Abstract
The bacterial cell division protein FtsZ polymerizes in a GTP-dependent manner to form a Z-ring that marks the plane of division. As a validated antimicrobial target, considerable efforts have been devoted to identify small molecule FtsZ inhibitors. We recently discovered the chrysophaentins, a novel suite of marine natural products that inhibit FtsZ activity in vitro. These natural products along with a synthetic hemi-chrysophaentin exhibit strong antimicrobial activity toward a broad spectrum of Gram-positive pathogens. To define their mechanisms of FtsZ inhibition and determine their in vivo effects in live bacteria, we used GTPase assays and fluorescence anisotropy to show that hemi-chrysophaentin competitively inhibits FtsZ activity. Furthermore, we developed a model system using a permeable E. coli strain, envA1, together with an inducible FtsZ-yellow fluorescent protein construct to show by fluorescence microscopy that both chrysophaentin A and hemi-chrysophaentin disrupt Z-rings in live bacteria. We tested the E. coli system further by reproducing phenotypes observed for zantrins Z1 and Z3, and demonstrate that the alkaloid berberine, a reported FtsZ inhibitor, exhibits auto-fluorescence, making it incompatible with systems that employ GFP or YFP tagged FtsZ. These studies describe unique examples of non-nucleotide, competitive FtsZ inhibitors that disrupt FtsZ in vivo, together with a model system that should be useful for in vivo testing of FtsZ inhibitor leads that have been identified through in vitro screens but are unable to penetrate the Gram-negative outer membrane.
Introduction
During the past decade, a steady occurrence of drug-resistant bacterial infections has drawn attention to a growing need for new antibiotics. More specifically, clinicians and researchers have emphasized the need for new classes of antibiotics that are effective against bacterial strains that are resistant toward clinically used antibiotics. This has led to the search for new bacterial targets,1–3 as well as the discovery or synthesis of new chemical classes of antibiotics.4,5 One prevalent example of late involves targeting the bacterial cell division machinery with an emphasis on the bacterial cell division protein FtsZ.6–9 FtsZ, the bacterial homolog of the eukaryotic protein tubulin, is a self-activating GTPase that assembles to form a so-called Z-ring at the bacterial plane of cell division. Previous studies have shown FtsZ to be essential for viability in the vast majority of bacteria.10,11 Accordingly, FtsZ inhibitors exhibit antibacterial activity by disrupting cell division, which ultimately leads to bacterial cell death.
In recent years, numerous studies describing FtsZ inhibitors have been published. As highlighted in several recent reviews, these studies have ranged from those describing the discovery of small molecule inhibitors and/or their in vitro modes of action, to those that have validated FtsZ as an antimicrobial target in vivo.6,7,12–14 Challenges for the field in general include the difficulties of working with this protein in vitro, tuning out promiscuity for the eukaryotic homolog tubulin, identifying non-nucleotide competitive inhibitors, and correlating the effects of in vitro and in vivo FtsZ inhibition.15–17
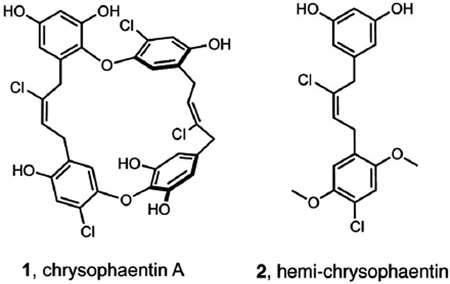
We recently described a new class of natural products called the chrysophaentins (exemplified by chrysophaentin A, 1) that we identified on the basis of their antibacterial activity toward drug sensitive and drug resistant Gram-positive bacteria.18 Using NMR and biochemical methods, we showed that these unusual halogenated bisbibenzyl ethers inhibit the in vitro GTPase activity and polymerization of FtsZ in a GTP-competitive manner. Further, we identified through chemical synthesis a hemi-chrysophaetin, 2, whose antibacterial profile is comparable to those of the natural products.19 Here, by developing a permeable Escherichia coli bacterial system employing FtsZ-YFP, together with fluorescence confocal microscopy and competitive binding studies, we describe a strategy for determining the modes of action of FtsZ inhibitors in vitro and in live bacteria.
Material and Methods
Protein expression and purification
The E. coli FtsZ expressing plasmid was a gift from William Margolin. The S. aureus FtsZ expressing plasmid was constructed by sub-cloning a synthetic gene encoding SaFtsZ into the same vector. Proteins were expressed and purified using standard procedures, detailed in the Supporting Information. The optimized 3-step purification included a 30% w/v ammonium precipitation step, followed by ion exchange chromatography using a ResQ column and gel filtration using a Superdex200 column. Recombinant FtsZ proteins were stored in buffer containing 50 mM Tris pH 7.4, 50 mM KCl, 1 mM EDTA, and 10% glycerol at −80 °C until use. Protein concentrations were determined by Bradford colorimetric assay.
