Summary
Cytosolic DNA arising from intracellular bacteria or viral infections is a powerful pathogen-associated molecular pattern (PAMP) that leads to innate immune host defense by the production of type I interferon and inflammatory cytokines. Recognition of cytosolic DNA by the recently discovered cyclic-GMP-AMP (cGA) synthase (cGAS) induces the production of cGA to activate the stimulator of interferon genes (STING). Here we report the crystal structure of cGAS alone and in complex with DNA, ATP and GTP along with functional studies. Our results explain cGAS’ broad specificity DNA sensing, show how cGAS catalyzes di-nucleotide formation and indicate activation by a DNA-induced structural switch. cGAS possesses a remarkable structural similarity to the antiviral cytosolic dsRNA sensor 2’-5’oligoadenylate synthase (OAS1), but contains a unique zinc-thumb that recognizes B-form dsDNA. Our results mechanistically unify dsRNA and dsDNA innate immune sensing by OAS1 and cGAS nucleotidyl transferases.
Introduction
Recognition of pathogen- or danger-associated molecular patterns (PAMPs or DAMPs) is crucial for host defense. Innate immunity ensures this recognition through germline-encoded pattern recognition receptors (PRRs) and triggers signaling cascades that result in production of proinflammatory cytokines and type I interferons (IFN-α and IFN-β) 1,2. Cytosolic DNA arising from intracellular bacteria or viral infections is a powerful PAMP and is also implicated as a DAMP in autoimmune diseases 1,3,4. Over the past years, a variety of PRRs for cytosolic DNA have been reported: DNA-dependent activator of IFN-regulatory factors (DAI) 5; absent in melanoma 2 (AIM2) 6–8; RNA polymerase III 9,10; leucine-rich repeat (in Flightless I) interacting protein-1 (LRRFIP) 11; DExD/H box helicases (DDX41, DHX9 and DHX36) 12,13 and IFN-inducible protein IFI16 14. However, these PRRs are either cell type or DNA sequence specific, possible accessory factors (DExD/H proteins) or trigger different pathways such as caspase-1 activation (AIM2) or a β-catenin dependent signaling pathway (LRRFIP1) 15.
Although the DNA sensor for type I IFN production with broad specificity and cell distribution was not identified until recently, it was known that IRF3 and NFκB activation in response to DNA requires STING (stimulator of interferon genes, also known as MITA/MPYS/ERIS), a transmembrane protein that is resident on the endoplasmic reticulum 16–18. STING co-localizes with DNA in vivo but binds DNA only with low affinity in vitro19, suggesting the presence of an additional sensor. Furthermore, STING is a direct PRR for cyclic di-nucleotides such as c-di-AMP and c-di-GMP 20, which are signaling molecules in prokaryotes and trigger IFN in response to e.g. intracellular bacteria 21,22.
Recent results identified human c-GMP-AMP (cGA) synthase (cGAS, also known as c6orf150, Male abnormal 21 domain containing-1 (Mb21d1)) as broad specificity cytosolic DNA sensor 23. In the presence of DNA cGAS produces cGA, which is an endogenous second messenger that activates STING 18, explaining how STING can stimulate IFN in response to both cyclic-di-nucleotides and DNA. To reveal the mechanism of DNA-stimulated cGA synthesis, we determined the crystal structure of porcine cGASMab21 (residues 135–497, comprising the highly conserved, DNA-stimulated nucleotidyl transferase (NTase) domain) with and without a 14mer dsDNA ligand and nucleotide substrates, along with functional studies in vitro and in living cells.
Crystal structure of cGASMab21
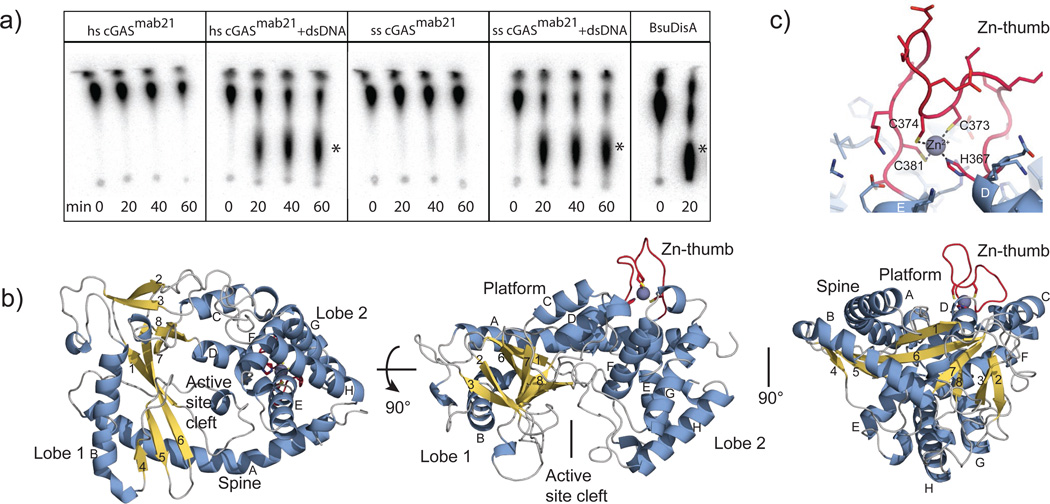
cGAS is a 60 kDA protein composed of an unstructured, not well conserved N-terminal stretch of approximately 130–150 residues followed by a highly conserved Mab21 domain that belongs to the nucleotidyl transferase (NTase) superfamily 24. In order to overproduce and crystallize cGAS, it was necessary to genetically remove the unstructured N-terminal tail. The resulting cGASMab21 used in this study (residues 155/161–522 for human cGAS and residues 135–497 for porcine cGAS) possesses DNA dependent di-nucleotide synthesis activity in the presence of a 50mer dsDNA that induces IFN in THP1 cells (Fig, 1a, suppl. Fig. S1a,b). While cGAS produces cGA also with a dsDNA 40mer, no activity was observed when we omitted either GTP or ATP from the reaction mixture or substituted dsDNA with ssDNA (suppl. Fig. S1a).
