2012.04.012. [DOI] [PubMed] [Google Scholar]