GTPase assays
The rates of GTP hydrolysis by EcFtsZ and SaFtsZ differ with SaFtsZ being a slower enzyme. To compare the effects of inhibitors on each of the proteins, experimental conditions were optimized so that the end-point production of inorganic phosphate was similar for both proteins keeping the initial amount of GTP in the reaction mixture constant. Inorganic phosphate production was detected using a malachite green-phosphomolybdate assay (Pi ColorLock Gold, Innova Biosciences) and standard curves for each of the FtsZ proteins were measured. Optimization conditions and specific activities of proteins are detailed in the Supporting Information. Conditions used in inhibition assays were as follows: assay buffer contained 50 mM 2-(N-morpholino)ethanesulfonic acid (MES), pH 6.0, 100 mM KCl, and 5 mM MgCl2. EcFtsZ: 2 µM EcFtsZ with 0.25 mM GTP for 10 min at 23 °C. SaFtsZ: 6 µM SaFtsZ with 0.25 mM GTP for 20 min at 30 °C. IC50 values for inhibition of FtsZ were determined by treating FtsZ with serially diluted inhibitor solutions under the same conditions described above. Inorganic phosphate was quantified by absorbance at 635 nm, and compared to a standard curve. Controls included DMSO, which had no effect on GTPase activity at concentrations up to 8%; FtsZ, buffer and inhibitor; FtsZ in buffer; and GTP in buffer. Last, GTPase assays were performed in the presence of 0.01% Triton X-10015 for compounds 1 and 2, with no change in inhibition observed.
Competition assays
The competition assays using mant-GTP (500 nM) and BsFtsZ (380 nM), or 3H-GTP (100 nM) and BsFtsZ (70 nM) were carried out exactly as described [19], adapting the method to BsFtsZ. For these types of competition assays that employ nucleotide-free FtsZ, BsFtsZ was chosen because EcFtsZ and SaFtsZ proteins are not stable when devoid of nucleotide. The sequence of SaFtsZ is 80% identical to BsFtsZ and 100% identical at the nucleotide binding site, which justifies the use of BsFtsZ for these assays.
Antibacterial assays
Antibacterial activities toward envA1 E. coli were determined using standard microbroth dilution assays as described in the CLSI guidelines. Inhibition curves were fit (Kaleidagraph 4.0) to a standard inhibition model using the equation y=100/[1+(concentration/MIC50)], where MIC50 is the concentration at which the growth of bacterial cultures are reduced by 50%. MIC90 values correspond to the lowest concentration of inhibitor at which bacterial growth could not be observed (relative to media alone).
Fluorescence microscopy
The arabinose inducible plasmid pFtsZ-YFP 20 (chloramphenicol resistance) was transformed into a permeable E. coli strain envA1 (E. coli K12 C600 leu thr lac (thi) galK lpxC::Tn10)21 via electroporation, and colonies (termed envA1/pFtsZ-YFP) were selected after plating onto LB agar containing 20 µg/mL chloramphenicol. Replicate plates were made, and clones that expressed yellow fluorescence at mid-cell after 15 min induction with 0.4% l-arabinose in LB containing 20 µg/mL chloramphenicol were harvested and stored frozen as glycerol stocks at −80 °C. Before use glycerol stocks were thawed and grown in 15 mL LB, 20 µg/mL chloramphenicol, for 3 hr at 37 °C with shaking at 200 rpm, or until the OD600 reached ~0.2. Fluorescent FtsZ-YFP expression was induced with 0.4% l-arabinose for 30 min, and cells were harvested by centrifugation at 3500 rpm, and resuspended in phosphate buffered saline (PBS) (pH 7.4) containing 50 µg/mL tetracycline to remove remaining arabinose, and maintain selection of the envA1 mutation. Cells were washed and centrifuged in PBS/tetracycline 3×, with a final wash in LB broth. Bacteria were then resuspended in 25 mL LB, 20 µg/mL chloramphenicol. One mL aliquots were removed and subjected either to 1% MeOH or 1% DMSO (as vehicle controls), or to each of inhibitors 1–5. Cells were visualized on polylysine coated slides with DIC and fluorescence microscopy using a Zeiss LSM700 confocal scanning laser unit with an inverted microscope (AXIO Observer.Z1), 100× objective, and 1.3 numerical aperture lens. FtsZ-YFP was visualized with excitation and emission at 488 nm and 558 nm, and FtsZ-RFP at 555 nm and 630 nm. Images were acquired using the Zeiss software package.