Figure 1. Crystal Structure of Mab21 cGASMab21.
a) Activity assays of human and porcine cGASMab21 alone or in presence of dsDNA. B. subtilis DisA, a c-di-AMP synthase is used as positive control. The di-nucleotide products are indicated with asterisks.
b) Side and top views of cGASMab21. The model is shown as ribbon representation with annotated domains and secondary structure (blue α-helices, yellow β-strands).
c) Close up view of the ‘zinc-thumb’.
We determined the crystal structure of porcine cGASMab21 by single-wavelength anomalous dispersion to 2.5Å resolution using a selenomethionine derivative. After density modification, we could build an initial model, which was completed and refined against the 2.0Å resolution native data, resulting in good R-factors and stereochemistry (suppl. Fig. S1c, suppl. Table S1).
The Mab21 domain of cGAS comprises two lobes, separated by a deep cleft (Fig. 1b). Lobe 1 possesses the NTase fold with a two-leaved highly twisted β-sheet (β1-β8) that is flanked on the outside by two long α-helices (αA and αB). At the inner side, lining the cleft, β1 and β6 harbor the signature catalytic site residues (E200, D202, D296) of the NTase superfamily that coordinate the catalytic Mg2+ ions and nucleotides. Lobe 2 is a bundle of four α-helices (αE-αH), connected to lobe 1 by a long “spine” (αA), two linker helices (αC, αD) and by a long active site loop connecting αA and β1.
The molecular surface opposite the active site is a fairly flat, slightly concave “platform”, formed predominantly by αA, αC, αD and the nucleotide-binding loop. An intriguing protrusion (residues 367–382) is situated at one end of the platform. This protrusion possesses highly conserved histidine and cysteines (H367, C373, C374 and C381), which together coordinate a Zn2+ ion (Fig. 1c). We denote this loop “Zn-thumb”. Its sequence is inserted between lobes 1 and 2 and is a highly conserved and characteristic feature of cGAS orthologs (suppl. Fig. S1d), suggesting an important functional role.
The cGAS:DNA:GTP:ATP complex
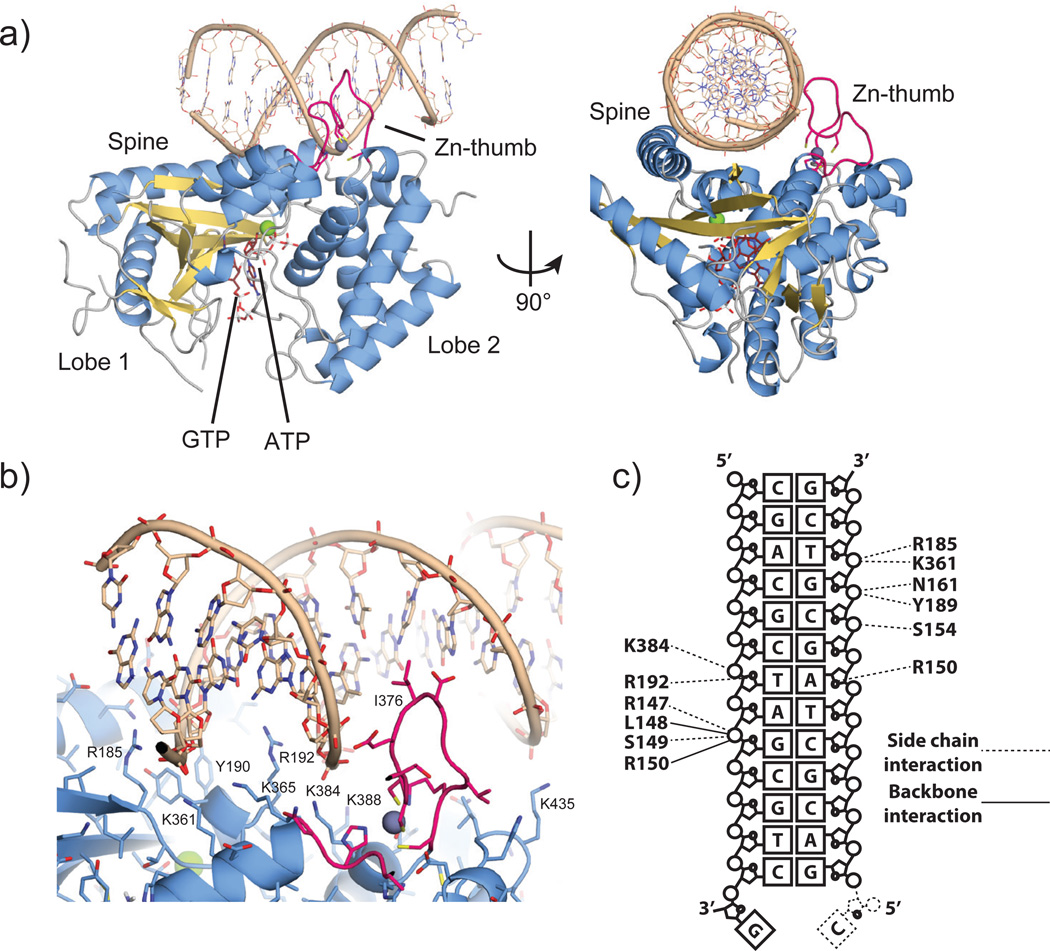
To reveal the structure of the activated conformation of cGAS, we co-crystallized cGASMab21(td) with a self-complementary 14mer oligonucleotide, ATP, GTP and MgCl2. In order to trap an activated conformation of cGASMab21 with DNA and bound nucleotides we mutated the NTase catalytic residues E200 and D202 to Q and N, respectively, thereby preventing catalysis during crystallization. The resulting transferase-deficient (td) variant is denoted cGASMab21(td). The structure of the cGASMab21(td):DNA:GTP:ATP complex was determined by molecular replacement using the coordinates for apo cGASMab21 as search model. 2Fo-Fc and Fo-Fc maps revealed interpretable density for 13 out of 14 base pairs of the dsDNA duplex and for both nucleotides bound at the active site (suppl. Fig. S2). The structure was refined at 3.1 Å resolution, resulting in a model with good R-factors and stereochemistry (suppl. Table S1).