Results
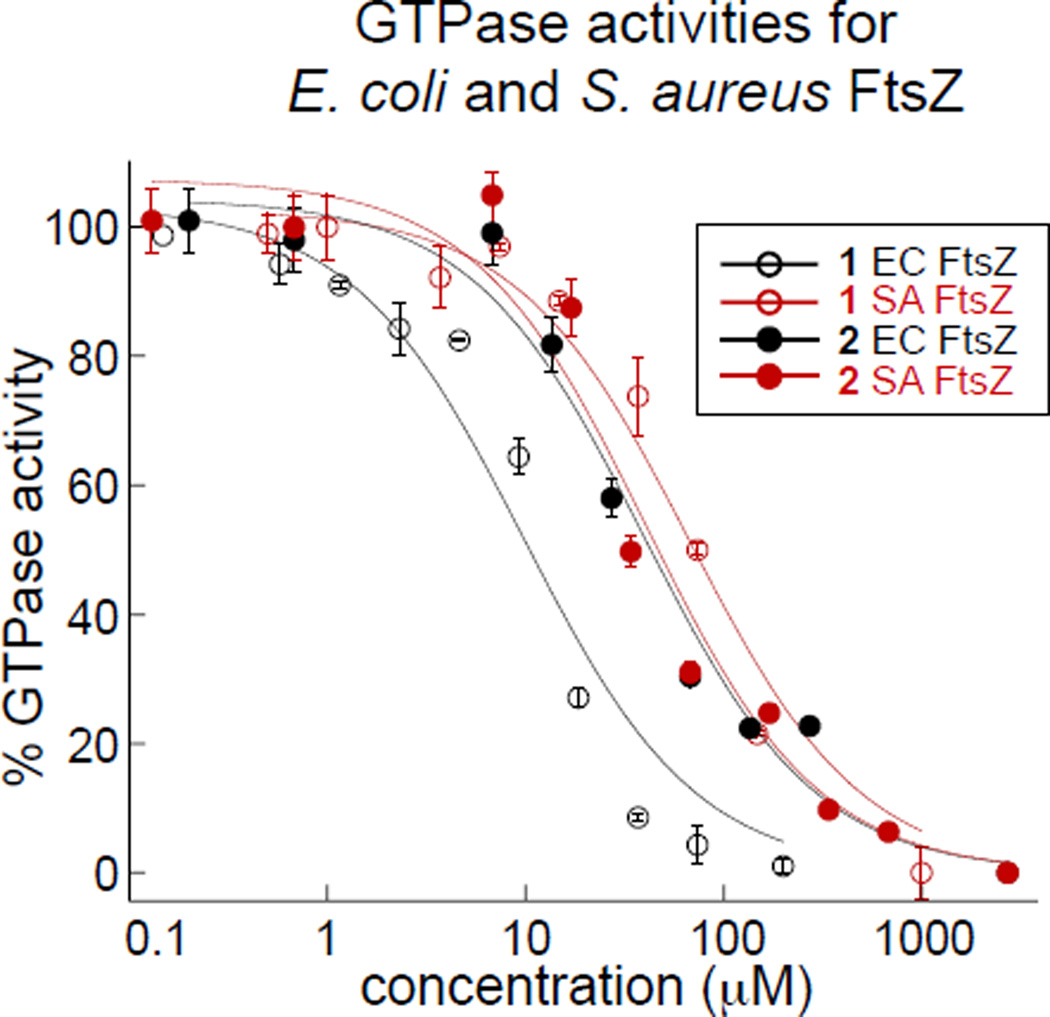
To date, the vast majority of studies describing in vitro inhibition of FtsZ polymerization or GTPase activity have been carried out with recombinant E. coli or Bacillus subtilis FtsZ constructs. Given the strong antimicrobial activity of the chrysophaentins toward drug-resistant S. aureus strains, and our desire to develop an E. coli model system for imaging Z-ring integrity in live bacteria, it was first necessary to determine the effect of 1 on S. aureus FtsZ (abbreviated SaFtsZ) and 2 on both S. aureus and E. coli FtsZ (abbreviated EcFtsZ). We produced recombinant EcFtsZ using a pET11a EcFtsZ expression plasmid (gift from W. Margolin),22 and to obtain SaFtsZ, we subcloned a synthetic gene encoding S. aureus FtsZ into the pET11a vector.23 Using a three-step purification scheme, we were able to obtain each recombinant protein in >95% purity and in good yield (see Methods). We tested SaFtsZ using an end-point GTPase assay, where the hydrolysis of GTP was measured as the production of inorganic phosphate (Pi) in a malachite green-phosphomolybdate colorometric assay. Using a similar approach, we showed previously that chrysophaentin A inhibited EcFtsZ in a dose-depended manner with an IC50 of 9.9±2.5 µM.18 Here we found that chrysophaentin A also inhibited SaFtsZ GTPase activity with an IC50 of 67±13 µM (Fig. 1), while hemi-chrysophaentin 2 was found to inhibit the hydrolysis activity of EcFtsZ with an IC50 of 37±7 µM and SaFtsZ with an IC50 of 38±9 µM (Fig. 1). Thus, compounds 1 and 2 each inhibit the GTPase activity of both EcFtsZ and SaFtsZ in vitro.
Figure 1.
Inhibitory activities of chrysophaentins 1 and 2. (A) Chrysophaentin A (1) and hemi-chrysophaentin (2) inhibit the in vitro GTPase activities of S. aureus FtsZ and E. coli FtsZ with IC50 values ranging from 9–67 µM; GTPase activity was measured using a colorimetric malachite green-phosphomolybdate assay.
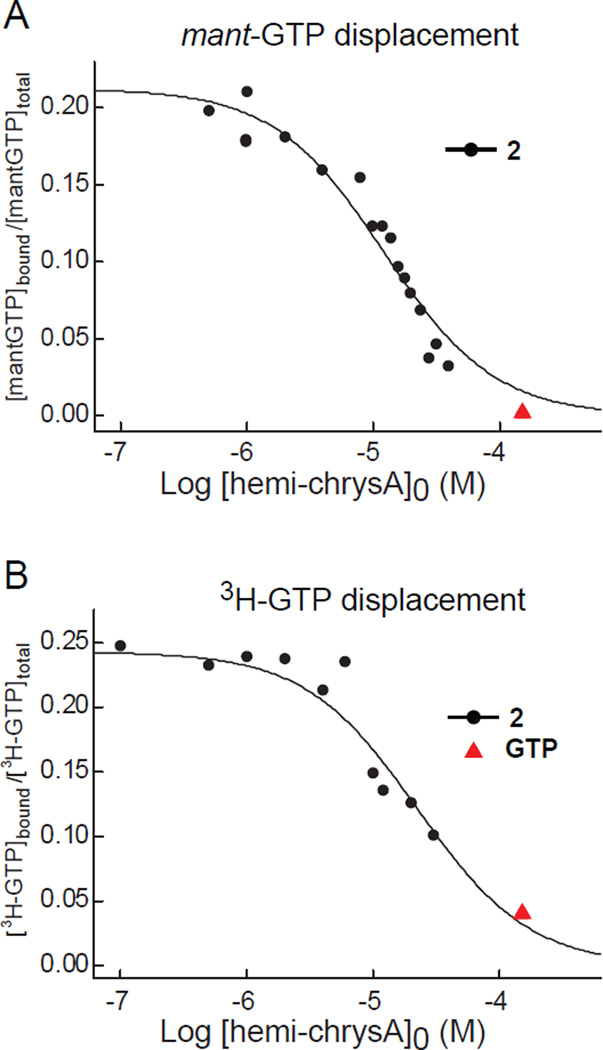
We next sought to determine the mode of inhibition of FtsZ by hemi-chrysophaentin. Previously, we demonstrated by STD-NMR that chrysophaentin A binds to the nucleotide binding pocket of FtsZ in a competitive manner with the non-hydrolyzable GTP analog GTPγS,18 and to date, this is the sole example of a competitive non-nucleotide inhibitor of FtsZ. To test whether hemi-chrysophaentin can also displace GTP in a competitive manner, we studied the binding of 2 to FtsZ by fluorescence anisotropy using a fluorescent GTP analog mant-GTP, and with a 3H-GTP competition assay.24 As shown in Figure 2, hemi-chrysophaentin competes with GTP for binding to the nucleotide binding pocket of FtsZ. Fitting of the data for 2 to a simple competition model where 2 binds to the same site as mant-GTP gives an equilibrium association constant, Ka, of 1.5 × 105 M−1. The mant-GTP probe was independently determined to bind with a Ka of (9.9 ± 0.5) × 105 M−1 by titrations with FtsZ. For displacement of 3H-GTP by compound 2, best fits to the data yield a Ka of 0.87 × 105 M−1. (3H-GTP was independently determined to bind to FtsZ with a Ka of (7.0 ± 0.7) × 106 M−1). Thus, despite its smaller size, 2 competes with GTP for binding to FtsZ like its parent molecule.
Figure 2.
Hemi-chrysophaentin 2 is a competitive inhibitor of FtsZ. (A) Curve showing the concentration dependent displacement of the fluorescent nucleotide mant-GTP (500 nM) by compound 2 in B. subtilis FtsZ (380 nM). Displacement is measured by the change in fluorescence anisotropy. The solid line represents the least squares best fit to the data (black circles) using a simple competition model.17 Control showing displacement of mant-GTP by GTP (150 µM) is included (red triangle). (B) Curve showing displacement of 3H-GTP (100 nM) from FtsZ (70 nM) by 2, determined by co-sedimentation and scintillation counting of 3H-GTP-bound FtsZ. The solid line shows the best fit to the data.
Both chrysophaentin A and hemi-chrysophaentin display broad spectrum antibacterial activity toward Gram-positive bacteria. However, neither compound inhibits the growth of Gram-negative bacteria, including E. coli. Because compounds 1 and 2 inhibit both EcFtsZ and SaFtsZ in vitro, we hypothesized that the compounds are unable to cross the outer membrane of Gram-negative organisms to exert their antimicrobial action. To test this, we used an E. coli strain, envA1, that contains a mutated UDP-3-O-acyl-GlcNAc deacetylase gene known to confer permeability to a wide variety of antibiotics.21,25 This approach has been used by pharmaceutical companies to screen for new antibiotics.25,26 Compounds 1 and 2 were tested against envA1 E. coli in microbroth dilution assays. We found that compound 1 inhibited the growth of permeable E. coli with an MIC50 of 27±9 µM, while compound 2 was also inhibitory with an MIC50 of 84 µM.27 The inhibition curve for 2 against envA1 E. coli was unusually steep. Although this type of curve can indicate aggregation of a small molecule inhibitor, we note that none of the in vitro inhibition curves for 2 toward FtsZ show this behavior. These results confirmed that the outer membrane of E. coli is preventing access to the conserved intracellular antibacterial target of compounds 1 and 2. When the outer membrane is defective, antibacterial activity is restored.
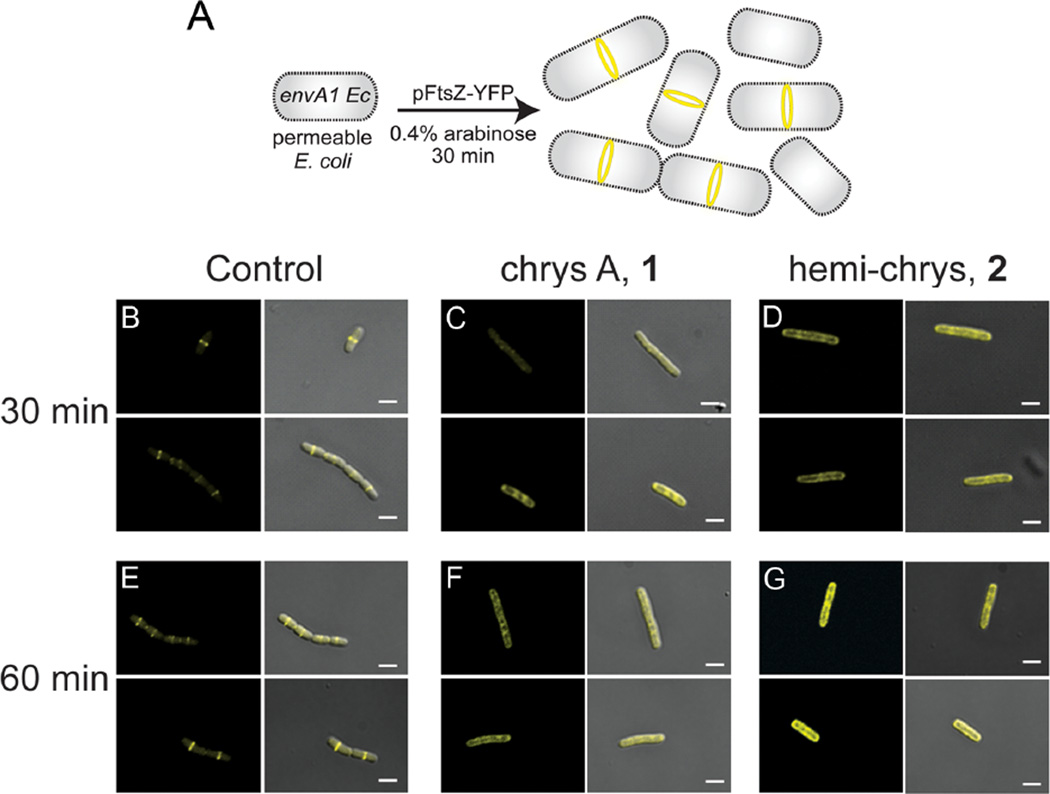
The above results demonstrated that compounds 1 and 2 inhibit the growth of Gram-positive and Gram-negative organisms through a conserved intracellular target, and that in cell-free systems both compounds competitively inhibit the biochemical activities of FtsZ. However, it remained to be shown whether these compounds could affect the physiological properties of FtsZ in bacteria, such as its ability to form a functional Z-ring during cell division. To address this question, we sought to develop a robust bacterial system wherein the presence or absence of Z-rings could be reliably detected by fluorescence microscopy, the antibacterial activity of the inhibitors is conserved, and the bacteria divide normally with clear Z-ring formation in the absence of inhibitor. After testing several bacterial strains in combination with different fluorescent FtsZ fusions, the bacterial system we chose (Fig. 3A) employed the envA1 E. coli strain transformed with an arabinose inducible plasmid that encodes full length E. coli FtsZ fused to a C-terminal yellow fluorescent protein (pFtsZ-YFP).20 Liew et al. described a S. aureus system for visualization of fluorescent Z-rings, but that system requires tight control of the expression of more than one plasmid making it more complex for screening purposes than an E. coli-based system.27 Experiments were conducted to determine times of induction that would allow for expression of FtsZ-YFP and co-localization of fluorescent and wild-type FtsZ at mid-cell during division (detailed in Methods). Fluorescence micrographs showed that prior to arabinose induction there was no observable background fluorescence (data not shown). Both before and after the washing step, almost all bacteria showed