DNA is bound along the platform between the spine on one side and the Zn-thumb on the other side (Fig. 2a). cGAS binds DNA predominantly by sequence independent interactions to both phosphate-sugar backbone strands along the minor groove (Fig. 2b,c). Hereby, cGAS binds seven nucleotides at the core of the platform, which are recognized by at least eleven residues via specific side and/or main chain contacts. In addition to the phosphate and sugar contacts, two arginine fingers (R150 and R192) are inserted into the minor groove, additionally stabilizing the interaction in a fairly sequence-independent manner. Besides binding to the array of conserved positively charged residues at the bottom of the platform, DNA is also bound by the spine and Zn-thumb. The continuous helix of the spine in apo-cGASMab21 is interrupted in the DNA complex and a DNA backbone phosphate is bound at the central kink of the spine helix. On the other side of the platform, the Zn-thumb contacts the DNA backbone near the major groove. We do not see close, direct polar contacts between Zn-thumb and DNA, but do not want to rule out water-mediated interactions here (suppl. Fig. S2a).
Figure 2. The cGASMab21:DNA:GTP:ATP complex.
a). Side and top views of cGASMab21 (color code of Fig. 1b) in complex with dsDNA (brown), GTP and ATP (ruby stick models). DNA binds along the platform between spine and Zn-thumb.
b) Close up view of the DNA binding site with selected annotated residues. DNA is bound mainly via the minor groove. A notable exception is the Zn-thumb near the major groove.
c) Schematic representation of DNA:cGAS contacts.
The Zn-thumb does not substantially change conformation or location between apo and DNA-bound cGAS. It seems to be a rather rigid element, in which the zinc ion serves as a structural stabilizer of the protruding loop, similar to Zn2+ in regulatory domains of RIG-I like receptors25. The location of the Zn-thumb at the backbone near the major groove suggests that it may assist in binding to B-form DNA. In support of this, we do not see a substantial perturbation of the bound DNA from canonical B-form DNA.
Altogether, our structure suggests a specific recognition of B-form dsDNA by cGAS via an extended B-DNA binding platform and flanking “Zn-thumb” across both lobes of the enzyme. The observed mode of binding is consistent with the key role of cGAS in sensing very different types of DNA in a sequence-independent manner18,23.
Structure-function analysis
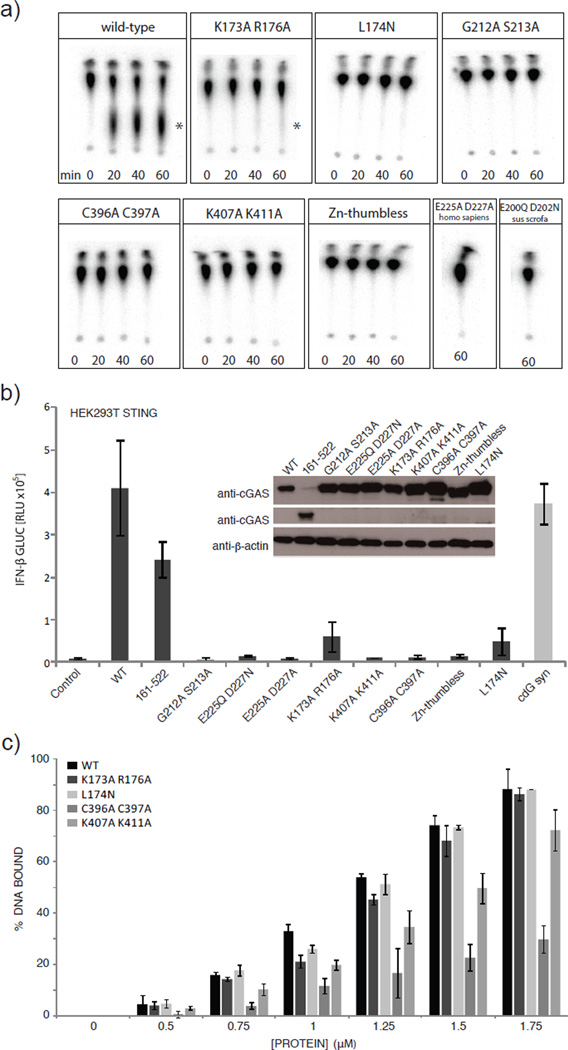
To validate the structural results, we mutated several conserved positively charged residues at the DNA binding platform of human cGAS, two active site residues and two zinc ligands in the Zn-thumb and tested for nucleotidyl-transferase activity in vitro by thin layer chromatography (TLC) (Fig. 3a). cGAS produces a product that migrates approximately in the range of c-di-AMP synthesized by DisA26, consistent with formation of a di-nucleotide. The conserved active site residues of NTases (human G212+S213 and E225+D227; porcine E200+D202) are essential for in vitro activity of both human and porcine cGASMab21. Moreover, mutation of conserved positively charged residues at the center and flanking regions of the platform (K173+R176 and K407+K411) either diminish or abolish activity, in accordance with this site being important for DNA sensing. Finally, disruption of the zinc-binding site of the thumb (human C396+C397, Zn-thumbless) abolishes DNA-induced NTase activity in vitro, highlighting the functional importance of the conserved Zn-thumb in DNA binding.
Figure 3. Platform and zinc thumb are involved in dsDNA-dependent activity.
a) NTase assays performed with different cGASMab21 mutants (2µM) in presence of 3µM dsDNA (50mer). Human WT cGASMab21 (positive control) synthesizes di-nucleotide, DNA binding site mutant K173A+R176A show reduced activity. K407A+K411A DNA-binding site mutant, C396A+C397A Zn-thumb mutant, Zn-thumbless, L174N structural switch mutant, active site mutants E200Q+D202N of porcine cGASMab21 and E225A+ D227A and G212A+S213A of human cGASMab21 are inactive. The asterisk indicates the di-nucleotide product.
b) IFN-β stimulation of cGAS mutants in HEK293T cells stably expressing murine STING. HEK293T cells were transfected with plasmids encoding indicated constructs along with the IFN-β promoter reporter plasmid pIFN-β-GLUC. Luciferase activity is plotted: mean ± sd (n=3). Both full-length and the crystallized region (cGASMab21 human 155–522) induce IFN-β promoter transactivation. Active site mutations (G212A+S213A and E225Q/A+D227N/A) abolish IFN-β stimulation. DNA-binding site mutants (K173A+R176A, K407A+K411A), Zn -thumb mutants (C396A+C397A, Zn-thumbless) and structural switch mutant (L174N) either reduce or abolish IFN-β stimulation. Empty vector was used as negative control while cyclic-di-GMP synthase (cdG syn) expressing vector was used as positive control. Inset: Western blot showing WT and mutant protein levels with β-actin as loading control.
c) Electrophoretic mobility shift analysis of 50mer dsDNA (0.2 µM) bound to cGASMab21 mutants at indicated concentrations. Plotted bars: mean±sd (n=3). While K407A+K411A DNA-binding site mutant and C396A+C397A Zn-thumb mutant show slightly reduced but not impaired affinity to dsDNA no detectable binding change was observed with the other mutants.
To test the effect of active site, platform and thumb mutations in living cells (Fig. 3b), we measured the transactivation of an IFN-β promoter reporter by transiently expressing human cGAS variants in HEK293T cells, which stably expressed murine STING. Induction of IFN-β by cGASMab21 (human cGAS155–552) in these cells is only moderately reduced compared to WT cGAS, showing that the Mab21 domain structurally addressed in this study is the catalytic active functional core of the sensor. The activity of full-length cGAS was abolished when residues of the NTase active site were mutated (E225Q/A+D227N/A or G212A+S213A) (Fig. 3b). Mutating charged platform residues (K173A+R176A; K407A+K411A) substantially reduced cGAS activity in living cells. Likewise, disrupting the zinc-binding site of the thumb (C396A+C397A, Zn-thumbless) severely compromised cGAS activity (Fig. 3b). These data validate the in vitro biochemical data and emphasize the importance of the structure-derived motifs and elements in living cells.
In order to see whether Zn-thumb and conserved platform surface residues are important for dsDNA binding and activity, we performed electrophoretic mobility shift assays (Fig. 3c). Both porcine and human wild-type cGASMab21 bind efficiently to dsDNA and – surprisingly – also dsRNA (suppl. Fig. S3a,c). The mutations in platform and thumb either did not affect DNA/RNA binding under these conditions, or reduced but did not abolish it (suppl. Fig S3b). However, both mutants fail to show DNA stimulated activity under conditions where they still bind DNA and dsRNA fails to stimulate activity under conditions where it robustly binds to the protein (suppl. Fig. S3c,d). Thus, while these analyses validate the functional relevance of the DNA binding platform and Zn-thumb on activating cGAS, they suggest that DNA or RNA interactions per se are not sufficient to activate the enzyme, indicating for instance the necessity for a precise DNA induced structural switch.