yellow fluorescence localized to mid-cell, indicating the presence of a Z-ring composed of both fluorescently-labeled and untagged wild-type FtsZ. Small volumes (~1 mL) of the resuspended bacteria were subjected to different treatments, including 1% MeOH (vehicle control), chrysophaentin A (1), and synthetic fragment 2. When treating envA1/pFtsZ-YFP with compounds 1 and 2, we used inhibitor concentrations slightly higher than their respective MIC90s (125 µM and 340 µM) to allow us to reproducibly observe the existence of different phenotypes in the majority of cells. The bacteria treated with 1% MeOH had normal Z-rings at 30 minutes (Fig. 3B). These bacteria looked very similar to those immediately after the wash step, indicating the presence of the fluorescent construct was not affecting the ability of the bacteria to grow and divide. This envA1 E. coli strain has been observed to undergo incomplete septation where the daughter cells do not fully separate after division.28 We also observed this phenotype which appears as short chains of bacteria, and can be seen in the lower panel of Fig. 3B. After 30 minutes of treatment with 1 (in 1% MeOH), the localization of fluorescent FtsZ differed from that of untreated bacteria (Fig. 3C). FtsZ-YFP was no longer localized to mid-cell and Z-rings were not observed; instead the protein appeared to be dispersed throughout the cytoplasm and was present in patches or foci in an estimated 80% of bacteria.23 Thirty minutes of treatment with 2 showed a similar phenotype to that observed in bacteria treated with 1 (Fig. 3D).
Figure 3.
Effects of chrysophaentins on Z-ring structure using fluorescence microscopy of envA1 E. coli/FtsZ-YFP. For each panel, micrographs showing fluorescence alone (ex 488, em 558) is shown on the left, and fluorescence merged with differential interference contrast (DIC) is shown on the right. Duplicate representative images of each experimental condition (including controls and inhibitor treatments at two time points) are shown. All images were acquired with a 100× lens; white scale bars, 2 µm. (A) Schematic showing the system used for visualizing fluorescent Z-rings in envA1/pFtsZ-YFP. Bacterial cells at different stages of cell division are present, as depicted in the cartoon. Microscopy showing treatment of envA1/pFtsZ-YFP with (B) 1% MeOH, 30 min; (C) 125 µM 1 in 1% MeOH, 30 min; (D) 340 µM 2 in 1% MeOH, 30 min; (E) 1% MeOH, 60 min; (F) 125 µM 1 in 1% MeOH, 60 min; (G) 340 µM 2 in 1% MeOH, 60 min. Micrographs taken at lower magnification that show numerous bacteria in a given field are available in Ref. 23.
An hour after treatment, the control bacteria containing the fluorescent construct still maintained single fluorescently-labeled Z-rings (Fig. 3E), and the number of short chains of bacteria in this group had increased, confirming that the bacteria were alive and dividing. Similarly, one hour after treatment with 1 or 2, we observed the same effects as those observed at 30 minutes where FtsZ had not localized to mid-cell and instead was present in patches throughout the cell (Figs. 3F,G). Background fluorescence at both time points was negligible in bacteria that were not induced with arabinose but treated with compounds (data not shown). While 1 and 2 do not appear to cause overt filamentation, the fact that the treated bacteria are larger in size and display visibly mis-localized fluorescent FtsZ, demonstrates that they are inhibiting division in envA1/pFtsZ-YFP. Last, we investigated the in vitro effect of hemi-chrysophaentin on B. subtilis FtsZ, and its in vivo effect on B. subtilis strain SU570 that contains Fts-GFP as the only FsZ protein.29 We found that 2 also inhibits the GTPase activity of B. subtilis FtsZ with an IC50 value of 70 µM, and perturbs Z-ring structure in this Gram-positive organism (Supporting Information).
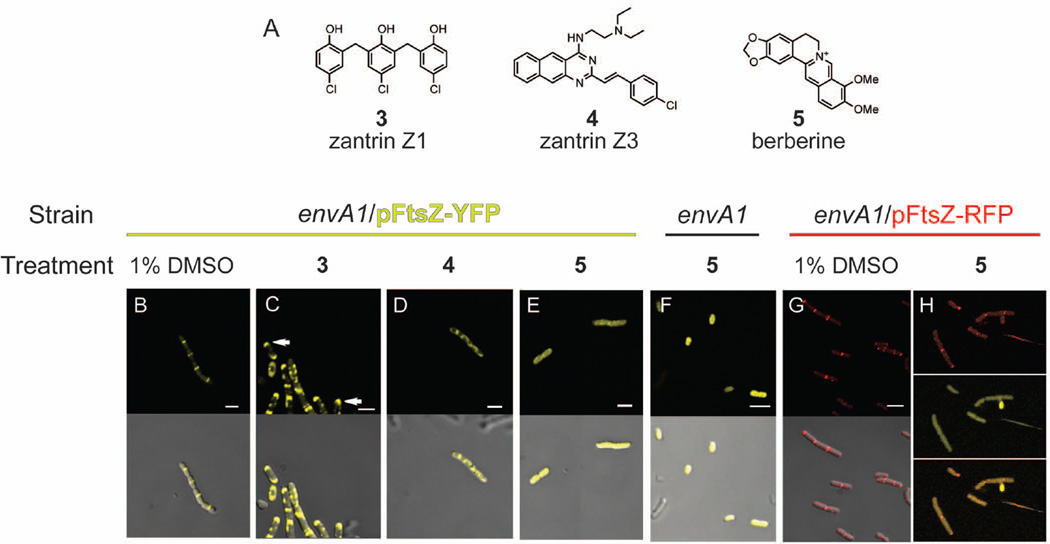
Our results show that compounds 1 and 2 disrupt FtsZ assembly and therefore the Z-ring in live bacteria in our permeable E.coli system. To test this system further, we wanted to compare the effects we observed with the chrysophaentins to those of other FtsZ inhibitors previously reported in the literature. To this end we chose the FtsZ inhibitors zantrin Z1, zantrin Z3, and berberine (Fig. 4A, 3–5). In experiments conducted with an anti-FtsZ antibody and a fluorescent secondary antibody, RayChaudhuri and co-workers found that treatment of E. coli with zantrin Z1 led to a mislocalization of FtsZ to the poles of the cell rather than mid-cell, and treatment with zantrin Z3 also led to perturbations of Z-ring formation and integrity.30 In two separate studies using the plant natural product berberine, a phenotype described as diffuse fluorescence throughout the interior of the cell was observed for bacteria expressing a FtsZ-GFP construct. This result, combined with an observed increase in cell size (filamentation), led the authors to conclude that berberine caused a disruption of Z-ring formation, and a decrease in the number of Z-rings per cell.31,32
Figure 4.
Effects of reported FtsZ inhibitors on Z-ring structure using envA1 E. coli and fluorescent FtsZ proteins. (A) Known FtsZ inhibitors zantrin Z1 (3), zantrin Z3 (4), and berberine (5). In panels B–H, the strain and treatment (inhibitor or control) are shown above the individual images, and fluorescence and merged fluorescence/DIC appear in the top and bottom panels, respectively. Inhibitor concentrations used approximate the MIC90 value of each compound toward the growth of envA1 E. coli. Treatment of envA1/pFtsZ-YFP with (B) 1% DMSO for 30 min; (C) 4.9 µM zantrin Z1 in 1% DMSO, 30 min, arrows indicate FtsZ polarization; (D) 120 µM zantrin Z3 in 1% DMSO, 30 min; (E) 680 µM berberine in 1% DMSO, 30 min. (F) Treatment of untransformed envA1 E. coli with 680 µM berberine in 1% DMSO, 30 min. Fluorescence is attributed solely to berberine. (G) Fluorescence micrographs of arabinose-induced envA1/pFtsZ-RFP in the presence of 1% DMSO, 30 min. (H) envA1/pFtsZ-RFP treated with 680 µM berberine, 30 min. Top panel, red fluorescence (ex 555, em 630); middle panel, yellow fluorescence (ex 488, em 558); and bottom panel, red/yellow fluorescence merged (channel 1: ex 488, channel 2: ex 555, em 605). All images were acquired with a 100× lens; white scale bars, 2 µm.
Using the envA1/pFtsZ-YFP system developed here, we investigated the effects of zantrin Z1 (3), zantrin Z3 (4), and berberine (5) on Z-ring integrity in live cells. Treatment with 4.9 µM zantrin Z1 caused mislocalization of the FtsZ fluorescence. At 30 minutes, FtsZ-YFP was no longer localized to mid-cell in a ring, as was observed for the DMSO-treated controls (Fig. 4B). Instead FtsZ-YFP appeared to be localizing to the cell poles (Fig. 4C) in the large majority of bacteria. By 60 minutes, most of the fluorescence was localized to the polar region of cells treated with zantrin Z1 (not shown). Treatment with 120 µM zantrin Z3 for 30 minutes showed FtsZ-YFP was no longer localized to mid-cell and instead showed foci of fluorescence throughout the cell (Fig. 4D), and a similar phenotype was observed at 60 minutes (not shown). We also visualized non-transformed envA1 cells treated with zantrin Z1 or zantrin Z3, and we did not observe instrinsic fluorescence for either compound at the wavelengths utilized for fluorescence microscopy.