NTase and DNA induced structural switch
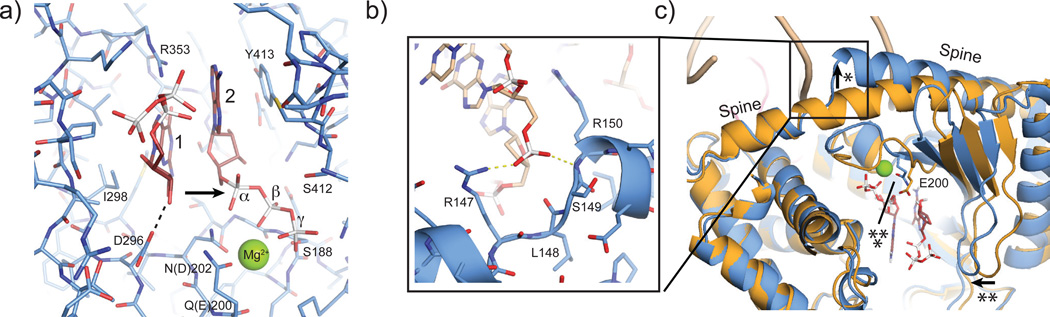
To reveal the mechanism of activation of cGAS by DNA, we first analyzed the NTase mechanism. We see clear electron density for two nucleotide triphosphate moieties (suppl. Fig. S2b). The two bases partially stack in an approx. 90° rotated conformation. They are sandwiched between I298 (lobe 1) and Y413 (lobe 2). The current resolution of the diffraction data does not allow us to unambiguously determine which base is adenine and which guanine. Binding of R353 at nucleobase 1 (the “receiving substrate” of NTases) near O6 and N7 would argue for this being guanine. In general, nucleobase 1 (interpreted as guanine here) is in hydrogen bonding distance to S355, S357 and T186, suggesting that this nucleotide is specifically recognized. In contrast, we do not observe direct hydrogen bonding contacts of the protein to nucleobase 2 (the “transferred” nucleotide in NTases; interpreted as adenine here). Nevertheless, this recognition might be mediated via water molecules such as in 3′ terminal uridylyl transferases 27.
The structure provides a mechanism for attack of nucleotide 1 on 2, consistent with the mechanism of other NTases, e.g. CCA adding enzyme28. The triphosphate chain of nucleotide 2 is well coordinated via S188 (lobe 1), S412 (lobe 2) and Mg2+ bound to E200 (Q in cGASMab21(td)) and D202 (N in cGAS Mab21(td)). As a consequence, the relative orientation of lobes 1 and 2 is important for the phosphate coordination of nucleotide 2. In our conformation, the α-phosphate of nucleotide 2 is well placed and oriented to promote nucleophilic attack of the sugar 2' OH from nucleotide 1 to form the 2'-5' linkage (Fig 4a, see accompanying manuscript by Ablasser et al. in this issue of Nature). The attacking OH of nucleotide 1 is polarized and activated by D296, consistent with the conserved features of NTases24. A second Mg2+ could be important for this catalytic step. However, distinct localization will require higher resolution.
Figure 4. NTase and DNA induced structural switch.
a) Close-up view of the NTase active site. Selected residues that are implicated in binding and catalysis are annotated. Both base moieties partially stack to each other and are further bound by stacking to Y413 and recognition by R353. E200 (mutated to Q in our structure) and D202 (mutated to N in our structure) bind an active site magnesium that coordinates phosphates of nucleotide 2. The attacking OH of nucleotide 1 is activated by D296 for nucleophilic attack on the α-phosphate of nucleotide 2 (arrow).
b) Close up view of DNA backbone phosphate binding at the spine.
c) This DNA phosphate binding triggers a change in the spine helix (*), which allows a closure of the active site cleft (**) and repositioning of the substrate binding loop for Mg2+-coordination of E200 (***).
cGAS is proposed to form a cyclic-di-nucleotide, which would require a second catalysis step and an additional attack of the OH of nucleotide 2 on the phosphate of nucleotide 1. Such an attack will require an almost 180° flip of the sugar moiety of nucleotide 2 to place its α-phosphate appropriately. In principle this is possible: in the course of our studies we determined the crystal structure of cGASMab21 bound to UTP in the absence of DNA and do observe an appropriate flip of the sugar moiety (suppl. Fig. S4). In any case, our structure satisfactorily explains the catalysis of formation of a specific (at present linear) di-nucleotide by cGAS, while formation of a cyclic di-nucleotide needs to be addressed in future studies.
To reveal a potential activation mechanism of cGAS, we superimposed apo-cGAS, cGASMab21:UTP and cGASMab21(td):DNA GTP:ATP complex (Fig. 4b,c; suppl. Fig S5a,b). We used cGASMab21:UTP because UTP binding orders the β-sheets on lobe 1 and we can also visualize conformational changes specifically induced by dsDNA rather than the nucleotides.
While UTP binding to cGAS ordered to some extend the nucleotide-binding loop in the active site, it does not substantially change the overall structure and active site geometry of cGAS (suppl. Fig S5b). In contrast, DNA phosphate binding to the spine (Fig. 4b) triggers a substantial structural switch in the spine helix (Fig. 4c) that closes lobes 1 and 2 and rearranges the active site loop, allowing magnesium coordinating of E200 to position and activate nucleotide 2.
To test the role of this DNA induced structural switch we mutated human L174 to N. L174 (porcine L148) is repositioned in response to DNA binding to stabilize the nucleotide-binding loop, but does not directly bind DNA or NTPs (suppl. Fig. S5c). While L174N shows fairly normal DNA binding (Fig. 3c, suppl. Fig. S3b), it lacks DNA stimulated cGA synthetase activity in vitro (Fig. 3a) and shows decreased interferon stimulation in cells (Fig. 3b). Thus, the structural and biochemical data suggest that cGAS is activated by a DNA induced structural switch that rearranges the NTase active site.
Conclusion
Here we provide the structure and mechanism of activation of the cytosolic DNA sensor cyclic-GMP-AMP synthase that readily explain the synthesis of a linear di-nucleotide intermediate by cGAS in response to DNA binding. The backbone binding of a canonical B-DNA by cGAS is consistent with a broad specificity innate immune PRR for cytosolic DNA and the structural elements of cGAS such as the positioning of residues involved in minor groove binding, arginine fingers and the Zn-thumb suggest that cGAS specifically responds to B-form DNA. This might explain the role for other innate immune DNA sensors to detect non-canonical DNA structures, such as DAI5. A structural switch transmitted by proper B-form DNA binding to the active site could also explain the lack of activation by dsRNA or in mutants that still bind DNA: slightly different conformations of RNA-bound, or DNA-bound mutant cGAS would not trigger robust cGA synthesis as even small differences in the active site geometry can strongly affect catalytic rates of enzymes.
In future, it will be important to address the specificity for other DNA structures in the activation of cGAS in more detail to see which types of DNA structures can activate cGAS. It will also be important to investigate additional requirements for efficient DNA sensing in vivo, because while shorter dsDNA molecules can stimulate cGASMab21 in vitro, DNA > 50mer is required for efficient IFN stimulation in vivo14,19. One possibility is that fraying of shorter DNA molecules prevents efficient stimulation or that the positively charged N-terminus contributes to sensing of longer DNA molecules. In addition, STING might play a direct role in DNA binding in a larger context in vivo19, although we do not see strong DNA binding in vitro and IFN stimulation in response to DNA in HEK293T cells in the absence of cGAS (suppl. Fig. S6).
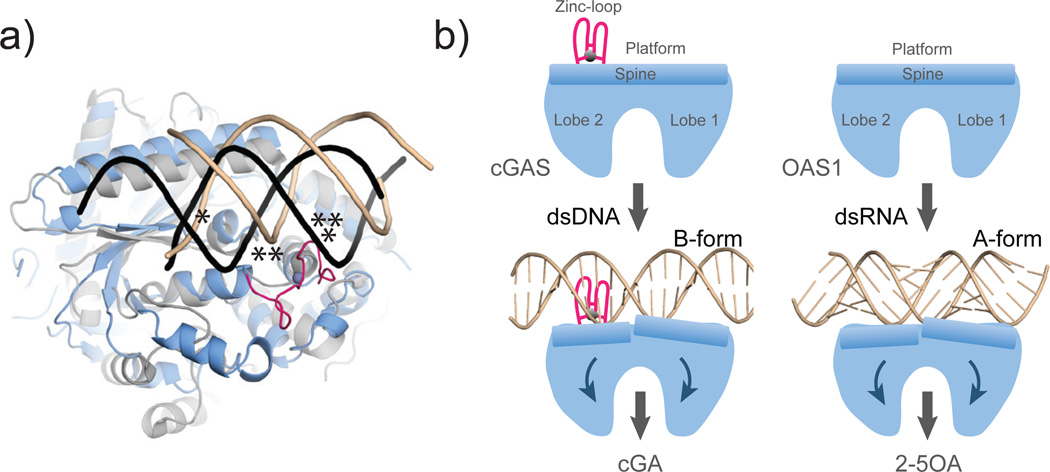
Interestingly, cGAS has remarkable fold similarity to the antiviral protein oligoadenylate synthase 1 (OAS1) 29,30 (Fig. 5). OAS1 synthesizes 2’-5’ linked oligoadenylate chains (2-5OA) in response to binding to cytosolic dsRNA. The structural similarity not only embraces the overall fold, several active site features, and arrangement of lobes 1 and 2, but also certain structural elements of the platform, including the long “spine” helix. Like cGAS, OAS1 binds dsRNA along the “platform” and triggers a structural change that is transmitted to the active site 30. However, while OAS1 is activated by A-form RNA, cGAS is activated by B-form DNA. The Zn-thumb in cGAS, missing in OAS1, likely acts a molecular “ruler” to specifically trigger activation in response to B-form but not A-form nucleic acids (Fig. 5). Despite these differences, cGAS shows a structural switch induced by dsDNA that is very similar to that of OAS1 induced by dsRNA30 (Fig. 5). Thus, our results structurally unify dsDNA and dsRNA sensing by cGAS and OAS1 NTases, respectively, in the innate immune system and suggests both processes are evolutionarily connected.
Figure 5. Model for DNA sensing by cGAS.
a) Superposition of cGAS�DNA (blue) with OAS1�RNA (grey) shows key elements for nucleic duplex selectivity. Both enzymes bind one DNA (brown) / RNA (black) backbone at the same protein site (*). The Zn-thumb specifically recognizes the position of the second DNA strand in B-form (**). However, it would clash with A-form RNA/DNA.
b) Unified activation model for cytosolic double-stranded nucleic acid sensing by cGAS and OAS1 NTases by a ligand induced structural switch.