We also tested the effect of berberine on Z-ring formation using this system. Upon treatment of envA1/pFtsZ-YFP with berberine (680 µM) we observed diffuse fluorescence throughout the cell (Fig. 4E) as reported previously. However, when envA1 E. coli that had not been transformed with pFtsZ-YFP was treated with berberine, we observed the same diffuse, intracellular fluorescence as we observed in the arabinose-induced FtsZ-YFP-expressing cells. Strikingly, our data revealed that in this system the diffuse fluorescence observed is due to the intrinsic auto-fluorescence of berberine and not to the presence of non-polymerized FtsZ (Fig. 4F). We were unable to view individually the fluorescence from YFP and berberine as the overlapping regions of their spectra are too large to filter. It remained possible that the strong fluorescence attributed to berberine was obscuring the presence of normal Z-rings. To be able to visualize Z-rings in the presence of berberine we constructed a plasmid encoding FtsZ with a C-terminal tdTomato fusion (pFtsZ-RFP) and carried out the same set of experiments using berberine and the arabinose inducible FtsZ-RFP. We note that the red fluorescent proteins such as mCherry and tdTomato are typically slower folding and require longer times to become fluorescent, making them non-ideal for use in most bacterial systems. Nonetheless, using a similar protocol and observing red fluorescence at 30 and 60 min post induction (Fig. 4G), we were able to observe intact red fluorescent Z-rings in cells treated with berberine, together with the diffuse intracellular fluorescence of berberine itself (Fig. 4H).
Discussion
These studies provide the first demonstration that chrysophaentins 1 and 2 inhibit FtsZ function in live bacteria. We have demonstrated that each compound inhibits the in vitro functions of E. coli, B. subtilis and S. aureus FtsZ; and for E. coli and B. subtilis FtsZ, 1 and 2 compete with the nucleotide ligand for binding to the GTP-binding site. This mode of inhibition therefore appears to represent both the in vitro and in vivo mechanisms of action. In vitro, the effects are seen as a decrease in the ability of FtsZ to hydrolyze GTP and to polymerize. In vivo, the effects are manifested as disruption in Z-ring localization and integrity, apparent from the absence of well resolved and properly located Z-rings in bacteria treated with 1 or 2. It is worth noting that in control experiments, we observed the presence of single fluorescently labeled Z-rings located at mid-cell in more than half the bacterial cells, both before and after addition of solvent-controls. In contrast, when treated with 1 or 2, virtually all of the bacterial cells exhibited the altered FtsZ phenotypes described above. Given this observation, the complete absence of normal Z-rings in bacteria treated with 1 or 2 suggests that these inhibitors are capable of disrupting previously formed Z-rings, in addition to preventing the formation of new Z-rings and causing FtsZ to mis-localize and remain in the cytoplasm, rather than self-associate at mid-cell in a Z-ring structure.
Our results with the known FtsZ inhibitors zantrin Z1 and Z3 using this system verified the phenotypes previously observed for these compounds, namely that zantrin Z1 leads to FtsZ localization at the cell poles rather than mid-cell,33 and zantrin Z3 disrupts proper FtsZ localization to mid cell. The intrinsic spectral properties of berberine on the other hand make it impossible to determine its mechanism of action and effect on Z-ring formation in systems that employ GFP or YFP tagged FtsZ. Overall, this model system can be used with a variety of inhibitors, provided that they do not possess intrinsic fluorescence. In closing, it has been proposed that non-nucleotide FtsZ inhibitors may show better selectivity for the active site of FtsZ over that of tubulin compared to the nucleotide-based competitive inhibitors reported to date. Thus our findings may have implications for future FtsZ inhibitor design.
Supplementary Material
Acknowledgements
We thank W. Margolin, H. Erikson, K. Young, and E. Harry for plasmids or bacterial strains; N. Dwyer (NIDDK) for assistance with confocal microscopy; and H. Erikson and A. Plaza for contributive discussions. This work was supported in part by the Intramural Research Program, National Institutes of Health (NIDDK), by NIGMS P50-GM067082 (PW), and by Plan Nacional de Investigación BFU 2011-23416 and CAM S2010/BMD-2353 (JMA). JLK acknowledges an Intramural AIDS Research Fellowship, Office of the Director, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary material
Complete Methods section and additional figures.