Full Methods
Constructs and cloning
The sequence encoding full-length or truncated Homo sapiens and Sus scrofa cGAS were amplified from total cDNA (courtesy of Dr. Stefan Bauersachs) and cloned into pIRESneo3 (Clontech) or a modified pET21 (Novagen), respectively. The mutants were generated by site directed mutagenesis using PfuUltra (Stratagene). Zn-thumbless mutant was created by replacing residues 390–405 (Homo sapiens) by three Gly-Ser replicates.
Protein production and purification
All proteins were produced in E.coli Rosetta(DE3) or B834 (DE3) strains for native or selenomethionine derivative proteins, respectively. Bacteria were grown until an OD600 of 0.6 to 0.8 and expression was induced at 18°C for 16 to 18h with 0.1mM IPTG. Proteins were purified by Ni-NTA agarose resin and incubated with TEV protease (ratio 1:50) at 4°C overnight to remove the 6xHis-MBP-tag. The proteins were further purified by cation exchange chromatography followed by size exclusion chromatography using a Superdex 200 column (GE Healthcare), equilibrated in 20mM TRIS pH 7.5, 150mM NaCl and 1mM DTT. Purified proteins were concentrated to 10mg/ml for crystallization. Human STING 139–379 was purified as described31. All purified proteins were frozen in liquid N2 and stored at −80°C.
Crystallization of cGASMab21
Purified porcine cGAS (10mg/ml) was crystallized by hanging drop vapor diffusion in 20% PEG3350 and 200mM sodium malonate. The crystals appeared after one day at 20°C and were flash frozen after addition of glycerol to a final concentration of 15% (v/v). The selenomethionine derivatized protein was crystallized in 100 mM BIS-TRIS propane pH 6.3, 18% PEG3350 and 200mM sodium malonate and cryo protected with 20% ethane-1,2-diol prior to flash freezing. UTP bound crystals were obtained by adding 20mM MgCl2 and 1:10 (v/v) of 50 mM of nucleotide in 100 mM TRIS pH 7.5 to the protein prior to crystallization.
For crystallizing the DNA-GTP-ATP-cGAS complex 20 mM MgCl2, 2 mM of both nucleotides and 14 bp ds DNA (5’-CGACGCTAGCGTCG-3’) in a molar ratio of 1:1.2 protein:DNA were added to the inactive porcine cGASMab21(td) (E200Q + D202N) (10mg/ml). Crystals were obtained by hanging drop vapor diffusion in 50 mM sodium cacodylate pH 7.0, 2.5 mM spermine, 60 mM MgCl2 and 3% (v/v) PEG 400 after one day at 20 °C. The crystals were soaked in reservoir solution containing 25 % (v/v) glycerol before flash freezing.
Data Collection and Refinement
X-ray diffraction data of cGAS and cGAS-UTP were collected at X06SA beamline (Swiss Light Source, Switzerland) and diffraction data of the cGASMab21(td)-GTP-ATP-DNA complex were collected at PetraIII beamline P14 (EMBL/DESY, Hamburg, Germany) at 100K. The selenomethionine derivative data were collected at the Selenium peak wavelength (λ=0.97961Å). Data processing was carried out with XDS 32. AutoSHARP was used to locate Se sites (SAD dataset) and to produce an initial solvent flattened map 33. An initial model was built using iterative cycles of Buccaneer 34 and ARP/wARP classic 35. The model was optimized by alternating manual building with Coot 36 and refinement using Phenix 37 against a 2.0Å native data set. The structure of UTP bound cGAS and the DNA-GTP-ATP-cGAS complex structure were determined using molecular replacement with Phaser 38 and optimized by manual building with Coot and refinement with Phenix or Autobuster 39. Data collection and refinement statistics are listed in suppl. Table S1.
NTase Assays
NTase Assays were performed like described in 26. Reaction mixtures with the indicated concentrations of protein and DNA (40mer: 5’GGATACGTAACAACGCTTATGCATCGC CGCCGCTACATCC3’/ 50mer: 5’GGATACGTAACAACGCTTATGCATCGCC GCC GCTACATCCCTGAGCTGAC3’ ) (unless indicated 50mer dsDNA is used) or RNA (sequence as 50mer DNA) in 0.1M NaCl, 40 mM TRIS pH7.5 and 10 mM MgCl2 were started by addition of 100 µM ATP and 100 µM GTP containing 1:600 α32P-ATP and/or α32P-GTP (3000 Ci/mmol, Hartmann Analytic GmbH, Germany). Analysis of the reaction products was done using thin layer chromatography (PEI-Cellulose F plates, Merck) with 1 M (NH4)2SO4/1.5 M KH2PO4 pH3.8 as running buffer for the TLC-plates. Assays were performed at 35°C. The dried TLC-plates were analyzed by phosphor imaging (GE Healthcare).
Electrophoretic Mobility Shift Assays
0.2 µM of dsDNA or dsRNA (same sequences used for NTase assays) was incubated with indicated amount of purified protein for 30 min on ice. As reaction buffer 20 mM TRIS pH 8.0 and 200 mM NaCl was used. Samples were separated by 1% Agarose gel prepared with Gel-Red (Biotium) as suggested by the manufacturer. The gel images were analyzed by ImageJ.