References
- 1.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Nat. Rev. Drug. Discov. 2007;6:29. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 2.Alksne LE, Dunman PM. Methods Mol. Biol. 2008;431:271. doi: 10.1007/978-1-60327-032-8_21. [DOI] [PubMed] [Google Scholar]
- 3.Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, Painter R, Parthasarathy G, Tang YS, Cummings R, Ha S, Dorso K, Motyl M, Jayasuriya H, Ondeyka J, Herath K, Zhang C, Hernandez L, Allocco J, Basilio A, Tormo JR, Genilloud O, Vicente F, Pelaez F, Colwell L, Lee SH, Michael B, Felcetto T, Gill C, Silver LL, Hermes JD, Bartizal K, Barrett J, Schmatz D, Becker JW, Cully D, Singh SB. Nature. 2006;441:358. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- 4.Fischbach MA, Walsh CT. Science. 2009;325:1089. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogers BL. Curr. Opin. Drug Discov. Devel. 2004;7:211. [PubMed] [Google Scholar]
- 6.Huang Q, Tonge PJ, Slayden RA, Kirikae T, Ojima I. Curr. Top. Med. Chem. 2007;7:527. doi: 10.2174/156802607780059790. [DOI] [PubMed] [Google Scholar]
- 7.Lock RL, Harry EJ. Nat. Rev. Drug Discov. 2008;7:324. doi: 10.1038/nrd2510. [DOI] [PubMed] [Google Scholar]
- 8.Kapoor S, Panda D. Expert Opin. Ther. Targets. 2009;13:1037. doi: 10.1517/14728220903173257. [DOI] [PubMed] [Google Scholar]
- 9.Kumar K, Awasthi D, Berger WT, Tonge PJ, Slayden RA, Ojima I. Future Med. Chem. 2010;2:19. doi: 10.4155/fmc.10.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai K, Lutkenhaus J. Journal of Bacteriology. 1991;173:3500. doi: 10.1128/jb.173.11.3500-3506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinho MG, Errington J. Mol. Microbiol. 2003;50:871. doi: 10.1046/j.1365-2958.2003.03719.x. [DOI] [PubMed] [Google Scholar]
- 12.Haydon DJ, Stokes NR, Ure R, Galbraith G, Bennett JM, Brown DR, Baker PJ, Barynin VV, Rice DW, Sedelnikova SE, Heal JR, Sheridan JM, Aiwale ST, Chauhan PK, Srivastava A, Taneja A, Collins I, Errington J, Czaplewski LG. Science. 2008;321:1673. doi: 10.1126/science.1159961. [DOI] [PubMed] [Google Scholar]
- 13.Ma S. ChemMedChem. 2012;7:1161. doi: 10.1002/cmdc.201200156. [DOI] [PubMed] [Google Scholar]
- 14.Schaffner-Barbero C, Martin-Fontecha M, Chacon P, Andreu JM. ACS Chem. Biol. 2012;7:269. doi: 10.1021/cb2003626. [DOI] [PubMed] [Google Scholar]
- 15.Anderson DE, Kim MB, Moore JT, O'Brien TE, Sorto NA, Grove CI, Lackner LL, Ames JB, Shaw JT. ACS Chem. Biol. 2012;7:1918. doi: 10.1021/cb300340j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foss MH, Eun YJ, Weibel DB. Biochemistry. 2011;50:7719. doi: 10.1021/bi200940d. [DOI] [PubMed] [Google Scholar]
- 17.Lappchen T, Pinas VA, Hartog AF, Koomen GJ, Schaffner-Barbero C, Andreu JM, Trambaiolo D, Lowe J, Juhem A, Popov AV, den Blaauwen T. Chem. Biol. 2008;15:189. doi: 10.1016/j.chembiol.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 18.Plaza A, Keffer JL, Bifulco G, Lloyd JR, Bewley CA. J. Am. Chem. Soc. 2010;132:9069. doi: 10.1021/ja102100h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keffer JL, Hammill JT, Lloyd JR, Plaza A, Wipf P, Bewley CA. Mar. Drugs. 2012;10:1103. doi: 10.3390/md10051103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osawa M, Erickson HP. J. Bacteriol. 2006;188:7132. doi: 10.1128/JB.00647-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young K, Silver LL. J. Bacteriol. 1991;173:3609. doi: 10.1128/jb.173.12.3609-3614.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corbin BD, Wang Y, Beuria TK, Margolin W. J. Bacteriol. 2007;189:3026. doi: 10.1128/JB.01581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keffer JL. Ph.D. Thesis. Georgetown University; 2012. Jul, [Google Scholar]
- 24.Schaffner-Barbero C, Gil-Redondo R, Ruiz-Avila LB, Huecas S, Lappchen T, den Blaauwen T, Diaz JF, Morreale A, Andreu JM. Biochemistry. 2010 doi: 10.1021/bi101577p. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Galgoci A, Kodali S, Herath KB, Jayasuriya H, Dorso K, Vicente F, González A, Cully D, Bramhill D, Singh S. J. Biol. Chem. 2003;278:44424. doi: 10.1074/jbc.M307625200. [DOI] [PubMed] [Google Scholar]
- 26.Cunningham ML, Kwan BP, Nelson KJ, Bensen DC, Shaw KJ. J. Biomol. Screen. 2013 doi: 10.1177/1087057113487208. [DOI] [PubMed] [Google Scholar]
- 27.Liew AT, Theis T, Jensen SO, Garcia-Lara J, Foster SJ, Firth N, Lewis PJ, Harry EJ. Microbiology. 2011;157:666. doi: 10.1099/mic.0.045146-0. [DOI] [PubMed] [Google Scholar]
- 28.Normark S, Boman HG, Matsson E. J. Bacteriol. 1969;97:1334. doi: 10.1128/jb.97.3.1334-1342.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strauss MP, Liew AT, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. PLoS Biol. 2012;10:e1001389. doi: 10.1371/journal.pbio.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margalit DN, Romberg L, Mets RB, Hebert AM, Mitchison TJ, Kirschner MW, RayChaudhuri D. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11821. doi: 10.1073/pnas.0404439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Domadia PN, Bhunia A, Sivaraman J, Swarup S, Dasgupta D. Biochemistry. 2008;47:3225. doi: 10.1021/bi7018546. [DOI] [PubMed] [Google Scholar]
- 32.Boberek JM, Stach J, Good L. PLoS One. 2010;5:e13745. doi: 10.1371/journal.pone.0013745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foss MH, Eun YJ, Grove CI, Pauw DA, Sorto NA, Rensvold JW, Pagliarini DJ, Shaw JT, Weibel DB. MedChemComm. 2013;4:112. doi: 10.1039/C2MD20127E. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.