Reporter Assays
HEK 293T cells stably expressing full-length murine STING (2×104 cells per 96-well) were transiently transfected with 25 ng IFN-β promoter reporter plasmid (pIFN-β-GLUC) in conjunction with 200 ng cGAS expression vectors using GeneJuice (Novagen) as indicated by the manufacturer. A codon-optimized version the diguanylate cyclase domain (AA83-248) of TM1788 (Thermotoga maritima MSB8) harboring a point mutation (R158A) to enhance c-di-GMP production was cloned into pEFBOS to contain a C-terminal HA tag (PMID: 19328769). This construct (c-di-GMP-synthase) was used to induce c-di-GMP production within 293T cells upon transient overexpression. 14h post transfection Luciferase activity was assessed.
THP-1 cells were stimulated with 200ng of either 50mer dsDNA (as in NTase assays) or tri-phosphate-RNA complexed with Lipofectamine 2000 (Life Technologies) according to the manufacturer´s instructions. Supernatants were collected 18h after stimulation and assayed for IP-10 production via ELISA. 90mer DNA used is as in ref. 19. CMA was purchased from Sigma Aldrich.
Immunoblotting
Cells were lysed in 1x Laemmli buffer and denatured at 95°C for 5 min. Probes were separated by 10% SDS–PAGE and transferred onto nitrocellulose membranes. Blots were incubated with anti-cGAS (Sigma Aldrich), anti-Phospho-IRF3 (Cell Signaling Technology) or anti-β-actin-IgG-HRP. Goat anti-rabbit-IgG-HRP was purchased from Santa Cruz Biotechnology.
Supplementary Material
Acknowledgements
We thank Agata Butryn for comments on the manuscript. We thank the Max-Planck-Crystallization facility for initial crystal screening and the Swiss Light Source, European Synchrotron Radiation Facility and the German electron synchrotron Petra III for beam time and excellent on site assistance. This work was funded by the National Institutes of Health (U19AI083025), the European Research Council Advanced Grant 322869, and the Center for Integrated Protein Science Munich (CIPSM) to K.-P.H., by DFG grant 3717/2-1 to G.W., by GRK1721 to KPH and G.W. by DFG grant SFB670 and ERC grant 243046 to V.H. C.M. is supported by GRK1721.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
F.C. crystallized and determined the structure of cGAS, performed biochemical assays, interpreted data and wrote the manuscript. T.D crystallized and refined the DNA complex. C.M., A.A., T.D., and M.M. performed biochemical assays. G.W. performed biochemical assays, interpreted data and helped with structure determination. V.H. supervised the cell-based experiments and interpreted data. K.-P.H designed the research, helped with structure determination, interpreted data and wrote the manuscript.
Author Information
Coordinates and structure factors have been deposited at the Protein Data Bank (4JLX, 4JLZ and 4KB6). Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Rathinam VA, Fitzgerald KA. Cytosolic surveillance and antiviral immunity. Current opinion in virology. 2011;1:455–462. doi: 10.1016/j.coviro.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Keating SE, Baran M, Bowie AG. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 2011;32:574–581. doi: 10.1016/j.it.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Krug A. Nucleic acid recognition receptors in autoimmunity. Handb Exp Pharmacol. 2008:129–151. doi: 10.1007/978-3-540-72167-3_7. [DOI] [PubMed] [Google Scholar]
- 5.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 6.Burckstummer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 7.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang P, et al. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- 12.Kim T, et al. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci U S A. 2010;107:15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, et al. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rathinam VA, Fitzgerald KA. Innate immune sensing of DNA viruses. Virology. 2011;411:153–162. doi: 10.1016/j.virol.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu J, et al. Cyclic GMP-AMP Is an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Science. 2012 doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abe T, et al. STING Recognition of Cytoplasmic DNA Instigates Cellular Defense. Mol Cell. 2013;50:5–15. doi: 10.1016/j.molcel.2013.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burdette DL, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McWhirter SM, et al. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J Exp Med. 2009;206:1899–911. doi: 10.1084/jem.20082874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science. 2012 doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuchta K, Knizewski L, Wyrwicz LS, Rychlewski L, Ginalski K. Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 2009;37:7701–7714. doi: 10.1093/nar/gkp854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui S, et al. The C-terminal regulatory domain is the RNA 5'-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–79. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 26.Witte G, Hartung S, Buttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 27.Stagno J, Aphasizheva I, Rosengarth A, Luecke H, Aphasizhev R. UTP-bound and Apo structures of a minimal RNA uridylyltransferase. J Mol Biol. 2007;366:882–899. doi: 10.1016/j.jmb.2006.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiong Y, Steitz TA. Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature. 2004;430:640–645. doi: 10.1038/nature02711. [DOI] [PubMed] [Google Scholar]
- 29.Hartmann R, Justesen J, Sarkar SN, Sen GC, Yee VC. Crystal structure of the 2'-specific and double-stranded RNA-activated interferon-induced antiviral protein 2'-5'-oligoadenylate synthetase. Mol Cell. 2003;12:1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- 30.Donovan J, Dufner M, Korennykh A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1218528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 31.Cavlar T, Deimling T, Ablasser A, Hopfner KP, Hornung V. Species-specific detection of the antiviral small-molecule compound CMA by STING. EMBO J. 2013 doi: 10.1038/emboj.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 34.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 35.Morris RJ, Perrakis A, Lamzin VS. ARP/wARP's model-building algorithms.I. The main chain. Acta Crystallogr D Biol Crystallogr. 2002;58:968–975. doi: 10.1107/s0907444902005462. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.